Abstract
High-level amplification of the protein phosphatase PPM1D (WIP1) is present in a subset of medulloblastomas (MBs) that have an expression profile consistent with active Sonic Hedgehog (SHH) signaling. We found that WIP1 overexpression increased expression of Shh target genes and cell proliferation in response to Shh stimulation in NIH3T3 and cerebellar granule neuron precursor (cGNP) cells in a p53-independent manner. Thus, we developed a mouse in which WIP1 is expressed in the developing brain under control of the Neurod2 promoter (ND2:WIP1). The external granule layer in early post-natal ND2:WIP1 mice exhibited increased proliferation and expression of Shh downstream targets. MB incidence increased and survival decreased when ND2:WIP1 mice were crossed with a Shh-activated MB mouse model. Conversely, Wip1 knock out significantly suppressed MB formation in two independent mouse models of Shh-activated MB. Furthermore, Wip1 knock-down or treatment with a WIP1 inhibitor suppressed the effects of Shh stimulation and potentiated the growth inhibitory effects of SHH pathway-inhibiting drugs in Shh-activated MB cells in vitro. This suggests an important cross-talk between SHH and WIP1 pathways that accelerates tumorigenesis and supports WIP1 inhibition as a potential treatment strategy for MB.
Keywords: WIP1, PPM1D, Hedgehog, p53, medulloblastoma
Introduction
Medulloblastoma (MB) is the most common malignant brain tumor of childhood. Unfortunately, current therapeutic regimens are associated with numerous long-term side effects.1 And, despite clinical advances, up to 30% of children with MB experience tumor progression or recurrence, for which no cure exists.2 Continued advances in therapy demand a better understanding of the molecular drivers of MB and treatment responsiveness.
Recent studies suggest that MB can be divided into at least four sub-groups based on gene expression and methylation profile: Wingless (WNT), Sonic Hedgehog (SHH), Group 3, and Group 4.3 One-quarter of MBs bear a signature of active SHH signaling. Clinically viable small molecule inhibitors of SHH signaling have been developed, and Phase I/II clinical trials have demonstrated efficacy of SHH-inhibiting drugs against SHH MBs.4 Unfortunately, resistance to SHH inhibitors develops quickly, and mechanisms of resistance are not fully understood.
Cytogenetics have previously shown that one-third of MBs exhibit gain of the long arm of chromosome 17 (17q) or isochromosome 17q (i17q), which is associated with poor disease-related survival.5 Surprisingly, the tumor suppressor gene, TP53, on chromosome 17p13, is usually wild-type in most MBs that exhibit loss of heterozygosity of 17p.6 Mutations of TP53 are constrained almost exclusively to WNT and SHH MBs and confer a dismal prognosis for survival in patients with SHH MBs. However, TP53 mutations are present in less than 10% of MBs.7 Yet, p53 function is compromised in a larger percentage of tumors, especially in aggressive histologic subtypes of MB.8 Northcott et al. have shown that a subset of MBs that have a signature of SHH pathway activation have high-level amplification of the chromosomal locus of WIP1 (wild-type p53-induced phosphatase 1; protein phosphatase, magnesium-dependent 1, delta, PPM1D) on chromosome 17q22-q23.9
We found that WIP1 overexpression in NIH/3T3 and cerebellar granule neuron precursor (cGNP) cells, a well-characterized MB cell of origin,10–13 increased expression of Shh target genes and cell proliferation in response to Shh stimulation in a p53-independent manner. WIP1 transgenic mice showed evidence of increased proliferation and expression of Shh downstream target genes in the external granule layer, where cGNPs arise and proliferate during early post-natal cerebellar development. When WIP1 transgenic mice were crossed with Shh-activated, SmoA1/SmoA1 MB-prone mice, MB incidence increased and MB-associated survival decreased. Conversely, Wip1 knock out significantly suppressed MB formation in SmoA1/SmoA1 and tamoxifen-induced Math1-creER; Ptc1 fl/fl mice. shRNA-mediated knock-down of Wip1 or treatment with a WIP1 inhibitor blocked the effects of Shh stimulation and potentiated the growth inhibitory effects of the SHH pathway-inhibiting drugs in SmoA1/SmoA1 or Math1-creER+; Ptc1 fl/fl MB cells under cell culture conditions. This suggests an important cross-talk between SHH and WIP1 signaling that accelerates MB tumorigenesis and that may be targetable with small molecules that inhibit WIP1 function.
