Abstract
After fertilization, rapid changes of the Caenorhabditis elegans cytoskeleton occur in the transition from meiosis to mitosis, requiring precise regulation. The MEI-1/MEI-2 katanin microtubule-severing complex is essential for meiotic spindle formation but must be quickly inactivated to allow for proper formation of the mitotic spindle. MEI-1/MEI-2 inactivation is dependent on multiple redundant pathways. The primary pathway employs the MEL-26 substrate adaptor for the CUL-3/cullin-based E3 ubiquitin ligase, which targets MEI-1 for proteosomal degradation. Here, we used quantitative antibody staining to measure MEI-1 levels to determine how other genes implicated in MEI-1 regulation act relative to CUL-3/MEL-26. The anaphase-promoting complex/cyclosome, APC/C, the DYRK (Dual-specificity tyrosine-regulated kinase), MBK-2, and the CUL-2-based E3 ubiquitin ligase act together to degrade MEI-1, in parallel to MEL-26/CUL-3. CUL-2 is known to keep MEL-26 low during meiosis, so CUL-2 apparently changes its target from MEL-26 in meiosis to MEI-1 in mitosis. RFL-1, an activator of cullin E3 ubiquitin ligases, activates CUL-2 but not CUL-3 for MEI-1 elimination. HECD-1 (HECT/Homologous to the E6AP carboxyl terminus domain) E3 ligase acts as a MEI-1 activator in meiosis but functions as an inhibitor during mitosis, without affecting levels of MEI-1 or MEI-2. Our results highlight the multiple layers of MEI-1 regulation that are required during the switch from the meiotic to mitotic modes of cell division.
Keywords: meiosis, spindle, microtubule severing, embryo, katanin
The cytoskeleton is a central player in maintaining cell structure, movement, shape, and cell division. Although the cytoskeleton itself has been extensively studied, less is known about what regulates cytoskeletal remodeling, especially during embryogenesis when cells rapidly adopt new forms and roles. Significant regulatory molecules can be in low abundance, short-lived, or transitory, making them difficult to identify and analyze. We use the nematode Caenorhabditis elegans as a model system to study the rapid transitions in the microtubule (MT) cytoskeleton in the early embryo. Fertilization triggers the completion of meiosis in the C. elegans zygote and within 15 min, the short, acentrosomal meiotic spindle must be disassembled and replaced by the astral mitotic spindle that fills most of the zygote’s volume (Albertson 1984; Kemphues et al. 1986; Albertson and Thomson 1993; McCarter et al. 1999; Yang et al. 2003). These dramatic changes require the precise regulation of molecules specific to each type of cell division. Protein degradation plays a key role in this meiosis-to-mitosis transition by eliminating oocyte-specific products that are no longer needed or would be detrimental to the developing embryo (Bowerman and Kurz 2006; Stitzel and Seydoux 2007; Verlhac et al. 2010; Robertson and Lin 2013).
The C. elegans genes mei-1 and mei-2 (meiosis defective) encode the two subunits of the katanin MT severing complex (Mains et al. 1990a; McNally and Vale 1993; Srayko et al. 2000; Roll-Mecak and McNally 2010; Sharp and Ross 2012). mei-1 and mei-2 are essential for meiotic spindle formation but must be rapidly inactivated prior to the first mitotic cleavage (Clark-Maguire and Mains 1994a). MEI-1/MEI-2 activity keeps meiotic spindles short, produces seeds for MT nucleation, and mediates the anaphase shortening of the spindle that is characteristic of the meiotic divisions (McNally et al. 2006, 2014; Srayko et al. 2006). Failure to eliminate katanin in mitosis results in lethality due to excess MT severing during the early cleavages (Mains et al. 1990a; Clark-Maguire and Mains 1994a; Srayko et al. 2006).
There are multiple redundant layers of regulation that ensure katanin MEI-1/MEI-2 MT severing is restricted to meiosis (Figure 1). The primary mode of MEI-1 regulation is through two independent protein degradation pathways. The major pathway involves MEL-26 (maternal effect lethal, Dow and Mains 1998) as a substrate recognition subunit that recruits MEI-1 to the CUL-3/cullin-based E3 ubiquitin ligase. This results in MEI-1 ubiquitylation and subsequent proteosomal degradation (Furukawa et al. 2003; Pintard et al. 2003b; Xu et al. 2003). Phosphorylation often precedes ubiquitylation and MBK-2 (mini-brain kinase), a member of the DYRK (Dual Specificity and Tyrosine Regulated Kinase) family of protein kinases, contributes to postmeiotic MEI-1 degradation (Pellettieri et al. 2003; Quintin et al. 2003; Ming Pang et al. 2004; Stitzel et al. 2006, 2007; Cheng et al. 2009; Wang et al. 2014). MBK-2 is not essential for MEL-26/CUL-3 degradation of MEI-1, but instead acts in a parallel protein degradation pathway with an unknown ubiquitin ligase (Lu and Mains 2007). RFL-1/ Uba3p (ectopic membrane ruffling) has also been implicated in MEI-1 degradation. rfl-1 encodes a subunit of an E1 activating enzyme that leads to the addition of a ubiquitin-like Nedd8 to cullins that activates E3 ubiquitin ligase activity (Kurz et al. 2002; Pintard et al. 2003a; Sarikas et al. 2011). Other modes of MEI-1 regulation include the PP4 phosphatase complex that increases MT severing activity at anaphase II (Han et al. 2009; Gomes et al. 2013) and postmeiotic inhibition of mei-1 mRNA translation by IFET-1/SPN-2 and OMA-1 (Li et al. 2009).
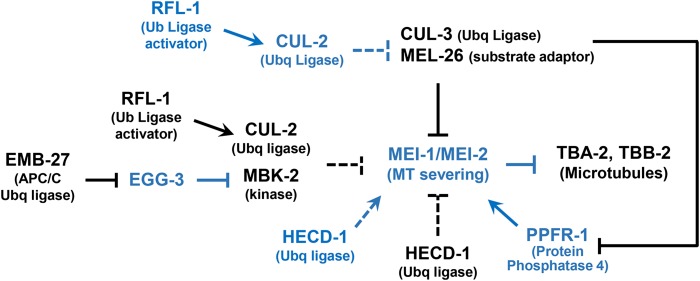
Figure 1.
The MEI-1 regulatory pathway. Proteins active in meiosis are in blue and those active in mitosis are in black. During meiosis, the katanin MEI-1/MEI-2 complex is activated by PPFR-1 and HECD-1 and so severs microtubules (MTs). When meiosis is completed, CUL-3/MEL-26 increases and EMB-27/APC/C activates CUL-2/MBK-2, leading to MEI-1 degradation. CUL-2 also functions during meiosis to prevent premature MEL-26 accumulation. RFL-1 and HECD-1 function at both divisions. Dashed lines indicate that interactions may be indirect. Not shown are mitotic translational inhibition of MEI-1 mRNA (messenger RNA) by IFET-1/SPN-2 and OMA-1 or the other PP4 subunits that act with PPFR-1.
MEI-1 degradation is under temporal control to ensure that sufficient MT severing activity remains until the completion of meiosis. Levels of the MEI-1 inhibitor MEL-26 remain low until the completion of meiosis due to the activity of RFL-1 and another E3 ubiquitin ligase, which contains CUL-2 (Johnson et al. 2009). The MBK-2-mediated MEI-1 degradation pathway is activated upon the completion of meiosis by the anaphase promoting complex/cyclosome (APC/C) that both drives meiotic anaphase progression and degrades the MBK-2 inhibitor EGG-3 (Lu and Mains 2007; Stitzel et al. 2007; Cheng et al. 2009; Parry et al. 2009). APC/C might also target the katanin MEI-2 subunit as the latter has an APC/C consensus “destruction box” motif RxxLxxxxN (RkkLthakN in MEI-2) (Harper et al. 2002; Lu and Mains 2007). MEI-1 and MEI-2 are each dependent on the other for their expression (Clark-Maguire and Mains 1994a; Srayko et al. 2000).
Previous work showing that APC/C and MBK-2 act in parallel to MEL-26 was based on the logic that if two genes act in the same pathway, their mutations will not enhance (exacerbate) one another’s phenotypes in doubly mutant animals; once a mutation in one gene inactivates a pathway, loss of a second gene in the same linear pathway is irrelevant. In contrast, if two genes act in parallel, the double mutant shows a more extreme (i.e., enhanced) phenotype than either of the single mutants. This analysis requires that at least one of the mutations must be a null; hypomorphic mutations acting sequentially may also show enhancement. Our previous work with MEL-26 and MBK-2 could rely on measuring embryonic viability because MEI-1 is the only essential substrate of MEL-26 (Lu et al. 2004; Lu and Mains 2005; Johnson et al. 2009) and so some mel-26(null) embryos survive at low temperatures due to the action of the MBK-2 pathway. In double mutants of mel-26(null) with a temperature-sensitive (ts) mbk-2 allele at a semipermissive temperature all embryos die, demonstrating that the genes work in parallel. However, we could not order MBK-2, APC/C, and RFL-1 relative to each other because these genes have other essential targets and so their null phenotypes are lethal, precluding assays of hatching rates (Davis et al. 2002; Stitzel et al. 2006; Dorfman et al. 2009; Wang et al. 2014). In this work, we quantitatively measured anti-MEI-1 staining levels in double mutant strains as an alternative metric to embryonic viability. We show that APC/C and RFL-1 act in the MBK-2 pathway. We identified CUL-2 as the E3 ubiquitin ligase acting with MBK-2. In addition, we found that HECD-1, a HECT domain ubiquitin ligase (homologous to the E6AP carboxyl terminus), also regulates MEI-1 without apparent effects on MEI-1 protein levels. Unexpectedly, HECD-1 acts as a MEI-1 activator in meiosis but as an inhibitor in mitosis. Thus, the regulation of MEI-1 MT severing includes unanticipated layers of regulation.
