Abstract
Samoans are a unique founder population with a high prevalence of obesity1–3, making them well suited for identifying new genetic contributors to obesity4. We conducted a genome-wide association study (GWAS) in 3,072 Samoans, discovered a variant, rs12513649, strongly associated with body mass index (BMI) (P = 5.3 × 10−14), and replicated the association in 2,102 additional Samoans (P = 1.2 × 10−9). Targeted sequencing identified a strongly associated missense variant, rs373863828 (p.Arg457Gln), in CREBRF (meta P = 1.4 × 10−20). Although this variant is extremely rare in other populations, it is common in Samoans (frequency of 0.259), with an effect size much larger than that of any other known common BMI risk variant (1.36–1.45 kg/m2 per copy of the risk-associated allele). In comparison to wild-type CREBRF, the Arg457Gln variant when overexpressed selectively decreased energy use and increased fat storage in an adipocyte cell model. These data, in combination with evidence of positive selection of the allele encoding p.Arg457Gln, support a ‘thrifty’ variant hypothesis as a factor in human obesity.
Obesity is essentially a disorder of energy homeostasis and has strong genetic and environmental components. As diets have modernized and physical activity has decreased, the prevalence of overweight and obesity in Samoa has escalated to be among the highest in the world. In 2003, 68% of men and 84% of women in Samoa were overweight or obese by Polynesian cutoffs (BMI >26 kg/m2)1; by 2010, prevalence had increased to 80% and 91%, respectively3. Although the contribution of environmental factors to this trend is clear, the estimated 45% heritability of BMI in Samoans remains largely unexplained1. Genetic susceptibility to obesity in the contemporary obesogenic environment may have resulted from putative selective advantages from efficient energy metabolism acquired during 3,000 years of Polynesian island discoveries, settlement, and population dynamics5–8 and/or from genetic drift due to founder effects, small population sizes, and population bottlenecks9–11.
To discover genes influencing BMI, we genotyped 659,492 markers across the genome in our discovery sample of 3,072 Samoans recruited from 33 villages across Samoa using the Affymetrix 6.0 chip (Supplementary Fig. 1 and Supplementary Table 1). We adjusted for population substructure and inferred relatedness using an empirical kinship matrix and then tested for association with BMI using linear mixed models. Quantile–quantile plots indicated that P-value inflation was well controlled (λGC = 1.07) (Supplementary Fig. 2).
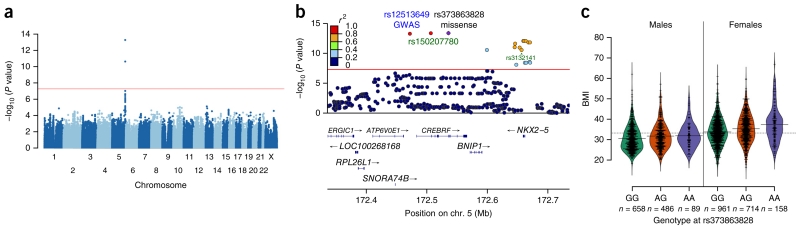
By far, the strongest association with BMI occurred at rs12513649 (P = 5.3 × 10−14) on chromosome 5q35.1 (Fig. 1a), and this association was strongly replicated (P = 1.2 × 10−9) in 2,102 adult Samoans from a 1990–1995 longitudinal study and a 2002–2003 family study, with participants of each study drawn from both American Samoa and Samoa (Table 1 and Supplementary Table 1). To fine-map the region encompassing this signal, we used the Affymetrix-based genotypes to select 96 individuals optimal for targeted sequencing of a 1.5-Mb region centered on rs12513649. The haplotypes generated from the sequencing data were used to impute genotypes for the rest of the discovery sample. Analyses of the imputed data highlighted two significantly associated variants in CREBRF (encoding CREB3 regulatory factor), rs150207780 and rs373863828 (Fig. 1b). Because of high linkage disequilibrium (LD) in the region, conditional analyses were not able to distinguish between the top variants on statistical grounds (Supplementary Fig. 3). Annotation indicated that neither rs12513649, located between ATP6V0E1 and CREBRF, nor rs150207780, located in intron 1 of CREBRF, had any predicted regulatory function, drawing our attention to rs373863828, which was the only strongly associated missense variant among the 775 variants with P ≤ 1 × 10−5 in the targeted sequencing region. The rs373863828 missense variant (c.1370G>A, p.Arg457Gln) is located at a highly conserved position (GERP score 5.49) with a high probability of being damaging (SIFT, 0.03; PolyPhen-2, 0.996). The BMI-increasing A allele of rs373863828 has an overall frequency of 0.259 in Samoans but is unobserved or extremely rare in other populations, with an allele count in the Exome Aggregation Consortium of only 5 among 121,362 measured alleles (Table 1)12. Bayesian fine-mapping with PAINTOR13 strongly supported following up the missense variant. The two variants in the region with the highest posterior probability (PP) of being causal were rs373863828 (PP = 0.80) and rs150207780 (PP = 0.22); when Encyclopedia of DNA Elements (ENCODE) functional annotation was included, these probabilities increased to 0.92 and 0.34, respectively.
Figure 1.
Association results from genome-wide and targeted sequencing and beanplots of BMI versus genotype in men and women from the discovery sample. (a) Manhattan plot of the genome-wide association scan for association with BMI. The red line corresponds to a P value of 5 × 10−8. (b) Association results using imputed data for the region encompassing CREBRF. The strength of LD, as measured by the squared correlation of genotype dosages, between each variant and the missense variant rs373863828 is represented by the color of each point. The red line corresponds to a P value of 5 × 10−8. The plot was generated using LocusZoom32. (c) Beanplots of BMI versus genotype in men (n = 1,233) and women (n = 1,833) from the discovery sample. Each bean consists of a mirrored density curve containing a one-dimensional scatterplot of the individual data. A solid line shows the average for each group, and the dashed line represents the overall average. The plot was generated using the R beanplot package33.
Table 1. Association details for rs12513649 and rs373863828.