Results
WIP1 promotes cell growth through sonic hedgehog signaling pathways
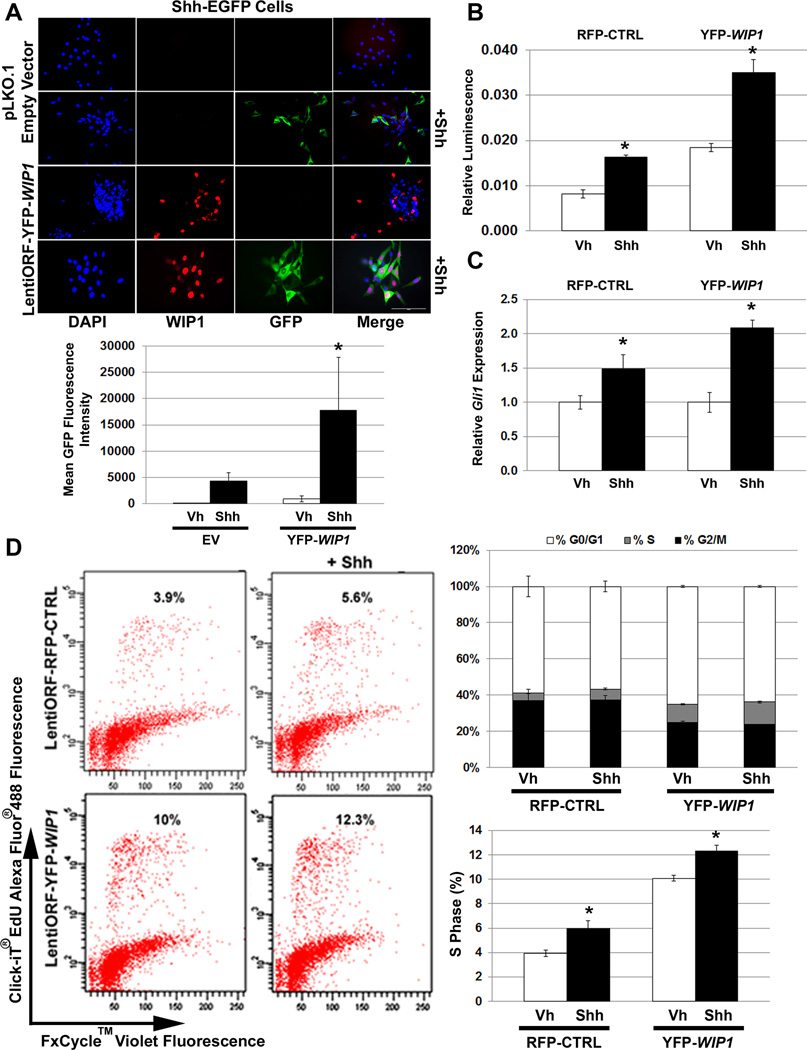
Previous studies support cross-talk between WIP1 and the SHH signaling pathway in multiple types of cancer, including MB.14, 15 To better understand this, we used NIH/3T3 cells stably transfected with a GLI-responsive Firefly luciferase reporter and a constitutive Renilla-luciferase expression vector (shh-LIGHT2) or with a Gli-dependent enhanced green fluorescent protein (EGFP) reporter (shh-EGFP), which provide downstream read-outs for activation of SHH signaling.16, 17 Immunofluorescence (IF) detected increased GFP in yellow fluorescent protein (YFP)-WIP1 (YFP-WIP1; LentiORF-YFP-WIP1)-transduced shh-EGFP cells, following stimulation with recombinant Shh (Shh) (Fig. 1A). We also found increased expression of Firefly luciferase in Shh-stimulated, YFP-WIP1-transduced shh-LIGHT2 cells, relative to controls (Fig. 1B). Quantitative real-time, RT-PCR (qRT-RT-PCR) confirmed increased expression of the downstream target of Shh signaling, Gli1, following Shh stimulation (Fig. 1C). Flow cytometry revealed a significant increase in the percentage of YFP-WIP1-transduced shh-EGFP cells in S phase following Shh stimulation (Fig. 1D). Overall, this shows that WIP1 increases proliferation through Shh-dependent and –independent mechanisms.
Figure 1. WIP1 promotes cell growth through hedgehog pathways.
