SUMMARY
Defective insulin signaling in hepatocytes is a key factor in type 2 diabetes. In obesity, activation of calcium/calmodulin-dependent protein kinase II (CaMKII) in hepatocytes suppresses ATF6, which triggers a PERK-ATF4-TRB3 pathway that disrupts insulin signaling. Elucidating how CaMKII suppresses ATF6 is therefore essential to understanding this insulin resistance pathway. We show that CaMKII phosphorylates and blocks nuclear translocation of histone deacetylase 4 (HDAC4). As a result, HDAC4-mediated SUMOylation of the corepressor DACH1 is decreased, which protects DACH1 from proteasomal degradation. DACH1, together with NCOR, represses Atf6 transcription, leading to activation of the PERK-TRB3 pathway and defective insulin signaling. DACH1 is increased in the livers of obese mice and humans, and treatment of obese mice with liver-targeted constitutively-nuclear HDAC4 or DACH1 shRNA increases ATF6, improves hepatocyte insulin signaling, and protects against hyperglycemia and hyperinsulinemia. Thus, DACH1-mediated corepression in hepatocytes emerges as an important link between obesity and insulin resistance.
INTRODUCTION
Excessive hepatic glucose production (HGP) and aberrant insulin receptor signaling are the major causes of hyperglycemia and insulin resistance in obesity-induced type 2 diabetes (T2D) (Pajvani and Accili, 2015). We have recently identified activation of hepatocyte (HC) CaMKII/p38 as a major contributor to aberrant HGP and perturbed insulin signaling in obesity (Ozcan et al., 2013; Ozcan et al., 2012). Treatment of obese mice with a drug inhibitor of the pathway lowers plasma glucose and corrects hyperinsulinemia and is additive in these benefits with the current leading T2D drug, metformin (Ozcan et al., 2015). The CaMKII pathway in HCs perturbs metabolism by lowering the transcription factor ATF6, which can function as a homeostatic regulator of the endoplasmic reticulum (ER) stress response by inducing the chaperone, p58IPK (Wu et al., 2007). As such, the decrease in ATF6 in obese HCs activates the PERK—ATF4 arm of the ER stress response, which induces an inhibitor of insulin receptor signaling called TRB3. In support of the key role of HC ATF6 in the CaMKII pathway as a driver of perturbed metabolism in obesity, both total and nuclear (cleaved) ATF6 and P58IPK are decreased in obese mice liver, and in human liver, ATF6 cleavage decreases in relation to HOMA-IR (Kumashiro et al., 2011; Wang et al., 2009). Moreover, ATF6 overexpression in cell culture models improves insulin signaling, although the underlying mechanisms have not been fully explored (Tang et al., 2011; Ye et al., 2010). ATF6 can also suppress HGP by disrupting the interaction of two gluconeogenic transcription factors, CREB and CRTC2, and restoration of ATF6 levels in the livers of obese mice lowers blood glucose (Wang et al., 2009).
Given the importance of CaMKII-mediated ATF6 suppression in HCs in linking obesity to T2D, a key question relates to the molecular mechanism through which CaMKII suppresses ATF6. We show here that the corepressor DACH1 is responsible for suppressing Atf6. DACH1 levels are increased in obesity by CaMKII-induced nuclear exclusion of HDAC4, which decreases HDAC4-mediated SUMOylation and degradation of DACH1. Accordingly, insulin sensitivity can be improved in obese mice by silencing either CaMKII or DACH1 or enforcing nuclear HDAC4 in HCs, without any change in food intake or body weight. These results identify DACH1 as a critical component of defective insulin action seen in obesity.
RESULTS
ATF6 Overexpression in Obese Mice Improves Insulin Sensitivity
While our previous work implicated suppression of ATF6 as a key mechanism of how CaMKII activation in HCs promotes insulin resistance in obesity (Ozcan et al., 2013), we asked whether genetically boosting hepatic ATF6 in obesity could improve metabolism. To this end, we treated DIO mice with adenoviral vectors encoding cleaved, nuclear ATF6 (ATF6N) or LacZ control. Adeno-ATF6N-treated mice had lower fasting blood glucose and lower plasma insulin levels and enhanced reduction of blood glucose in response to insulin stimulation (Figure S1A and S1B) in the absence of any change in body weight. Adeno-ATF6N treatment also improved acute insulin-induced p-Akt (Figure S1C); which is a dynamic measure of improved insulin signaling.
CaMKII Suppresses HC Atf6 mRNA in Obesity
Given the critical role of ATF6 in CaMKII-mediated improvement in insulin signaling, we next investigated whether CaMKII regulates ATF6 at the transcriptional level. For this purpose, we first analyzed the livers of Camk2gfl/fl obese mice treated with control AAV-TBG-LacZ vs. AAV-TBG-Cre, which specifically deletes CaMKII in HCs and thereby increases insulin sensitivity (Ozcan et al., 2013). The livers of obese mice with HC-deleted CaMKII had higher Atf6 mRNA levels than the livers of the control obese mice (Figure S1D). We then isolated primary HCs from Camk2gfl/fl mice and treated them ex vivo with adeno-Cre to delete CaMKII, or adeno-LacZ control and incubated with BSA (control) or palmitate, which mimics features of HC pathophysiology seen in obesity (Boden et al., 2005), including defective insulin signaling (Ozcan et al., 2013). Using this model, we confirmed the increase in Atf6 mRNA in palmitate-treated CaMKII deficient HCs (Figure S1E). We then conducted Atf6 promoter ChIP assays using this model. CaMKII-deficient HCs showed increased Atf6 promoter occupancy of both RNA polymerase II (Pol II) and H3K27ac, a histone mark associated with active gene transcription (Wang et al., 2008) (Figure S1F). In summary, hepatic CaMKII activation in obese mice suppresses Atf6 transcription, and restoring hepatic ATF6 through genetic engineering improves metabolic health.
Obesity Induces HDAC4 Phosphorylation and Regulates Its Nuclear Localization
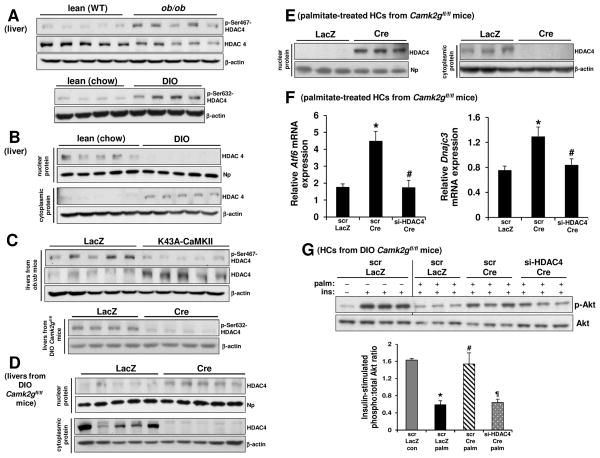
The data above raise the key issue of how hepatic CaMKII lowers Atf6 transcription in obesity. Previous work has implicated CaMKII in transcriptional regulation through its ability to phosphorylate and promote the nuclear export of the class IIa histone deacetylase HDAC4 (Backs et al., 2006; Zhang et al., 2007). The possible relevance of CaMKII-induced phosphorylation and nuclear export of HDAC4 in obese mouse liver was suggested by the results of several experiments. First, phosphorylation of HDAC4 on Ser467 and Ser632, two CaMKII phosphorylation sites (Backs et al., 2006), were significantly increased in the livers of obese mice compared with lean mice (Figure 1A). Second, in the livers of obese mice, nuclear HDAC4 levels were lower and cytoplasmic levels were higher (Figure 1B). Third and most importantly, liver-CaMKIIγ deficiency or CaMKII inhibition markedly decreased p-Ser467 and p-Ser632-HDAC4 and promoted HDAC4 nuclear localization in the livers of obese mice (Figure 1C–D). Palmitate-treated CaMKII-deficient HCs also showed increased nuclear HDAC4 compared to HCs from control mice (Figure 1E). To determine causation, we silenced HDAC4 in CaMKII-deficient HCs using siRNA and found that HDAC4 deficiency lowered Atf6 and its downstream target, Dnajc3 (P58IPK), and abrogated the improvement in insulin-Akt signaling in CaMKII-deficient HCs (Figure 1F and 1G) upon palmitate treatment. We then tested causation in vivo by silencing HDAC4 in DIO mice and found increased fasting blood glucose and plasma insulin levels and reduced glucose-disposal curves post-insulin (Figure S2) without a change in body weight. These data provide initial support for the role of an obesity-driven CaMKII-HDAC4 nuclear exclusion pathway in ATF6 suppression and defective insulin signaling in HCs.
