Abstract
The expansion of species ranges frequently necessitates responses to novel environments. In plants, the ability of seeds to disperse to marginal areas relies in part to its ability to germinate under stressful conditions. Here we examine the genetic architecture of Arabidopsis thaliana germination speed under a novel, saline environment, using an Extreme QTL (X-QTL) mapping platform we previously developed. We find that early germination in normal and salt conditions both rely on a QTL on the distal arm of chromosome 4, but we also find unique QTL on chromosomes 1, 2, 4, and 5 that are specific to salt stress environments. Moreover, different QTLs are responsible for early vs. late germination, suggesting a temporal component to the expression of life history under these stress conditions. Our results indicate that cryptic genetic variation exists for responses to a novel abiotic stress, which may suggest a role of such variation in adaptation to new climactic conditions or growth environments.
Keywords: salinity tolerance, salt stress, QTL, bulk segregant analysis, abiotic stress
Germination is a key life history stage in plants. In the model species Arabidopsis thaliana, germination has been shown to determine key aspects of organismal fitness (Donohue 2002; Donohue et al. 2010, 2005b). Seasonal cueing of germination, for example, has been shown to have major consequences both for shaping life history strategies in this species and for local adaptation (Donohue 2002; Donohue et al. 2005a). One key germination trait is germination speed (Yuan et al. 2016), i.e., the time lapse for a nondormant seed to germinate upon the reception of cue, or the time lapse for an isogenic seed lot to reach 50% germination. Though little is known about germination speed in A. thaliana, comparative studies with various other species correlates this trait with fitness under fluctuating environments (Parsons 2012), competitiveness (Miller et al. 1994; Weinig 2000; Dubois and Cheptou. 2012), and establishment during range expansion (Graebner et al. 2012; Beckman et al. 2011). In a previous study, we demonstrated that germination speed is variable among A. thaliana accessions, and identified QTL for this trait using an Extreme QTL (X-QTL) mapping strategy (Yuan et al. 2016).
As sessile organisms, plants rely on seed dispersal to alleviate resource competition and seed predation in local environments. Seed dispersal also enables migration to novel resource patches. Given the ability of plants to disperse their seeds, they can presumably encounter novel environments to which they have had no previous adaptive history. Should a seed manage to germinate in these novel environments, the habitat can determine the strength and types of selection the plant must experience throughout all stages of its life cycle.
Response to novel environments underlies the process of local adaptation, range expansion, and ecological speciation (Atwell et al. 2014). Recent studies have focused on characterizing the types of traits that serve as the primary targets of selection under novel environments (Atwell et al. 2014; Dutilleul et al. 2014; Handelsman et al. 2013). Life history traits and the genotype-by-environment (G×E) interactions in the expression of life history stages are believed to be crucial in determining the likely path of adaptation and range expansion (Handelsman et al. 2013; Seiter and Kingsolver 2013; Cameron et al. 2013). However, relatively little is known regarding the genetics of life history traits in response to novel environments, including the types of genes that are likely under selection. Understanding the genetics of novel environmental response is important in examining how selection acts when organisms disperse to new environments at the edges of species ranges, or experience new climactic regimes.
The ability to respond to novel environment differs within natural populations, depending on the amount of standing genetic variation for the trait under selection (Volis et al. 2016; Etterson and Shaw 2001). Standing genetic variation can underlie phenotypic diversity, but recently it has been shown that cryptic genetic variation can be important under certain environments or in genetic backgrounds (Paaby and Rockman 2014; McGuigan et al. 2011; Jarosz and Lindquist 2010; Hayden et al. 2011). The lack of selection on cryptic genetic variation in native environments allows them to accumulate within populations (Paaby and Rockman 2014), and enable them to respond rapidly to selective pressures under novel environments.
Salinity is an abiotic soil condition caused by natural processes such as mineral weathering, past or present proximity to coastal waters, or through anthropogenic processes such as irrigation (Rhoades and Loveday 1990). Worldwide, saline soils are found over ∼323 million hectares, or ∼7% of global land area (Lynch and Clair 2004). Soil salinity is rapidly becoming a major agricultural issue, but for wild species it also represents a marginal soil environment, which can exert strong adaptive pressure (Assman 2013; Lefebvre et al. 2009). The model wild species A. thaliana, for example, is not known to have widespread adaptation to saline environment, though as a colonizing species, there is a possibility for an A. thaliana individual and/or population to encounter a saline environment. Moreover, local adaptation in coastal regions was shown to be possible (Rus et al. 2005; Baxter et al. 2010). Salinity thus represents a novel but relevant environment for most Arabidopsis populations.
Relatively little is known about salinity tolerance during the germination stage in A. thaliana, although evidence suggests that the genetic mechanism of salt tolerance during germination is quite different from the vegetative growth stage (Quesada et al. 2002; Lefebvre et al. 2009). Several studies have identified different genomic regions responsible for salt tolerance in both germination and vegetative growth stages (for example, Clerkx et al. 2004; DeRose-Wilson and Gaut 2011; Joosen et al. 2010; Vallejo et al. 2010; Guo et al. 2016; Galpaz and Reymond 2010; Quesada et al. 2002; Rus et al. 2005; Baxter et al. 2010). Moreover, RAS1, a large-effect negative regulator of germination salt tolerance, was cloned in A. thaliana (Ren et al. 2010). This gene is distinct from the AtHKT1 locus, which is a salinity tolerance gene during vegetative growth (Rus et al. 2005).
In this study, we examine cryptic genetic variation for germination speed under novel high-salt environments by mapping germination speed QTL in A. thaliana using the X-QTL approach (Ehrenreich et al. 2010; Yuan et al. 2016). X-QTL mapping applies bulk segregant analysis and high-throughput genotyping to very large mapping populations (∼105–106 individuals), and can provide greater power in detecting moderate- to small-effect QTL (Ehrenreich et al. 2010; Yuan et al. 2016). By identifying germination speed QTL under saline environments and comparing them to loci found in normal growth conditions (Yuan et al. 2016), we can begin to dissect the genetic architecture of A. thaliana in response to a novel and ecologically important environment. We can then catalog the extent to which cryptic genetic variation contributes to potential life history adaptation to a stressful environment, and uncover these variants through induced artificial selection.
Materials and Methods
Plant materials
The mapping population was generated using Bs-2 (CS6628) and Col-0 (CS6673) accessions of A. thaliana as parental accessions. For plant cultural conditions, seed storage condition, and a detailed description of the mapping population see Yuan et al. (2016).