(A) Twenty-four hours after 1×105 Shh-EGFP cells were seeded on poly-L ornithine-coated plates, cells were transduced with empty vector (pLKO.1) or yellow (YFP) fluorescent protein-tagged WIP1 (YFP-WIP1) lentivirus and stimulated with vehicle (Vh) or Shh (Shh-N recombinant protein, 3µg/mL) for another 24 hours, followed by fixation and incubation with α-WIP1 primary and RFP-tagged secondary antibodies. Representative photomicrographs show fluorescence for DAPI, RFP, and GFP (top panel). Total fluorescence was measured using CellProfiler software (bottom panel), *p<0.0001. Scale bar, 100µm. (B) Shh-LIGHT2 cells were transduced with control red (RFP) fluorescent protein-tagged (RFP-CTRL) or YFP-WIP1 lentivirus and stimulated with Vh or Shh for 24 hours. Cells were subsequently lysed and assayed for expression of Firefly luciferase, relative to Renilla luciferase, *p<0.005. (C) Shh-EGFP cells, transduced with RFP-CTRL or YFP-WIP1 lentivirus and stimulated with Vh or Shh for 24 hours, were harvested and lysed for total RNA. mRNA was used to determine expression of Gli1, relative to Gapdh and normalized to expression in Vh-treated cells, by real-time, RT-PCR, *p<0.005. (D) Shh-EGFP cells were treated as in (C), then incubated with 10µM Click-iT EdU for 2 hours, followed by detection of cell cycle by flow cytometry. Error bars, standard deviation (SD) among replicates of at least three per treatment. All experiments were repeated at least three times.
WIP1 promotes hedgehog signaling through p53-independent pathways
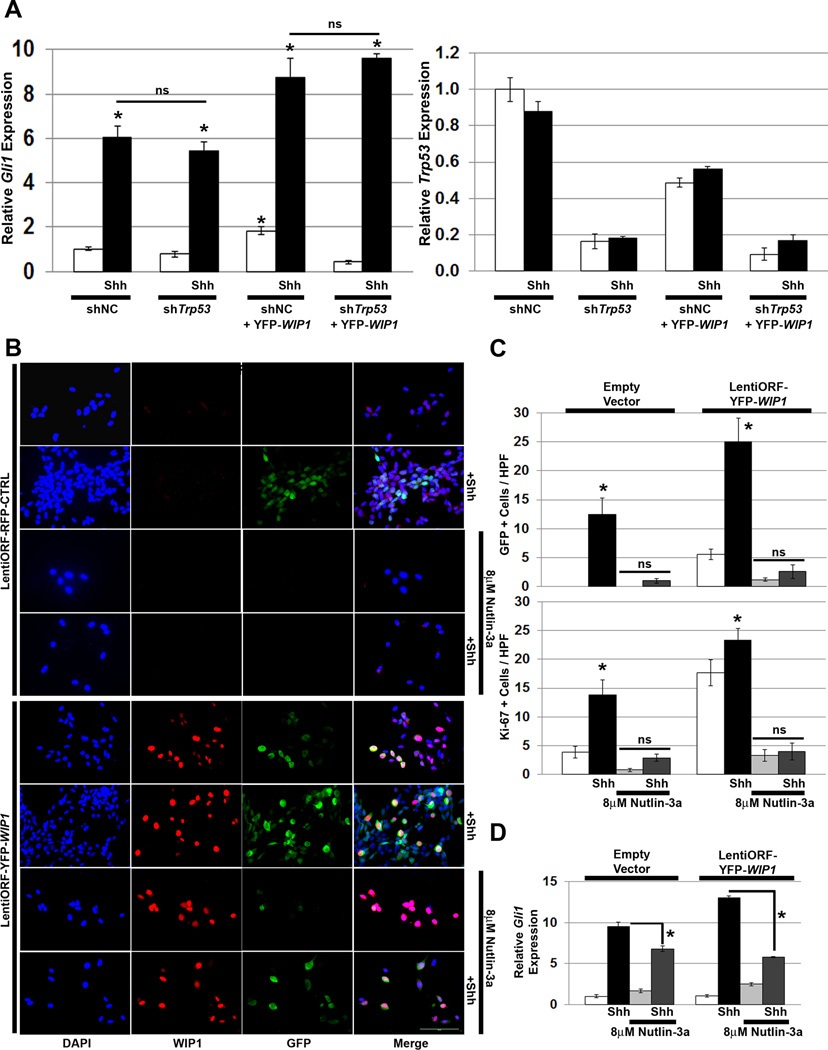
Although earlier studies have shown that WIP1 promotes growth primarily through p53 signaling pathways, recent publications suggest that the interaction between WIP1 and SHH signaling occurs independent of p53.15 To validate this prior finding, we transduced shh-EGFP cells with either Trp53 shRNA or YFP-WIP1. In contrast to the prior report in which TP53 knockdown enhanced Shh signaling, in shh-EGFP cells, knock-down of Trp53 did not affect Shh-stimulated expression of Gli1 in the presence or absence of WIP1 (Fig. 2A, Fig. S1).