Materials and Methods
Nematode culture and strains
All strains, including the wild type (N2, var. Bristol) were maintained under standard conditions at 15° unless otherwise stated (Brenner 1974). Hatching rates of complete broods of at least four hermaphrodites and > 400 offspring for each strain are reported as previously described (Mains et al. 1990b). ts strains were upshifted to their nonpermissive temperatures (20° or 25°) at the last larval stage (L4). The following alleles were used and detailed descriptions are found at http://www.wormbase.org. Linkage group I: mei-1(ct46ts, ct46ct101, and or1178ts), mei-2(ct98 and sb39ts), and mel-26(ct61sb4 and or184ts). Linkage group II: zyg-11(b2ts) and emb-27(g48ts). Linkage group III: tbb-2(sb26), cul-2(or209ts and ek1), zer-1(gk165593), rfl-1(or198ts), and fem-2(b245ts). Linkage group IV: hecd-1(ok1437), mbk-2(dd5ts and pk1427), and fem-1(hc17ts and e1965). mei-1(or1178ts) included the transgenes ruIs57[pAZ147 pie-1::GFP::tbb-2 unc-119(+)] III; ltIs37[Paa64:pie-1p::mCherry::his-58 unc-119(+)]. zer-1(gk165593) is a nonsense allele isolated in the Million Mutant Project (Thompson et al. 2013). zer-1 and hecd-1 were outcrossed eight times before use and genotypes for strains that included these alleles were confirmed by PCR or sequencing. Alleles were often marked with closely-linked morphological markers to facilitate strain construction. The tbb-2(sb26)/β tubulin allele, which is refractory to MEI-1 severing, was included in all strains that were measured by anti-MEI-1 staining to restore normal embryo morphology for easier comparisons of cell stages (Lu et al. 2004).
RNAi
RNAi feeding followed the protocol described by Kamath and Ahringer (2003). A pair of L4 hermaphrodites were placed together on RNAi plates and transferred every 24 hr or until they ceased to lay embryos. The first brood was not counted due to RNAi not being fully effective. The exception was for the mbk-2(RNAi) experiment, where RNAi effects take place earlier and so the first brood was included in the analysis. Control worms were fed with Escherichia coli carrying the L4440 vector plasmid.
Antibody staining
Young adult or L4 hermaphrodites were upshifted to the appropriate temperature 24 hr before fixation. Mutant and wild-type gravid hermaphrodites were mounted on the same polylysine slide, far enough apart to accurately distinguish the two populations. Embryos were extruded from hermaphrodites with gentle pressure on the cover slip. The embryos were fixed with methanol-acetone and immediately used for antibody staining as described (Kemphues et al. 1986; Clark-Maguire and Mains 1994a). The same antibody solution covered both the wild-type and mutant samples on the same slide. Rabbit anti-MEI-1 (Clark-Maguire and Mains 1994a) and rabbit anti-PPFR-1 (Gomes et al. 2013) were used at a 1/100 dilution, rat anti-MEL-26 (Johnson et al. 2009) and rabbit anti-MEI-2 (Srayko et al. 2000) were used at a 1/50 dilution, and mouse anti-α-tubulin (M1A, Sigma-Aldrich Inc.) was used at a 1/200 dilution. FITC-conjugated goat anti-rabbit (Sigma-Aldrich Inc.) and Texas Red-conjugated donkey anti-mouse (Jackson ImmunoResearch Inc.) were used at a 1/100 dilution. Slides were mounted in an antifade solution (Roche Diagnostics) that contained 1 µg/ml DAPI (4’, 6-diamidine-2’phenylindole dihydrochloride, Roche) to visualize DNA.
Measurement and analysis of anti-MEI-1 staining
Photomicrographs through a focal plane that included both centrosomes were taken on a Zeiss Axioplan microscope at 40 × magnification equipped with a Hamatsu Orca ER digital camera using constant settings and exposure time controlled with Zeiss Axiovision (4.7.1) software. Embryos were measured using ImageJ (http://imagej.nih.gov/ij/) by circling the perimeter of the embryo (or individual cells) and determining the mean pixel intensity. The polar body was excluded because it retains high levels of MEI-1 (Clark-Maguire and Mains 1994a). The traced area was superimposed to a nearby area in the photomicrograph free of embryos or worm debris and the pixel value was subtracted from the embryo value. All images presented here were cropped without further editing. Data for each experimental day was normalized to the average of the wild type for that day. Cells were scored as prophase up to the point where DAPI-stained chromosomes aligned at the middle of the tubulin-stained spindle, metaphase during the period of alignment, anaphase from the time when chromosomes separated until they reached the centrosome, and telophase/interphase until completion of cytokinesis. Statistical significance was assessed with the Mann–Whitney Rank Sum test using Sigma Plot software as some data sets did not show the normal distribution required for t-tests (paired t-tests led to the same conclusions in all cases).
Cell cycle timing
Embryos were mounted on a 2% agarose pads (Sulston et al. 1983). Images were taken using the Zeiss Axioplan microscope equipped with Nomarski optics and a Hamatsu OrcaER digital camera with Axiovision software. The cell cycle times from the completion of one cytokinesis to the next for the first two mitotic cell divisions were determined from images taken every 20 sec in the central focal plane. P0 cycle length was measured from pronuclear fusion to when the cleavage furrow fully bisected the daughter cells.
Data availability
Strains and reagents are available upon request. Additional immunostaining data are included in supplemental files.
Results
Quantification of anti-MEI-1 levels
Our previous work, which placed both APC/C and mbk-2 in parallel to mel-26, took advantage of the viability of the null truncation allele mel-26(ct61sb4) (hereafter referred to as mel-26(null)) at 15° but not 25°. This allele behaves as a null in genetic tests at all temperatures but is inherently ts, indicating that the parallel MEI-1 degradation pathway is sufficient at 15° but not 25° (Dow and Mains 1998; Lu and Mains 2007). Thus, we could use hatching rates in double mutants with mel-26(null) to order genes in the pathway. However, we could not determine how APC/C and mbk-2 acted relative to one another because their null alleles showed complete lethality due to their activity in other pathways. Therefore, we developed a quantitative method to measure anti-MEI-1 staining as a substitute for measuring enhancement based on viability. When MEI-1 is ectopically expressed in mitosis it localizes to the cytoplasm, with higher expression on the spindle and yet higher levels in the center of the centrosomes and on the chromosomes (Clark-Maguire and Mains 1994a). Previous work scored increases in ectopic MEI-1 as the ratio of centrosomal MEI-1 to that in adjacent cytoplasm free of MTs (Lu and Mains 2007). However, as centrosomal MEI-1 increases in mutants, MEI-1 also increases in the cytoplasm but they may not do so in parallel. Other reports have used a translation MEI-1::GFP fusion protein to measure MEI-1 expression (Pellettieri et al. 2003; Pintard et al. 2003b; McNally et al. 2006; Parry et al. 2009). However, the transgene used in those studies rescues mei-1(null) poorly (P. E. Mains, unpublished results) and crossing the transgene into many mutant backgrounds, each with multiple mutations, would be difficult. Therefore, we used antibody staining to measure MEI-1 levels.