Figure 1. Obesity Promotes CaMKII-Mediated Phosphorylation and Nuclear Exclusion of HDAC4, Leading to Defective Insulin Signaling in HCs.
(A) Upper blot, p-Ser467-HDAC4, HDAC4, and β-actin were assayed in liver extracts of lean (WT) and ob/ob mice. Lower blot, p-Ser632-HDAC4 and β-actin were assayed in liver extracts of lean (chow-fed) and DIO mice.
(B) Liver extracts of lean (chow-fed) and DIO mice were assayed by immunoblot for nuclear HDAC4 and nucleophosmin (Np) (upper blot) and for cytoplasmic HDAC4 and β-actin (lower blot).
(C) p-Ser467-HDAC4, HDAC4 and β-actin were assayed in liver extracts of ob/ob mice treated with adeno-LacZ or adeno-K43A-CaMKII (upper blot), and p-Ser632-HDAC4 and β-actin were assayed in liver extracts of DIO Camk2gfl/fl mice treated with AAV8-TBG-LacZ or AAV8-TBG-Cre (lower blot).
(D) Liver extracts of DIO Camk2gfl/fl mice treated with AAV8-TBG-LacZ or AAV8-TBG-Cre were assayed for nuclear HDAC4 and nucleophosmin (Np) (upper blot) and for cytoplasmic HDAC4 and β-actin (lower blot).
(E) Primary HCs from Camk2gfl/fl mice transduced with adeno-LacZ or adeno-Cre were incubated with 0.3 mM palmitate for 4 h, and then nuclear and cytoplasmic lysates were assayed for HDAC4, nucleophosmin (Np), and β-actin.
(F) HCs from Camk2gfl/fl mice were pretreated with either scrambled RNA (scr) or siRNA targeting HDAC4 (si-HDAC4) for 12 h and then transduced with adeno-LacZ or adeno-Cre. After an additional 24 h, the cells were incubated with 0.3 mM palmitate for 11 h, with the last 5 h in serum-free media. The cells were then assayed for Atf6 and Dnajc3 mRNA by RT-qPCR (n = 3; bars with different symbols are different from each other and control, mean ± SEM, p < 0.05).
(G) As in (F), except that some of the cells received BSA control instead of palmitate and were then treated with PBS control or 100 nM insulin for 5 min, as indicated by the minus and plus symbols. Cell lysates were then assayed for p-Akt and total Akt. Densitometric quanti cation of the immunoblot data is shown in the bar graph (n = 3; bars with different symbols are different from each other and control, mean ± SEM, p < 0.05). See also Figure S1.
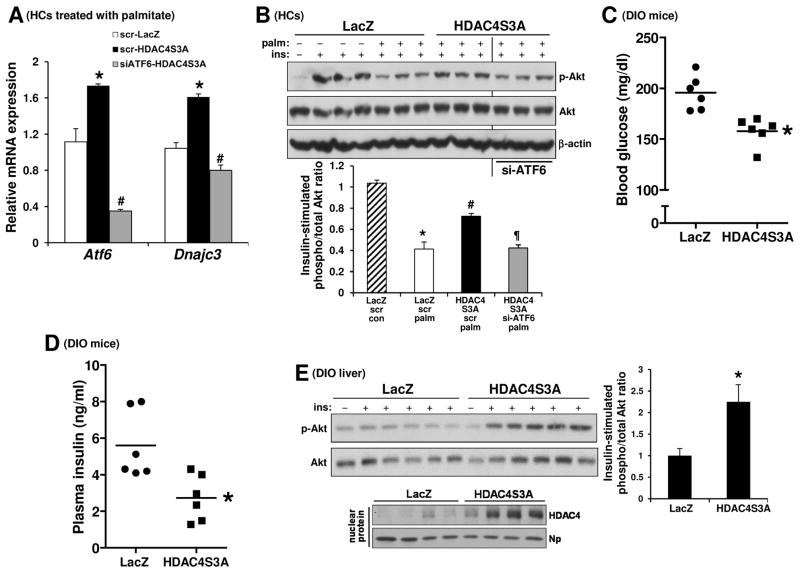
To establish causation, we took advantage of a phosphorylation-defective, constitutively nuclear HDAC4 mutant, HDAC4S3A (S246A/S467A/S632A) (Backs et al., 2006). Transfection of primary HCs with a relatively low titer of this mutant increased Atf6 and Dnajc3 mRNA levels and improved insulin-induced p-Akt after palmitate treatment (Figure 2A and 2B—compare white and black bars). To test the functional role of ATF6 in this improvement, we silenced Atf6 in palmitate-treated HDAC4S3A-overexpressing HCs using siRNA and found that this treatment lowered Dnajc3 and reduced insulin-induced p-Akt to the level of palmitate-treated control HCs (Figure 2A and 2B; compare black and grey bars). We then treated DIO mice with adenoviral vectors encoding the mutant HDAC4 vs. LacZ control. Adeno-HDAC4S3A-treated mice had lower fasting blood glucose and lower plasma insulin levels in the absence of any change in body weight (Figure 2C–D). To link these findings to hepatic insulin signaling, we assayed p-Akt in the livers of mice injected with insulin through the portal vein. Similar to the case with palmitate-treated HCs, there was enhanced insulin-stimulated Akt phosphorylation in the livers of adeno-HDAC4S3A-treated DIO mice (Figure 2E). Finally, acetylation of the key gluconeogenic transcription factor FoxO1, which suppresses its binding to DNA target sites, was shown to be increased in mice with combined silencing of HDAC4, HDAC5, and HDAC7 (Mihaylova et al., 2011). However, we found that when hepatic HDAC4 alone was either silenced or rendered constitutively nuclear (HDAC4S3A), the level of acetylated FoxO1 was not altered (Figure S2F). These collective data indicate the presence of CaMKII → HDAC4 phosphorylation/nuclear exclusion → ATF6 suppression pathway in obese liver, resulting in perturbed hepatic insulin signaling and disturbed glucose homeostasis.
Figure 2. Constitutively Nuclear HDAC4S3A Increases Atf6, Dnajc3, and Insulin-Stimulated p-Akt and Improves Metabolism in DIO Mice.