Figure 2. WIP1 enhances hedgehog signaling independent of p53.
(A) Twenty-four hours after plating 1×105 Shh-EGFP cells, media was changed to serum-free media containing vehicle or Shh (3µg/mL). Cells were transduced with lentivirus containing negative control (shNC) or Trp53 shRNA (shTrp53) in combination with lentivirus containing an empty vector or YFP-tagged WIP1 cDNA (LentiORF-YFP-WIP1), and incubated for another 48 hours. Shown is real-time, RT-PCR for Gli1 and Trp53, relative to Gapdh and normalized to vehicle-treated, shNC and empty vector-transduced controls, *p<0.005. (B) Twenty-four hours after plating 1×105 Shh-EGFP cells, media was changed to serum-free media containing vehicle, 8µM Nutlin-3a, and/or Shh (3µg/mL). Cells were also transduced with lentivirus containing pLKO.1 empty vector or LentiORF-YFP-WIP1. 48 hours later, cells were fixed in 4% paraformaldehyde, permeabilized, incubated with α-WIP1 or α-Ki-67 antibody, and mounted using media containing DAPI. Scale bar, 100µm. (C) Quantitation of cells from (B) that express green fluorescent protein (GFP) or Ki-67, per high power field (HPF). Bars, mean cell counts from 10 representative fields for each condition, *p<0.005. (D) mRNA was used to determine Gli1, relative to Gapdh and normalized to expression in empty vector-transduced, vehicle-treated cells, by real-time, RT-PCR, *p<0.005. Error bars, standard deviation (SD) among replicates of at least three per treatment. All experiments were repeated at least three times. ns, not significant.
Next, we treated shh-EGFP cells with Nutlin-3a, the active enantiomer of Nutlin-3, which disrupts the interaction between p53 and Mdm2, preventing proteasome-mediated p53 degradation.18 Nutlin-3a and p53 shRNA alter p53 activity in opposite ways. shTrp53 reduces p53 activity by blocking p53 expression, while Nutlin-3a activates p53, by stabilizing the p53 protein. Treatment with Nutlin-3a suppressed activation of the Gli promoter in shh-EGFP cells, as evident by suppression of GFP as well as of expression of the proliferation marker Ki-67 in Shh-stimulated, empty vector-transduced shh-EGFP cells. Nutlin-3a treatment of YFP-WIP1-transduced shh-EGFP cells similarly suppressed activation of the Gli promoter and Ki-67 following Shh stimulation (Fig. 2B–C). And, Nutlin-3a suppressed Shh-stimulated expression of Gli in shh-EGFP cells transduced with empty vector or YFP-WIP1 (Fig. 2D). This suggests that high p53 levels override growth support from Shh or transduced WIP1.