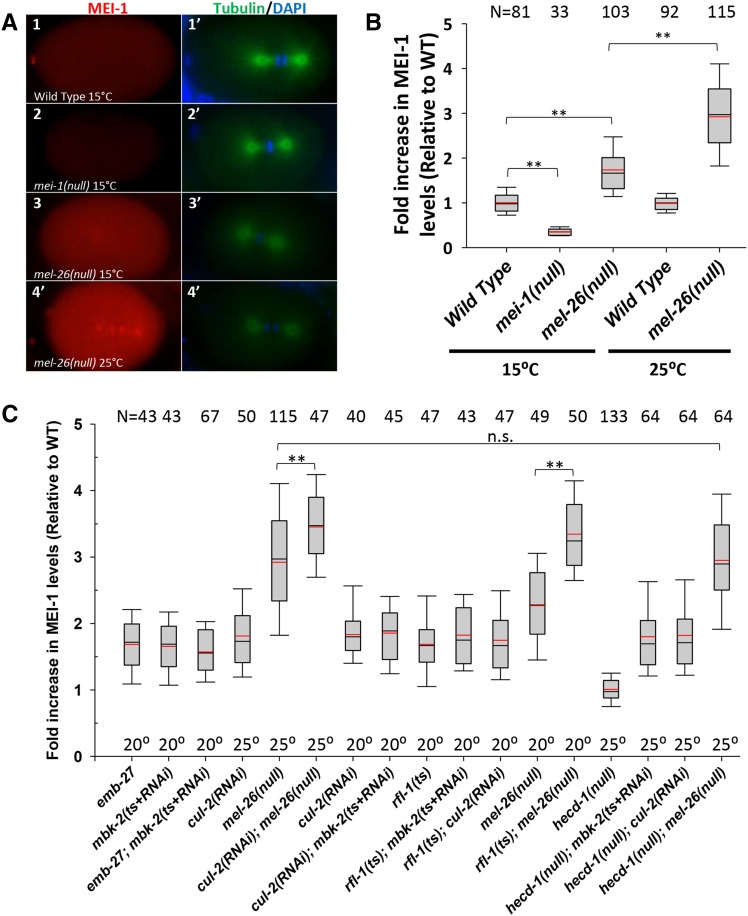
To optimize and standardize antibody staining methods, we measured the mean pixel intensity of MEI-1 staining within the entire embryo subtracted from the background. To minimize experimental variation, we normalized to the wild type processed on the same slide (see Materials and Methods). At 15°, mel-26(null) embryos showed approximately 1.5-fold higher anti-MEI-1 staining compared to wild-type embryos (P < 0.001) and this increased to 3 fold at 25° (P < 0.001) (Figure 2, A and B, Mann–Whitney Rank Sum tests are used throughout). Thus, our assay recapitulates the previous (more qualitative) results, showing that mel-26(null) results in ectopic MEI-1 and that this increases with temperature. The experiment was repeated on five different days, which demonstrated reproducibility (Supplemental Material, Figure S1A). Anti-MEI-1 staining was consistent for one cell (from the point when the female pronucleus became visible after meiosis II), two cell, and four cell embryos and did not differ with cell cycle (Figure S1, B and C), so the data we present pools all one-to-four-cell embryos. mei-1(null) embryos had significantly lower MEI-1 than wild-type (P < 0.001), confirming specificity of the antibody signal (Figure 2B). All measurements of ectopic MEI-1 staining were done in strains that included the tbb-2(sb26) allele (Lu et al. 2004; Lu and Mains 2007). This mutation encodes a β-tubulin that is refractory to MT severing and so restores normal embryo morphology to mutant strains with ectopic MEI-1, which otherwise have short, misoriented spindles and abnormal cytokinesis (Mains et al. 1990a). For simplicity, tbb-2(sb26) is left out of genotypes in most cases in the text and figures. This allele is not included in strains where embryo viability is scored, unless otherwise noted.
Figure 2.
Quantitative anti-MEI-1 indirect immunofluorescence in wild-type and mutant embryos. All strains (including “Wild Type”) contained tbb-2(sb26) to restore normal morphology to embryos that expressed ectopic MEI-1 for easier comparisons. (A) Embryos at the first mitotic cleavage. Anti-MEI-1 (red) is shown in the left column with the corresponding anti-tubulin (green) and DAPI (blue) shown in the right column. The low levels of MEI-1 present in the wild type (A1 and A1’) were above levels shown in mei-1(null) embryos (A2 and A2’) at 15°, demonstrating antibody specificity. Higher than wild-type levels of MEI-1 were present in mel-26(null) at 15° (A3 and A3′) and levels further increased at 25° (A4 and A4’). (B) Anti-MEI-1 mean pixel intensity measurements were subtracted from the nearby background and divided by the average of the wild type processed on the same slide (1.0 = average wild-type level). Data are pooled for one-to-four-cell cell embryos and for all stages of the cell cycle (see Figure S1 for unpooled data). Boxes indicate the 25th to the 75th percentiles and whiskers show the 10th and 90th percentiles. Black lines indicate the median and red lines the mean. **: P < 0.001. N values are shown at the top of the chart. (C) Quantification of anti-MEI-1 levels for mutant strains. emb-27; mbk-2 had similar levels of ectopic MEI-1 to the individual mutations, indicating the genes act in the same pathway. cul-2; mel-26 had higher MEI-1 than the singles, indicating the genes act in parallel. cul-2; mbk-2 was similar to the individual mutations, indicating these genes act in the same pathway. rfl-1 double mutants with mbk-2 or cul-2 had MEI-1 levels similar to single mutants, and so these genes act in the same pathway. rfl-1; mel-26 had elevated levels of MEI-1, and so those genes act in parallel. hecd-1 had wild-type MEI-1 levels and hecd-1 did not affect the levels found in mbk-2, cul-2, or mel-26, and so hecd-1 does not appear to be involved in the degradation pathways. **, P ≤ 0.001; n.s., not significant; P ≥ 0.13. All other double mutant combinations did not differ significantly (0.13 > P > 0.90) from the higher of the corresponding single mutants, but this is not shown for to simplify this figure. N values are shown at the top of the chart and temperature are along the bottom. DAPI, 4’, 6-diamidine-2’phenylindole dihydrochloride; RNAi, RNA interference; WT, wild-type.
The APC/C and MBK-2 act in the same pathway to regulate MEI-1 levels
Stitzel et al. (2007) demonstrated that the APC/C acts as an upstream activator of MBK-2 leading to MEI-1 degradation (Figure 1). It is possible that the APC/C also directly targets the destruction box consensus motif found in MEI-2, which in turn could lead to MEI-1 degradation. Indeed, mei-2 mutations lead to the loss of MEI-1 staining (Clark-Maguire and Mains 1994a). This is also seen with the corresponding subunits of Drosophila katanin (Grode and Rogers 2015).
We asked if the APC-6/CDC16 subunit gene emb-27 acts in parallel or strictly sequentially with mbk-2. This analysis requires double mutants, one of which must be null. If two genes act in parallel, they enhance one another’s phenotypes. The ts allele emb-27(g48ts) arrests embryos in meiosis I anaphase at the restrictive temperature (Golden et al. 2000), precluding later analysis of mitotically-expressed MEI-1. The null allele mbk-2(pk1427) in combination with tbb-2(sb26) shows defects in germline development and so hermaphrodites produce few oocytes (C. Lu and P. E. Mains, unpublished results), again preventing examination of MEI-1 levels during mitosis in early embryos. Therefore, we mimicked a null situation by combining the ts allele mbk-2(dd5ts) and mbk-2(RNAi), both of which partly compromise mbk-2 activity at the semipermissive temperature of 20°. mbk-2(dd5ts); tbb-2(sb26) hermaphrodites were upshifted from 15° to 20° and placed on mbk-2(RNAi) bacteria as young adults, bypassing earlier mbk-2; tbb-2(sb26) defects in germline development. mbk-2(dd5ts+RNAi) had a hatching rate of 1/411 (the hatched embryo arrested without further growth), compared to 58% hatching for mbk-2(dd5ts) and 12% for mbk-2(RNAi) alone. Thus mbk-2(dd5ts+RNAi) represents a strong loss of function (lf) and was used to approximate a null. The emb-27(g48ts) allele is also partially compromised at 20°, showing 56% hatching (Lu and Mains 2007).
Ectopic MEI-1 was 1.7-fold higher in both mbk-2(dd5ts+RNAi) and emb-27(ts) embryos at 20° Compared to the wild type (Figure 2C). emb-27; mbk-2(dd5ts+RNAi) double mutants had ectopic MEI-1 staining levels of 1.6-fold above the wild type, not significantly different from the levels seen in each of the single mutants (P > 0.13). This lack of enhancement suggests that emb-27/APC/C only acts within the mbk-2 pathway to induce MEI-1 degradation and does not have an additional function in parallel by targeting MEI-2.
CUL-2 acts in parallel to MEL-26 to regulate MEI-1 levels
As we and others have reported previously (Stitzel et al. 2006; Johnson et al. 2009), loss of cul-2 leads to ectopic MEI-1 expression in mitosis. Indeed, Kurz et al. (2002) observed that 2/9 cul-2(RNAi) embryos had mel-26-like phenotypes (short, misoriented spindles with abnormal cytokinesis). If CUL-2 is the E3 ubiquitin ligase acting in concert with MBK-2, in parallel to MEL-26 (Figure 1), then mel-26; cul-2 would have higher MEI-1 levels than either of the single mutants. In contrast, no changes would be seen in cul-2; mbk-2. If CUL-2 is instead acting in the MEL-26/CUL-3 rather than the MBK-2 pathway, the cul-2; mbk-2 genotype would show higher MEI-1 levels.
We initially used the cul-2(ek1) null allele and found ectopic MEI-1 staining in mitotic embryos; however, the null allele produced too few embryos for our analysis because of its earlier role in gonad development (Feng et al. 1999; Burger et al. 2013). As an alternative, we used cul-2 RNAi on young adult worms or a cul-2(ts) allele to bypass the earlier defects in our cul-2 experiments. MEI-1 levels in cul-2(RNAi) worms were 1.8-fold higher than the wild type at 25° (Figure 2C). mel-26(null) was 2.9-fold above the wild type. mel-26(null); cul-2(RNAi) showed significantly higher (P < 0.001) ectopic MEI-1 staining than mel-26(null) alone, with levels reaching about 3.5-fold higher than the wild type. In contrast, at 20° there was no significant difference (P > 0.6) between the ectopic MEI-1 levels of cul-2(RNAi) (the higher of the two single mutants) and cul-2(RNAi); mbk-2(dd5ts+RNAi) (Figure 2C). Thus, CUL-2 acts in parallel to CUL-3/MEL-26 and so could be the E3 ubiquitin ligase acting in concert with MBK-2.
cul-2 genetic interactions indicate differing roles in meiosis and mitosis
We next asked if we could detect genetic interactions based on embryo viability that would reflect ectopic MEI-1 function in cul-2 mutants. Interpreting cul-2 double mutants with other mei-1 pathway genes is complicated by the dual roles of cul-2 in regulating both MEI-1 and delaying the build-up of the MEI-1 inhibitor MEL-26 until meiosis is complete (Johnson et al. 2009). However, analysis of cul-2; mel-26(null) is straightforward as this genotype eliminates effects on MEI-1 caused by premature increases in MEL-26. There was a strong genetic interaction between the ts allele of cul-2(or209ts) and the mel-26(null) allele at temperatures semipermissive for cul-2. At 15°, 64% of cul-2(or209ts) embryos hatched compared to 12% of mel-26(null) (Table 1, lines 1 and 2). If the two genes did not interact, then 64% × 12% = 7.7% of double mutant embryos should hatch. Instead, only 0.7% hatched, an 11-fold enhancement (line 3). This genetic interaction supports the conclusion from the anti-MEI-1 staining in Figure 2C that cul-2 and mel-26 work in parallel to eliminate MEI-1.