(A) Primary HCs from WT mice were pretreated with either scrambled RNA (scr) or siRNA targeting ATF6 (siATF6) for 12 h followed by transduction with adeno-LacZ or adeno-HDAC4S3A. After an additional 24 h, the cells were incubated with 0.3 mM palmitate for 11 h, with the last 5 h in serum-free media. The cells were then assayed for Atf6 and Dnajc3 mRNA by RT-qPCR (n = 3; bars with different symbols are different from each other and control, mean ± SEM, p < 0.05).
(B) As in (A), except that some of the cells received BSA control instead of palmitate and were then treated with PBS control or 100 nM insulin for 5 min, as indicated by the minus and plus symbols. Cell lysates were then assayed by immunoblot for p-Akt and total Akt and β-actin. Densitometric quanti cation of the immunoblot data is shown in the bar graph (n = 3; bars with different symbols are different from each other and control, mean ± SEM, p < 0.05).
(C–D) Fasting blood glucose and plasma insulin in DIO mice treated with adeno-LacZ or adeno-HDAC4S3A (n = 6 mice/group; mean ± SEM, *p <0.05).
(E) DIO mice treated with adeno-LacZ or adeno-HDAC4S3A were fasted for 5 h and then injected with 1.5 IU/kg insulin through the portal vein for 3 min. Liver extracts were assayed for p-Akt and total Akt (upper blot), with densitometric quantification of the data shown in the bar graph (n = 5; mean ± SEM, *p < 0.05). The lower blot shows the expression level of nuclear HDAC4 in the two groups of mice. See also Figure S2.
Nuclear Exclusion of HC HDAC4 in Obesity Elevates Nuclear DACH1, a Corepressor That Lowers Atf6 and Promotes Insulin Resistance
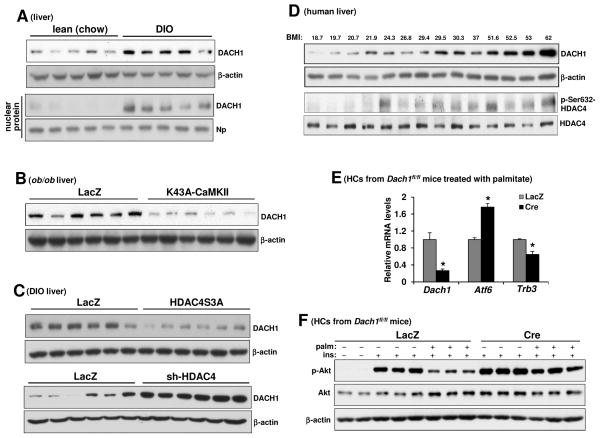
We next turned our attention to the mechanism of how HDAC4 could induce Atf6 transcription. Because HDAC4 usually represses gene transcription (Parra, 2015), we considered the possibility that HDAC4 repressed an Atf6 gene repressor. Indeed, previous studies in skeletal muscle identified a transcriptional repressor called Dachshund homolog (DACH) as a target of HDAC4-mediated repression (Tang and Goldman, 2006; Tang et al., 2009). Thus, we hypothesized that nuclear exclusion of HDAC4 in HCs in obesity would result in the de-repression of DACH1, which would then repress Atf6. We first asked whether hepatic DACH1 is increased in obesity. DACH1 levels were indeed increased in the livers of both DIO and ob/ob mice compared with lean controls (Figure 3A upper panel and Figure S3A). Because DACH1 protein is predominantly nuclear (Mardon et al., 1994), we next checked nuclear DACH1 and found that nuclear DACH1 was also increased in obese vs. lean mouse liver (Figure 3A lower panel). Moreover, this increase in hepatic DACH1 was significantly suppressed in obese mice in which liver CaMKII was deleted or inhibited (Figure 3B and Figure S3B) or in obese mice subjected to treatment with adeno-HDAC4S3A to enforce nuclear HDAC4 (Figure 3C upper panel). Conversely, silencing hepatic HDAC4 in obese mice further increased hepatic DACH1, suggesting that DACH1 lies downstream of HDAC4 (Figure 3C lower panel). We also tested the effect of CaMKII inhibition in metabolism-qualified human HCs and found that DACH1 was increased by palmitate treatment, and this increase was prevented by inhibition of CaMKII using adeno-K43A-CaMKII (Figure S3C). Furthermore, analysis of 14 human liver biopsy specimens spanning body-mass indexes (BMIs) from 19 to 62 showed an overall trend for higher levels of DACH1 and p-Ser632-HDAC4 in the livers of obese vs. leaner subjects (Figure 3D).
Figure 3. Hepatic DACH1 is Increased in Obesity Via CaMKII Activation and Nuclear Exclusion of HDAC4, Leading to Defective Insulin Signaling in HCs.
(A) Extracts of total liver (upper blot) and liver nuclei (lower blot) from WT mice fed a chow or DIO diet for 12 wks were assayed for DACH1, β-actin, and nucleophosmin (Np) by immunoblot.
(B) DACH1 and β-actin were assayed in liver extracts from ob/ob mice treated with adeno-LacZ or adeno-K43A-CaMKII.
(C) DACH1 and β-actin were assayed in livers from DIO mice treated with adeno-LacZ or adeno-HDAC4S3A (upper blot) or DIO mice treated with adeno-LacZ or adeno-sh-HDAC4 (lower blot).
(D) DACH1, β-actin, p-Ser467-HDAC4 and HDAC4 were assayed in liver extracts from human subjects with the indicated BMIs.
(E) HCs from Dach1fl/fl mice were transduced with adeno-LacZ or adeno-Cre. After 36 h, the cells incubated with 0.3 mM palmitate for 11 h, with the last 5 h in serum-free media. The cells were then assayed for Dach1, Atf6 and Trb3 mRNA (n = 3; mean ± SEM, *p < 0.05).
(F) As in (E), except that some of the cells received BSA control instead of palmitate and were then treated with PBS control or 100 nM insulin for 5 min, as indicated by the minus and plus symbols. Cell lysates were then assayed for p-Akt and total Akt and β-actin. See also Figure S3.
We next tested the importance of DACH1 in the regulation of the ATF6–TRB3-insulin signaling pathway using WT vs. DACH1-deficient HCs treated in the absence or presence of palmitate. For this experiment, we used primary HCs isolated from Dach1fl/fl mice and then treated the cells ex vivo with adeno-Cre vs. control adeno-LacZ. The data show that Atf6 mRNA was significantly higher and Trb3 mRNA was significantly lower in DACH1-deficient HCs (Figure 3E). Most importantly, DACH1 deficiency resulted in prevention of palmitate-induced suppression of insulin-induced p-Akt (Figure 3F).
