Abstract
Although hepatotropic viruses are important causes of human disease, the intrahepatic immune response to hepatitis viruses is poorly understood due to a lack of tractable small animal models. Here we describe a murine model of hepatitis A virus (HAV) infection that recapitulates critical features of type A hepatitis in humans. We demonstrate that the capacity of HAV to evade MAVS-mediated type I interferon responses defines its host species range. HAV-induced liver injury was associated with interferon-independent intrinsic hepatocellular apoptosis and hepatic inflammation that unexpectedly results from MAVS and IRF3/7 signaling. This murine model thus reveals a previously undefined link between innate immune responses to virus infection and acute liver injury, providing a new paradigm for viral pathogenesis in the liver.
Although viral hepatitis is an important cause of human morbidity worldwide, there are no small animal models that accurately recapitulate liver disease caused by any of the five responsible viruses (1, 2). Previous studies have relied heavily on nonhuman primates, especially chimpanzees (3, 4), to investigate pathogenesis and immune responses to hepatitis viruses. This has handicapped efforts to understand host responses within the unique immunologic environment of the liver (5, 6). Recent NIH policies effectively eliminate the use of chimpanzees in such studies (7), intensifying the need for alternative models. Here, we report a murine model that recapitulates many features of human infection with hepatitis A virus (HAV), an hepatotropic picornavirus (genus Hepatovirus) that circulates in blood as quasi-enveloped, membrane-cloaked virions and is shed in feces as naked, nonenveloped particles (8).
Like hepatitis B (HBV) and hepatitis C (HCV) viruses, the host range of HAV is considered restricted to humans and nonhuman primates (2, 9). However, successful adaptation to growth in murine and guinea pig cells suggests a broader host range (10, 11). Closely related viruses have also been discovered recently in bats, rodents, shrews and hedgehogs, with phylogenetic evidence suggesting past shifts among host species (12). HAV replication is strongly suppressed by type I interferon (IFN) (13), but HAV, like HCV, blunts interferon responses in human cells by expressing proteinases that degrade MAVS and TRIF, adaptor molecules involved in induction of IFN (13, 14). As a result, infected chimpanzees demonstrate limited type I IFN responses (4). Since the sequences targeted in human MAVS and TRIF are not conserved in small mammals (fig. S1A), the inability of HAV to infect these species could stem from a failure to disrupt IFN responses.
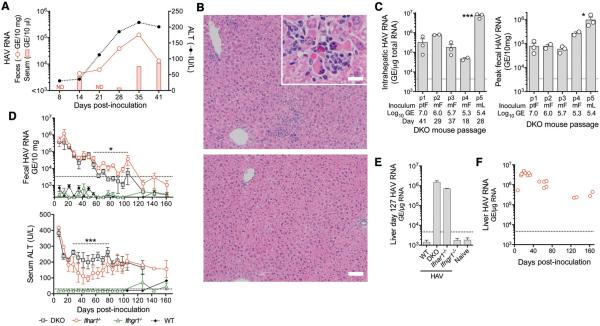
To test this hypothesis, we intravenously inoculated Ifnar1−/−Ifngr1−/− (DKO) mice that lack receptors for both type I and type II IFN with wild-type human HAV (15). These mice proved highly permissive for infection, developing multiple features of acute hepatitis A in humans (4, 16): fecal HAV shedding, low-grade viremia, and elevated serum alanine aminotransferase (ALT) activity (Fig. 1A). Multifocal inflammatory cell infiltrates, often surrounding necrotic or apoptotic hepatocytes, were present in liver 37–41 days post-inoculation (d.p.i) and associated with HAV RNA (Fig. 1B, fig. S2A). Fecal shedding of infectious virus was confirmed by 3 subsequent passages in DKO mice, each leading to intrahepatic HAV RNA, fecal virus shedding, and elevated ALT (Fig. 1C, fig. S2B). Anti-HAV antibodies were detectable 28 d.p.i. (fig. S3A). A fifth serial passage used 4th passage liver extract as inoculum. Unlike non-enveloped virions present in feces (density ~1.23 gm/cm3) (8), ~65% of liver-derived virus was membrane-associated (~1.11 gm/cm3) (fig. S4). This inoculum rapidly induced ALT elevation with impressive fecal shedding and intrahepatic HAV RNA abundance (Fig. 1C,D).