Table 1. Genetic interactions of MEI-1 pathway components.
| Temperature | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 15° | 20° | 25° | ||||||||
| Maternal Genotypea | % Hatch | Fold Change | % Male | % Hatch | Fold Change | % Male | % Hatch | Fold Change | % Male | |
| 1 | cul-2(ts) | 64 | 0.5 | 51 | 2.6 | |||||
| 2 | mel-26(null) | 12 | ||||||||
| 3 | cul-2(ts); mel-26(null) | 0.7 | 11 × ↓ | |||||||
| 4 | tbb-2b | 100 | 0 | |||||||
| 5 | cul-2(ts) tbb-2 | 40 | 1.6 × ↓ | 2.6 | 27 | 1.9 × ↓ | 6.0 | |||
| 6 | rfl-1(ts) | 89 | 72 | |||||||
| 7 | rfl-1(ts); mel-26(null) | 0.9 | 12 × ↓ | |||||||
| 8 | mel-26(null); tbb-2 | 68 | 7.8 × ↑ | |||||||
| 9 | rfl-1(ts); tbb-2 | 92 | ||||||||
| 10 | rfl-1(ts); mel-26(null); tbb-2 | 65 | 77 × ↓ | |||||||
| 11 | mbk-2(ts) | 87 | 59 | |||||||
| 12 | rfl-1(ts); mbk-2(ts) | 35 | 2.2 × ↓ | 0.5 | 85 × ↓ | |||||
| 13 | mbk-2(ts); tbb-2 | 78 | ||||||||
| 14 | rfl-1(ts); mbk-2(ts); tbb-2 | 1.5 | ||||||||
| 15 | hecd-1 | 89 | 96 | 0 | 77 | 0 | ||||
| 16 | hecd-1; mel-26(null) | 0c | > 190 × ↓ | |||||||
| 17 | hecd-1(RNAi) | 99 | 99 | |||||||
| 18 | hecd-1(RNAi); mel-26(null) | 0c | > 150 × ↓ | |||||||
| 19 | hecd-1; tbb-2 | 91 | 85 | 0.4 | ||||||
| 20 | hecd-1(RNAi); mel-26(null); tbb-2 | 50 | ||||||||
| 21 | mei-1(gf) | 12 | ||||||||
| 22 | hecd-1(RNAi); mei-1(gf) | 0c | > 60 × ↓ | |||||||
| 23 | mbk-2(ts) | 74 | ||||||||
| 24 | hecd-1; mbk-2(ts) | 81 | ||||||||
| 25 | mei-2(sb39ts) | 78 | 1.3 | 41 | 2.2 | |||||
| 26 | hecd-1; mei-2(sb39ts) | 28 | 2.5 × ↓ | 3.4 | 8 | 4.9 × ↓ | 13 | |||
| 27 | mei-2(ct98) | 71 | 2.6 | |||||||
| 28 | hecd-1; mei-2(ct98) | 19 | 2.9 × ↓ | 8.9 | ||||||
| 29 | cul-2(ts); hecd-1 | 16 | 3.5 × ↓ | 2.3 | 11 | 4.4 × ↓ | 12 | |||
ts alleles are presumed hypomorphs at intermediate temperatures. hecd-1 = ok1437, cul-2(ts) = or209, mbk-2(ts) = dd5, mei-1(ts) = or1178, mei-1(gf) = ct46, mel-26(null) = ct61sb4, mel-26(ts) = or184, rfl-1(ts) = or198, and tbb-2 = sb26.
Data from Lu et al. (2004).
N values for lines 16, 18, and 22 are 1587, 1290, and 501, respectively. Fold enhancement is calculated based on if one embryo had hatched.
We examined the role of cul-2 in meiosis and asked if premature MEL-26 expression in cul-2(or209ts) could in turn lower the amount of MEI-1 available for proper meiotic spindle formation and so cause meiotic defects. This situation would be equivalent to mei-1 or mei-2 hypomorphs, which result in meiotic nondisjunction due to spindle defects. While nondisjunction of autosomes results in lethality, if only the X chromosome is lost, viable XO males result (Him phenotype, high incidence of males). Thus, male progeny serve as an easily-scored metric of meiotic spindle defects (Mains et al. 1990a; Clandinin and Mains 1993; Lu and Mains 2005). The wild-type baseline frequency of XO males resulting from the selfing of XX hermaphrodites is 0.2% (Hodgkin et al. 1979). cul-2(or209ts) produced over 10-fold (2.6%) more males at 20°, indicating meiotic defects (Table 1, line 1). To determine if decreased MT severing was the cause of this Him phenotype, we compromised MEI-1 activity using the tbb-2(sb26) allele that encodes a β-tubulin refractory to MEI-1 severing. In addition to decreasing MT severing at mitosis (which restores normal morphology to embryos with ectopic mitotic MEI-1 as seen in Figure 2) (Lu et al. 2004; Lu and Mains 2005), this tubulin allele also decreases the normal MT severing that is required during meiosis: while tbb-2(sb26) alone does not increase the frequency of XO males, it does so in combination with mei-2 hypomorphs (Lu et al. 2004). Indeed, the Him phenotype of cul-2(or209ts) was increased from the 2.6% described above to 6.0% with the addition of the tubulin allele at 20° (lines 1 and 5). At 15°, where cul-2 alone had 0.5% males, cul-2(or209ts) tbb-2(sb26) hermaphrodites had 2.6% male offspring (line 5). These data with tbb-2 indicate that MT severing is the relevant target for the cul-2 Him phenotype. This is consistent with the loss of cul-2 resulting in increased MEL-26, which in turn decreases MEI-1 activity during meiosis.
rfl-1 acts upstream of CUL-2, but not MEL-26, to regulate MEI-1
RFL-1 is a component of an E1 complex that leads to ubiquitin ligase activation by coupling the Nedd8 ubiquitin-like molecule to cullins (Kurz et al. 2002; Pintard et al. 2003a; Petroski and Deshaies 2005; Sarikas et al. 2011). Like mel-26 and cul-2, rfl-1 has ectopic MEI-1 in mitosis (Kurz et al. 2002; Pintard et al. 2003a). To determine where RFL-1 is acting in MEI-1 regulation, we used the ts allele of rfl-1(or198) (Dorfman et al. 2009). rfl-1 embryos showed 1.8-fold more MEI-1 staining than the wild type at the semipermissive temperature of 20° (Figure 2C). We found that there was no significant difference (P > 0.13) in ectopic MEI-1 staining levels between the single knockdowns, rfl-1 and mbk-2(ts+RNAi), and the double rfl-1; mbk-2(ts+RNAi). In addition, rfl-1; cul-2(RNAi) showed no significant increase (P > 0.15) in ectopic MEI-1 staining compared to each single at 20°. This suggests that RFL-1 is acting in the same pathway as MBK-2 and CUL-2. In contrast, at 20°, rfl-1; mel-26(null) embryos did show significantly higher (P = 0.001) ectopic MEI-1 staining than mel-26(null) embryos (Figure 2C), indicating that rfl-1 and mel-26 likely act in parallel. Thus, the data indicate that RFL-1 appears to contribute to CUL-2-, but not CUL-3, -mediated MEI-1 degradation (Figure 1).
We examined hatching rates of double mutants between rfl-1 and mei-1 pathway mutations as another way to determine in which degradation pathway rfl-1 functions. rfl-1 strongly enhanced mel-26(null) lethality, leading to 12-fold less hatching than predicted if the genes acted independently (Table 1, lines 2, 6, and 7), supporting our conclusion that rfl-1 acts in parallel to MEL-26. To determine if MT severing was the underlying cause of the enhanced lethality, rather than some other target shared by MEL-26 and RFL-1, we examined the effect of the tbb-2(sb26) allele that inhibits ectopic MEI-1 activity. As reported previously (Lu et al. 2004), tbb-2(sb26) suppresses mel-26 lethality (lines 2 and 8). tbb-2(sb26) had no effect on rfl-1 alone (line 9, Johnson et al. 2009), but tbb-2(sb26) did block the enhancement between mel-26 and rfl-1 (line 10). Thus, MEI-1/MT severing was the relevant target of the rfl-1 genetic interaction with mel-26.
We next examined rfl-1 genetic interactions with the mbk-2 pathway. Unexpectedly, rfl-1 also strongly enhanced mbk-2 (85-fold at 20°; Table 1, lines 11 and 12). However, this lethality was not blocked by tbb-2(sb26) (lines 13 and 14) indicating that these genes likely share essential targets other than MEI-1. Indeed, while CUL-3 and MEL-26 may have only MEI-1 as their major target (Lu and Mains 2005, 2007), MBK-2, CUL-2, and RFL-1 have many other targets in the C. elegans germline and early embryo (Feng et al. 1999; DeRenzo et al. 2003; Pellettieri et al. 2003; Liu et al. 2004; Sonneville and Gonczy 2004; Sasagawa et al. 2005; Stitzel et al. 2006; Lu and Mains 2007; Dorfman et al. 2009; Guven-Ozkan et al. 2010; Starostina et al. 2010; Burger et al. 2013; Brockway et al. 2014; Wang et al. 2014).