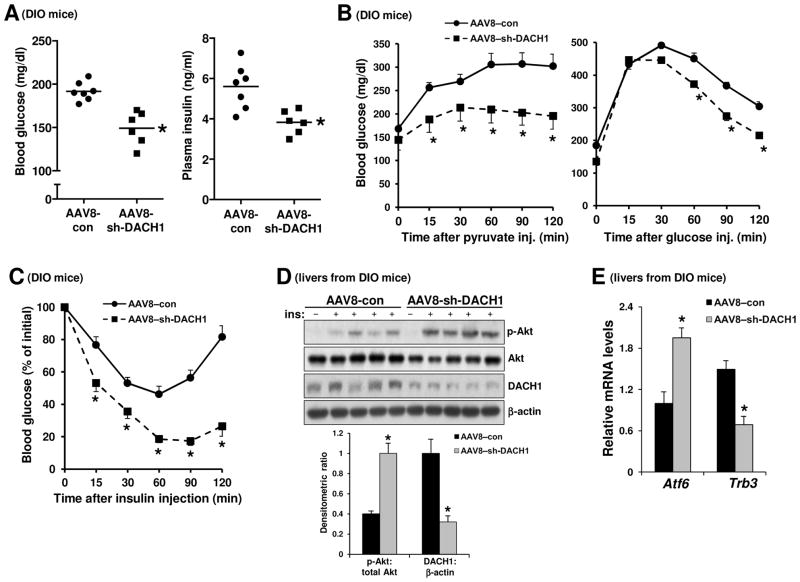
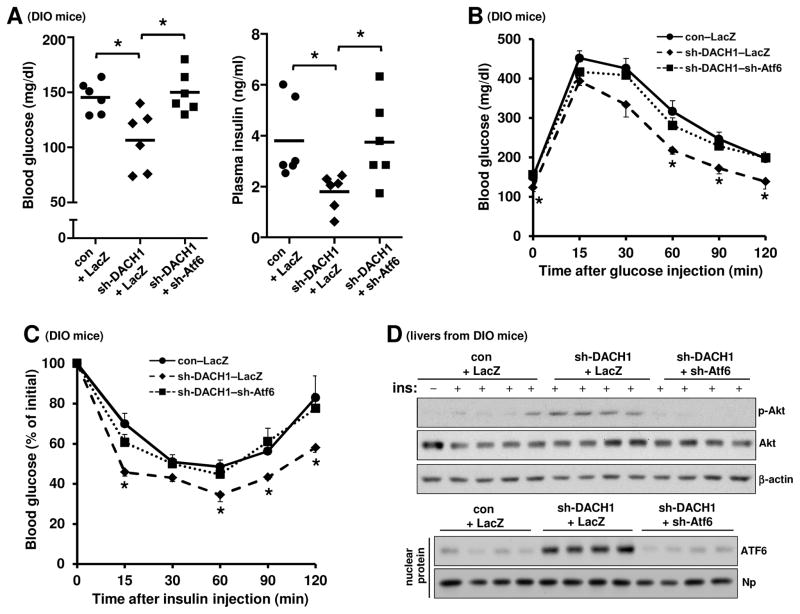
To test the functional importance of DACH1 in glucose metabolism in obesity, we silenced hepatic DACH1 in DIO mice using adeno-associated virus-8 (AAV8) mediated shRNA (Lisowski et al., 2014). This treatment, which lowered hepatic DACH1 levels by approximately 70% (Figure 4D) without a change in body weight, significantly lowered fasting blood glucose and plasma insulin (Figure 4A). DACH1 silencing also lowered plasma glucose in response to pyruvate challenge, which is a measure of HGP; improved the blood glucose response to glucose challenge; and enhanced reduction of blood glucose in response to insulin stimulation (Figure 4B and 4C). Consistent with our in vitro data, we observed an increase in both Atf6 mRNA and insulin-stimulated Akt phosphorylation and a significant decrease in Trb3 mRNA levels in the DACH1-silenced DIO cohort (Figure 4D and 4E). Similar results were found in DIO Dach1fl/fl mice injected with AAV-8 encoding Cre recombinase driven by the HC-specific thyroxin binding globulin promoter (TBG-Cre) (Figure S4). These data are consistent with the hypothesis that hepatic DACH1 depletion improves glucose metabolism in obese mice by increasing hepatic insulin sensitivity.
Figure 4. Silencing Hepatic DACH1 in Obese Mice Improves Glucose and Insulin Metabolism.
(A–C) Fasting blood glucose and plasma insulin (A) and pyruvate, glucose, and insulin tolerance tests (B–C) in DIO mice after treatment with AAV8–sh-DACH1 or empty AAV (AAV8–con) (n = 6–7 mice/group; mean ± SEM, *p < 0.05).
(D) DIO mice were treated with AAV8-sh-DACH1 or empty AAV (AAV8-con). After 14 days, the mice were fasted for 5 h and injected with insulin through the portal vein, followed 3 mins later by harvesting of liver. Liver extracts were assayed for p-Akt, total Akt, DACH1, and β-actin by immunoblot. Densitometric quanti cation of the immunoblot data is shown in the graph (n =4–5; mean ± SEM, *p < 0.05).
(E) As in (D), except that Atf6 and Trb3 mRNA levels were assayed by RT-qPCR (n = 5; mean ± SEM, *p < 0.05). See also Figures S4–6.
DACH1 has a highly conserved N-terminal Dach Box domain, which shares approximately 35% amino acid identity to the Ski/Sno proteins and is therefore known as the Dach Ski/Sno (DS) domain (Li et al., 2002; Wu et al., 2003). The DS domain mediates the interaction of DACH1 with both DNA and other corepressors and is required for the ability of DACH1 to repress a number of genes (Sundaram et al., 2008; Wu et al., 2008; Wu et al., 2003). Accordingly, an engineered mutant of DACH1 in which the DS domain is deleted (ΔDS-DACH1) has been shown to act as dominant-negative in blocking DACH1 gene repression (Sunde et al., 2006). As a further test of the importance of DACH1 in liver metabolism, we treated obese mice with adeno-ΔDS-DACH1 vs. adeno-LacZ control. The ΔDS-DACH1 cohort had lower fasting blood glucose, lower plasma insulin levels, improved blood glucose response to glucose challenge, and enhanced reduction of blood glucose upon insulin stimulation, all without a change in body weight (Figure S5A–D). Adeno-ΔDS-DACH1 treatment also improved insulin-induced p-Akt and raised the level of P58IPK (Figure S5E). Similar results were obtained using ob/ob mice (Figure S6). These results further support the role of DACH1 in glucose homeostasis in obesity and indicate that the DS domain of DACH1 is required for this effect.
DACH1 Silencing Improves Glucose Metabolism in Obese Mice By Increasing ATF6
To test whether the metabolic improvement seen with hepatic DACH1 deficiency was dependent on the increase in ATF6, we silenced hepatic ATF6 in liver-DACH1-deficient obese mice using adeno-sh-ATF6 (Wang et al., 2009). The results show that all of the beneficial effects of DACH1 silencing—decreased blood glucose and plasma insulin, improved blood glucose response to glucose challenge, enhanced reduction of blood glucose in response to insulin, and increased insulin-induced p-Akt in liver—were abrogated by also silencing hepatic ATF6 (Figure 5A–D). These data provide further evidence that hepatic DACH1 depletion improves insulin sensitivity and glucose metabolism in obese mice by de-repressing ATF6.
Figure 5. Improvement in Glucose Homeostasis by Hepatic DACH1 Silencing is Abrogated by Also Silencing ATF6.
(A–C) WT DIO mice were treated with AAV8–sh-DACH1 or AAV8–con. After 3 days, half of the AAV8–sh-DACH1 mice received adeno–sh-Atf6, while the other half received adeno-LacZ. After an additional 5 days, fasting blood glucose and plasma insulin were assayed, and glucose and insulin tolerance tests were conducted (n = 6 mice/group; mean ± SEM, p< 0.05).
(D) WT DIO mice were treated as in (A–C). After 14 days, the mice were fasted for 5 h and injected with insulin through the portal vein, followed 3 mins later by harvesting of liver. Total liver extracts were assayed for p-Akt, total Akt, and β-actin (top blot), and nuclear extracts were assayed for ATF6 and nucleophosmin (Np) (bottom blot).