Figure 1.
HAV infection in DKO (Ifnar1−/−Ifngr1−/−), Ifnar1−/−, and Ifngr1−/− mice. (A) Representative course of infection in a DKO mouse inoculated i.v. with 107 GE human HAV (chimpanzee fecal extract). (B) H&E-stained liver from representative (top) infected and (bottom) control DKO mice 41 d.p.i. showing inflammatory infiltrates and (inset) apoptotic hepatocytes (bar = 50 μm, inset bar = 12.5 μm). (C) Summary of serial passage of HAV in DKO mice showing (left) intrahepatic HAV RNA and (right) fecal HAV RNA, source and magnitude of HAV inocula (Pt-F, chimpanzee feces; M-F, DKO mouse feces; M-L, DKO mouse liver), and day of harvest. Data are mean ± SEM or range, n=2–3 animals as shown. *p<0.05, ***p<0.001 p1 vs. p5 by 1-way ANOVA. (D) (top) Fecal HAV RNA and (bottom) serum ALT in DKO, Ifnar1−/−, Ifngr1−/− and wild-type (WT) BL6 mice challenged with 4th passage DKO liver extract (2.6 ×108 GE). Shown are means ± SEM, n=4. *p<0.05, *** p<0.001 for Ifnar1−/− vs. DKO by ANOVA. (E) Intrahepatic HAV RNA in WT, DKO, Ifnar1−/− and Ifngr1−/− mice 127 d.p.i. (mean ± range, n=2). (F) Intrahepatic HAV RNA in Ifnar1−/− mice infected with 4th passage liver extract. Symbols represent individual mice. Dotted horizontal lines in panels indicate level of detection (RNA) or upper limit of normal (ALT).
Like DKO mice, Ifnar1−/− animals shed virus and developed ALT elevation when challenged with liver-derived virus, whereas type II IFN receptor Ifngr1−/− knockouts and wild-type (WT) mice showed no evidence of infection (Fig. 1D,E). The rapid induction of disease in this experiment, compared with slower onset in early DKO passages (Fig. 1A), resulted from a higher inoculum titer rather than viral adaptation to mice. Only a single nonsynonymous nucleotide substitution occurred in the viral sequence over 4 mouse passages (table S1). Infection persisted in Ifnar1−/− and DKO mice for over 3 months (Fig. 1D). Declining serum ALT and fecal virus shedding over this period of time suggested slow immune control in both types of mice, but histopathologic lesions persisted throughout (fig. S2C). As in chimpanzees (4), HAV RNA copy numbers remained high in liver after fecal virus shedding had terminated (Fig. 1E,F).
These data suggest that the capacity of HAV to evade type I IFN responses defines its host range. However, DKO mice were resistant to challenge with either fecal or liver-derived virus administered by oral gavage, possibly reflecting a greater role for type III IFN in the gut (17, 18), or absence of an essential receptor. Rag1−/− and NSG mice lacking adaptive immunity were resistant to intravenous virus challenge (Fig. 2A and fig. S5A), further highlighting the importance of innate immunity in control of HAV. Since HAV-encoded proteinases disrupt IFN responses by degrading human MAVS and TRIF (13, 14), we challenged Mavs−/− and Trif−/− mice to ascertain whether signaling through these adaptor molecules restricts replication. Mavs−/− mice were highly permissive for HAV, shedding 10-fold more virus than DKO or Ifnar1−/− mice (Fig. 2A), whereas Trif−/− mice were nonpermissive (Fig. 2A and fig. S5B,C). Thus, MAVS-mediated type I IFN responses block HAV replication in WT mice. Consistent with this, HAV 3ABC, a proteinase that degrades MAVS in human cells (13), does not cleave murine MAVS (fig. S1B).