Phenotyping the germination stages
Germination sensu stricto is defined as the moment when the radicle (embryonic root) ruptures the endosperm and protrudes through the seed coat. However, for the convenience of our study, we used a phase early in seedling growth, cotyledon expansion, as our marker for germination. All our ensuing usage of the word “germination” therefore refers to cotyledon expansion. For each parental accession, 50 mg (∼2000) dry seeds were evenly spread onto an 80-cm2 gridded square petri dish (Fisher brand) containing Murashige–Skoog basal solid media with or without 250 mM NaCl supplement. A sterilized 500-μm nylon mesh was applied to the interface of the medium and the seeds to prevent seed clumping. Three square grid units (1.74 cm2 each) were randomly chosen from each plate, and the germination stage of individual seeds within the chosen grid units were closely examined under a stereoscopic microscope (Stereozoom3; Bausch & Lomb) using 10× magnification at 6-hr time intervals. The number of seeds reaching the testa rupture, radicle protrusion, cotyledon greening, and cotyledon expansion phases at each time point were recorded.
Phenotyping germination speed
Triplicate scoring was carried out for each accession under both no-salt and salt (250 mM NaCl) conditions, and each replicate contained 50 mg (∼2000) dry seeds. Pretreatment, plating, and stratification followed Yuan et al. (2016). Germination was scored by visually identifying, counting, and removing germinants that reached the cotyledon expansion phase. For the no-salt condition the scoring lasted for 5 d at 6-hr intervals, and for saline condition, daily for 15 consecutive days.
Selection experiment and mapping
Selection for salt-tolerant germinants was carried out in duplicate of ∼100,000 F3 populations. Seeds were harvested, after-ripen, sterilized, plated, stratified, and placed into the chamber conditions as described in Yuan et al. (2016). Germinants reaching the cotyledon expansion phase were visually identified, counted, and collected from the plate daily. The sampling process lasted 15 d for each replicate, until very few (n < 100) individuals germinated each day. Approximately 23% of F3 seeds (23,000 individuals) had germinated by the end of the experiment.
The germinants were grouped into early [0–7 d after stratification (DAS); ∼0-5th percentile of total seed], medium (8–12 DAS; ∼5th–20th percentile of total seed), and late (13–15 DAS; ∼20th–23rd percentile of total seed) cohorts, each corresponding to the lagging, linear, and plateau phase of the sigmoidal cumulative germination curve. DNA was extracted from each of the pooled cohorts, and a fourth pool consisting of total DNA of all ∼23,000 F3 salt-tolerant germinants was made by proportionally mixing together three DNA pools according to their DNA concentration and the number of individuals in each pool.
We mapped for salt-tolerance germination QTL using the X-QTL method (Ehrenreich et al. 2010) as developed for A. thaliana (Yuan et al. 2016). We determined allele frequency for 30,389 single nucleotide polymorphisms (SNPs) using a custom-designed isothermal Agilent microarray DNA chip we previously developed for A. thaliana (Yuan et al. 2016). Dye-swap hybridization was carried out for each of the four pools, with pool-extracted DNA from ∼2000 random F2 recombinants (equal in size to the founder population, and at the same germination stage) as reference. Random F2 individuals were used as reference to control for the possible segregation distortion in the founder population that might have biased our allele frequency estimates at specific SNP markers.
The microarray-based allele frequency estimate of each salt-tolerant germinant DNA pool was compared against that of a control DNA pool of ∼100,000 F3 germinants, sampled under no-salt conditions, using a sliding window, three-way nested ANOVA model described previously (Yuan et al. 2016). Statistical significance for QTL peaks was established via permutation. We then identified SNPs above the significance threshold, and defined QTL as regions with consecutive SNPs that exceed the threshold. In cases (e.g., peak cluster on chromosome 4) where two or more groups of consecutively significant SNPs were <1 Mbp apart, the entire region was considered as one large QTL.
Germination curve fitting
Raw cumulative germination fraction of the parental accessions and the F3 mapping populations at each time point were used to fit a four-parameter Hill function (4PHF) models (El-Kassaby et al. 2008; Joosen et al. 2010). The parameters for each independent experiment were solved using the nls() function in R (R Core Team 2014) package chemCal (Ranke 2014), and with the least sum-of-squares method. Germination speed for each accession was calculated by averaging the final estimates of time to reach 50% germination from all three replicates.
Data availability
Bulk-segregant microarray genotyping data of early, medium and late salt-tolerant germination cohorts, and the entire salt germinant pool are available in Supplemental Material, File S1.
Results
Salinity increases the difference in germination speed between parental accessions
An F3 mapping population was created by crossing two natural accessions: Col-0 (CS6673) and Bs-2 (CS6628). Neither of the parental accessions originated from a coastal area or a known inland saline area; Bs-2 being from Basel, Switzerland and Col-0 being genetically closest to populations from Gückingen, Germany (Nordborg et al. 2005). Therefore it is unlikely that either accession came from a population that had prior adaptation history to salinity, and salinity likely represents a novel abiotic environment for these two accessions. Despite this, Quesada et al. (2002) reported significant difference in germination salt tolerance between the accessions, with Bs-2 being tolerant and Col-0 being sensitive to salt (250 mM NaCl) treatment on soil at the germination stage.
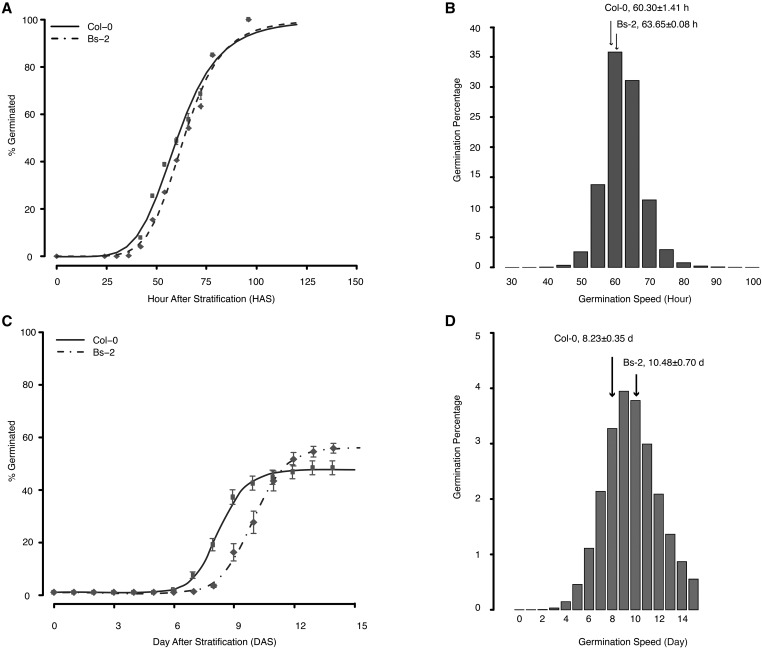
We measured the germination speed of these two accessions under no-salt (Murashige–Skoog basal solid media) and extremely saline (Murashige–Skoog basal solid media supplemented with 250 mM NaCl) conditions. The latter is equivalent to 42% of seawater, and would be considered a highly stressful abiotic environment. As reported in our previous publication, the accessions exhibited significant yet small differences in their germination speed under no-salt conditions (difference: 3.35 hr, P < 0.05; Figure 1A, data from Yuan et al. 2016).