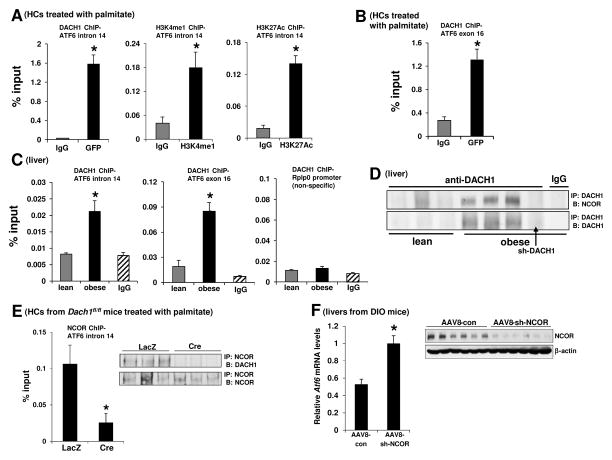
Recent work has identified a consensus DACH1 DNA-binding sequence using genome-wide in silico promoter analysis together with cyclic amplification and selection of targets (Zhou et al., 2010). Using this information, we identified an intronic region (intron 14) and an exon (exon 16) in the Atf6 gene that contain DACH1 consensus sequences. ChIP analysis in GFP-DACH1-transfected primary HCs showed significantly increased recruitment of DACH1 to both of these regions (Figure 6A left graph and Figure 6B), whereas a control segment at the 3′ end of the Atf6 gene gave no signal (not shown). Moreover, in view of the fact that corepressors bind to enhancer sites to suppress gene transcription, we found through additional ChIP experiments that the intronic site was enriched for two enhancer marks, histone H3 lysine 4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27Ac) (Heintzman et al., 2007) (Figure 6A, middle and right graphs). We also observed significantly increased recruitment of DACH1 to the identified intronic and exonic regions in obese liver compared with lean liver, whereas a control segment (Rplp0) gave no signal (Figure 6C).
Figure 6. DACH1-NCOR Complex Represses Atf6.
(A) Primary HCs from WT mice were transfected with an expression plasmid encoding GFP-DACH1. After 48 h, the cells were incubated with 0.3 mM palmitate for 3 h, and then ChIP was performed using anti-GFP, anti-H3K4me1, anti-H3K27Ac, or IgG control antibodies. The region spanning a specific intron site containing a predicted DACH1-binding sequence (intron 14) was amplified by RT-qPCR and normalized to the values obtained from the input DNA (n = 3; mean ± SEM, *p < 0.05).
(B) Same as (A), except that region spanning a specific exon site containing a predicted DACH1-binding sequence (exon 16) was amplified by RT-qPCR and normalized to the values obtained from the input DNA (n = 3; mean ± SEM, *p < 0.05).
(C) ChIP was performed from liver extracts of lean and DIO mice with anti-DACH1 or IgG control antibodies. The region spanning specific intron and exon sites containing a predicted DACH1-binding sequence (intron 14 and exon 16, respectively) and a non-specific region (Rplp0) were amplified by RT-qPCR and normalized to the values obtained from the input DNA ( n= 3; mean ± SEM, *p < 0.05).
(D) DACH1 was immunoprecipitated (IP:) from liver extracts and then probed for NCOR or DACH1 by immunoblot (B:). Lanes 1–3 are lean mice, and lanes 4–8 are DIO mice. All DIO mice were treated with AAV8-con, except for lane 7, which was treated with AAV8-sh-DACH1. Lane 8 used control IgG instead if anti-DACH1 for the IP step.
(E) HCs from Dach1fl/fl mice were transduced with adeno-LacZ or adeno-Cre. After 48 h, the cells were incubated with 0.3 mM palmitate for 3 h, and then ChIP was performed using an antibody against NCOR. The region spanning the aforementioned Atf6 intronic site was amplified by RT-qPCR and normalized to the values obtained from the input DNA (n = 3; mean ± SEM, *p < 0.05). Inset, ChIP from a parallel set of cells that was treated the same as above, up until the final post-IP wash. The beads were then boiled in loading buffer and assayed by immunoblot for NCOR and DACH1.
(F) Liver extracts from DIO mice treated with AAV8–con or AAV8–sh-NCOR were assayed for Atf6 mRNA levels by RT-qPCR (n = 6, mean ± SEM, *p < 0.05). Inset, protein extracts were probed for NCOR and β-actin by immunoblot to document NCOR silencing.
In view of previous work showing that DACH1 can interact with nuclear receptor corepressor (NCOR) to repress genes (Wu et al., 2003), we tested the interaction between DACH1 and NCOR. We observed that NCOR could be coimmunoprecipitated with DACH1 in liver from obese but not lean mice, suggesting that DACH1 and NCOR physically interact in the setting of obesity (Figure 6D). Consistent with the data in Figure 3A, very little DACH1 could be immunoprecipitated from lean liver. Moreover, ChIP analysis revealed the presence of NCOR on the intronic region where DACH1 binds Atf6, and this ChIP signal was markedly diminished in DACH1-deficient cells (Figure 6E). As further evidence, we showed that AAV8-sh-NCOR-treated obese mice had higher hepatic Atf6 mRNA levels compared with control mice (Figure 6F). These combined data are consistent with a model in which DACH1-NCOR complex binds to DACH1 consensus sites in the Atf6 gene in HCs in obesity and represses Atf6.
HDAC4-Mediated DACH1 SUMOylation Leads to Its Degradation
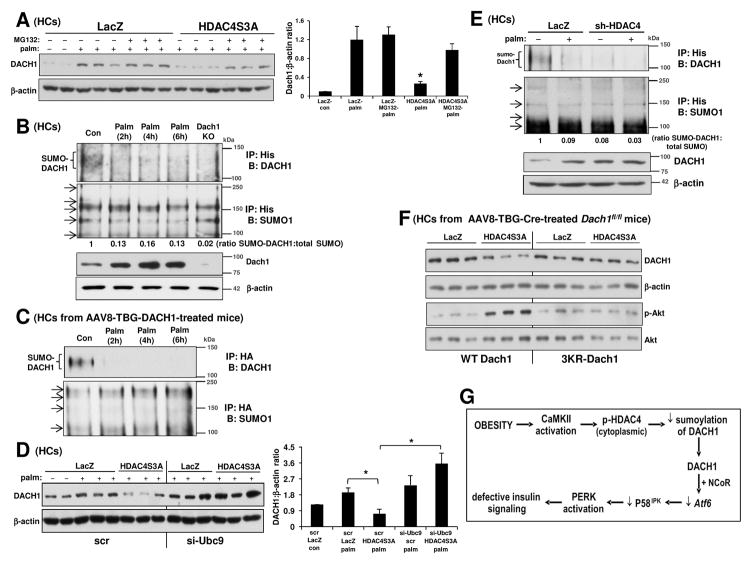
To elucidate the molecular mechanism of DACH1 suppression by nuclear HDAC4, we first analyzed the livers of obese mice treated with adeno-LacZ vs. adeno-HDAC4S3A and found no decrease in Dach1 mRNA in the HDAC4S3A cohort (Figure S7A). Similar results were obtained using palmitate-treated control vs. HDAC4S3A-transfected primary HCs (Figure S7B). Using the HC model, we then explored the possibility that nuclear HDAC4 increases the proteasomal degradation of DACH1 protein. Consistent with this idea, the proteasome inhibitor MG132 partially prevented the decrease in DACH1 protein conferred by HDAC4S3A transfection (Figure 7A).
Figure 7. HDAC4-Mediated SUMOylation of DACH1 Leads to Proteasomal Degradation of DACH1 and Defective Insulin Signaling in HCs.