WIP1 enhances Shh signaling in cerebellar GNPs
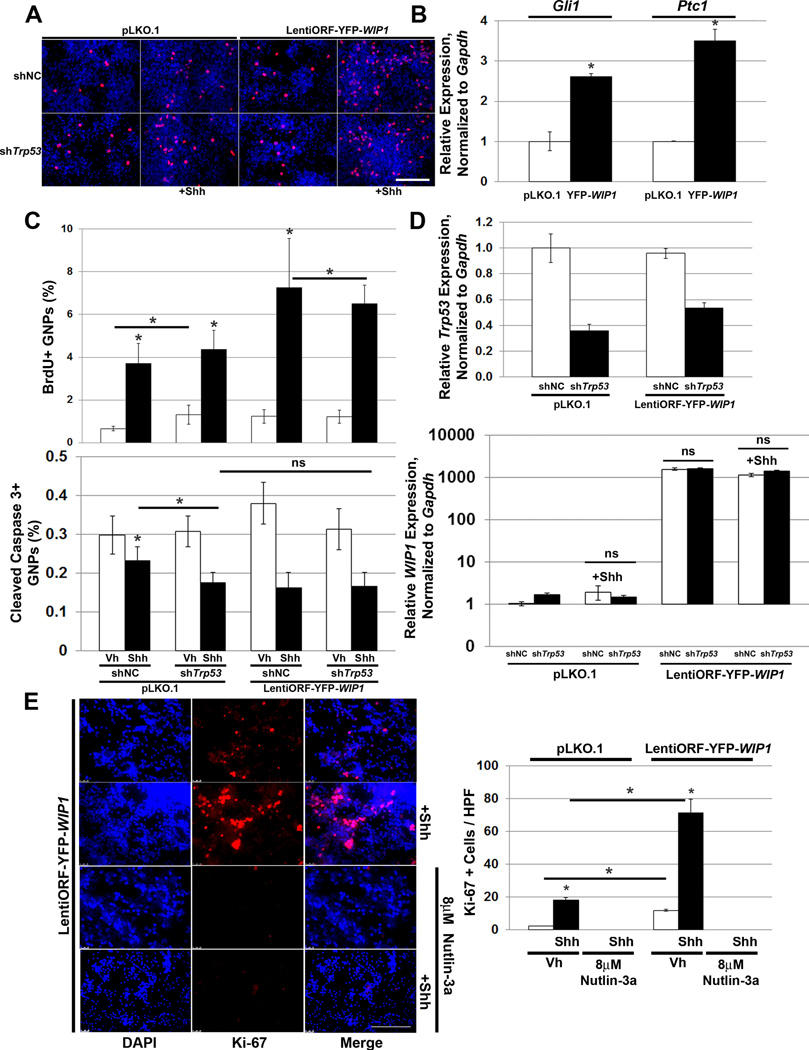
To understand the significance of high expression or amplification of WIP1 in MB tumorigenesis, we transduced cGNPs from post-natal day seven (P7) wild-type mice with empty vector or YFP-WIP1. This resulted in increased expression of the downstream Shh target genes Gli1 and Ptc1 (Fig. 3B).
Figure 3. WIP1 enhances hedgehog signaling and proliferation of cerebellar granule neuron precursor cells.
(A) Three hours after plating 1×106 post-natal day five (P5) cerebellar granule neuron precursor cells (GNPs), media was changed to serum-free media containing vehicle or Shh (3µg/mL) and cells were transduced with lentivirus containing shNC or shTrp53 in combination with lentivirus containing empty vector or LentiORF-YFP-WIP1. 48 hours later, cells were incubated with 3µg/mL BrdU for four hours, fixed in 4% paraformaldehyde (PFA), permeabilized, and incubated with α-BrdU antibody, followed by Alexa Fluor 594-conjugated secondary antibody. Shown are representative photomicrographs 48 hours after viral transduction. BrdU, red; DAPI, blue. Scale bar, 100µm. (B) Lentivirus-transduced GNPs from (A) were examined by real-time, RT-PCR for Gli1 and Ptc1, relative to Gapdh and normalized to empty vector-transduced controls, *p<0.005. (C) Quantitation of GNPs, from (A), that fluoresce positive for BrdU (top panel) or Cleaved Caspase 3 (bottom panel), relative to DAPI, using CellProfiler software, *p<0.005. (D) Real-time, RT-PCR for Trp53 (top panel) or WIP1 (bottom panel), relative to Gapdh and normalized to vehicle-treated, shNC-transduced controls, *p<0.005. (E) Twenty-four hours after plating 1×106 P5 GNPs, media was changed to serum-free media containing vehicle, 8µM Nutlin-3a, and/or Shh (3µg/mL). Cells were also transduced with lentivirus containing pLKO.1 or LentiORF-YFP-WIP1. 48 hours later, cells were fixed in 4% PFA, permeabilized, incubated with α-Ki-67 antibody, and mounted using media containing DAPI (left-hand panel). Scale bar, 100µm. Quantitation of cells from (C) that express Ki-67, per high power field (HPF) (right-hand panel), *p<0.005. Bars, mean of cell counts from 10 representative fields for each experimental condition. Error bars, standard deviation (SD) among replicates of at least three per treatment. All experiments were repeated at least three times.