| Discovery variant | Missense variant | |
|---|---|---|
| SNP rs ID | rs12513649 | rs373863828 |
| Chromosome | 5 | 5 |
| Physical position (GRCh37.p13) (bp) | 172,472,052 | 172,535,774 |
| Effect allele | G | A |
| Other allele | C | G |
| Nearest gene upstream of the SNP | ATP6V0E1 | CREBRF |
| Distance to nearest upstream gene (bp) | 10,152 | 0 |
| Nearest gene downstream of the SNP | CREBRF | CREBRF |
| Distance to nearest downstream gene (bp) | 11,302 | 0 |
| Sample sizes (phenotyped and genotyped) | ||
| GWAS Samoans from the 2010s (discovery) | 3,072 | 3,066 |
| Samoans from the 1990s (replication) | 1,020 | 1,020 |
| Samoans from the 2000s (replication) | 1,082 | 1,083 |
| Meta-analysis of the 1990s and 2000s samples | 2,102 | 2,103 |
| Meta-analysis of the 1990s, 2000s, and 2010s samples | 5,174 | 5,169 |
| Samoan children from the 2000s | 409 | 409 |
| P values for log-transformed BMI | ||
| GWAS Samoans from the 2010s (discovery) | 5.3 × 10−14 | 7.0 × 10−13 |
| Samoans from the 1990s (replication) | 5.8 × 10−4 | 8.0 × 10−4 |
| Samoans from the 2000s (replication) | 3.0 × 10−7 | 6.5 × 10−7 |
| Meta-analysis of the 1990s and 2000s samples | 1.2 × 10−9 | 3.5 × 10−9 |
| Meta-analysis of the 1990s, 2000s, and 2010s samples | 4.0 × 10−22 | 1.4 × 10−20 |
| Samoan children from the 2000s | 4.1 × 10−3 | 1.1 × 10−3 |
| Effect sizes (β (s.e.)) for log-transformed BMI | ||
| GWAS Samoans from the 2010s (discovery) | 0.041 (0.005) | 0.039 (0.005) |
| Samoans from the 1990s (replication) | 0.029 (0.008) | 0.028 (0.008) |
| Samoans from the 2000s (replication) | 0.056 (0.011) | 0.054 (0.011) |
| Samoan children from the 2000s | 0.031 (0.011) | 0.035 (0.011) |
| Effect allele frequencies | ||
| GWAS Samoans from the 2010s | 0.276 | 0.276 |
| Samoans from the 1990s | 0.251 | 0.251 |
| Samoan adults from the 2000s | 0.224 | 0.225 |
| Samoan children from the 2000s | 0.236 | 0.235 |
| All of the 1990s, 2000s and 2010s samples | 0.258 | 0.259 |
| Individuals of East Asian descent from 1000G | 0.063 | 0.000 |
| Individuals of South Asian descent from 1000G | 0.003 | 0.000 |
| Individuals of European descent from 1000G | 0.000 | 0.000 |
| Individuals of admixed American descent from 1000G | 0.059 | 0.000 |
| Individuals of African descent from 1000G | 0.001 | 0.000 |
| Individuals of East Asian descent from ExAC | NA | <0.001a |
| Individuals of South Asian descent from ExAC | NA | 0.000 |
| Individuals of European descent from ExAC | NA | <0.001b |
| Individuals of Latino descent from ExAC | NA | 0.000 |
| Individuals of African descent from ExAC | NA | 0.000 |
| Individuals of other descent from ExAC | NA | 0.001c |
This table provides detailed results for rs12513649 and rs373863828. 1000G, 1000 Genomes Project; ExAC, Exome Aggregation Consortium12; s.e., standard error; NA, not available.
Two A alleles in 8,636 measured alleles.
Two A alleles in 73,328 measured alleles.
One A allele in 908 measured alleles.
We then genotyped the missense variant rs373863828 in the discovery and replication samples, obtaining very significant evidence of association with BMI in adults (P = 7.0 × 10−13 and P = 3.5 × 10−9, respectively), with a combined meta-analysis P value of 1.4 × 10−20 (Table 1). The meta-analysis showed no evidence of heterogeneity (I2 = 0%; Q = 1.12; P = 0.571). In our discovery sample, each copy of the A allele increased BMI by 1.36 kg/m2 (Fig. 1c). In our adult replication sample, each copy of the A allele increased BMI by 1.45 kg/m2. There was a strong effect on BMI at this locus even after stratifying by sex and cohort (Supplementary Fig. 4; however, sex–genotype interactions were not significant (discovery P = 0.060; replication P = 0.555)). There was also suggestive evidence (P = 1.1 × 10−3) that this variant increased BMI in our sample of 409 Samoan children (Table 1). The rs373863828 variant (encoding p.Arg457Gln) accounted for 1.93% of the variance in BMI in our discovery sample and 1.08% of the variance in BMI in our replication sample. In comparison, rs1558902, the main risk-associated variant in FTO, increases BMI by 0.39 kg/m2 per copy of the risk-associated allele and accounts for only 0.34% of the variance in BMI in Europeans14,15. In searches of the literature and databases (including GRASP16,17), we were unable to identify any significant associations with BMI in the CREBRF region in other human studies.
In addition to BMI, the A allele of rs373863828 was also positively associated with obesity risk (odds ratio (OR) = 1.305 and 1.441 in the discovery and replication cohorts, respectively) as well as measures of total and regional adiposity, including percent body fat, abdominal circumference, and hip circumference, in both cohorts (Table 2 and Supplementary Table 2). The A allele was also positively associated with serum leptin levels in women (both cohorts) and men (replication cohort) before but not after adjusting for BMI. These data indicate that the association between the missense variant and BMI is indeed due to an association with adiposity.
Table 2. Association of rs373863828 with untransformed adiposity, metabolic, and lipid traits in the discovery sample.
| Quantitative trait | n | β (s.e.) | P | Covariatesa |
|---|---|---|---|---|
| Adiposity traits | ||||
| BMI (kg/m2) | 3,066 | 1.356 (0.183) | 1.12 × 10−13 | A, A2, S, A × S |
| Body fat (%) | 2,893 | 2.199 (0.345) | 1.78 × 10−10 | A, A2, S, A × S |
| Abdominal circumference (cm) | 3,057 | 2.842 (0.404) | 2.05 × 10−12 | A, A2, S, A × S, A2 × S |
| Hip circumference (cm) | 3,058 | 2.361 (0.332) | 1.19 × 10−12 | A, A2, S, A2 × S |
| Abdominal–hip ratio | 3,056 | 0.005 (0.002) | 2.23 × 10−3 | A, A2, S, A × S, A2 × S |
| Metabolic traits | ||||
| Fasting glucose (mg/dl)b | 2,393 | −1.652 (0.423) | 9.52 × 10−5 | A, A2, S |
| Fasting insulin (μU/ml)b | 2,392 | 1.342 (0.449) | 2.83 × 10−3 | A, S, A × S |
| HOMA-IRb | 2,392 | 0.241 (0.114) | 0.035 | A, S, A × S |
| Adiponectin (μg/ml) | 2,858 | −0.228 (0.083) | 0.006 | A, A2, S, A × S |
| Leptin in men (ng/ml)c | 1,151 | 0.719 (0.326) | 0.027 | A |
| Leptin in women (ng/ml)c | 1,707 | 1.888 (0.525) | 3.25 × 10−4 | |
| Metabolic traits adjusted for BMI | ||||
| Fasting glucose (mg/dl)b | 2,383 | −2.248 (0.417) | 6.89 × 10−8 | A, A2, S, B |
| Fasting insulin (μU/ml)b | 2,382 | 0.225 (0.420) | 0.592 | A, A2, S, B, A × S, A2 × S |
| HOMA-IRb | 2,382 | −0.034 (0.107) | 0.754 | A, B |
| Adiponectin (μg/ml) | 2,844 | −0.066 (0.080) | 0.412 | A, A2, S, B, A × S |
| Leptin in men (ng/ml)c | 1,143 | −0.262 (0.210) | 0.213 | A, A2, B |
| Leptin in women (ng/ml)c | 1,701 | −0.516 (0.366) | 0.159 | A, A2, B |
| Serum lipid levels | ||||
| Total cholesterol (mg/dl) | 2,858 | −3.203 (1.029) | 1.84 × 10−3 | A, A2, S, A × S, A2 × S |
| Triglycerides (mg/dl) | 2,858 | 0.349 (2.769) | 0.900 | A, S, A × S |
| HDL cholesterol (mg/dl) | 2,858 | −0.322 (0.321) | 0.317 | A, A2, S |
| LDL cholesterol (mg/dl) | 2,851 | −2.347 (0.945) | 0.013 | A, A2, S, A2 × S |
|
| ||||
| Dichotomous traits | n | OR (95% CI) | P | Covariatesa |
|
| ||||
| Obesity (>32 kg/m2) | 3,066 | 1.305 (1.159–1.470) | 1.12 × 10−5 | A, A2, S, A × S |
| Diabetes | 2,876 | 0.637 (0.536–0.758) | 3.86 × 10−7 | A |
| Diabetes adjusted for BMI | 2,861 | 0.586 (0.489–0.702) | 6.68 × 10−9 | A, B |
| Hypertension | 3,041 | 1.014 (0.898–1.145) | 0.818 | A, S |
Boldface represents a P value <2.17 × 10−3. s.e., standard error; OR, odds ratio; 95% CI, 95% confidence interval.