(A) Primary HCs from WT mice were transduced with adeno-LacZ or adeno-HDAC4S3A. After 24 h, the cells were pretreated with either vehicle or MG132 for 1 h followed by incubation with either BSA or 0.3 mM palmitate (palm) for 6 h. Lysates were probed for DACH1 and β-actin by immunoblot. Densitometric quanti cation of the immunoblot data is shown in the bar graph (n = 3; mean ± SEM, *p < 0.05 vs. all other groups except LacZ-con).
(B) Lysates from WT or DACH1 KO HCs transfected with poly-His-tagged SUMO1 and treated with BSA (Con) or palmitate (palm) were immunoprecipitated using anti-poly-His and blotted for DACH1 or SUMO1. Arrows indicate SUMOylated proteins. The numbers below the SUMO1 immunoblot are the densitometric ratios of SUMO-DACH1:total SUMO. The lower blot shows DACH1 and β-actin in whole cell lysates.
(C) Primary HCs from WT mice treated with AAV8-TBG-DACH1 were transfected with HA-tagged SUMO1 and treated with BSA (Con) or palmitate (palm) as indicated. Lysates were immunoprecipitated using anti-HA and blotted for DACH1 or SUMO1. Arrows indicate SUMOylated proteins.
(D) HCs were pretreated with either scrambled RNA (scr) or siRNA targeting Ubc9 (si-Ubc9). After 12 h, the cells were transduced with adeno-LacZ or adeno-HDAC4S3A. After an additional 24 h, the cells were incubated with BSA control or 0.3 mM palmitate (palm) for 5 h as indicated by the minus and plus symbols. Lysates were then probed for DACH1 and β-actin by immunoblot. Densitometric quanti cation of the immunoblot data is shown in the bar graph (n = 3; mean ± SEM; *p < 0.05; bars 4 and 5 are not significantly different).
(E) Similar to (C) except that the cells were transduced with adeno-LacZ or adeno-sh-HDAC4 and lysates were immunoprecipitated using anti-His antibody.
(F) HCs from Dach1fl/fl mice treated with AAV-TBG-Cre were transduced with plasmids encoding WT-DACH1 or 3KR-mutant DACH1. After 12 h, the cells were transduced with adeno-LacZ or adeno-HDAC4S3A. After an additional 24 h, the cells were incubated with BSA control or 0.3 mM palmitate for 11 h, followed by insulin stimulation for 5 mins. Lysates were probed for DACH1, β-actin, p-Akt, and Akt.
(G) Summary scheme of CaMKII-HDAC4-DACH1 pathway linking obesity to defective insulin signaling in HCs. Based on the data in this report and our previous publications (Ozcan et al., 2013; Ozcan et al., 2012), obesity-induced activation of CaMKII in HCs phosphorylates HDAC4, which promotes its exit from the nucleus. As a result, nuclear SUMOylation of the corepressor DACH1 is decreased, which prolongs its half-life and increases its level in the nucleus. DACH1, together with NCOR, decreases the transcription of Atf6, leading to activation of the PERK-TRB3 pathway and defective insulin signaling, as described previously (Ozcan et al., 2013). See also Figure S7.
HDAC4 can act as an E3 ligase that affects protein SUMOylation (Gregoire and Yang, 2005; Zhao et al., 2005), which in turn can promote ubiquitination and proteasomal degradation of SUMOylated proteins (Miteva et al., 2010). To address whether a SUMOylation-dependent pathway was involved in the HDAC4-mediated suppression of DACH1, we first asked whether endogenous DACH1 gets SUMOylated. In palmitate-treated HCs, immunoprecipitation-Western blot (IP:WB) analysis revealed that DACH1 SUMOylation occurs and is diminished upon palmitate treatment, concomitant with an increase in DACH1 protein (Figure 7B). We obtained similar results when we overexpressed DACH1 in HCs (Figure 7C and S7C). Moreover, when we silenced the E2-conjugating enzyme Ubc9, which transfers the activated SUMO to protein substrates (Ghisletti et al., 2007; Hay, 2005), HDAC4-mediated DACH1 degradation was abrogated (Figure 7D). As a direct evidence that HDAC4 promotes DACH1 SUMOylation, we found that silencing HDAC4 abrogated DACH1 SUMOylation in primary HCs (Figure 7E).
Based on the consensus SUMO-acceptor site sequence (Rodriguez et al., 2001) (http://www.abgent.com/doc/sumoplot), we identified three high-probability sites for SUMOylation in murine DACH1: K341, K644, K676. In order to determine whether these lysine residues are targets for SUMOylation and degradation of DACH1, we created a mutant of DACH1 with lysine to arginine substitutions at these three sites (DACH1-3KR mutant) and transfected DACH1-deficient HCs with this construct. In non-palmitate-treated HCs, wild-type DACH1 was SUMOylated as before, but this was not seen with DACH1-3KR (Figure S7D). Furthermore, HDAC4S3A was unable to decrease DACH1 levels in the DACH1-3KR-transfected cells, in contrast to the situation with HCs expressing wild-type DACH1 (Figure 7F—top two blots). Most importantly, the improvement in insulin signaling conferred by HDAC4S3A in palmitate-treated HCs was abrogated when the cells expressed 3KR-DACH1 instead of WT DACH1 (Fig 7F—bottom two blots). These combined results show that nuclear HDAC4 decreases DACH1 through SUMOylation and subsequent proteasome-mediated degradation. With this finding, we present a summary of the pathway in which CaMKII activation in HCs in obesity leads to defective insulin signaling (Figure 7G).
DISCUSSION
Understanding how ATF6 is suppressed in HCs in obesity is a critical issue in view of its importance in both insulin signaling, supported by our previous study (Ozcan et al., 2013) and bolstered here, and HGP (Wang et al., 2009). Additionally, two independent studies reported associations between variants of the ATF6 gene in humans and disturbed glucose homeostasis and T2D (Meex et al., 2007; Thameem et al., 2006). The mechanism of ATF6 suppression revealed here involves a very interesting pathway in which the half-life of the corepressor DACH1 is prolonged, leading to decreased transcription of the Atf6 gene. Even though the key step in ATF6 activation is its post-translational proteolytic cleavage and nuclear translocation (Ye et al., 2000), the data here imply that the level of Atf6 transcription expression becomes rate-limiting in HCs in the setting of obesity.
Our work shows that decreased levels of nuclear HDAC4 in HCs in obesity indirectly lowers Atf6 transcription by increasing the level of DACH1. In this context, a recent paper using an unbiased proteomics approach found that obesity in humans is associated with a decrease in HDAC4, which improves with physical exercise, suggesting the possibility that nuclear HDAC4 is protective in obesity (Abu-Farha et al., 2013). In contrast, Mihaylova et al. reported that combined silencing of hepatic HDAC4, 5, and 7 increases FoxO1 acetylation in fasted lean mice and lowers HGP in lean and obese mice (Mihaylova et al., 2011). In this regard, we found no difference in the level of acetylated FoxO1 in the livers of obese mice treated with constitutively nuclear HDAC4 or HDAC4 shRNA. Moreover, while the major endpoint of the Mihaylova et al. study was blood glucose levels, the study here focused on liver insulin signaling and whole body insulin sensitivity.