Figure 1.
Germination speed of the founder accessions and their F3 populations under no-salt condition and extreme salinity. (A) Cumulative germination percentage of the founder accessions (Bs-2 and Col-0). Gray dots show the percentage germinated at each sampling time point, with the center of the dot indicating mean germination percentage, and the error bar showing the SE. Where error bars cannot be seen, they are because the size of the error bars at those time points are smaller than the size of the dots. The black lines show the 4PHF fitting result. Raw data were fitted to 4PHF with 100 maximum iteration and least sum-of-square method. Data from Yuan et al. (2016). (B) Histogram showing the distribution of germination speed in ∼100,000 Bs-2 × Col-0 F3 population. Data for plotting the histogram was derived from fitting raw F3 germination data to 4PHF. The germination speed of Bs-2 and Col-0 were indicated by the arrows. The F3 population exhibits transgressive segregation in germination speed Data from Yuan et al. (2016). (C) Cumulative germination percentage of the founder accessions under extreme salinity (250 mM NaCl). Compared to germination under no-salt condition (Figure 1A), both accessions showed significant delay in onset of germination (P < 2.2 × 10−16), reduction in final germination percentage (P = 5.3 × 10−10), and a greater difference between their germination speed (P = 2.7 × 10−9). (D) Histogram showing the distribution of germination speed in ∼100,000 F3 population under extreme salinity. The higher tail was cut off at the end of the experiment. The transgressive segregation of germination speed under salt is evident in the F3 population.
The addition of salt dramatically delayed the onset of germination of both parental accessions from ∼30 hr after stratification to ∼5–6 DAS, and drastically reduced the final germination percentage to ∼40–50%. More importantly, the saline environment amplified the difference in germination speeds between the accessions, from ∼3 hr to 2.25 d (Figure 1C). Both genotype effect and G×E effect are significant (Pgenotype = 2.71 × 10−9; PG×E = 1.95 × 10−9, two-way ANOVA) in explaining the difference in germination speed observed between parents under different treatments.
To examine the germination behavior in the F3 population, ∼100,000 F3 seeds were evenly spread onto the surface of Murashige–Skoog basal solid media supplemented with 250 mM NaCl at a density of ∼40 seeds/cm2. Germinants reaching the cotyledon expansion phase were counted daily and removed from the plate. The germination speed of the F3 population exhibited transgressive segregation (Figure 1D), with sporadic germinants observed as early as 3 DAS. Approximately 23,000 germinants (23%) were collected in a 15-d period, and scoring of germination stopped at 15 d as germination reached a plateau phase.
The increase in germination speed variation is via differential timing of each developmental stage
For the convenience of our study, we use a phase early in seedling growth, cotyledon expansion, as our marker for germination. According to our expansive definition, the germination process thus consists of several physiological/developmental stages: testa rupture, radicle protrusion, cotyledon greening, and cotyledon expansion.
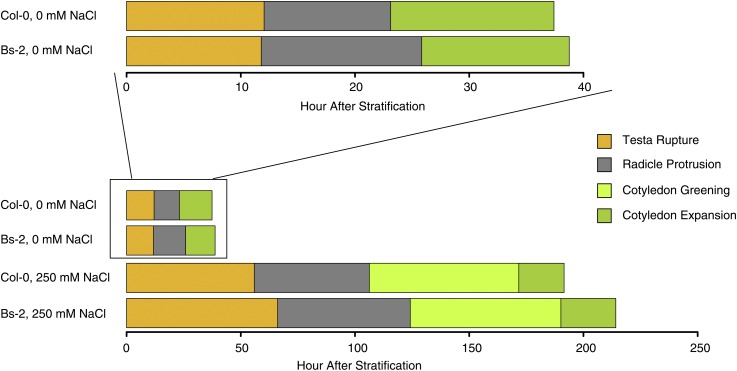
The timing for each of the four stages during the germination process for the parental accessions is measured in both no-salt and saline conditions (Figure 2). In no-salt conditions, both accessions reached 100% germination at the end of the experiment, and the schedule of testa rupture was the same between Bs-2 and Col-0 (Student’s t-test, P > 0.62; see Table 1). The Bs-2 accession, however, showed later progression into radicle protrusion and cotyledon expansion stages (Student’s t-test, P < 0.005). The delay in radicle protrusion and cotyledon expansion in Bs-2 likely explains the overall slower germination for this accession compared to Col-0.
Figure 2.
Schedule and duration of four stages during germination in the founder populations. The mean time for Bs-2 and Col-0 seeds sown onto no-salt or salt (250 mM NaCl) media plates to reach testa rupture (gold), radicle protrusion (gray), cotyledon greening (light green), and cotyledon expansion (dark green) are shown. The cotyledon greening phase was too transient to record with our time interval under no-salt conditions. The top panel shows a magnified view of the no-salt condition, under which only the schedules of radicle protrusion and cotyledon expansion were significantly different between the accessions (P < 0.005), while schedules of all four stages were significantly different between the accessions under saline condition (P < 0.05).