Candidate substrate recognition subunits used by CUL-2 leading to MEL-26 or MEI-1 inactivation
The CUL-2-containing E3 ubiquitin ligase apparently switches from targeting MEL-26 during meiosis to MEI-1 during mitosis (Figure 1). However, we do not know if either interaction reflects direct degradation or involves intermediates. Identification of the substrate recognition subunits used by CUL-2 for MEL-26 and MEI-1 regulation would help address these questions. Thus, we tested candidates for the substrate recognition subunits using the criteria that depleting a substrate adaptor targeting MEL-26 would suppress a mel-26 hypomorph and enhance mei-1 or mei-2 hypomorphs. A substrate adaptor for MEI-1 (or MEI-2) would show the opposite interactions. In addition, these adaptors should alter the levels of MEI-1 or MEL-26. Follow-up testing for physical interactions between the substrate recognition subunits and MEI-1 pathway genes could then determine if CUL-2 targets each protein directly. However, none of the tested genes described below fulfilled our criteria.
In C. elegans, five proteins have been shown to bind CUL-2 as known or candidate substrate adaptors: FEM-1, ZYG-11, ZER-1, VHL-1, and LRR-1 (Starostina et al. 2007; Vasudevan et al. 2007; Merlet et al. 2010; Burger et al. 2013). We tested the first four for genetic interactions with mel-26 and mei-1 (lrr-1 was not tested due to its sterility).
ZYG-11 acts with CUL-2 during meiotic progression (Liu et al. 2004; Sonneville and Gonczy 2004; Vasudevan et al. 2007). Double mutants between zyg-11 and gain of function alleles of mel-26 or mei-1 have been shown previously to have minimal enhancement of embryonic lethality (Mains et al. 1990a). We also found no interactions between zyg-11(ts) and lf alleles of mei-1, mei-2, or mel-26 (Table S1, lines 1–9). We previously reported that zyg-11 does not affect MEL-26 accumulation (Johnson et al. 2009).
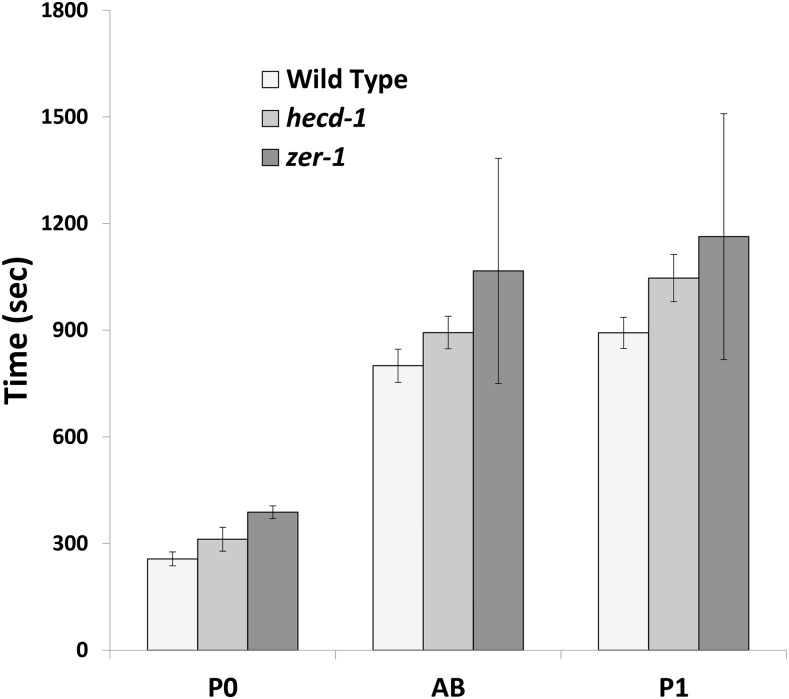
ZER-1 is a ZYG-11-related gene that binds CUL-2, but zer-1 has no phenotype other than weakly enhancing zyg-11 (Vasudevan et al. 2007). The zer-1(gk165593) allele (Q336stop in the 860 amino acid protein) suppressed mel-26(null) lethality. Additionally, zer-1 also suppressed mbk-2(ts) (Table S1, lines 10–18). The suppression of both mel-26(null) and mbk-2 by zer-1 argue that zer-1 is an activator of MEI-1, likely independent of those two pathways. However, zer-1 showed little or no enhancement of mei-1 or mei-2 hypomorphs. MEI-1, MEI-2, and PPFR-1 staining of zer-1 embryos did not show any obvious changes (Figure S2, A–L). Since previous work showed that mbk-2(ts) is suppressed by mutations that extend the cell cycle (Wang et al. 2014), we measured the lengths of the first two rounds of cleavage of zer-1 embryos. Indeed, zer-1 embryos have significantly longer cell cycle times (Figure 3). It is not clear if this alone explains the suppression of mel-26: as described below, hecd-1 also has longer cell cycle times, but hecd-1 enhanced rather than suppressed mel-26 and mei-1 defects.
Figure 3.
Cell cycles of hecd-1 and zer-1. Embryos were observed by Nomarski microscopy at room temperature (∼23°) and the length of the cell cycles were noted. P0 cycle length was measured from pronuclear fusion to when the cleavage furrow fully bisected the daughter cells. The AB and P1 cell divisions were scored from one cleavage to the next. hecd-1 and zer-1 both lengthened cell cycles compared to wild-type for all three divisions (P < 0.04, unpaired t-tests, N ≥ 5 for each division of each genotype).
FEM-1 acts with CUL-2 to target the TRA-1 sex determination transcription factor (Starostina et al. 2007) and the PAR-6 polarity protein in the early embryo (Pacquelet et al. 2008). The ts allele fem-1(hc17ts) showed the anticipated pattern of the substrate adapter for MEL-26 suppressing mel-26(ts) while enhancing mei-1(ts), although both interactions were modest (Table S1, lines 19–27). However, the allele used, hc17ts, is an unusual fem-1 mutation in affecting only the gonad and not the soma (Hodgkin 1986). The fem-1 null allele, e1965, failed to recapitulate the genetic interactions of hc17ts. Furthermore, while FEM-1 acts with FEM-2 in regulating TRA-1 degradation, we found no genetic interactions with ts or null alleles of fem-2 (Table S1, lines 28–33). Finally, fem-1 did not affect MEL-26 accumulation as would be anticipated for a MEL-26 substrate adapter (Figure S2, M–P).
vhl-1 is involved in C. elegans DNA repair and longevity (Muller et al. 2009; Sendoel et al. 2010). However, vhl-1(RNAi) had no genetic interactions with mel-26(null) or mel-26(ts) (Table S1, lines 34–36).
hecd-1 enhances mel-26(null) but does not affect MEI-1 levels
In an RNAi screen for ubiquitin ligases acting in parallel to MEL-26, we identified HECD-1, a HECT domain (Homologous to the E6-AP Carboxyl Terminus) E3 ubiquitin ligase, the ortholog of human Hectd1 (Chen and Greenwald 2014). A null allele hecd-1(ok1437) (Chen and Greenwald 2014) and hecd-1(RNAi) showed little effect on viability on their own but both decreased embryonic viability of mel-26(null) mutants, from 12 to 0% (N = 1587 and 1290, respectively), and so enhancement was at least 190-fold (Table 1, lines 2 and 15–18). tbb-2(sb26) rescued mel-26; hecd-1(RNAi) lethality (lines 19 and 20), indicating that MTs are the relevant target for the hecd-1 genetic interactions. This interpretation was confirmed by showing that a mei-1(ct46gf) allele, which is refractory to MEL-26-mediated degradation (Clark-Maguire and Mains 1994a; Pintard et al. 2003b), was also enhanced by hecd-1(RNAi) (lines 21 and 22). In contrast, hecd-1 did not affect mbk-2 lethality (lines 23 and 24). Together, these results indicate that HECD-1 is an inhibitor of MEI-1 function, acting in parallel to MEL-26.
Loss of a MEI-1 inhibitor could suppress hypomorphic MEI-1 activity. Unexpectedly, hecd-1 instead showed a two-to-five-fold enhancement of the lethality of two different mei-2 hypomorphs. This lethality was accompanied by an increase in the percentage of XO males among the surviving progeny (Table 1, lines 15 and 25–28): hecd-1 alone did not increase the level of spontaneous males, but there was a sixfold increase in male progeny for mei-2(sb39ts); hecd-1 at 20° and 3.5-fold for mei-2(ct98); hecd-1 at 25°. The increase in male progeny (likely via meiotic nondisjunction) suggests that the enhancement is due to meiotic spindle defects rather than nonspecific interactions leading to lethality. Thus, HECD-1 is acting as an activator of MEI-1 function at meiosis but as an inhibitor in mitosis (Figure 1). Finally, hecd-1 also enhanced cul-2 lethality, and this was accompanied by a fivefold increase in male progeny (Table 1, lines 1, 15, and 29).