A, age; A2, age2; S, sex; A × S, age × sex interaction; A2 × S = age2 × sex interaction, B, log(BMI).
Analysis was conducted only in non-diabetics.
Leptin was not analyzed in men and women together because the distributions were very different for the sexes.
Higher BMI and adiposity are usually associated with greater insulin resistance (higher fasting insulin levels and homeostatic model assessment–insulin resistance (HOMA-IR)), an atherogenic lipid profile (especially higher serum triglyceride and lower HDL cholesterol levels), and lower adiponectin levels. We therefore expected the BMI-increasing A allele of rs373863828 to also be associated with these metabolic variables. However, even though the A allele was consistently associated with higher BMI and adiposity in both the discovery and replication cohorts, the expected associations with the above obesity-related comorbidities were not observed and, in some cases, were even in the opposite direction to that expected (Table 2 and Supplementary Table 2). Notably, when considering all subjects, the risk of diabetes was actually lower (OR = 0.586 for the discovery cohort, P = 6.68 × 10−9) or trended lower (0.742 for the replication cohorts, P = 0.029) in carriers of the A allele. Likewise, even in non-diabetic subjects, the variant was associated with moderately but significantly lower fasting glucose levels in both the discovery and replication cohorts (1.65 mg/dl (P = 9.5 × 10−5) and 1.54 mg/dl (P = 8.8 × 10−4) lower for each copy of the A allele, respectively). These effects became even more significant after adjusting for BMI (2.25 mg/dl, P = 6.9 × 10−8 and 2.09 mg/dl, P = 7.6 × 10−6), suggesting an independent effect of the variant on glucose homeostasis and diabetes risk. Such effects are unlikely to be due to survival bias, as no correlation between age and genotype was observed (linear regression P = 0.849). These effects seem to be independent of obesity-associated insulin resistance, as associations with fasting insulin levels and HOMA-IR were not consistently observed across the cohorts (associations were stronger only in the replication cohort before adjusting for BMI). Furthermore, although the variant was associated with lower total cholesterol levels in the discovery cohort, consistent effects on serum lipid or adiponectin levels were likewise not observed. Together, these data suggest that the missense variant does not promote, and may even protect against, obesity-associated comorbidities; however, additional studies will be required to confirm these findings and directly test this hypothesis.
Although the majority of genes contributing to obesity do so by influencing the central regulation of energy balance18, emerging evidence highlights the contribution of altered cellular metabolism to obesity19. Therefore, we examined the impact of rs373863828 on cellular bioenergetics. To do so, we selected the established 3T3-L1 mouse adipocyte model for two reasons: (i) CREBRF is widely expressed in virtually all tissues, including adipose tissue (Supplementary Fig. 5), suggesting a fundamental cellular function, and (ii) several CREB family proteins have been linked to mitochondrial function and metabolic phenotypes in adipocytes20–23. Thus, this model is well suited to assess multiple potentially relevant metabolic phenotypes.
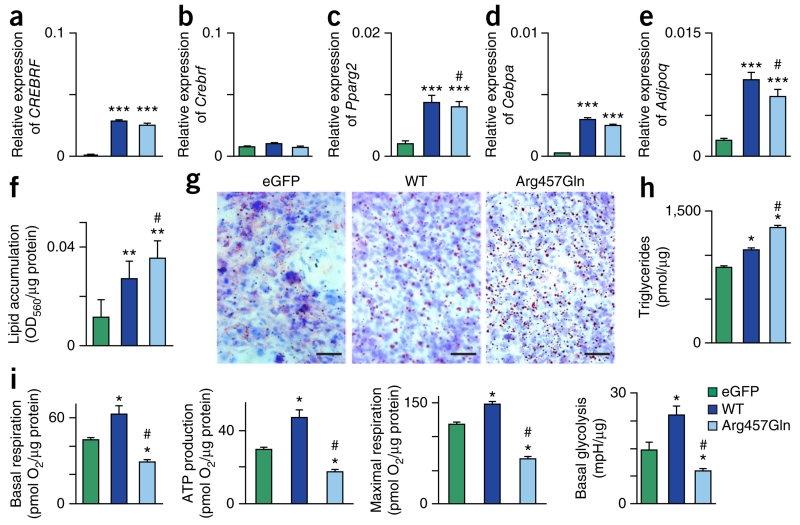
We first characterized the effects of adipogenic differentiation and ectopic overexpression of human wild-type or Arg457Gln CREBRF on endogenous Crebrf expression in 3T3-L1 cells. Crebrf expression was induced during adipogenesis in conjunction with that of adipogenic markers (Cebpa, Pparg, and Adipoq), suggesting a role for CREBRF in this process (Supplementary Fig. 6). Indeed, comparable stable overexpression of the transcripts for human wild-type and Arg457Gln CREBRF (Fig. 2a), without changing endogenous Crebrf levels (Fig. 2b), was sufficient to induce the expression of adipogenic markers (Fig. 2c–e) and promote lipid and triglyceride accumulation (Fig. 2f–h) in the absence of standard hormonal induction of adipogenesis. Although Arg457Gln CREBRF resulted in slightly weaker induction of adipogenic markers than wild-type protein (Fig. 2c,e), it promoted significantly (P < 0.02) greater lipid and triglyceride accumulation (Fig. 2f–h). To determine whether this increased energy storage was associated with decreased energy use, we next assessed glycolysis, mitochondrial respiration, and ATP production. Consistent with published data24,25, glycolysis was suppressed and mitochondrial respiration and ATP production were enhanced by hormonally induced adipogenic differentiation (Supplementary Fig. 7). Stable overexpression of wild-type CREBRF increased whereas Arg457Gln CREBRF decreased multiple measures of cellular energy use, including basal and maximal mitochondrial respiration, mitochondrial ATP production, and basal glycolysis (Fig. 2i). These data indicate that the Arg457Gln CREBRF variant promotes more lipid storage while using less energy than wild-type CREBRF.
Figure 2.
CREBRF variants, adipogenic differentiation, lipid accumulation, and energy homeostasis. 3T3-L1 mouse preadipocytes overexpressing enhanced GFP–only negative control (eGFP), wild-type human CREBRF (WT), or Arg457Gln human CREBRF were collected at 8 d after confluence in the absence of hormonal stimulation of adipogenic differentiation. (a–e) mRNA levels of human CREBRF (a) and endogenous mouse Crebrf (b), Pparg2 (c), Cebpa (d), and Adipoq (e) relative to those of the β-actin (Actb) reference gene determined using quantitative RT–PCR. Values are given as means ± s.e.m. from three biological replicates with four technical replicates each (n = 3 × 4 = 12). Representative results from one of four experiments are shown. (f) Quantification of lipid accumulation with Oil Red O staining normalized to protein content (OD560/μg protein). Data are shown as means ± s.e.m. from three transfection replicates with eight wells for each transfection (n = 3 × 8 = 24). (g) Representative photomicrographs of Oil Red O staining to visualize lipid droplets (red) with counterstaining of nuclei with hematoxylin (blue). Scale bars, 50 μm. (h) Biochemical assay for triglycerides. Data are shown as means ± s.e.m., n = 2 biological replicates. (i) Key bioenergenic variables as determined on the basis of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) normalized to protein content. Values are given as means ± s.e.m. (n = 6 biological replicates). mpH, 0.01 pH unit. Statistical analysis: one-way analysis of variance (ANOVA), two-sided Games–Howell post-hoc test. *P < 0.03, **P < 1 × 10−3, ***P < 1 × 10−4 compared to 3T3-L1 cells transfected with eGFP control construct; #P < 0.05 compared to 3T3-L1 cells transfected with construct for wild-type CREBRF.