Table 1. Cumulative time (in hours) for the founder populations to reach four distinct phases during germination under no-salt and saline conditions.
| Condition | Stage | Accession | Average/hr | SD | P Value |
|---|---|---|---|---|---|
| 0 mM NaCl (no-salt) | Testa rupture | Bs-2 | 11.789 | 0.227 | 0.6211 |
| Col-0 | 12.043 | 0.79 | |||
| Radicle protrusion | Bs-2 | 25.836 | 0.5 | 0.0011** | |
| Col-0 | 23.116 | 0.25 | |||
| Cotyledon expansion | Bs-2 | 38.761 | 0.348 | 0.0036** | |
| Col-0 | 37.419 | 0.152 | |||
| 250 mM NaCl (salt) | Testa rupture | Bs-2 | 66.133 | 1.62 | 0.0004*** |
| Col-0 | 55.907 | 0.235 | |||
| Radicle protrusion | Bs-2 | 124.202 | 1.687 | 0.0085* | |
| Col-0 | 106.282 | 6.209 | |||
| Cotyledon greening | Bs-2 | 190.125 | 1.957 | 0.0177* | |
| Col-0 | 171.707 | 8.846 | |||
| Cotyledon expansion | Bs-2 | 214.168 | 0.59 | 0.0016** | |
| Col-0 | 191.542 | 5.125 |
P < 0.05; ** P < 0.005; *** P < 0.0005.
The salinity-induced difference in germination speed between accessions is likely the combined effects of timing differences through all the developmental stages. In saline conditions, the difference in timing of every single stage of development was amplified between the two parental accessions, with the Bs-2 accession consistently exhibiting slower progress through the germination stages (Student’s t-test, P < 0.05 for all stages; Table 1). Notably, under extreme salt stress, the germination of many plants in both accessions was arrested at the cotyledon greening phase without ever progressing into cotyledon expansion. The ratio of individuals at cotyledon greening phase vs. cotyledon expansion phase approached a constant value for Col-0 at 6 d (∼40% greening vs. 45% expansion), and for Bs-2 at 9 d (∼30% greening vs. 60% expansion). Also, the slower germinating Bs-2 accession exhibited acceleration in cotyledon greening after 6 d, exceeding Col-0 in the fraction of seeds that reached this germination stage, and later on exhibited a greater percentage of plants reaching cotyledon expansion.
Germination speed QTL under salinity stress
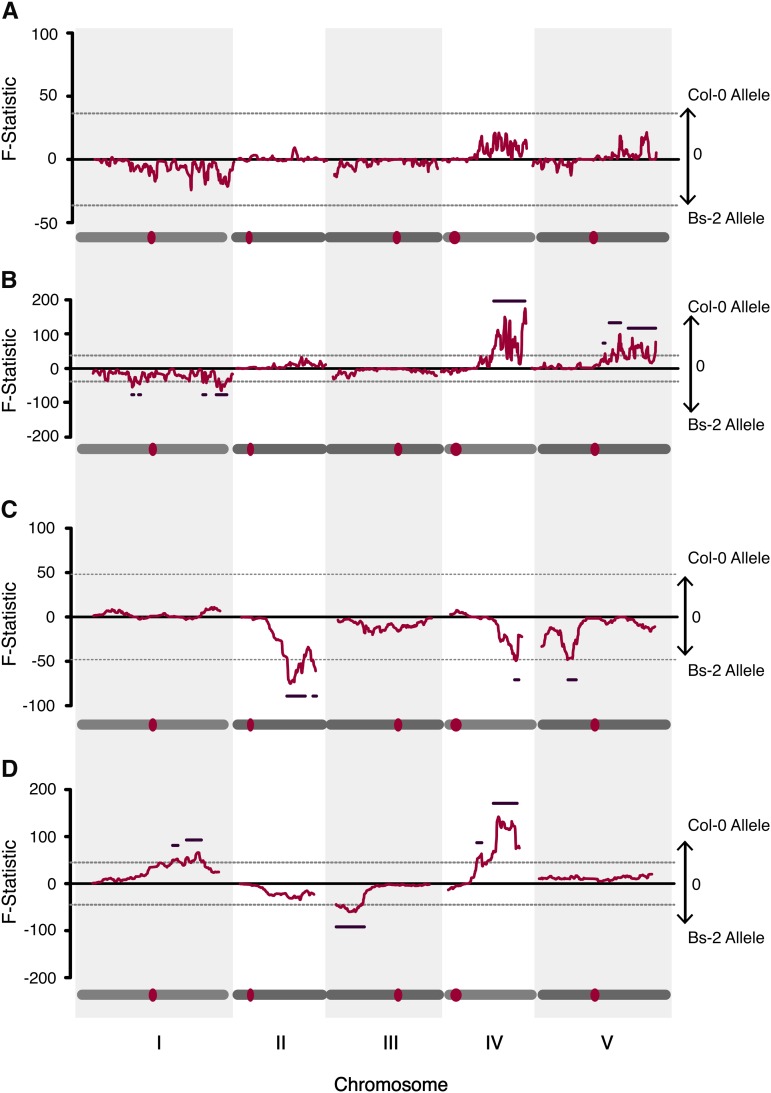
In a previous study, we developed an X-QTL mapping platform for A. thaliana and used this to identify QTL for germination speed under no-salt conditions. To compare germination speed QTL between no-salt and salt stress conditions, we collected the early germinants (first 5% of F3 individuals) under salt conditions in our experiment. The DNA of this early germinant pool was extracted and hybridized to a custom isothermal SNP microarray (Yuan et al. 2016) to quantify allele frequency changes that are associated with QTL for germination speed under saline conditions (Figure 3B).
Figure 3.
Genetic architecture of germination speed under extreme salinity in Arabidopsis thaliana. Allele frequency of (A) all germinants, (B) early germinants, (C) late salt germinants, and (D) early germinants under no-salt condition. Figure 3D is generated with data from Yuan et al. (2016). Data were fitted to a sliding window, three-way nested ANOVA model. The F-statistic was plotted along the genome, with positive values indicating bias for the Col-0 allele and negative values for the Bs-2. Significance threshold (shown as gray dashed lines) and optimal window size were established via permutation. The dark thin bars within each plot indicate the QTL regions, and the gray bars underneath each panel indicate the chromosomes, and the red dots indicate the position of the centromere.
Using this X-QTL method, we were able to identify one peak on chromosomes 4 and three on chromosome 5, all with allele frequency biased toward the Col-0 allele for early germination. Four peaks were also identified on chromosome 1, biased toward the Bs-2 allele. In addition, multiple regions on chromosomes 2 and 3 were identified that just reached the significance threshold. The position and size of the QTL are shown in Table 2.