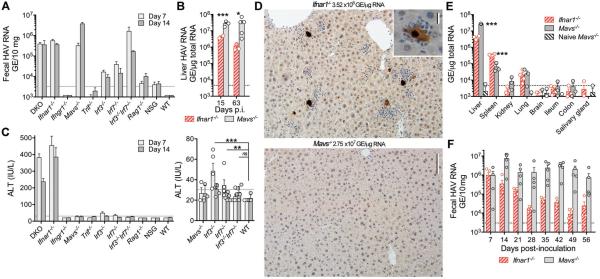
Figure 2.
HAV infection in Ifnar1−/− vs. Mavs−/− mice. (A) Fecal HAV RNA on day 7 and 14 after i.v. challenge of different genetically-deficient mice. Data are mean ± SEM, n=3-5. (B) Viral RNA in livers of Ifnar1−/− vs. Mavs−/− mice 15 and 63 d.p.i. Data are mean ± SEM, n=2–5 as shown. *p<0.05, ***p<0.001 by two-sided t test. (C) Serum ALT 7 and 14 d.p.i. in genetically-deficient mice, with expanded low ALT range on the right. Data are mean ± SEM, n= 5. **p<0.01, ***p<0.001 for combined day 7 and 15 data by two-sided Mann-Whitney test. (D) Immunohistochemical staining of cleaved caspase 3 in liver from representative (top) Ifnar1−/− vs. (bottom) Mavs−/− mice 15 d.p.i. Bar = 100μm (inset, 12.5μm). (E) Tissue distribution of HAV RNA in infected Ifnar1−/− vs. Mavs−/− mice. Data are mean ± SEM, n=3–4. ***p<0.001 by multiple t-test with false discovery rate 1%. (F) Fecal virus shedding in infected Ifnar1−/− or Mavs−/− mice over 56 days of infection. Data are mean ± SEM, n=3-5. ***p<0.001 by multiple t-test with false discovery rate 1%.
Intrahepatic HAV RNA copy numbers were 10-fold higher in Mavs−/− mice than Ifnar1−/− mice (Fig. 2B), with the majority of Mavs−/− hepatocytes containing HAV RNA (fig. S5D–E, table S2). Nonetheless, Mavs−/− mice developed neither ALT elevation (Fig. 2C) nor hepatic inflammation (Fig. 2D). Immunohistochemical staining for activated caspase 3 (Fig. 2D) and TUNEL assays (fig. S6A) revealed numerous apoptotic hepatocytes in infected Ifnar1−/− liver, but none in Mavs−/− tissue. Apoptotic cells in Ifnar1−/− and DKO mice were surrounded by inflammatory infiltrates in proximity to cells containing HAV RNA (Fig. 2D, fig. S6B). Both caspase 8 and 9 (and caspase 3) activities were slightly increased in infected DKO and Ifnar1−/− liver (fig. S6C), but cleaved caspase was not detected in immunoblots as only ~1% of hepatocytes were apoptotic (fig. S6D). These data show that apoptosis and inflammation results from a MAVS-dependent but IFN-independent mechanism. MAVS-mediated apoptosis has been recognized previously, but its role in vivo is uncertain (19, 20).
Virus was largely restricted to the liver in Mavs−/− mice: HAV genomes were 400-fold less abundant in spleen and 1000-fold less in lung (Fig. 2E). Viral RNA was more abundant in spleens of Ifnar1−/− mice, possibly reflecting sequestration of virus released from damaged hepatocytes. Little virus was present in ileum or colon of either knockout, indicating that fecal shedding originates in the liver, as in primates (21, 22). Thus, HAV is highly hepatotropic in mice. Viral shedding persisted unabated for 56 days in Mavs−/− mice with only minimal ALT increases (Fig. 2F, fig. S6E). Rare, isolated apoptotic hepatocytes were observed in only 2 of 5 mice 63 d.p.i. The appearance of anti-HAV antibody was delayed in Mavs−/− mice (fig. S3B), but virus neutralizing activities were comparable to Ifnar1−/− mice 63 d.p.i..