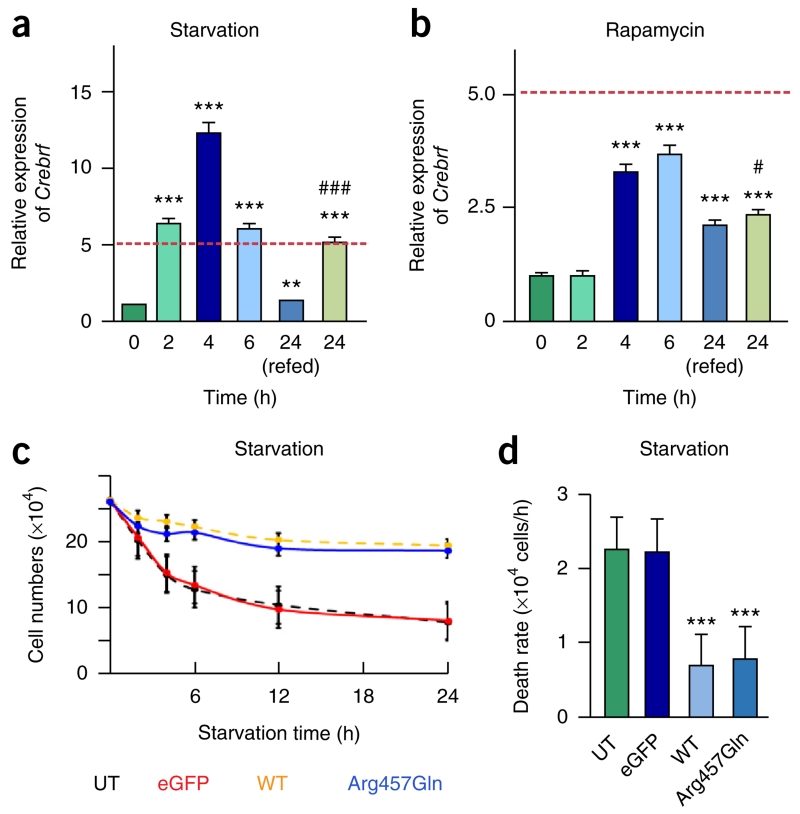
In addition to having a role in cellular energy storage and use, the Drosophila melanogaster CREBRF ortholog REPTOR has recently been implicated in both cellular and organismal adaptation to nutritional stress by mediating the downstream transcriptional response to the cellular energy sensor TORC1 (refs. 26,27). In support of this hypothesis, expression of CREBRF orthologs is highly induced by starvation in all tissues of Drosophila26,27 as well as in human lymphoblasts28,29. Moreover, REPTOR-knockout flies26 and Crebrf-knockout mice30 have lower total energy storage and body weight, respectively. Similarly, we found that nutrient starvation of 3T3-L1 preadipocytes rapidly increased Crebrf mRNA levels, which peaked by 4 h at levels 13-fold higher than those seen at 0 h (P = 1.1 × 10−16) and remained elevated by 5-fold at 24 h after the start of starvation (P = 4.1 × 10−14) (Fig. 3a). Treatment with rapamycin, a TORC1 inhibitor, also rapidly increased Crebrf mRNA levels, but did so to a lesser extent than starvation (Fig. 3b), indicating that additional TORC1-independent signals converge on Crebrf. Furthermore, overexpression of wild-type and Arg457Gln human CREBRF equivalently reduced the cell death rate to approximately one-third of that in controls within the first 6 h of nutrient starvation in 3T3-L1 preadipocytes (P = 5 × 10−6 and P = 4 × 10−5, respectively; Fig. 3c,d). These data indicate that CREBRF is a starvation-responsive factor and that wild-type and Arg457Gln CREBRF when overexpressed confer similar protection against cellular nutritional stress.
Figure 3.
Induction of Crebrf expression by nutritional stress and protection against starvation. (a,b) 3T3-L1 preadipocytes were starved (a) or treated with 20 ng/ml rapamycin (b) for 0, 2, 4, 12, or 24 h. A set of cells was starved or treated with rapamycin for 12 h and then refed with fresh growth medium for an additional 12 h (24 h (refed)). Crebrf mRNA levels were determined relative to Actb levels and normalized to baseline levels (0 h). Values are given as means ± s.e.m. from three biological replicates with four technical replicates each (n = 3 × 4 = 12). Statistical analysis: one-way ANOVA and two-sided Bonferroni post-hoc tests. **P = 0.002, ***P < 1 × 10−11 compared to cells at 0 h; #P = 0.02, ###P = 8.8 × 10−13 compared to cells at 24 h (refed). (c,d) 3T3-L1 preadipocytes were either untransfected (UT) or transfected with plasmid encoding eGFP-only negative control, wild-type human CREBRF, or Arg457Gln CREBRF and starved. (c) Time course of 3T3-L1 cell survival upon starvation up to 24 h. (d) Cell death rates after 0–6 h of starvation. Values are given as means ± s.e.m. from two transfection replicates with six wells for each transfection and three technical (cell counting) replicates (n = 2 × 6 × 3 = 36). This experiment was performed once following a pilot experiment with fewer time points showing similar results. Statistical analysis: one-way ANOVA and two-sided Games–Howell post-hoc tests. ***P < 5 × 10−5 compared to cells transfected with control eGFP construct.
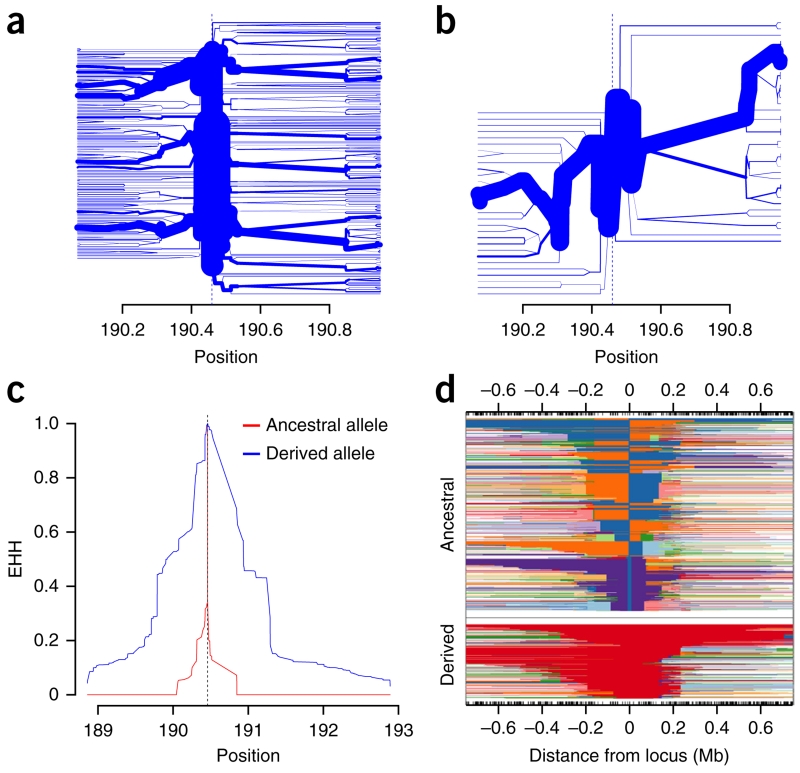
Complementing the functional evidence of ‘thriftiness’, we identified evidence of positive selection at the missense variant in Samoan genomes. The core haplotype carrying the derived BMI-increasing allele exhibited long-range LD (corresponding to the single thick branch in Fig. 4b versus Fig. 4a) and had elevated extended haplotype homozygosity (EHH) relative to haplotypes carrying the ancestral allele (Fig. 4c). Haplotypes carrying the derived allele were longer than haplotypes carrying the ancestral allele (Fig. 4d). Evidence of positive selection was provided by an integrated haplotype score (iHS) of 2.94 (P ≈ 0.003) and a number of segregation sites by length (nSL) score of 2.63 (P ≈ 0.008) (Supplementary Fig. 8).