Table 2. Positions of germination speed under salt X-QTL, and their overlap with previously identified salt germination QTL.
| QTL | Chromosome | Range/Mbp | Size/Mbp | Overlap | Trait |
|---|---|---|---|---|---|
| SaltEarlyQ1.1 | 1 | 8.58–10.78 | 2.2 | Clerkx et al. 2004 | Ler/Sha RIL, 150 mM NaCl, % germination |
| DeRose-Wilson and Gaut 2011 | Ler/Cvi RIL, % germination, 150 mM | ||||
| DeRose-Wilson and Gaut 2011 | Ler0/Col4 RIL, % cotyledon expansion | ||||
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, Gmax | ||||
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, AUC | ||||
| Vallejo et al. 2010 | Bay0/Sha RIL, 50 mM NaCl, % germination at 4 d | ||||
| Guo et al. 2016 | Tsu1/Sha F2, 150 mM NaCl, cotyledon greening | ||||
| Galpaz and Reymond 2010 | Sha/Ler RIL, 175 mM NaCl, % germinated at 10 d after sowing | ||||
| SaltEarlyQ1.2 | 1 | 11.92–12.20 | 0.18 | Quesada et al. 2002 | Ler0/Col4 RIL, 250 mM NaCl, % germination at 15 d |
| Guo et al. 2016 | Tsu1/Sha F2, 150 mM NaCl, cotyledon greening | ||||
| SaltEarlyQ1.3 | 1 | 23.60–24.59 | 0.99 | Guo et al. 2016 | Tsu1/Sha F2, 150 mM NaCl, cotyledon greening |
| SaltEarlyQ1.4 | 1 | 26.29–28.75 | 2.46 | ||
| SaltEarlyQ4.1 | 4 | 10.87–18.08 | 7.21 | Quesada et al. 2002 | Ler0/Col4 RIL, 250 mM NaCl, % germination at 15 d |
| DeRose-Wilson and Gaut 2011 | Ler/Cvi RIL, % germination, 150 mM | ||||
| DeRose-Wilson and Gaut 2011 | Ler0/Cvi RIL, % cotyledon expansion | ||||
| SaltEarlyQ5.1 | 5 | 15.88–16.09 | 0.21 | Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, AUC |
| SaltEarlyQ5.2 | 5 | 17.33–25.18 | 7.85 | Clerkx et al. 2004 | Ler/Sha RIL, 150 mM NaCl, % germination |
| DeRose-Wilson and Gaut 2011 | Ler/Cvi RIL, % germination, 150 mM | ||||
| DeRose-Wilson and Gaut 2011 | Ler0/Col4 RIL, T50 | ||||
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, Gmax | ||||
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, T50 | ||||
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, AUC | ||||
| Vallejo et al. 2010 | Bay0/Sha RIL, 50 mM NaCl germination at 4 d | ||||
| Galpaz and Reymond 2010 | Sha/Col RIL, 175 mM NaCl, % germinated at 10 d after sowing | ||||
| SaltEarlyQ5.3 | 5 | 26.34–26.66 | 0.32 | ||
| SaltLateQ2.1 | 2 | 10.76–15.01 | 4.25 | Guo et al. 2016 | Tsu1/Sha F2, 150 mM NaCl, cotyledon greening |
| SaltLateQ2.2 | 2 | 16.32–17.47 | 1.15 | ||
| SaltLateQ4.1 | 4 | 14.89–15.29 | 0.4 | DeRose-Wilson and Gaut 2011 | Ler0/Cvi RIL, % cotyledon expansion |
| SaltLateQ5.1 | 5 | 6.90–6.98 | 0.08 | Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, T50 |
| Joosen et al. 2010 | Bay0/Sha RIL, 100 mM NaCl, AUC | ||||
| Guo et al. 2016 | Tsu1/Sha F2, 150 mM NaCl, cotyledon greening |
Three rows are empty for Overlap and Trait because no overlap with previous publications were identified for those QTLs. Gmax, maximum germination percentage; AUC, area under cumulative germination curve.
We compared the X-QTL mapping results of the early cohorts (the initial 5000 germinants) in the saline conditions vs. no-salt conditions (Figure 3, B and D). While only three peak clusters on chromosomes 1, 3, and 4 were significant in no-salt conditions, multiple peaks on chromosomes 1, 4, and 5 were identified for rapid germination under salt. Among all peak regions that were significant, only the peaks on the right arm of chromosome 4 were shared between the rapid germinating cohorts under both conditions, and the Col-0 allele was favored in both cases.
Time-cohort germination QTL under salinity stress
Unlike under no-salt conditions, germination under salinity stress occurred over a prolonged time period that spanned >12 d. Given this prolonged period, we could examine what QTL were associated with germination under different time periods during salinity stress.
We identified two germination cohorts for analysis. The first 5% that germinated of the entire F3 population (see above) were designated the early cohort, and corresponds to the lagging phase of the cumulative germination curve. The late cohort corresponds to the plateau phase of germination curve, and constitutes the 20th–23rd percentiles that germinated of the same F3 populations.
Three genomic regions exhibited a response to selection in the late cohort, on chromosome 2, 4, and 5 respectively, and all three regions favor the Bs-2 allele (Figure 3C). In this late cohort, the peak on chromosome 2 passed the permutation threshold, while the ones on chromosome 4 and 5 narrowly reached significance. Interestingly, overlapping regions on chromosome 2 and 4 also showed response to selection in the early cohort, although the allele frequency bias on chromosome 2 is not significant; however the allele favored for rapid germination in the early cohort was Col-0.
Finally, we examined whether there was a genetic basis for the ability to germinate under salt stress (as opposed to germination speed). We pooled DNA isolated from all time cohorts, representing the ∼23% of F3 recombinants that germinated under salt across the 15 d of the experiment. In this population, we observed allele frequency skews on chromosomes 1, 4, and 5. However, none of these were significant as determined by permutation (Figure 3A). This could be attributable to the different QTL associated with germination at different time points (e.g., early vs. late cohorts), some of which are attributed to QTL alleles with different directions in phenotypic effect.
Discussion
Salinity is a novel environment for the majority of A. thaliana populations, and previous studies have examined natural variation for salt tolerance in this species in both germination and vegetative growth stages. Salt tolerance during vegetative growth, for example, has been shown to be under control of few loci of large effect (e.g., Rus et al. 2005; Quesada et al. 2002; Katori et al. 2010). The RAS1 locus was cloned in a Ler × Sha recombinant inbred line (RIL) mapping population, and a naturally occurring premature stop codon in the Sha allele leads to decreased salt tolerance (Ren et al. 2010). Also, a weak allele of the sodium transporter AtHKT1 gene shows altered expression patterns in two coastal populations of A. thaliana that confers salt tolerance (>30% variation explained) during vegetative growth. A broader survey of >300 natural populations shows an adaptive cline of salt tolerance that was driven by the distribution of the weak AtHKT1 allele.