Irf3−/− and Irf7−/− mice lack transcription factors downstream of MAVS that drive type I IFN expression (23). These mice supported only limited HAV replication, whereas Irf3−/−Irf7−/− double knockouts shed virus and accumulated intrahepatic HAV RNA levels equivalent to Mavs−/− or Ifnar1−/− mice (Fig. 2A, fig. S5A, and table S2). This is consistent with redundant roles for IRF3 and IRF7 in control of flaviviruses (24, 25). Serum ALT elevations were minimal in infected Irf3−/−, Irf7−/−, and Irf3−/−Irf7−/− mice (Fig. 2C and fig. S7A). Irf7−/− and Irf3−/−Irf7−/− livers contained rare apoptotic hepatocytes (fig. S7B), possibly reflecting activation of IRF5 (25). However, a general lack of pathology in infected Irf3−/−Irf7−/− animals mirrored the absence of disease in Mavs−/− mice.
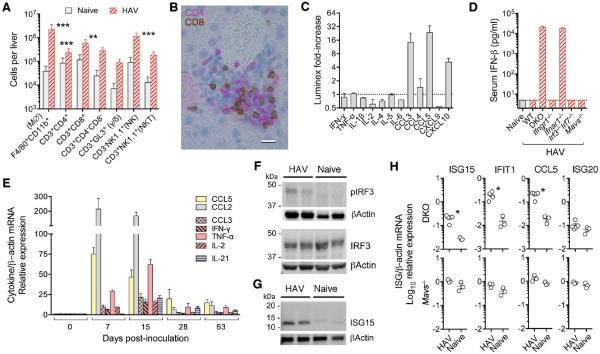
Hepatocyte apoptosis is known to drive inflammation within the liver (26). The livers of 7 d.p.i Ifnar1−/− mice showed 55- and 13-fold increases in F4/80+CD11b+ macrophages and NK1.1+ NK cells, whereas CD4+ and CD8+ T cells were increased only 3- to 5-fold (Fig. 3A). Increases in CD3+CD4−CD8− and γ/δ T cells did not achieve statistical significance. Immunohistochemistry confirmed a mixed cellular infiltrate (Fig. 3B). Luminex assays showed increased hepatic CCL3 (MIP-α), CCL5 (RANTES), and CXCL10 (IP-10) protein, but not IFN-γ, TNF-α, IL-1β, IL-2 or IL-6 (Fig. 3C). Similarly, serum IFN-β was markedly elevated (>10 ng/ml) in infected DKO and Ifnar1−/− mice (Fig. 3D), but ELISA assays for IFN-γ, TNF-α, IL-1β, and IL6 were negative. Nonetheless, RT-PCR demonstrated HAV-induced intrahepatic transcripts for multiple cytokines and chemokines in DKO and Ifnar1−/−, but not Mavs−/− mice (Fig. 3E, fig. S8A). CCL2 (MCP-1) and CCL5 mRNA responses were maximal 7 d.p.i, whereas CCL3, IFN-γ, and TNF-α mRNAs peaked 15 d.p.i. (Fig. 3E) despite the absence of detectable protein in serum or liver. Diminishing chemokine and cytokine responses at 28 d.p.i. (Fig. 3E) correlated temporally with a 100-fold decline in fecal virus shedding. NLRP3 inflammasome-related transcripts were not increased (fig. S8B).
Figure 3.