Figure 4.
Evidence of positive selection centered on the missense variant rs373863828. Findings are shown for 626 Samoans who are not closely related. (a,b) Haplotype bifurcation plots for haplotypes carrying the ancestral allele (a) and the derived allele (b) at rs373863828 show that haplotypes carrying the derived allele have unusual long-range homozygosity. (c,d) Haplotypes carrying the derived allele have elevated EHH values as one moves away from rs373863828 (vertical dashed line) (c) and are longer than those carrying the ancestral allele (d).
In 1962, James Neel posited the existence of a thrifty gene that provides a metabolic advantage in times of famine but promotes metabolic disease in times of nutritional excess31. By carrying out a genome-wide association analysis of BMI in Samoans, we discovered and replicated a strong association with a missense variant in CREBRF that has a much larger effect size than any other known common risk-associated variant for BMI18. Functional evidence from an adipocyte model further demonstrated that CREBRF with this missense variant promotes cellular energy conservation by increasing fat storage and decreasing energy use in comparison to the wild-type protein.
The potential importance of this variant in organismal energy homeostasis is further supported by the ‘lean’ phenotype of mice30 and flies26 lacking the ortholog for this gene. These data, in combination with evidence of positive selection, support a thrifty variant hypothesis for human obesity and underscore the value of examining unique populations to identify new genetic contributions to complex traits.
However, many questions remain unanswered. More detailed studies in animal models and humans are required to define the systemic and tissue-specific (particularly central) contributions of the missense variant to overall energy balance. Such studies would also help confirm and clarify the mechanism by which this missense variant might protect against obesity-associated metabolic disease, which perhaps involves preferential promotion of more metabolically ‘safe’ or efficient energy storage and use. Studies that consider potential modifying and mediating environmental influences of this variant as well as gene–gene interactions might illuminate additional new factors contributing to these complex traits. Finally, additional anthropological genetic studies might determine the evolutionary origin of this variant or the potential role of drift in determining its frequency. Such research is urgently needed to inform decisions about how to use knowledge of this obesity risk variant to benefit Samoans at both individual and population health levels and to determine how this discovery might contribute to the understanding and treatment of more common obesity in general.
ONLINE METHODS
Participants
The participants in this study are derived from the populations of the Independent State of Samoa and the US territory of American Samoa. We used two samples in this study: a discovery sample of 3,072 phenotyped and genotyped Samoans and a replication sample of 2,103 phenotyped and genotyped Samoans and American Samoans (Supplementary Table 1). An additional sample of 409 phenotyped and genotyped Samoan children was not included in the main analyses, but analyses with our associated variants were also conducted in this sample. Details about participant recruitment can be found in the Supplementary Note. The parent GWAS, sample selection and data collection methods, and phenotype levels, including those of lipids and lipoproteins, have been reported3. This study has been approved by the Health Research Committee of the Samoa Ministry of Health and the institutional review boards of Brown University, the University of Cincinnati, and the University of Pittsburgh. All participants gave informed consent.
In the original GWAS study design, our goal of a discovery sample size of 2,500 (which we exceeded) was chosen so as to have high power to detect risk-associated SNPs with realistic effect sizes. Power was estimated as follows: we used Quanto34,35 to estimate the power to detect the rs9930506 SNP in FTO, which in the Sardinia study36 explained 1.34% of variance in BMI. If we assume that this SNP has the same allele frequencies and that BMI has the same overall mean values and standard deviation as in Scuteri et al.36, then at a significance level of 1 × 10−5 power is ≥80% when the risk-associated SNP explains at least 1.1% of the variance (and power is 90% when the SNP explains 1.3% of the variance). If we instead test at a threshold of 1 × 10−7, power is ≥80% if the SNP explains at least 1.5% of the variance.
Anthropometric and biochemical measurements
Height, weight, and BMI were measured as previously described3,37,38. Polynesian cutoffs were used to classify adults as normal weight, overweight, or obese on the basis of BMI of <26 kg/m2, 26–32 kg/m2, and >32 kg/m2, respectively39. Obesity in children was categorized from BMI using the international age- and sex-specific classifications developed by Cole et al.40.
In the discovery sample, abdominal (at the level of the umbilicus) and hip circumferences were measured in duplicate, and the measures were averaged (Supplementary Table 1). Bioelectrical impedance measures of resistance and reactance (RJL BIA-101Q device, RJL Systems) were used to estimate percent body fat on the basis of Polynesian-specific equations38,39. Serum separated from whole-blood samples, collected after a 10-h overnight fast, was assayed for cholesterol (total, HDL, and LDL), triglycerides, glucose, and insulin. The assay techniques for these metabolic markers have been described previously1. Individuals were classified as having type 2 diabetes on the basis of fasting serum glucose levels ≥126 mg/dl or the current use of diabetes medication41. Hypertensives either had systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg, or were currently taking hypertension medication. Additionally, serum levels of leptin and adiponectin were obtained by using commercially available radioimmunoassay kits (EMD Millipore). HOMA-IR was calculated as glucose (mg/dl) × insulin (μU/ml)/405, as recommended42.
Genotyping
Genotyping of the discovery sample was performed using Genome-Wide Human SNP 6.0 arrays (Affymetrix). Extensive quality control was conducted on the basis of a pipeline developed by Laurie et al.43. Additional details for sample genotyping and genotype quality control can be found in the Supplementary Note.
Statistical analysis
During quality control, significant relatedness was observed among the discovery sample participants, so empirical kinship coefficients were estimated using genotyped markers, in two iterations. In the first iteration, we selected 10,000 independent autosomal markers using PLINK44 and used them to generate empirical kinship coefficients with GenABEL45. Individuals with kinship coefficients less than 0.0625 (corresponding to first cousins) were considered unrelated. A maximal set of 1,891 unrelated individuals was then determined using previously published methods46. In the second iteration, the kinship matrix for all participants was estimated using a new set of 10,000 independent autosomal markers that had been selected using the set of unrelated individuals.
We tested for association between autosomal marker genotypes and BMI residuals while using the empirical kinship matrix to adjust for population substructure and subject relatedness. The tests were conducted using a score test as implemented in the mmscore function in GenABEL47. The statistics for association of X-chromosome genotypes with BMI residuals were calculated in GenABEL without adjusting for the empirical kinship estimates.