Germination of dispersed seeds is crucial for the range expansion and establishment of plants in novel environments, and germination speed affects the fitness of plants under fluctuating environments. Much less is known about the genetic basis of salt tolerance during germination. We identified nine QTL studies (see Table 2 for the full list) that examined the genetic architecture of A. thaliana germination salt tolerance. The number of QTL in each mapping population ranges from one to six, and most of these mapped loci are of large phenotypic effect, explaining 5–27% of the total trait variance. In general, QTL for germination salt tolerance and those for the vegetative stage do not overlap, indicating different mechanisms are involved in salt tolerance for these the two stages. The most common phenotype measured was the percentage of germination/cotyledon greening, although two studies (DeRose-Wilson and Gaut 2011; Joosen et al. 2010) scored the time to reach 50% germination (i.e., germination speed). DeRose-Wilson and Gaut (2011) mapped three QTL for germination speed under salt from Col × Ler RILs, on chromosomes 3 and 5, and they only partially overlap with the percentage germination/cotyledon greening QTL. Likewise, Joosen et al. (2010) mapped four germination speed QTL under salt from Bay × Sha RILs, also on chromosomes 3 and 5, partially overlapping QTL they mapped for other salt-tolerant germination traits. These findings indicate that germination speed may contribute to germination salt tolerance trait.
We used the X-QTL mapping platform to examine the genetics of germination speed. Unlike previous QTL studies, this approach potentially has higher power (with a sample size of ∼100,000 F3 recombinants), and can provide a more comprehensive catalog of loci compared to previous mapping studies. We observed increased phenotypic variation compared to the no-salt condition (i.e., a range in germination time of 15 d instead of 72 hr), and changes in the schedules of each developmental stage (testa rupture, radicle protrusion, cotyledon greening, and cotyledon expansion) during germination.
We identified eight QTL for early germination under extreme salinity in our mapping population, and four QTL for late germination. We compared the QTL maps of germination speed under saline conditions to that observed in no-salt conditions (Yuan et al. 2016). We identified that the QTL on the distal arm of chromosome 4 in saline conditions (SaltEarlyq4.1) overlaps with a germination speed QTL we previously identified under no-salt germination conditions (Yuan et al. 2016). We also found that SaltEarlyq5.2 and SaltLateq5.1 overlap with a salt germination speed QTL previously mapped by other groups (DeRose-Wilson and Gaut 2011; Joosen et al. 2010). This suggests that some of the early germination response to salt stress may simply be due to genes associated with a general acceleration in germination timing.
However, most peaks do not overlap between the salt and normal conditions, and we identified more QTL in salt compared to no-salt conditions (12 vs. five loci). These results suggest there are polymorphic genes between the Bs-2 and Col-0 accessions that remain unexpressed under no-salt conditions, but are observed in the novel saline environment. This release of cryptic genetic variation leads to increased variation in germination speed between the founder accessions.
When we attempted to map the loci underlying germination ability under salt stress (by pooling all germinants from all time cohorts), we detected few regions that exhibited an allele frequency bias, and none of them were statistically significant. This might be due to the contrasting genes/alleles at different time points in the germination process.
This appears to be the case, as we found that the genetic architecture of germination under salt stress differs in early vs. late germinating cohorts, suggesting multiple pathways to salt tolerance during the developmental period between testa rupture and cotyledon expansion. Moreover, QTL on chromosomes 2 and 4 exhibit opposite allele frequency biases in early and late germination cohorts. There are two possible reasons for this observation: either these are due to different genes with opposite effects in early vs. late germination, or time-dependent allelic effects of the same genes. Establishing the genetic mechanism for this phenomenon will have to await isolation of the causal genes.
We found that some of the QTL we identified appear to overlap with QTL identified in previously published A. thaliana germination salt tolerance QTL studies. Determining exact overlap is difficult. Given that positions of most of the previously identified QTL were reported on a genetic (rather than physical) map, we are only able to approximate the extent of overlap between studies. Nevertheless, it appears that the SaltEarlyQ1.1 and SaltEarlyQ5.2 overlaps with eight previously identified QTL from various studies, while SaltEarlyQ4.1 and SaltLateQ5.1 overlaps with three QTL each (see Table 2 for a full list of overlapping QTLs). However, many of other QTL we detected by X-QTL mapping do not overlap with any known QTL.
It is noteworthy to compare our approach with another bulk segregant mapping study of germination salt tolerance in A. thaliana (Guo et al. 2016). Although the approaches behind our two studies are similar, we used different founder accessions, a much larger sample size, and more stringent selection threshold. Nevertheless, of the nine QTL they reported, five colocalize with QTL on chromosomes 1, 2, and 5 identified in our study. The three QTL on chromosome 1 are in close proximity, and other classical QTL studies cannot separate these three loci; in both our and the Guo et al. (2016) study, however, these QTL are well-resolved, highlighting the power and resolution of high-throughput selection/bulk segregant analysis in mapping QTL.
A major goal of QTL analysis is to identify specific genes that underlie complex traits. Although functional identification of specific loci is beyond the scope of this study, we attempted to identify plausible positional candidate genes by examining the annotations of genes that fall within the QTL peaks (Table 2). We considered a gene to be a strong candidate if it had annotation attributed to salt/dehydration stress response, germination, ABA metabolic/signaling pathways, and GA metabolic/signaling pathways. The choice of relevant gene annotation was based on the osmotic and ionic stress components of extreme salinity (Munns and Tester 2008), as well as the key roles of the hormones ABA and GA in both germination (Bewley 1997) and stress response (Nakashima and Yamaguchi-Shinozaki 2013). Eighteen genes met these criteria. We then looked for tissue/stage-specific gene expression pattern for these genes using ePlant (http://bar.utoronto.ca/~dev/eplant). With these criteria, we identified 10 genes in four QTL regions that had appropriate functional annotation and had either high expression levels in mature seeds, or a dramatic change in expression level from dry to imbibed seeds (see Table 3). Future functional validation by quantitative complementation of the alleles for these genes will be necessary to conclusively demonstrate whether any of these loci indeed drives the phenotypic difference between the parental accessions.