Cellular and cytokine response to HAV infection in DKO and Ifnar1−/− mice. (A) Estimated intrahepatic leukocyte numbers in naïve versus infected Ifnar1−/− mice 7 d.p.i. Data are mean ± SD, n=5 (mean ALT=372 IU/L). **p<0.01, ***p<0.001 by two-way ANOVA with Tukey's multiple comparison test. (B) Dual immunohistochemical staining of infected Ifnar1−/− liver for CD4 (magenta) and CD8 (brown) showing a mixed cellular infiltrate14 d.p.i. Bar = 10 μm. (C) Fold-increase in liver cytokine levels in HAV-infected DKO mice (Luminex assay) with ALT >200 IU/L. Mean ± range, n=2. (D) Serum IFNβ measured by ELISA 7 d.p.i. Data are mean ± SD, n=4. (E) Fold-increase in intrahepatic cytokine and chemokine mRNA abundance in Ifnar1−/− mice. Data are mean ± SEM, n=4–5. Immunoblots of (F) phospho-Ser-396 and total IRF3, and (G) ISG15 in livers from HAV-infected vs. naïve DKO mice. β-actin included as a loading control. (H) Intrahepatic transcripts of IRF3-regulated ISGs, ISG15, IFIT1 (ISG56), CCL5 (RANTES), and ISG20 (not directly regulated by IRF3), in HAV-infected DKO (n=4) and Mavs−/− (n=3) mice vs. naïve animals 18–28 d.p.i. *p<0.05 by t test.
IFN-β transcription is coordinately regulated by IRF3/7 and NF-κB (27). Phospho-IRF3 confirmed IRF3 activation in infected Ifnar1−/− mice (Fig. 3F), and interferon-stimulated genes (ISGs) such as ISG15, IFIT1, and CXCL10 that are directly regulated by IRF3 (28) were induced (Figs. 3C,G,H). IRF3 similarly regulates CCL5 transcription (29), explaining prominent and early CCL5 expression by HAV-infected hepatocytes in Ifnar1−/− but not Mavs−/− mice (Fig. 3C,E, fig. S8C). The phospho-p65 component of NF-κB was not measurably increased (fig. S8D).
Several possible mechanisms could account for apoptosis induced through a MAVS-IRF3/7 pathway (fig. S8E). First, CCL5 expression could recruit cytotoxic lymphocytes to the liver, resulting in death receptor-mediated apoptosis (30, 31). However, depletion of CD4+ or CD8+ T cells had no impact on acute (7 d.p.i) disease in Ifnar1−/− mice (fig. S9). Moreover, virus-specific T cell responses were minimal in Ifnar1−/− and Mavs−/− mice (fig. S10). Depletion of NK1.1+ NK cells similarly failed to reduce liver injury (fig. S11). This argues against primary death receptor-mediated apoptosis. Clodronate depletion of macrophages prior to infection also had no effect on viral replication or inflammation (fig. S12).
Alternatively, apoptosis could be induced by ISGs that are directly regulated by IRF3 (28). The functions of these proteins are only partly understood, but IFIT2, an ISG that is transcriptionally regulated by IRF3, is known to trigger mitochondrial apoptosis in human cells (28, 32). IRF3 similarly regulates PMAIP1, a pro-apoptotic BH3-only protein (33). Importantly, both Ifit2 and Pmaip1 transcripts were induced early and to a greater extent in Ifnar1−/− than Mavs−/− mice (fig. S13). IRF3 can also induce apoptosis through a transcription-independent mechanism involving a direct interaction with mitochondrial Bax (34). However, this would not explain the rare apoptotic hepatocytes observed in Irf3-deficient mice (fig. S7B).
Although many details remain to be resolved, our data show that Ifnar1−/− mice provide a useful model that recapitulates many aspects of type A hepatitis in humans. Despite heroic efforts, such a model has proved elusive for HBV or HCV infection (2). Our results suggest that HAV host species range is dictated largely by its capacity to evade MAVS-mediated type I IFN responses, and reveal an unexpected role for MAVS signaling in virus-mediated liver injury. Such signaling leads to IRF3/7-dependent, but IFNα/β- and IFNγ-independent hepatocellular apoptosis with a secondary inflammatory response (fig. S8E). This may explain why HAV and HCV have evolved independently to target MAVS for degradation. Disrupting innate immune signaling upstream of IRF3/7 not only limits IFN-mediated antiviral responses, but also restricts inflammation within the liver, delays anti-viral antibody responses, and slows viral clearance (Figs. 2D,E fig. S3B, S6E). IRF3, activated through STING as a result of endoplasmic reticulum stress, has been implicated recently in acute ethanol-induced hepatitis (35), suggesting a common final pathway for toxin- and virus-induced liver injury. Altogether, our findings establish the critical importance of innate immune responses in control of viral infection in the liver, and provide a paradigm for HAV pathogenesis that is likely relevant to other hepatotropic human viruses.