Meta-analysis of the adult samples was performed using METAL48 to generate two replication P values: one for the adult replication samples and one for the adult replication samples and the discovery sample together (Table 1). Additional details of the statistical analyses, including ancestry principal components (Supplementary Fig. 1 and Supplementary Video 1), can be found in the Supplementary Note.
Targeted sequencing
Before undertaking targeted sequencing, we first used SHAPEIT49–53 and IMPUTE2 (refs. 54–56) for imputation in our region of interest centered on rs12513649 with the December 2013 1000 Genomes Project Phase I integrated variant set release haplotype reference panel. The approach implicated only one strongly associated variant (with a predicted allele frequency of 0.075), but when we genotyped this variant in a pilot sample it turned out to be monomorphic (as it was in the subsequent targeted sequencing experiment). On the basis of this experience, as well as what we would expect given the unique population history of Samoans, we believe that the best way to perform accurate imputation in Samoans is by using a Samoan-specific reference panel. This idea is in agreement with recent recommendations for optimal fine-mapping in populations with unique ancestry not found in a cosmopolitan reference panel57. A panel of 1,295 Samoans from the discovery sample is currently undergoing whole-genome sequencing by the National Heart, Lung, and Blood Institute (NHLBI) TOPMed Consortium. Additional details for targeted sequencing can be found in the Supplementary Note.
Imputation
We prephased the targeted sequencing sample using SHAPEIT49–53 and then imputed into our discovery sample using IMPUTE2 (refs. 54–56). Association testing was carried out using ProbABEL58, adjusting for relatedness with the empirical kinship matrix generated by GenABEL. Three variants had nearly equivalent P values (rs12513649, rs150207780, and rs373863828) because of nearly perfect LD between them (r2 ≥0.988); imputation was very good for rs150207780 and rs373863828 (IMPUTE2 info metric = 0.954 for both variants). To determine which of these variants might be the most likely causal candidate, we tested for association in the targeted sequencing region with conditioning on each of these variants as well as the next most significant variant (rs3095870; info metric = 0.957), using ProbABEL and adjusting for relatedness. As expected for variants in such high LD, the signals in the region were eliminated after conditioning (Supplementary Fig. 3).
Bayesian fine mapping
Details can be found in the Supplementary Note.
Confirmatory genotyping
Genotyping was attempted for both rs150207780 and rs373863828 using TaqMan technology in all discovery and replication sample participants. The assay for rs150207780 failed; genotyping was not reat-tempted because this SNP showed no residual association signal in the analyses of the imputed data with conditioning on the missense variant rs373863828 (Supplementary Fig. 3). The replication plates included the 96 samples that had been sequenced in the targeted sequencing experiment. Laboratory personnel were blinded to the sequence-derived genotypes of these 96 samples, as well as to the phenotypes for all the samples. Association analysis was performed using the same regression models and meta-analysis as for the GWAS and replication analyses above. Effect size estimates were calculated using untransformed BMI separately for men and women from the discovery sample with age and age2 as covariates.
Association analyses of additional phenotypes
rs373863828 genotype was examined for association with the additional adiposity-related phenotypes listed in Table 2. Association was assessed in both the discovery sample (Table 2 and Supplementary Table 2a) and a mega-analysis of the adults from the replication sample (Supplementary Table 2b). Although meta-analysis of properly transformed phenotypes generates more accurate P values (as in Table 1), we chose instead to carry out mega-analyses here because we were primarily interested in estimating effect sizes on the natural scale for each trait. Sex-stratified analyses were also conducted in both samples (Supplementary Table 2). Diabetics were excluded from analyses of glucose, insulin, and HOMA-IR. Because the distributions of leptin levels varied greatly for women and men, a combined-sex analysis was not conducted for this trait. Residuals for quantitative traits were generated using linear regression. Age, age2, sex, and the interactions between age and sex and between age2 and sex were initially included in sex-combined models. For glucose, insulin, HOMA-IR, adiponectin, leptin, and diabetes status, a second set of models was used that included log-transformed BMI as a covariate. Sex and age × sex interactions were not included in the sex-stratified models. In the replication mega-analysis models, polity (Samoa or American Samoa) and cohort (1990s or 2000s) were initially included in the models as well. Stepwise regression was used to reduce the number of covariates for each trait separately. For quantitative traits, residuals were tested for association using the mmscore function of GenABEL45, adjusted for the empirical kinship matrix as above. Dichotomous traits were analyzed using the palogist function of ProbABEL58 while adjusting for covariates and empirical kinship. A Bonferroni-corrected P-value threshold of 2.17 × 10−3 was used to assess significance; this threshold is conservative, as it adjusts for 23 tests even though some traits are correlated with each other. To assess a possible survivor effect as the cause of the association between the BMI-increasing allele and decreased fasting glucose levels and risk of diabetes, we conducted linear regression of age by genotype. In the discovery sample, in regard to the association of rs373863828 with BMI, fasting glucose, fasting insulin, obesity risk, and diabetes risk, addition of the first ten ‘local’ principal components from Supplementary Figure 1b into the statistical models had a negligible effect on the effect estimates and statistical significance (data not shown).
Expression of CREBRF in human and mouse tissues
For human gene expression analysis, a Human Normal cDNA Array was obtained from Origene Technologies (HMRT103 and HBRT101). The human standard curve was prepared from Control Human Total RNA (Thermo Fisher Scientific, 4307281). For mouse gene expression analysis, mouse tissues were collected from 8–10 a.m. from littermate-matched, ad libitum–fed male C56BL/6J mice at 10 weeks of age (n = 6 mice/group). The mouse standard curve was prepared from pooled kidney RNA from the above mice. mRNA was prepared using the RNeasy Lipid Tissue Mini kit with on-column DNase treatment (Qiagen) followed by reverse transcription to cDNA using qScript cDNA Supermix (Quanta Biosciences). Gene expression was determined by qPCR (Quanta PerfeCTa SYBR Green FastMix or PerfeCTa qPCR FastMix) using an Eppendorf Realplex System. Human CREBRF was amplified using species-specific primers (Supplementary Table 3). Mouse Crebrf was amplified using a species-specific primer–probe set (Thermo Fisher Scientific, Mm00661538_m1). CREBRF expression was normalized to species-specific peptidylprolyl isomerase A or cyclophilin A as the endogenous control gene (Thermo Fisher Scientific, 4333763T and Mm02342430_g1 for human and mouse, respectively). Mouse data are expressed as means plus s.e.m. Data are relative expression values, and so randomization, blinding, and statistical comparisons were not indicated. Gene expression analysis was performed in accordance with Minimum Information for Publication of Quantitative Real–Time PCR Experiments (MIQE) guidelines. Mouse experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee and conducted in conformity with the Public Health Service Policy for Care and Use of Laboratory Animals. Human samples from Origene Technologies conform to federal policies for the protection of human subjects (45 CDR 46) and are HIPAA compliant. Additional information and documentation can be obtained by contacting the company.
Plasmid construction and mutagenesis
Expression plasmids with ORFs for eGFP and human CREBRF (NM_153607.2) were obtained from GeneCopoeia (EX-EGFP-M10, EX-E3374-M10). The backbone vector was pReceiver-M10, which has a cytomegalovirus promoter and encodes a C-terminal Myc-(His)6 tag. A rare missense variant, c.1447A>G, p.Thr483Ala (rs17854147), affecting a conserved residue was present in the CREBRF ORF. To avoid using this potentially function-altering variant, we converted CREBRF to the wild-type sequence and introduced the BMI risk-associated mutation c.1370G>A, p.Arg457Gln (rs373863828), using PCR mutagenesis. The segments obtained by PCR in each plasmid were verified by sequencing before large-scale plasmid purification for transfection.