Table 3. Candidate genes within germination speed under salt X-QTL regions.
| AGI | Gene Name | QTL | Description | Citation |
|---|---|---|---|---|
| AT1G72770 | HAB1 | SaltEarlyQ1.4 | Protein phosphatase 2C, mutant has ABA hypersensitive inhibition of seed germination | Kuhn et al. 2006 |
| AT2G29090 | CYP707A2 | SaltLateQ2.1 | Protein involved in ABA catabolism, plays a major role in the rapid decrease in ABA levels during early seed imbibition | Liu et al. 2009 |
| AT2G29380 | HAI3 | SaltLateQ2.1 | Highly ABA-induced PP2C gene 3 | Schweighofer et al. 2004 |
| AT2G31660 | SAD2 | SaltLateQ2.1 | Encodes an importin β-domain family protein likely to be involved in nuclear transport in ABA signaling | Verslues et al. 2006 |
| AT4G28520 | CRU3 | SaltEarlyQ4.1 | Encodes a 12S seed storage protein whose phosphorylation state is modulated in response to ABA in Arabidopsis thaliana seeds | Lin et al. 2013 |
| AT4G30660 | N/A | SaltEarlyQ4.1 | Low temperature and salt responsive protein family | www.arabidopsis.org |
| AT5G45830 | DOG1 | SaltEarlyQ5.2 | DELAY OF GERMINATION 1. Causal gene of a quantitative trait locus involved in the control of seed dormancy | Bentsink et al. 2006 |
| AT5G51760 | AHG1 | SaltEarlyQ5.2 | ABA-hypersensitive germination 1, a putative PP2C. Expressed in seeds and functions in seed development and germination | Nishimura et al. 2007 |
| AT5G52300 | LTI65 | SaltEarlyQ5.2 | Encodes a protein that is induced in expression in response to water deprivation such as cold, high-salt, and desiccation via ABA | Nakashima et al. 2006 |
| AT5G54390 | AHL | SaltEarlyQ5.2 | Encodes a 3′-phosphoadenosine-5′-phosphate (PAP) phosphatase that is sensitive to physiological concentrations of Na+ | Gil-Mascarell et al. 1999 |
AGI, Arabidopsis Genome Initiative.
In this study, we used the X-QTL mapping platform and a time series experimental design to map germination speed QTL under a novel abiotic environment. The mapping population size (∼100,000 F3 recombinants) is by far the largest used in mapping studies for multicellular organisms, which can potentially lead to higher mapping power and precision (Ehrenreich et al. 2010; Beavis 1997). The combination of salt stress that increased the phenotypic range of germination speed variation, the large mapping population, and the time-series sampling design enabled us to isolate three temporal germination cohorts. Unlike previous studies, the isolation of these germination cohorts allowed us to examine dynamic germination behavior under salinity, and to focus our attention on how intrinsic variation of germination speed responds to salinity, rather than simply mapping whether or not seeds germinate under salinity.
Our findings suggest that the range of phenotypic variation in germination speed increases upon saline treatment, consistent with previous studies that point to a decrease in genetic canalization under novel (stressful) environments (Waddington 1942; Waddington 1953; Paaby and Rockman 2014). We also observe intraspecific standing genetic variation that is observed only in saline conditions, indicating cryptic genetic variation that exhibits a clear G×E interaction. Our results suggest that cryptic genetic variation may play a crucial adaptive role in facilitating species range expansions, exploitation of new niches or response to climate change. Identification of the causal genes underlying this cryptic genetic variation in abiotic stress tolerance may illuminate the mechanisms that can underlie response to novel environments, and lead to greater understanding of adaptation that may accompany species dispersal. Moreover, many of these genes may also prove useful in an agricultural context, providing new genes that can be used to improve crop performance in marginal environments.
Supplementary Material
Acknowledgments
We would like to thank Ian Ehrenreich for his help in developing the X-QTL platform in Arabidopsis. This work was supported in part by grants from the National Science Foundation Plant Genome Research Program, and the New York University Abu Dhabi Research Institute to M.D.P.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.033944/-/DC1
Communicating editor: S. A. Jackson
Literature Cited
- Assman S. M., 2013. Natural variation in abiotic stress and climate change responses in Arabidopsis: implications for twenty-first-century agriculture. Int. J. Plant Sci. 174: 3–26. [Google Scholar]
- Atwell J. W., Cardoso G. C., Whittaker D. J., Price T. D., Ketterson E. D., 2014. Hormonal, behavioral, and life-history traits exhibit correlated shifts in relation to population establishment in a novel environment. Am. Nat. 184(6): E147–E160. [DOI] [PubMed] [Google Scholar]
- Baxter I., Brazelton J. N., Yu D., Huang Y. S., Lahner B., et al. , 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W. D., 1997. QTL analysis: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by Patterson A. H. CRC, Boca Raton, New York. [Google Scholar]
- Beckman M., Bruelheide H., Erfmeier A., 2011. Germination responses of three grassland species differ between native and invasive origins. Ecol. Res. 26: 763–771. [Google Scholar]
- Bentsink L., Jowett J., Hanhart C. J., Koornneef M., 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J. D., 1997. Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron T. C., O’Sullivan D., Reynolds A., Piertney S. B., Benton T. G., 2013. Eco-evolutionary dynamics in response to selection on life-history. Ecol. Lett. 16: 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx E. J. M., El-Lithy M. E., Vierling E., Ruys G. J., Blankestijn-De Vries H., et al. , 2004. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Ler and Shakdara, using a new recombinant inbred line population. Plant Physiol. 135: 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose-Wilson L., Gaut B. S., 2011. Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS One 6(8): 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K., 2002. Germination timing influences natural selection on life-history characters in Arabidopsis thaliana. Ecology 83: 1006–1016. [Google Scholar]
- Donohue K., Dorn L., Griffith C., Kim E., Aguilera A., et al. , 2005a Niche construction through germination cueing: life-history responses to timing of germination in Arabidopsis thaliana. Evolution 59: 771–785. [PubMed] [Google Scholar]
- Donohue K., Dorn L., Griffith C., Kim E., Aguilera A., et al. , 2005b The evolutionary ecology of seed germination of Arabidopsis thaliana: variable natural selection on germination timing. Evolution 59: 758–770. [PubMed] [Google Scholar]
- Donohue K., de Casas R., Burghardt L., Kovach K., Willis C. G., 2010. Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 41: 293–319. [Google Scholar]