Supplementary Material
Acknowledgements
The authors thank Daisuke Yamane, Kevin McKnight, Tiffany Benzine, Lily Wai, Dawud Hilliard, and Michael Chua for helpful discussions and technical assistance, Robert Lanford (Texas Biomedical Research Institute) for chimpanzee-passaged HAV, and Christopher Walker (Research Institute of Nationwide Children's Hospital) for the generous gift of HAV peptides. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. DNA sequences are deposited in GenBank with the accession numbers: KX343014, KX343015, KX343016, KX343017, and KX343018. This work was supported in part by NIH grants R01-AI103083 (SML), U19-AI109965 (SML), R01-AI074862 (JKW), R21-AI117575 (JKW), R56-AI110682 (JKW), and an NCI Center Core Support Grant to the Lineberger Comprehensive Cancer Center, P30-CA016086.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Dembek C, Protzer U. Mouse models for therapeutic vaccination against hepatitis B virus. Med Microbiol Immunol. 2015;204:95–102. doi: 10.1007/s00430-014-0378-6. [DOI] [PubMed] [Google Scholar]
- 2.Winer BY, Ding Q, Gaska J, Ploss A. In vivo models of hepatitis B and C virus infection. FEBS Lett. 2016 doi: 10.1002/1873-3468.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanford RE, Lemon SM, Walker C. In: Hepatitis C Antiviral Drug Discovery & Development. He Y, Tan T, editors. Horizons Scientific Press; Norwich: 2011. pp. 99–132. [Google Scholar]
- 4.Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, Yamane D, Perelson AS, Walker CM, Lemon SM. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Nat'l Acad Sci USA. 2011;108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 6.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser J. Biomedical Research: An end to U.S. chimp research. Science. 2015;350:1013. doi: 10.1126/science.350.6264.1013. [DOI] [PubMed] [Google Scholar]
- 8.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong S-H, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deinhardt F, Deinhardt JB. In: Hepatitis A. Gerety RJ, editor. Academic Press, Inc; Orlando: 1984. pp. 185–204. [Google Scholar]
- 10.Hornei B, Kammerer R, Moubayed P, Frings W, Gauss-Muller V, Dotzauer A. Experimental hepatitis A virus infection in guinea pigs. J Med Virol. 2001;64:402–409. doi: 10.1002/jmv.1065. [DOI] [PubMed] [Google Scholar]
- 11.Feigelstock DA, Thompson P, Kaplan GG. Growth of hepatitis A virus in a mouse liver cell line. J Virol. 2005;79:2950–2955. doi: 10.1128/JVI.79.5.2950-2955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drexler JF, Corman VM, Lukashev AN, van den Brand JMA, Gmyl A, Brunink S, Rasche A, Seggewiss N, Feng H, Leijten LM, Vallo P, Kuiken T, Dotzauer A, Ulrich RG, Lemon SM, Drosten C. Hepatovirus Ecology Consortium, Evolutionary origins of hepatitis A virus in small mammals. Proc Nat'l Acad Sci U S A. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Nat'l Acad Sci USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu L, Feng Z, Yamane D, Liang Y, Lanford RE, Li K, Lemon SM. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7:e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supplementary materials at the Science website.