We hypothesized that HECD-1 contributes to the degradation of mitotic MEI-1 in parallel to MEL-26. However, quantitative staining of hecd-1 embryos at 25° showed wild-type MEI-1 in mitosis (Figure 2C and Figure S3, A–D). Furthermore, hecd-1 did not affect MEI-1 levels (P > 0.4) in sensitized backgrounds that included mbk-2(dd5ts+RNAi), cul-2, or mel-26 (Figure 2C). HECD-1 could also influence regulators of MEI-1 activity, but we found no obvious changes in MEI-2, MEL-26, or the MEI-1 activator PPFR-1 by immunostaining (Figure S3, E–P).
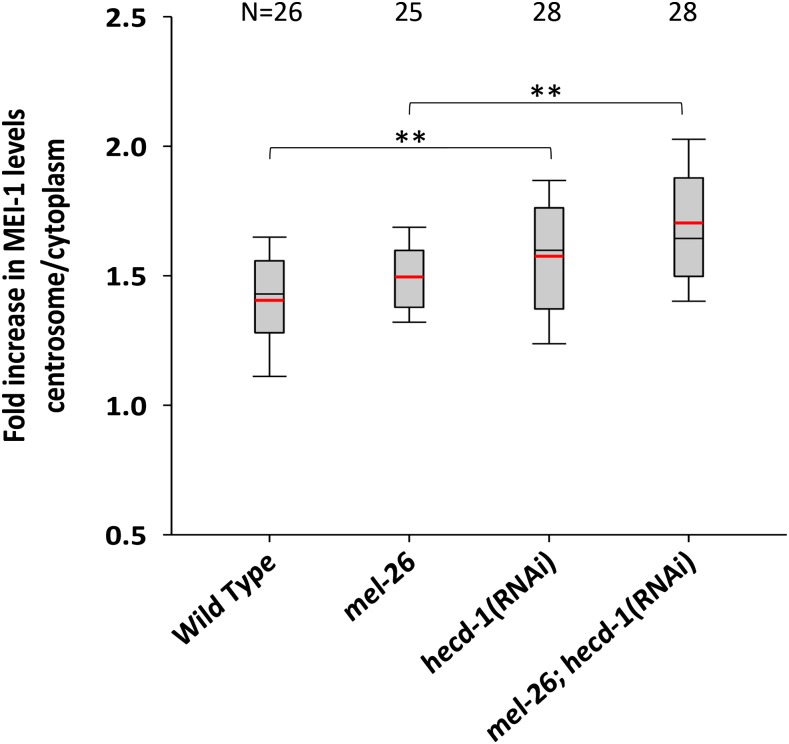
HECT domain proteins often alter the localization or trafficking of their targets rather than leading to proteosomal degradation (Mukhopadhyay and Riezman 2007; Metzger et al. 2012; Sarkar and Zohn 2012; Sato et al. 2014). The immunostaining results imply that HECD-1 is not influencing MEI-1 degradation. To test the effect of HECD-1 on MEI-1 location, we measured the ratio of anti-MEI-1 staining at the centrosome compared to the embryo as a whole. The ratio increased from an average of 1.5 in mel-26 to 1.7 in mel-26; hecd-1 (Figure 4). Although modest, this was statistically significant (P < 0.001). However, it is not clear that this small change alone accounts for the strong genetic interactions.
Figure 4.
Quantification of centrosomal anti-MEI-1 levels. The MEI-1 level at the centrosome was compared to the embryo as a whole in wild-type, mel-26, and hecd-1 embryos at 25°. Boxes indicate the 25th to the 75th percentiles and whiskers show the 10th and 90th percentiles. Black lines are at the median and red lines at the mean. hecd-1(RNAi) increased the ratio of centrosome/embryo relative to wild-type and also increased the ratio in the presence of mel-26(null). mel-26 strains included the suppressor mutation tbb-2(sb26) to restore normal embryo morphology. **: P ≤ 0.003. N values are shown at the top of the figure. Note that unlike Figure 2, 1.0 does not represent the wild-type measurement. RNAi, RNA interference.
We examined the effects of hecd-1 on the cell cycle. One possibility is that hecd-1 shortens the cell cycle to decrease the ability of other compromised MEI-1 regulators to complete their function, which might explain the genetic enhancement with MEI-1 pathway genes. However, hecd-1 had the opposite effect, lengthening the cell cycle (Figure 3). Both hecd-1 and zer-1 lengthen the cell cycle, but have opposite genetic interactions with mel-26.
Discussion
At fertilization, the C. elegans embryo rapidly transitions from the meiotic to mitotic divisions (Albertson 1984; Kemphues et al. 1986; McCarter et al. 1999; Yang et al. 2003). Meiosis-specific products, such as the MEI-1/MEI-2 katanin MT severing complex, must be downregulated or eliminated as their persistence would be detrimental to later development (Clark-Maguire and Mains 1994a; Bowerman and Kurz 2006; Stitzel and Seydoux 2007; Verlhac et al. 2010; Robertson and Lin 2013). By measuring genetic interactions using quantitative antibody staining and embryo hatching rates, we determined how APC/C, MBK-2, CUL-2, RFL-1, and HECD-1 function relative to one another (Figure 1). Some genetic interactions as measured by embryo viability were modest, sometimes in the range of only twofold from that predicted assuming independence. These could be misleading and might represent nonspecific effects on general viability. Therefore, we focused on genetic interactions likely to be specific to the MEI-1 pathway: those that also increase meiotic nondisjunction, a characteristic of hypomorphic MEI-1 function, or interactions that were modified by a tubulin allele that specifically affects MEI-1-mediated MT severing.
APC/C and MBK-2 act sequentially to degrade MEI-1 in mitosis
APC/C and MBK-2 were previously shown to act in parallel to CUL-3/MEL-26 for MEI-1 degradation (Lu and Mains 2007). While APC/C is required for the activation of MBK-2 to trigger MEI-1 degradation at meiotic exit (Maruyama et al. 2007; Stitzel et al. 2007), it was possible that APC/C could also act directly on the MEI-1/MEI-2 complex in a third pathway. Indeed, MEI-2 has an APC/C consensus destruction box (D-box). Here, we have demonstrated that APC/C and MBK-2 act strictly sequentially with one another and in parallel with MEL-26: double mutants of emb-27 (APC-6/CDC16) and mbk-2 showed the same level of anti-MEI-1 staining as the single mutants, demonstrating that the genes act in the same pathway. In contrast, mutations of the two genes increased MEI-1 levels when each were combined with mel-26, demonstrating that emb-27 and mbk-2 act in parallel with mel-26 (Figure 2).
Gomes et al. (2013) showed that mutating the MBK-2 phosphorylation site on MEI-1 affects MEL-26 interactions with MEI-1 in yeast two-hybrid assays. This may imply that MBK-2 is also acting within the MEL-26/CUL-3 pathway in addition to our evidence that MBK-2 and MEL-26 act in parallel. However, this predicts that the mbk-2 single mutant would have MEI-1 levels higher than the loss of mel-26 alone, which was not observed (Figure 2). Possibilities to explain the lack of a genetic requirement of mbk-2 for mel-26 function is that another kinase phosphorylates the same MEI-1 residue as does MBK-2, or that in vivo interactions between unphosphorylated MEI-1 and MEL-26 are sufficiently strong to mediate MEI-1 degradation.
CUL-2 regulates MT severing by multiple pathways
Given that MBK-2 is acting in parallel to the MEL-26/CUL-3 E3 ubiquitin ligase, we asked if the CUL-2 E3 ubiquitin ligase was acting in concert with MBK-2. Kurz et al. (2002) reported a weakly penetrant cul-2(RNAi) phenotype of small, misoriented spindles typical of cul-3 and mel-26. Previous work found that MEI-1 persists into mitosis when CUL-2 was inactivated (Stitzel et al. 2006; Johnson et al. 2009). Our quantitative staining showed that CUL-2 acts with MBK-2, in parallel to MEL-26: cul-2(RNAi) increases MEI-1 levels in mel-26 but not mbk-2 backgrounds (Figure 2). The genetic enhancement of mel-26 lethality by cul-2 also suggests that the two genes act in parallel (Table 1).
CUL-2 acts at two points of MEI-1 regulation, having opposite effects at meiosis and mitosis (Figure 1). MEL-26 levels are low during meiosis but then increase. When cul-2 is lost, MEL-26 accumulates prematurely to high levels during meiosis (although mitotic levels are not increased) (Johnson et al. 2009). We asked if high meiotic MEL-26 in cul-2 mutants resulted in the meiotic defects due to lowered MEI-activity. Meiotic spindle defects stemming from suboptimal MEI-1 activity result in a high incidence of males (Him) phenotype due to nondisjunction generating XO males from XX hermaphrodite oocytes (Hodgkin et al. 1979; Clandinin and Mains 1993). Notably, at semipermissive temperatures, cul-2(ts) has the Him phenotype. It is possible that this nondisjunction phenotype could arise from the delayed meiotic progression seen in cul-2 mutants (Liu et al. 2004; Sonneville and Gonczy 2004) rather than MEI-1 misregulation. However, the cul-2 Him phenotype was likely mediated through compromised MEI-1 function as males were further increased when cul-2 was combined with the tubulin mutation tbb-2(sb26) (Lu et al. 2004) that hinders MEI-1 activity (Table 1). The allele mei-1(ct46ct103) shows a similar mixture of both meiotic and mitotic defects to those of cul-2. This mutation decreases but does not eliminate MEI-1 enzymatic function, resulting in a meiotic Him phenotype, but the product persists into mitosis with sufficient activity to cause mitotic phenotypes (Clandinin and Mains 1993; Clark-Maguire and Mains 1994b; McNally and McNally 2011; McNally et al. 2014).