- Dubois J., Cheptou P. O., 2012. Competition/colonization syndrome mediated by early germination in non-dispersing achenes in the heteromorphic species Crepis sancta. Ann. Bot. (Lond.) 110: 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul M., Bonzom J.-M., Lecomte C., Goussen B., Daian F., et al. , 2014. Rapid evolutionary responses of life history traits to different experimentally-induced pollutions in Caenorhabditis elegans. BMC Evol. Biol. 14: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kassaby Y. A., Moss I., Kolotelo D., Stoehr M., 2008. Seed germination: mathematical representation and parameters extraction. For. Sci. 54: 220–227. [Google Scholar]
- Etterson J. R., Shaw R. G., 2001. Constraint to adaptive evolution in response to global warming. Science 294: 151–154. [DOI] [PubMed] [Google Scholar]
- Galpaz N., Reymond M., 2010. Natural variation in Arabidopsis thaliana revealed a genetic network controlling germination under salt stress. PLoS One 5: e15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Mascarell R., López-Coronado J. M., Bellés J. M., Serrano R., Rodríguez P. L., 1999. The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 17: 373–383. [DOI] [PubMed] [Google Scholar]
- Graebner R. C., Callaway R. M., Montesinos D., 2012. Invasive species grows faster, competes better, and shows greater evolution toward increased seed size and growth than exotic non-invasive congeners. Plant Ecol. 213: 545–553. [Google Scholar]
- Guo J., Fan J., Hauser B. A., Rhee S. Y., 2016. Target enrichment improves mapping of complex traits by deep sequencing. G3 (Bethesda) 6: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman C. A., Broder E. D., Dalton C. M., Ruell E. W., Myrick C. A., et al. , 2013. Predator-induced phenotypic plasticity in metabolism and rate of growth: rapid adaptation to a novel environment. Integr. Comp. Biol. 53: 975–988. [DOI] [PubMed] [Google Scholar]
- Hayden E. J., Ferrada E., Wagner A., 2011. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474: 92–95. [DOI] [PubMed] [Google Scholar]
- Jarosz D. F., Lindquist S., 2010. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330: 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen R. V., Kodde J., Willems L. A., Ligterink W., van der Plas L. H., et al. , 2010. GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 62: 148–159. [DOI] [PubMed] [Google Scholar]
- Katori T., Ikeda A., Iuchi S., Kobayashi M., Shinozaki K., et al. , 2010. Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 61: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. M., Boisson-Dernier A., Dizon M. B., Maktabi M. H., Schroeder J. I., 2006. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140: 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Kiani S. P., Durand-Tardif M., 2009. A focus on natural variation for abiotic constraints response in the model species Arabidopsis thaliana. Int. J. Mol. Sci. 10: 3547–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Pajak A., Marsolais F., McCourt P., Riggs C. D., 2013. Characterization of a cruciferin deficient mutant of Arabidopsis and its utility for overexpression of foreign proteins in plants. PLoS One 8: e65980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shi L., Ye N., Liu R., Jia W., et al. , 2009. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183: 1030–1042. [DOI] [PubMed] [Google Scholar]
- Lynch J. P., Clair S. B. S., 2004. Mineral stress: the missing link in understanding how global climate change will affect plants in real world soils. Field Crops Res. 90: 101–115. [Google Scholar]
- McGuigan K., Nishimura N., Currey M., Hurwit D., Cresko W. A., 2011. Cryptic genetic variation and body size evolution in threespine stickleback. Evolution 65: 1203–1211. [DOI] [PubMed] [Google Scholar]
- Miller T. E., Winn A., Schemske D., 1994. The effects of density and spatial distribution on selection for emergence time in Prunella vulgaris. Am. J. Bot. 81: 1–6. [Google Scholar]
- Munns R., Tester M., 2008. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K., 2013. ABA signaling in stress-response and seed development. Plant Cell Rep. 32: 959–970. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., et al. , 2006. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., et al. , 2007. ABA-Hypersensitive Germination 1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50: 935–949. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2014. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R. F., 2012. Incidence and ecology of very fast germination. Seed Sci. Res. 22: 161–167. [Google Scholar]
- Quesada V., Garcia-Martinez S., Piqueras P., Ponce M. R., Micol J. L., 2002. Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol. 130: 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ranke, J., 2014 chemCal: calibration functions for analytical chemistry. R package version 0.1–34. Available at: http://chem.uft.uni-bremen.de/ranke/?page=chemCal. Accessed June 4, 2015.
- Ren Z. H., Zheng Z. M., Chinnusamy V., Zhu J. H., Cui X. P., et al. , 2010. RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, J. D., and J. Loveday, 1990 Salinity in irrigated agriculture, pp. 1089–1142 in Irrigation of Agricultural Crops (Monograph 30), edited by B. A. Stewart, D. R. Nielsen. American Society of Agronomists, Madison, WI. [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., et al. , 2006. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I., 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9: 236–243. [DOI] [PubMed] [Google Scholar]
- Seiter S., Kingsolver J., 2013. Environmental determinants of population divergence in life-history traits for an invasive species: climate, seasonality and natural enemies. J. Evol. Biol. 26: 1634–1645. [DOI] [PubMed] [Google Scholar]
- Vallejo A. J., Yanovsky M. J., Botto J. F., 2010. Germination variation in Arabidopsis thaliana accessions under moderate osmotic and salt stresses. Ann. Bot. (Lond.) 106: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P. E., Guo Y., Dong C. H., Ma W., Zhu J. K., 2006. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 47: 776–787. [DOI] [PubMed] [Google Scholar]
- Volis S., Ormanbekova D., Yermekbayev K., Song M., Shulgina I., 2016. The conservation value of peripheral populations and a relationship between quantitative trait and molecular variation. Evol. Biol. 43(1): 26–36. [Google Scholar]
- Waddington C. H., 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Waddington C. H., 1953. Genetic assimilation of an acquired character. Evolution 7: 118–126. [Google Scholar]
- Weinig C., 2000. Differing selection in alternative competitive environments: shade-avoidance responses and germination timing. Evolution 54: 124–136. [DOI] [PubMed] [Google Scholar]
- Yuan W., Flowers J. M., Sahraie D. J., Ehrenreich I. M., Purugganan M. D., 2016. Extreme QTL mapping of germination speed in Arabidopsis thaliana. Mol. Ecol. DOI: 10.1111/mec.13768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk-segregant microarray genotyping data of early, medium and late salt-tolerant germination cohorts, and the entire salt germinant pool are available in Supplemental Material, File S1.