Cell culture and transfection, adipocyte differentiation, Oil Red O plate assays, microscopy, triglyceride assays, and quantitative RT–PCR
These methods are described in detail in the Supplementary Note.
Bioenergetic profiling
OCR, a measure of mitochondrial respiration, and ECAR, a measure of glycolysis, were determined using an XF96 extracellular flux analyzer (Seahorse Bioscience). Transfected 3T3-L1 cells were seeded in a 96-well XF96 cell culture microplate (Seahorse Bioscience) at a density of 7,000 cells per well in 200 μl of DMEM (4.5 g/l glucose) supplemented with 10% FBS (Sigma) 36 h before measurement. Six replicates per cell type were included in the experiments, and four wells were chosen evenly in the plate to correct for temperature variation. On the day of the assay, the growth medium was exchanged for assay medium (unbuffered DMEM with 4.5 g/l glucose). Oligomycin at a final concentration of 2.0 μM, FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone) at 1.0 μM, 2-deoxyglucose at 100 mM, and rotenone at 15.0 μM were sequentially injected into each well in accordance with the manufacturer’s protocol. Basal mitochondrial respiration, maximal respiration, ATP production, and basal glycolysis were determined according to the manufacturer’s instructions. At the conclusion of the assay, cells in the analysis plate were lysed using CelLytic M (Sigma). Protein concentration was measured using the Bradford assay59 and used to normalize the bioenergetic profile data.
Starvation and rapamycin treatment
3T3-L1 preadipocytes were subjected to starvation for 0, 2, 4, 12, and 24 h by culturing cells in Hank’s balanced salt solution (HBSS). To investigate the response to refeeding starving cells, a set of cells undergoing 12 h of starvation was fed with fresh growth medium for an additional 12 h (Fig. 3a). For rapamycin stimulation, preadipocytes were treated with 20 ng/ml rapamycin (Sigma), for 2, 4, 12, and 24 h. A set of cells kept in rapamycin for 12 h was cultured in fresh growth medium for the following 12 h (Fig. 3b). To quantify cell survival, 3T3-L1 cells and transfected cells were seeded in six-well plates at 86,000 cells per well. Two days later, the cells were starved in HBSS. At 0, 2, 4, 6, 12, and 24 h, the cells were collected and 100 μl of the cell suspension samples was added to an equal volume of trypan blue (Life Technologies). The mixture was loaded into an automated cell counter (Cellometer Mini, Nexcelom Bioscience), and viable cell numbers were measured. Cell death rates were calculated by subtracting the number of viable cells at 6 h from cell numbers at 0 h and dividing the result by the cell numbers at 6 h.
Cell studies statistical analysis
For the cell studies, adequate sample sizes were determined on the basis of publications using similar methods and pilot experiments. No blinding was used. Each experiment was performed twice with similar results unless otherwise stated in the corresponding figure legend. The data were initially evaluated by one-way ANOVA implemented in SPSS (IBM). The homogeneity of variances was examined using Levene’s test. Two-sided Bonferroni and Games–Howell post-hoc tests were used to compare data with equal and unequal variance, respectively. Alternatively, pairwise two-sided t tests for unequal variance were used. P < 0.05 was considered to be statistically significant. SPSS analyses were verified using the same tests as implemented in R (ref. 60).
Selection analyses
On the basis of the genome-wide Affymetrix 6.0 SNP genotype data, we used Primus61,62 to select 626 individuals from the discovery sample using a kinship threshold (0.039) halfway between the values expected for first and second cousins, so that first cousins and more closely related relatives were excluded. These ‘unrelated’ individuals were then haplotyped using SHAPEIT49–53 and were annotated with ancestral allele information using the selectionTools pipeline63. Haplotype bifurcation diagrams and EHH plots were drawn using the rehh R package64. The haplotype bifurcation diagram65 visualizes the breakdown of LD as one moves away from the core allele at the focal SNP; each branch reflects the creation of new haplotypes, and the thickness of the line reflects the number of samples with the haplotype. EHH represents the probability that two randomly chosen chromosomes are identical by descent from the focal SNP to the current position of interest65. Selection at the core allele is expected to result in EHH values close to 1 in an extended region centered on the focal SNP. To measure the deviation, we used selscan66 to compute the iHS67, which is defined as the log of the ratio of the integrated EHH for the derived allele over the integrated EHH for the ancestral allele. These values are then normalized in frequency bins across the whole genome (we used 25 bins). Note that selscan’s definition of iHS differs from earlier definitions where the ancestral allele was in the numerator of the ratio66,67. In our case, a large positive iHS indicates that a derived allele has had its frequency increase owing to selection. We computed an approximate two-sided P value under the assumption that after normalization the iHS is approximately distributed as a standard normal. We also used selscan to compute nSL scores (the number of segregation sites by length)68. The nSL is similar to the iHS, but instead of integrating over genetic distance the nSL uses the number of segregating sites as a measure of ‘distance’. Thus, the nSL is more robust to demographic assumptions than the iHS, as it does not depend on a genetic map. As with the iHS, we normalized the nSL scores in 25 frequency bins across the whole genome and computed approximate two-sided P values assuming a standard normal distribution. The selscan program was run using its assumed default values. As we were focused on testing whether there is positive selection at the missense variant, we did not adjust the P values for multiple testing.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the Samoan participants of the study, and local village authorities and the many Samoan and other field workers over the years. We acknowledge the Samoan Ministry of Health and the Samoan Bureau of Statistics, and the American Samoan Department of Health for their support of this research. We also acknowledge S.S. Shiva and C.G. Corey at the University of Pittsburgh Center for Metabolism and Mitochondrial Biology for assistance with cellular bioenergetic profiling. This work was funded by NIH grants R01-HL093093 (S.T.M.), R01-AG09375 (S.T.M.), R01-HL52611 (I. Kamboh), R01-DK59642 (S.T.M.), P30 ES006096 (S.M. Ho), R01-DK55406. (R.D.), R01-HL090648 (Z.U.), and R01-DK090166 (E.E.K.) and by Brown University student research funds. Genotyping was performed in the Core Genotyping Laboratory at the University of Cincinnati, funded by NIH grant P30 ES006096 (S.M. Ho). Illumina sequencing was conducted at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine (Baltimore).
Footnotes
URLs. BGTEx portal, http://www.gtexportal.org/; BioGPS portal, http://biogps.org/.