- 16.Lemon SM. Type A viral hepatitis: new developments in an old disease. N Engl J Med. 1985;313:1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- 17.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei Y, Moore CB, Liesman RM, O'Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan K, Zheng Z, Song T, He X, Xu C, Zhang Y, Ma S, Wang Y, Xu Q, Cao Y, Li J, Yang X, Ge X, Wei C, Zhong H. MAVS regulates apoptotic cell death by decreasing K48-linked ubiquitination of voltage-dependent anion channel 1. Mol Cell Biol. 2013;33:3137–3149. doi: 10.1128/MCB.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulman AN, Dienstag JL, Jackson DR, Hoofnagle JH, Gerety RJ, Purcell RH, Barker LF. Hepatitis A antigen particles in liver, bile, and stool of chimpanzees. J Infect Dis. 1976;134:80–84. doi: 10.1093/infdis/134.1.80. [DOI] [PubMed] [Google Scholar]
- 22.Walker CM, Feng Z, Lemon SM. Reassessing immune control of hepatitis A virus. Curr Opin Virol. 2015;11:7–13. doi: 10.1016/j.coviro.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 24.Chen HW, King K, Tu J, Sanchez M, Luster AD, Shresta S. The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J Immunol. 2013;191:4194–4201. doi: 10.4049/jimmunol.1300799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, Nikolich-Zugich J, Moses AV, Gale M, Jr., Fruh K, Diamond MS. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faouzi S, Burckhardt BE, Hanson JC, Campe CB, Schrum LW, Rippe RA, Maher JJ. Anti-Fas induces hepatic chemokines and promotes inflammation by an NF-kappa B-independent, caspase-3-dependent pathway. J Biol Chem. 2001;276:49077–49082. doi: 10.1074/jbc.M109791200. [DOI] [PubMed] [Google Scholar]
- 27.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol.Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 28.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin R, Heylbroeck C, Genin P, Pitha PM, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehermann B. Natural killer cells in viral hepatitis. Cell Mol Gastroenterol Hepatol. 2015;1:578–588. doi: 10.1016/j.jcmgh.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014;147:577–594.e571. doi: 10.1053/j.gastro.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 32.Reich NC. A death-promoting role for ISG54/IFIT2. J Interferon Cytokine Res. 2013;33:199–205. doi: 10.1089/jir.2012.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity. 2016;44:1151–1161. doi: 10.1016/j.immuni.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, Fitzgerald KA, Szabo G. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A. 2013;110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 37.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 38.Althof N, Harkins S, Kemball CC, Flynn CT, Alirezaei M, Whitton JL. In vivo ablation of type I interferon receptor from cardiomyocytes delays coxsackieviral clearance and accelerates myocardial disease. J Virol. 2014;88:5087–5099. doi: 10.1128/JVI.00184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M., Jr. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity. 2012;37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 41.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima A, Nishimura K, Nakaima Y, Oh T, Noguchi S, Taniguchi T, Tamura T. Cell type-dependent proapoptotic role of Bcl2L12 revealed by a mutation concomitant with the disruption of the juxtaposed Irf3 gene. Proc Natl Acad Sci U S A. 2009;106:12448–12452. doi: 10.1073/pnas.0905702106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 44.Daffis S, Suthar MS, Szretter KJ, Gale M, Jr., Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gust ID, Lehmann NI, Crowe S, McCrorie M, Locarnini SA, Lucas CR. The origin of the HM175 strain of hepatitis A virus. The Journal of Infectious Diseases. 1985;151:365–367. doi: 10.1093/infdis/151.2.365. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C, Gillette DD, Li X, Zhang Z, Wen H. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J Biol Chem. 2014;289:17020–17029. doi: 10.1074/jbc.M114.563114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. Journal of Virology. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Counihan NA, Daniel LM, Chojnacki J, Anderson DA. Infrared fluorescent immunofocus assay (IR-FIFA) for the quantitation of non-cytopathic and minimally cytopathic viruses. J Virol Methods. 2006;133:62–69. doi: 10.1016/j.jviromet.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 50.Samuel MA, Morrey JD, Diamond MS. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J Virol. 2007;81:2614–2623. doi: 10.1128/JVI.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D, Li C, Chen Y, Burnett C, Liu XY, Downs S, Collins RD, Hawiger J. Nuclear import of proinflammatory transcription factors is required for massive liver apoptosis induced by bacterial lipopolysaccharide. J Biol Chem. 2004;279:48434–48442. doi: 10.1074/jbc.M407190200. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Callendret B, Xu D, Brasky KM, Feng Z, Hensley LL, Guedj J, Perelson AS, Lemon SM, Lanford RE, Walker CM. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med. 2012;209:1481–1492. doi: 10.1084/jem.20111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.