HDAC4, like other class IIa HDACs (5, 7, and 9), exhibit a very low deacetylase activity because of the substitution of a catalytic Tyr with His (Lahm et al., 2007). As such, the role of HDAC4 shown here is related its ability to SUMOylate and thereby promote the proteasomal degradation of DACH1 when HDAC4 is nuclear, e.g., in the lean state. Previous studies have shown that HDAC4 can serve as SUMO E3 ligase for a number of target proteins in various cell types (Ghisletti et al., 2007; Gregoire and Yang, 2005) and that SUMOylation can serve as a priming process for ubiquitination, leading to protein ubiquitination and proteasomal degradation of the target proteins (Lee et al., 2014; Liu et al., 2013; Miteva et al., 2010). Consistent with the findings here, only the nuclear form of HDAC4 possesses SUMO E3 ligase activity (Lee et al., 2009; Yao and Yang, 2011). Of note, DACH1 can be affected by other post-translational modifications that affect its function, including acetylation and phosphorylation (Chen et al., 2013; Wu et al., 2014), and thus it will be interesting to examine in the future whether these modifications of DACH1 occur in HCs in obesity.
Class IIa HDACs can affect gene transcription by other mechanisms, but none of these appear to be involved in the pathway described here. For example, HDAC4 can effect gene repression by recruiting HDAC3 and its coregulators to the nucleus, which repress gene transcription (Fischle et al., 2002). However, we found that siRNA mediated-silencing of HDAC3 did not suppress Atf6 transcription in palmitate-treated CaMKII-deficient HCs (data not shown). Consistent with the lack of a role of HDAC3 in the pathway presented here, a recent study showed that when HDAC3 is silenced in HCs in obese mice, metabolic precursors are rerouted away from glucose production into lipid synthesis, causing steatosis and improved insulin sensitivity (Sun et al., 2012).
Most studies on DACH1 have focused on its roles in development and tumorigenesis (Popov et al., 2010). Ironically, one of the developmental roles is related to perinatal pancreatic beta cell proliferation, which is associated with DACH1-mediated repression of p27Kip1 (Kalousova et al., 2010). The hypothesis that DACH1 is required for normal insulin secretion was suggested as a mechanism to explain a link between a DACH1 polymorphism and diabetes in a Chinese population (Ma et al., 2014), but replication in additional populations and direct links to beta cell function are needed to substantiate this claim. In contrast, the findings here show a non-developmental role of liver DACH1 as a detrimental factor in obesity-associated insulin resistance. Accordingly, silencing of DACH1 in the livers of obese mice, by improving insulin signaling in HCs and improving overall metabolism, corrects hyperinsulinemia. Moreover, we found a striking correlation between hepatic DACH1 levels and BMI in humans.
We provide evidence that suppression of Atf6 in HCs in obesity involves DACH1-NCOR complex. However, this finding does not imply that hepatic NCOR silencing in obese mice would mimic the metabolic improvement seen with hepatic DACH1 silencing, because NCOR has functions in the liver that are independent of DACH1. For example, NCOR in complex with HDAC3 represses genes involved in hepatic lipogenesis, and thus one of the effects of HC-targeted NCOR deletion is steatosis (Sun et al., 2013). In contrast, hepatic steatosis is improved by liver-targeted silencing of DACH1 (not shown) or its upstream effector, CaMKII (Ozcan et al., 2013).
In summary, our study reveals a role for DACH1 in obesity-induced glucose intolerance and insulin resistance in mice, with a striking correlation between liver DACH1 level and obesity in humans. While DACH1 upregulation in obesity may affect a number of different mechanisms, its ability to suppress insulin signaling by repressing Atf6 transcription appears to be dominant. Nonetheless, future studies will likely reveal additional DACH1 targets in HCs that may shed additional light on gene expression changes in obesity. Finally, we recently published that drug-mediated inhibition of MK2, an enzyme linked to CaMKII in HCs in obesity, improves metabolism in obese mice (Ozcan et al., 2015), which is consistent with genetic studies conducted by our group (Ozcan et al., 2013) and an independent laboratory (Ruiz et al., 2015). Given that the upstream kinase pathway promotes diabetes by suppressing ATF6, the findings in this report provide important mechanistic underpinnings for future therapeutic strategies that attempt to improve metabolism in obese T2D subjects by targeting this pathway.
EXPERIMENTAL PROCEDURES
Mouse Experiments
Ob/ob and diet-induced obese (DIO) mice were obtained from Jackson Labs. DIO mice were fed a high-fat diet with 60% kcal from fat (Research Diets) and maintained on a 12 h-light-dark cycle. Dach1fl/fl mice were generated as described previously (Chen et al., 2015). The detailed procedures are shown in the Supplemental Information.
Primary HCs
Primary mouse HCs were isolated from 8- to 12-week-old mice as described previously (Ozcan et al., 2012). Unless indicated otherwise, the cells were cultured in DMEM containing 10% fetal bovine serum, treated as described in the figure legends, and then incubated for 5 h in serum-free DMEM before harvesting. The detailed procedures are shown in the Supplemental Information.
Statistical Analysis
All results are presented as mean ± SEM. P values were calculated using the Student’s t-test for normally distributed data and the Mann-Whitney rank sum test for non-normally distributed data. One-way ANOVA with post-hoc Tukey test was used to evaluate differences among groups when 3 or more groups were analyzed.
Supplementary Material
Acknowledgments
We thank Dr. Harold A. Singer (Albany Medical College) for adeno-LacZ and K43A-CaMKII; Dr. Marc Montminy (Salk Institute for Biological Studies) for adeno-sh-ATF6 and -ATF6-N; Dr. Eric Olson (UT Southwestern) for adeno-HDAC4S3A; Dr. Reuben Shaw (Salk Institute for Biological Studies) for adeno-sh-HDAC4; and Dr. Tso-Pang Yao (Duke University) for anti-pSer-467-HDAC4 antibody. This work was supported by an American Heart Association Scientist Development Grant (11SDG5300022) and a NYONRC Pilot and Feasibility Grant (DK26687) to L.O.; by a São Paulo Research Foundation grant (FAPESP/BEPE 2012/21290-4) to J.C.S; and by NIH grants CA132115-05A1 to R.P. and HL087123 and HL075662 to I.T. Authors L.O. and I.T. are in the group of cofounders of Tabomedex Biosciences LLC, which is developing inhibitors of the upstream kinases in the pathway described in this report for the treatment of T2D.
Footnotes
Supplemental Information includes seven figures.