Accession codes. The discovery data set is available from the database of Genotypes and Phenotypes (dbGaP) under accession phs000914.v1.p1.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
R.L.M. performed the genotype quality control and association analyses, with guidance from D.E.W. and assistance from O.D.B. and J.L.; D.E.W. and R.L.M. wrote the relevant sections of the manuscript. N.L.H. led the field work data collection and phenotype analyses with guidance from S.T.M. G.S. led and directed genotyping experiments (using the Affymetrix 6.0 chip) and assay development for validation and replication (using the TaqMan platform) with guidance from R.D. H.C. participated extensively in DNA extraction, genotyping, and quality control of the data under the supervision of G.S. and R.D. Z.U. and C.-T.S. designed and performed the CREBRF overexpression, lipid accumulation, and adipocyte differentiation and starvation experiments, analyzed the data, and wrote the relevant sections of the manuscript. E.E.K. contributed mouse and human gene expression profiling data as well as contributed to the design and analysis of the functional studies. M.S.R., S.V., and J.T. facilitated fieldwork in Samoa and American Samoa. T.N. contributed to the discussion of the public health implications of the findings. All authors contributed to this work, discussed the results, and critically reviewed and revised the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Åberg K, et al. Susceptibility loci for adiposity phenotypes on 8p, 9p, and 16q in American Samoa and Samoa. Obesity (Silver Spring) 2009;17:518–524. doi: 10.1038/oby.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGarvey ST. Obesity in Samoans and a perspective on its etiology in Polynesians. Am. J. Clin. Nutr. 1991;53(Suppl. 6):1586S–1594S. doi: 10.1093/ajcn/53.6.1586S. [DOI] [PubMed] [Google Scholar]
- 3.Hawley NL, et al. Prevalence of adiposity and associated cardiometabolic risk factors in the Samoan genome-wide association study. Am. J. Hum. Biol. 2014;26:491–501. doi: 10.1002/ajhb.22553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tishkoff S. Strength in small numbers. Science. 2015;349:1282–1283. doi: 10.1126/science.aad0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGarvey ST, Bindon JR, Crews DE, Schendel DE. In: Human Population Biology: A Transdisciplinary Science. Little MA, Haas JD, editors. Academic Press; 1989. pp. 263–279. [Google Scholar]
- 6.McGarvey ST. The thrifty gene concept and adiposity studies in biological anthropology. J. Polyn. Soc. 1994;103:29–42. [Google Scholar]
- 7.Zimmet P, Dowse G, Finch C, Serjeantson S, King H. The epidemiology and natural history of NIDDM—lessons from the South Pacific. Diabetes Metab. Rev. 1990;6:91–124. doi: 10.1002/dmr.5610060203. [DOI] [PubMed] [Google Scholar]
- 8.Kirch PV, Rallu J-L. In: The Growth and Collapse of Pacific Island Societies. Kirch PV, Rallu J-L, editors. University of Hawaii Press; 2007. pp. 1–14. [Google Scholar]
- 9.Friedlaender JS, et al. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai H-J, et al. Distribution of genome-wide linkage disequilibrium based on microsatellite loci in the Samoan population. Hum. Genomics. 2004;1:327–334. doi: 10.1186/1479-7364-1-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green RC. In: The Growth and Collapse of Pacific Island Societies. Kirch PV, Rallu J-L, editors. University of Hawaii Press; 2007. pp. 203–231. [Google Scholar]
- 12.Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. 2016 doi: 10.1038/nature19057. Preprint at bioRxiv http://dx.doi.org/10.1101/030338. [DOI] [PMC free article] [PubMed]
- 13.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eicher JD, et al. GRASP v2.0: an update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–D804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie R, O’Donnell CJ, Johnson AD. GRASP: analysis of genotype–phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearce LR, et al. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155:765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vankoningsloo S, et al. CREB activation induced by mitochondrial dysfunction triggers triglyceride accumulation in 3T3-L1 preadipocytes. J. Cell Sci. 2006;119:1266–1282. doi: 10.1242/jcs.02848. [DOI] [PubMed] [Google Scholar]
- 21.Reusch JE, Colton LA, Klemm DJ. CREB activation induces adipogenesis in 3T3-L1 cells. Mol. Cell. Biol. 2000;20:1008–1020. doi: 10.1128/mcb.20.3.1008-1020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, et al. CREBL2, interacting with CREB, induces adipogenesis in 3T3-L1 adipocytes. Biochem. J. 2011;439:27–38. doi: 10.1042/BJ20101475. [DOI] [PubMed] [Google Scholar]
- 23.Kim TH, et al. Identification of Creb3l4 as an essential negative regulator of adipogenesis. Cell Death Dis. 2014;5:e1527. doi: 10.1038/cddis.2014.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson-Fritch L, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keuper M, et al. Spare mitochondrial respiratory capacity permits human adipocytes to maintain ATP homeostasis under hypoglycemic conditions. FASEB J. 2014;28:761–770. doi: 10.1096/fj.13-238725. [DOI] [PubMed] [Google Scholar]
- 26.Tiebe M, et al. REPTOR and REPTOR-BP regulate organismal metabolism and transcription downstream of TORC1. Dev. Cell. 2015;33:272–284. doi: 10.1016/j.devcel.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stocker H. Stress relief downstream of TOR. Dev. Cell. 2015;33:245–246. doi: 10.1016/j.devcel.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Chen R, Mallelwar R, Thosar A, Venkatasubrahmanyam S, Butte AJ. GeneChaser: identifying all biological and clinical conditions in which genes of interest are differentially expressed. BMC Bioinformatics. 2008;9:548. doi: 10.1186/1471-2105-9-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dengjel J, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martyn AC, et al. Luman/CREB3 recruitment factor regulates glucocorticoid receptor activity and is essential for prolactin-mediated maternal instinct. Mol. Cell. Biol. 2012;32:5140–5150. doi: 10.1128/MCB.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 32.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kampstra P. Beanplot: a boxplot alternative for visual comparison of distributions. J. Stat. Softw. 2008;28:1–9. [Google Scholar]
- 34.Gauderman WJ. Sample size requirements for association studies of gene–gene interaction. Am. J. Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 35.Gauderman WJ. Sample size requirements for matched case–control studies of gene–environment interaction. Stat. Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 36.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGarvey ST, Levinson PD, Bausserman L, Galanis DJ, Hornick CA. Population-change in adult obesity and blood-lipids in American-Samoa from 1976–1978 to 1990. Am. J. Hum. Biol. 1993;5:17–30. doi: 10.1002/ajhb.1310050106. [DOI] [PubMed] [Google Scholar]
- 38.Keighley ED, McGarvey ST, Turituri P, Viali S. Farming and adiposity in Samoan adults. Am. J. Hum. Biol. 2006;18:112–122. doi: 10.1002/ajhb.20469. [DOI] [PubMed] [Google Scholar]
- 39.Swinburn BA, Ley SJ, Carmichael HE, Plank LD. Body size and composition in Polynesians. Int. J. Obes. Relat. Metab. Disord. 1999;23:1178–1183. doi: 10.1038/sj.ijo.0801053. [DOI] [PubMed] [Google Scholar]
- 40.Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. Br. Med. J. 2000;320:1240–1243. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2012;35(Suppl. 1):S64–S71. doi: 10.2337/dc12-s064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews DR, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 43.Laurie CC, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 46.Heath SC, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur. J. Hum. Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 47.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 50.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 2013;93:687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 52.O’Connell J, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delaneau O, Marchini J, 1000 Genomes Project Consortium. 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 55.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, et al. Evaluation of transethnic fine mapping with population-specific and cosmopolitan imputation reference panels in diverse Asian populations. Eur. J. Hum. Genet. 2016;24:592–599. doi: 10.1038/ejhg.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genomewide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 60.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2004. [Google Scholar]
- 61.Staples J, Nickerson DA, Below JE. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol. 2013;37:136–141. doi: 10.1002/gepi.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staples J, et al. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cadzow M, et al. A bioinformatics workflow for detecting signatures of selection in genomic data. Front. Genet. 2014;5:293. doi: 10.3389/fgene.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 65.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 66.Szpiech ZA, Hernandez RD. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014;31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 2014;31:1275–1291. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.