CUL-2 targets many substrates in the germline and acquires additional substrates in the newly fertilized embryo (Feng et al. 1999; DeRenzo et al. 2003; Liu et al. 2004; Sonneville and Gonczy 2004; Pacquelet et al. 2008; Merlet et al. 2010; Starostina et al. 2010). We do not know if CUL-2 regulation of MEL-26 or MEI-1 is direct and so we tried to identify the substrate adapters that could mediate the genetic interactions. Unlike mutations of cul-2 that both increase MEI-1 activity in mitosis while decreasing MEI-1 in meiosis (via ectopic MEL-26), the loss of a substrate adapter would most likely affect meiosis or mitosis, but not both. Thus an adaptor would likely have reciprocal interactions with hypomorphs of mel-26 vs. mei-1 or mei-2. Unfortunately, none of the candidates tested met these genetic criteria (Table S1) or altered levels of MEI-1, MEI-2, MEL-26, and/or PPFR-1 (Figure S2).
RFL-1 activates the MBK-2/CUL-2-mediated MEI-1 degradation pathway
RFL-1 leads to the neddylation of cullins, activating these E3 ubiquitin ligases (Kurz et al. 2002; Pintard et al. 2003a; Bosu et al. 2010; Sarikas et al. 2011; Pierce et al. 2013). RFL-1 is involved in neddylation of both C. elegans CUL-2 and CUL-3 (Luke-Glaser et al. 2007; Dorfman et al. 2009; Bosu et al. 2010). RFL-1 appears to function with CUL-2 to temporally regulate MEL-26 meiotic levels (Johnson et al. 2009). During mitosis, we found that RFL-1 acts sequentially with the MBK-2/CUL-2 pathway and in parallel to MEL-26-mediated MEI-1 degradation; loss of rfl-1 further increased MEI-1 levels when combined with mel-26 but did not do so in combination with mbk-2 or cul-2(RNAi) (Figure 2). rfl-1 enhancement of mel-26 lethality also indicates that the genes act in parallel and this was ameliorated by tbb-2(sb26), demonstrating that MEI-1 is a relevant target (Table 1). rfl-1 also enhanced lethality in the parallel mbk-2 pathway, which would imply that RFL-1 acts in both MEI-1 degradation pathways. However, rfl-1; mbk-2 enhanced lethality did not stem from MEI-1 misregulation since MEI-1 levels were not increased in the double mutant and lethality was not suppressed by tbb-2(sb26) (Figure 2 and Table 1). Furthermore, if RFL-1 acted in both pathways, rfl-1 mutants alone would have very high levels of ectopic MEI-1, which was not the case (Figure 2). Thus, mbk-2 and rfl-1 must act in parallel on other targets to explain the enhancement of lethality. Many targets other than MEI-1 have been reported for MBK-2 and RFL-1 (Feng et al. 1999; DeRenzo et al. 2003; Pellettieri et al. 2003; Liu et al. 2004; Sonneville and Gonczy 2004; Sasagawa et al. 2005; Stitzel et al. 2006; Lu and Mains 2007; Dorfman et al. 2009; Guven-Ozkan et al. 2010; Starostina et al. 2010; Burger et al. 2013; Brockway et al. 2014; Wang et al. 2014).
Previous reports assumed that the effects of RFL-1 on MEI-1 levels were mediated through CUL-3/MEL-26 rather than CUL-2 (Kurz et al. 2002, 2005; Pintard et al. 2003a; Dorfman et al. 2009). rfl-1 mutants affect neddylation of both CUL-2 and CUL-3 (Luke-Glaser et al. 2007; Dorfman et al. 2009; Bosu et al. 2010). Neddylation/deneddylation cycles are proposed to activate Cullin complexes by mediating the exchange of substrate adaptors (Pierce et al. 2013). Since most cul-3(RNAi) lethality is suppressed by tbb-2(sb26) (Johnson et al. 2009), MEI-1 may be the major essential CUL-3 target and so dynamic exchange of CUL-3 substrate adapters might not be critical to the C. elegans embryo. Meanwhile, CUL-2 may require RFL-1 for MEI-1 degradation to facilitate exchange of the substrate adaptors for other CUL-2 targets in the germline and embryo (Feng et al. 1999; DeRenzo et al. 2003; Liu et al. 2004; Sonneville and Gonczy 2004; Sasagawa et al. 2005; Dorfman et al. 2009; Guven-Ozkan et al. 2010; Starostina et al. 2010; Burger et al. 2013; Brockway et al. 2014). In addition to MEI-1 degradation, RFL-1 appears to also function with CUL-2 by maintaining low meiotic MEL-26 levels for timely progression through meiosis (Johnson et al. 2009).
HECD-1 acts in MEI-1 regulation without affecting protein levels
We found that HECD-1 genetically acts as both a MEI-1 activator in meiosis (hecd-1 enhanced mei-2 hypomorphs) and a MEI-1 inhibitor in mitosis (hecd-1 enhanced mutations that result in ectopic MEI-1, Table 1). Both interactions appeared specific to MEI-1 regulation, as hecd-1 increased mei-2 meiotic nondisjunction (as indicated by XO male offspring) and hecd-1 enhancement of mel-26 was suppressed by tbb-2(sb26). hecd-1 increased mel-26 lethality by > 190-fold, the strongest enhancement we have observed in the mei-1 system. However, hecd-1 had no discernible effects on protein levels of MEI-1, MEI-2, or PPFR-1 (Figure 2 and Figure S3).
HECTD E3 ubiquitin ligases differ from cullin-based E3 ligases (e.g., worm CUL-2/3) in that they directly bind the substrate without an adapter subunit and can build ubiquitin chains via K63 instead of K48 linkages (Mukhopadhyay and Riezman 2007; Metzger et al. 2012). K63 ubiquitin modifies protein location, protein–protein interactions, and/or trafficking, although cases of target degradation are known (Li et al. 2013; Segref et al. 2014). HECTD E3 ligases function in mammalian neural tube formation, regulate heat shock protein 90 secretion and subsequent cell migration (Sarkar and Zohn 2012), placental development (Sarkar et al. 2014), and Wnt signaling (Tran et al. 2013). C. elegans hecd-1 acts in Notch signaling (Chen and Greenwald 2014), vulva development (Cui et al. 2006), mitochondrial metabolism (Segref et al. 2014), and in membrane protein endocytosis after fertilization (Sato et al. 2014).
Although we did not observe any change in MEI-1, MEI-2, or PPFR-1 levels in hecd-1 mutants, effects could be subtle. A speculative model is that HECD-1 modifies MEI-1 or MEI-2 to increase function in the meiotic relative to the mitotic spindles. Thus, hecd-1 loss would shift the balance back toward mitotic MEI-1 function. This would explain why hecd-1 enhances mutants resulting in both meiotic loss and mitotic gain of MEI-1. We did find that hecd-1 increased MEI-1 centrosomal localization relative to the cytoplasm (Figure 4), but the effects were modest and it is not clear if they are sufficient to explain our genetic interactions. Further studies will focus on identifying HECD-1 targets.
In conclusion, our results indicate that many levels of regulation are involved in ensuring that MEI-1 is quickly inactivated before mitosis to allow for proper embryogenesis. We showed that the APC/C acts strictly sequentially with MBK-2. We demonstrated a role for CUL-2 acting in concert with MBK-2 to degrade MEI-1. The neddylation pathway gene rfl-1 regulates MEI-1 through CUL-2 rather than CUL-3. Finally, we identified a role for HECD-1 in regulating MEI-1 at both meiosis and mitosis, apparently independent of protein levels. Further analysis of these players are needed to decipher their exact roles in the process.