Author Contributions: L.O. and I.T. designed the research. L.O., D.S.G., Z.Z., and J.C.S. conducted the research. L.O., D.S.G., Z.Z., J.C.S., K.C., M.Bessler, M.Bagloo., B.S., R.P. and I.T. analyzed the data. L.O., R.P., and I.T. wrote the manuscript. K.C. and R.P. contributed new reagents and analytical tools. M.Bessler, M.Bagloo, and B.S. organized patient recruitment and human liver sample collection.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Farha M, Tiss A, Abubaker J, Khadir A, Al-Ghimlas F, Al-Khairi I, Baturcam E, Cherian P, Elkum N, Hammad M, et al. Proteomics analysis of human obesity reveals the epigenetic factor HDAC4 as a potential target for obesity. PLoS ONE. 2013;8:e75342. doi: 10.1371/journal.pone.0075342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, She P, Mozzoli M, Cheung P, Gumireddy K, Reddy P, Xiang X, Luo Z, Ruderman N. Free fatty acids produce insulin resistance and activate the proinflammatory nuclear factor-kappaB pathway in rat liver. Diabetes. 2005;54:3458–3465. doi: 10.2337/diabetes.54.12.3458. [DOI] [PubMed] [Google Scholar]
- Chen K, Wu K, Gormley M, Ertel A, Wang J, Zhang W, Zhou J, Disante G, Li Z, Rui H, et al. Acetylation of the cell-fate factor dachshund determines p53 binding and signaling modules in breast cancer. Oncotarget. 2013;4:923–935. doi: 10.18632/oncotarget.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wu K, Jiao X, Wang L, Ju X, Wang M, Di Sante G, Xu S, Wang Q, Li K, et al. The endogenous cell-fate factor dachshund restrains prostate epithelial cell migration via repression of cytokine secretion via a cxcl signaling module. Cancer Res. 2015;75:1992–2004. doi: 10.1158/0008-5472.CAN-14-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Kalousova A, Mavropoulos A, Adams BA, Nekrep N, Li Z, Krauss S, Stainier DY, German MS. Dachshund homologues play a conserved role in islet cell development. Dev Biol. 2010;348:143–152. doi: 10.1016/j.ydbio.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumashiro N, Erion DM, Zhang D, Kahn M, Beddow SA, Chu X, Still CD, Gerhard GS, Han X, Dziura J, et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GY, Jang H, Lee JH, Huh JY, Choi S, Chung J, Kim JB. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol Cell Biol. 2014;34:926–938. doi: 10.1128/MCB.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, Joe EH, Jou I. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell. 2009;35:806–817. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang YD, Guo L, Huang HY, Zhu H, Huang JX, Liu Y, Zhou SR, Dang YJ, Li X, et al. Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein beta (C/EBPbeta) during adipogenesis. Mol Cell Biol. 2013;33:4606–4617. doi: 10.1128/MCB.00723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma RC, Lee HM, Lam VK, Tam CH, Ho JS, Zhao HL, Guan J, Kong AP, Lau E, Zhang G, et al. Familial young-onset diabetes, pre-diabetes and cardiovascular disease are associated with genetic variants of DACH1 in Chinese. PLoS ONE. 2014;9:e84770. doi: 10.1371/journal.pone.0084770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Meex SJ, van Greevenbroek MM, Ayoubi TA, Vlietinck R, van Vliet-Ostaptchouk JV, Hofker MH, Vermeulen VM, Schalkwijk CG, Feskens EJ, Boer JM, et al. Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch Caucasians. J Clin Endocrinol Metab. 2007;92:2720–2725. doi: 10.1210/jc.2006-2280. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva M, Keusekotten K, Hofmann K, Praefcke GJ, Dohmen RJ. Sumoylation as a signal for polyubiquitylation and proteasomal degradation. Subcell Biochem. 2010;54:195–214. doi: 10.1007/978-1-4419-6676-6_16. [DOI] [PubMed] [Google Scholar]
- Ozcan L, Cristina de Souza J, Harari AA, Backs J, Olson EN, Tabas I. Activation of calcium/calmodulin-dependent protein kinase II in obesity mediates suppression of hepatic insulin signaling. Cell Metab. 2013;18:803–815. doi: 10.1016/j.cmet.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan L, Wong CC, Li G, Xu T, Pajvani U, Park SK, Wronska A, Chen BX, Marks AR, Fukamizu A, et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15:739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan L, Xu X, Deng SX, Ghorpade DS, Thomas T, Cremers S, Hubbard B, Serrano-Wu MH, Gaestel M, Landry DW, et al. Treatment of Obese Insulin-Resistant Mice With an Allosteric MAPKAPK2/3 Inhibitor Lowers Blood Glucose and Improves Insulin Sensitivity. Diabetes. 2015;64:3396–3405. doi: 10.2337/db14-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajvani UB, Accili D. The new biology of diabetes. Diabetologia. 2015;58:2459–2468. doi: 10.1007/s00125-015-3722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra M. Class IIa HDACs - new insights into their functions in physiology and pathology. FEBS J. 2015;282:1736–1744. doi: 10.1111/febs.13061. [DOI] [PubMed] [Google Scholar]
- Popov VM, Wu K, Zhou J, Powell MJ, Mardon G, Wang C, Pestell RG. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol Metab. 2010;21:41–49. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Ruiz M, Coderre L, Lachance D, Houde V, Martel C, Legault JT, Gillis MA, Bouchard B, Daneault C, Carpentier AC, et al. Mk2 Deletion In Mice Prevents Diabetes-Induced Perturbations In Lipid Metabolism And Cardiac Dysfunction. Diabetes. 2015 doi: 10.2337/db15-0238. [DOI] [PubMed] [Google Scholar]
- Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram K, Mani SK, Kitatani K, Wu K, Pestell RG, Reddy SV. DACH1 negatively regulates the human RANK ligand gene expression in stromal/preosteoblast cells. J Cell Biochem. 2008;103:1747–1759. doi: 10.1002/jcb.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde JS, Donninger H, Wu K, Johnson ME, Pestell RG, Rose GS, Mok SC, Brady J, Bonome T, Birrer MJ. Expression profiling identifies altered expression of genes that contribute to the inhibition of transforming growth factor-beta signaling in ovarian cancer. Cancer Res. 2006;66:8404–8412. doi: 10.1158/0008-5472.CAN-06-0683. [DOI] [PubMed] [Google Scholar]
- Tang H, Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc Natl Acad Sci USA. 2006;103:16977–16982. doi: 10.1073/pnas.0601565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Macpherson P, Marvin M, Meadows E, Klein WH, Yang XJ, Goldman D. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol Biol Cell. 2009;20:1120–1131. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Shen H, Chen J, Wang X, Zhang Y, Chen LL, Rukachaisirikul V, Jiang HL, Shen X. Activating transcription factor 6 protects insulin receptor from ER stress-stimulated desensitization via p42/44 ERK pathway. Acta Pharmacol Sin. 2011;32:1138–1147. doi: 10.1038/aps.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thameem F, Farook VS, Bogardus C, Prochazka M. Association of amino acid variants in the activating transcription factor 6 gene (ATF6) on 1q21-q23 with type 2 diabetes in Pima Indians. Diabetes. 2006;55:839–842. doi: 10.2337/diabetes.55.03.06.db05-1002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wu K, Chen K, Wang C, Jiao X, Wang L, Zhou J, Wang J, Li Z, Addya S, Sorensen PH, et al. Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. 2014;74:829–839. doi: 10.1158/0008-5472.CAN-13-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Katiyar S, Li A, Liu M, Ju X, Popov VM, Jiao X, Lisanti MP, Casola A, Pestell RG. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc Natl Acad Sci USA. 2008;105:6924–6929. doi: 10.1073/pnas.0802085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Yang Y, Wang C, Davoli MA, D’Amico M, Li A, Cveklova K, Kozmik Z, Lisanti MP, Russell RG, et al. DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J Biol Chem. 2003;278:51673–51684. doi: 10.1074/jbc.M310021200. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM. Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J Biomed Biotechnol. 2011;2011:146493. doi: 10.1155/2011/146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kohlhaas M, Backs J, Mishra S, Phillips W, Dybkova N, Chang S, Ling H, Bers DM, Maier LS, et al. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem. 2007;282:35078–35087. doi: 10.1074/jbc.M707083200. [DOI] [PubMed] [Google Scholar]
- Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang C, Wang Z, Dampier W, Wu K, Casimiro MC, Chepelev I, Popov VM, Quong A, Tozeren A, et al. Attenuation of Forkhead signaling by the retinal determination factor DACH1. Proc Natl Acad Sci USA. 2010;107:6864–6869. doi: 10.1073/pnas.1002746107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.