Supplementary Material
Acknowledgments
We would like to thank J. D. McGhee and D. Hansen and members of their labs for their input during the course of this project. We thank L. Pintard, E. Kipreos, and M. Labouesse for antibodies, plasmids, and strains. C. Wang, P. Gupta, D. Hansen, and B. Gavinolla are thanked for technical assistance. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a grant from the Canadian Institute of Health Research to P.E.M.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.031666/-/DC1
Communicating editor: M. C. Zetka
Literature Cited
- Albertson D. G., 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101: 61–72. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Bosu D. R., Feng H., Min K., Kim Y., Wallenfang M. R., et al. , 2010. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Dev. Biol. 346: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Kurz T., 2006. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development 133: 773–784. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway H., Balukoff N., Dean M., Alleva B., Smolikove S., 2014. The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans. PLoS Genet. 10: e1004757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J., Merlet J., Tavernier N., Richaudeau B., Arnold A., et al. , 2013. CRL(2LRR-1) E3-ligase regulates proliferation and progression through meiosis in the Caenorhabditis elegans germline. PLoS Genet. 9: e1003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Greenwald I., 2014. hecd-1 Modulates Notch Activity in Caenorhabditis elegans. G3 (Bethesda) 5: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Klancer R., Singson A., Seydoux G., 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139: 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T. R., Mains P. E., 1993. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics 134: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P. E., 1994a Localization of the mei-1 gene product of Caenorhabditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P. E., 1994b mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136: 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Kim E. B., Han M., 2006. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. S., Wille L., Chestnut B. A., Sadler P. L., Shakes D. C., et al. , 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C., Reese K. J., Seydoux G., 2003. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman M., Gomes J. E., O’Rourke S., Bowerman B., 2009. Using RNA interference to identify specific modifiers of a temperature-sensitive, embryonic-lethal mutation in the Caenorhabditis elegans ubiquitin-like Nedd8 protein modification pathway E1-activating gene rfl-1. Genetics 182: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow M. R., Mains P. E., 1998. Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics 150: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., et al. , 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1: 486–492. [DOI] [PubMed] [Google Scholar]
- Furukawa M., He Y. J., Borchers C., Xiong Y., 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J. E., Tavernier N., Richaudeau B., Formstecher E., Boulin T., et al. , 2013. Microtubule severing by the katanin complex is activated by PPFR-1-dependent MEI-1 dephosphorylation. J. Cell Biol. 202: 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grode K. D., Rogers S. L., 2015. The non-catalytic domains of Drosophila katanin regulate its abundance and microtubule-disassembly activity. PLoS One 10: e0123912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Robertson S. M., Nishi Y., Lin R., 2010. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development 137: 3373–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Gomes J. E., Birmingham C. L., Pintard L., Sugimoto A., et al. , 2009. The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo. Genetics 181: 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Burton J. L., Solomon M. J., 2002. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 16: 2179–2206. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., 1986. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114: 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction Mutants of the Nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Lu C., Raharjo E., McNally K., McNally F. J., et al. , 2009. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis. Dev. Biol. 330: 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Wolf N., Wood W. B., Hirsh D., 1986. Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. Dev. Biol. 113: 449–460. [DOI] [PubMed] [Google Scholar]
- Kurz T., Pintard L., Willis J. H., Hamill D. R., Gonczy P., et al. , 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295: 1294–1298. [DOI] [PubMed] [Google Scholar]
- Kurz T., Ozlu N., Rudolf F., O’Rourke S. M., Luke B., et al. , 2005. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435: 1257–1261. [DOI] [PubMed] [Google Scholar]
- Li W., DeBella L. R., Guven-Ozkan T., Lin R., Rose L. S., 2009. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J. Cell Biol. 187: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhou Q., Sunkara M., Kutys M. L., Wu Z., et al. , 2013. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J. Cell Sci. 126: 2617–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Vasudevan S., Kipreos E. T., 2004. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development 131: 3513–3525. [DOI] [PubMed] [Google Scholar]
- Lu C., Mains P. E., 2005. Mutations of a Redundant α-Tubulin Gene Affect Caenorhabditis elegans Early Embryonic Cleavage via MEI-1/Katanin-Dependent and -Independent Pathways. Genetics 170: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Mains P. E., 2007. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev. Biol. 302: 438–447. [DOI] [PubMed] [Google Scholar]
- Lu C., Srayko M., Mains P. E., 2004. The Caenorhabditis elegans Microtubule-severing Complex MEI-1/MEI-2 Katanin Interacts Differently with Two Superficially Redundant beta-Tubulin Isotypes. Mol. Biol. Cell 15: 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S., Roy M., Larsen B., Bihan T. L., Metalnikov P., et al. , 2007. CIF-1, a shared subunit of the COP9/Signalosome and eIF3 complexes regulates MEL-26 levels in the C. elegans embryo. Mol. Cell. Biol. 27: 4526–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains P. E., Kemphues K. J., Sprunger S. A., Sulston I. A., Wood W. B., 1990a Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 126: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains P. E., Sulston I. A., Wood W. B., 1990b Dominant maternal-effect mutations causing embryonic lethality in Caenorhabditis elegans. Genetics 125: 351–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R., Velarde N. V., Klancer R., Gordon S., Kadandale P., et al. , 2007. EGG-3 Regulates Cell-Surface and Cortex Rearrangements during Egg Activation in Caenorhabditis elegans. Curr. Biol. 17: 1555–1560. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McNally F. J., Vale R. D., 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75: 419–429. [DOI] [PubMed] [Google Scholar]
- McNally K. P., McNally F. J., 2011. The spindle assembly function of C. elegans katanin does not require microtubule-severing activity. Mol. Biol. Cell 22: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J., 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K., Berg E., Cortes D. B., Hernandez V., Mains P. E., et al. , 2014. Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol. Biol. Cell 25: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlet J., Burger J., Tavernier N., Richaudeau B., Gomes J. E., et al. , 2010. The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137: 3857–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M. B., Hristova V. A., Weissman A. M., 2012. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Pang K., Ishidate T., Nakamura K., Shirayama M., Trzepacz C., et al. , 2004. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev. Biol. 265: 127–139. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H., 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205. [DOI] [PubMed] [Google Scholar]
- Muller R. U., Fabretti F., Zank S., Burst V., Benzing T., et al. , 2009. The von Hippel Lindau tumor suppressor limits longevity. J. Am. Soc. Nephrol. 20: 2513–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A., Zanin E., Ashiono C., Gotta M., 2008. PAR-6 levels are regulated by NOS-3 in a CUL-2 dependent manner in Caenorhabditis elegans. Dev. Biol. 319: 267–272. [DOI] [PubMed] [Google Scholar]
- Parry J. M., Velarde N. V., Lefkovith A. J., Zegarek M. H., Hang J. S., et al. , 2009. EGG-4 and EGG-5 Link Events of the Oocyte-to-Embryo Transition with Meiotic Progression in C. elegans. Curr. Biol. 19: 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G., 2003. Coordinate Activation of Maternal Protein Degradation during the Egg-to-Embryo Transition in C. elegans. Dev. Cell 5: 451–462. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J., 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., et al. , 2013. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., Kurz T., Glaser S., Willis J. H., Peter M., et al. , 2003a Neddylation and Deneddylation of CUL-3 Is Required to Target MEI-1/Katanin for Degradation at the Meiosis-to-Mitosis Transition in C. elegans. Curr. Biol. 13: 911–921. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J. H., Willems A., Johnson J. L., Srayko M., et al. , 2003b The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Quintin S., Mains P. E., Zinke A., Hyman A. A., 2003. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 4: 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S., Lin R., 2013. The oocyte-to-embryo transition. Adv. Exp. Med. Biol. 757: 351–372. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., McNally F. J., 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z. Q., 2011. The cullin protein family. Genome Biol. 12: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. A., Zohn I. E., 2012. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J. Cell Biol. 196: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. A., Nuwayhid S. J., Maynard T., Ghandchi F., Hill J. T., et al. , 2014. Hectd1 is required for development of the junctional zone of the placenta. Dev. Biol. 392: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y., Kikuchi K., Dazai K., Higashitani A., 2005. Caenorhabditis elegans Elongin BC complex is essential for cell proliferation and chromosome condensation and segregation during mitosis and meiotic division II. Chromosome Res. 13: 357–375. [DOI] [PubMed] [Google Scholar]
- Sato M., Konuma R., Sato K., Tomura K., 2014. Fertilization-induced K63-linked ubiquitylation mediates clearance of maternal membrane proteins. Development 141: 1324–1331. [DOI] [PubMed] [Google Scholar]
- Segref A., Kevei E., Pokrzywa W., Schmeisser K., Mansfeld J., et al. , 2014. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab. 19: 642–652. [DOI] [PubMed] [Google Scholar]
- Sendoel A., Kohler I., Fellmann C., Lowe S. W., Hengartner M. O., 2010. HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature 465: 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Ross J. L., 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125: 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Gonczy P., 2004. zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131: 3527–3543. [DOI] [PubMed] [Google Scholar]
- Srayko M., Buster D. W., Bazirgan O. A., McNally F. J., Mains P. E., 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14: 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko M., E. T. O’Toole, A. A. Hyman, and T. Müller-Reichert, 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16: 1944–1949. [DOI] [PubMed] [Google Scholar]
- Starostina N. G., Lim J. M., Schvarzstein M., Wells L., Spence A. M., et al. , 2007. A CUL-2 Ubiquitin Ligase Containing Three FEM Proteins Degrades TRA-1 to Regulate C. elegans Sex Determination. Dev. Cell 13: 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina N. G., Simpliciano J. M., McGuirk M. A., Kipreos E. T., 2010. CRL2(LRR-1) targets a CDK inhibitor for cell cycle control in C. elegans and actin-based motility regulation in human cells. Dev. Cell 19: 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Seydoux G., 2007. Regulation of the oocyte-to-zygote transition. Science 316: 407–408. [DOI] [PubMed] [Google Scholar]
- Stitzel M. L., Pellettieri J., Seydoux G., 2006. The C. elegans DYRK Kinase MBK-2 Marks Oocyte Proteins for Degradation in Response to Meiotic Maturation. Curr. Biol. 16: 56–62. [DOI] [PubMed] [Google Scholar]
- Stitzel M. L., Cheng K. C., Seydoux G., 2007. Regulation of MBK-2/Dyrk Kinase by Dynamic Cortical Anchoring during the Oocyte-to-Zygote Transition. Curr. Biol. 17: 1545–1554. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The Million Mutation Project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H., Bustos D., Yeh R., Rubinfeld B., Lam C., et al. , 2013. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J. Biol. Chem. 288: 3753–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Starostina N. G., Kipreos E. T., 2007. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac M. H., Terret M. E., Pintard L., 2010. Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr. Opin. Cell Biol. 22: 758–763. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang J. T., Rasoloson D., Stitzel M. L., O’Connell K. F., et al. , 2014. Identification of Suppressors of mbk-2/DYRK by Whole-Genome Sequencing. G3 (Bethesda) 4: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., et al. , 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321. [DOI] [PubMed] [Google Scholar]
- Yang H., McNally K., McNally F. J., 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans. Dev. Biol. 260: 245–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and reagents are available upon request. Additional immunostaining data are included in supplemental files.