To determine dependence on p53 signaling, we transduced wild-type P5 cGNPs shNC or shTrp53 in combination with empty vector or YFP-WIP1 (Fig. 3A, D). WIP1 overexpression enhanced proliferation, while stimulation with Shh inhibited apoptosis. Furthermore, knock-down of Trp53 did not significantly affect Shh-stimulated proliferation in the presence or absence of overexpressed WIP1 (Fig. 3A, C). This also suggests that WIP1 augments Gli1 through p53-independent pathways. Nutlin-3a significantly suppressed Shh-stimulated proliferation of cGNPs transduced with empty vector or YFP-WIP1, which again suggests that high levels of p53 override growth support provided by Shh or transduced WIP1 (Fig. 3E).
WIP1 enhances Shh signaling and proliferation in the neonatal cerebellum
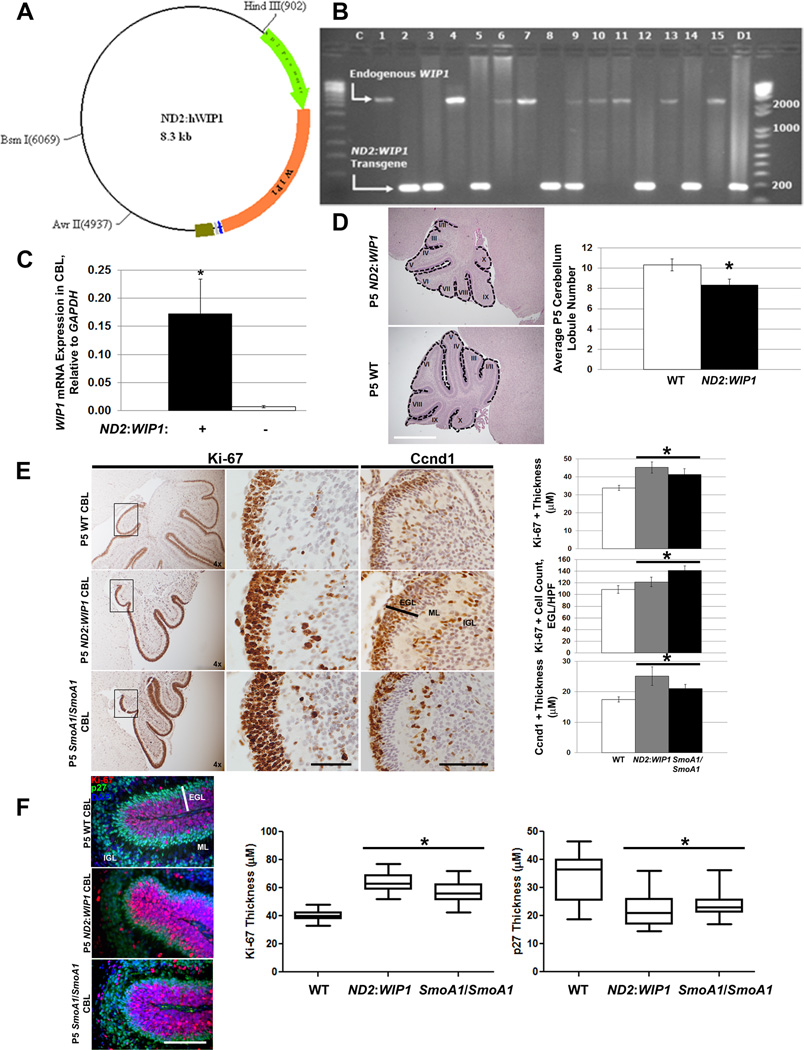
To understand the role of WIP1 in MB tumorigenesis, we developed a mouse in which human WIP1 is expressed under the control of the Neurod2 promoter (ND2:WIP1), which has previously been used to drive constitutive expression of activated Smoothened in the developing cerebellum (Fig. 4A).19 Embryos were screened for ND2:WIP1 transgene expression by conventional PCR (Fig. 4B). WIP1 expression was significantly higher in the P7 cerebellum of ND2:WIP1 transgenic mice, compared to age-matched, wild-type controls (Fig. 4C). ND2:WIP1 transgenic founder #12 was used in subsequent experiments since WIP1 expression was highest in the cerebellum of these mice by qRT-RT-PCR.
Figure 4. WIP1 promotes increased proliferation and downstream hedgehog signaling in the neonatal cerebellum.
(A) Schematic of construct used to generate ND2: WIP1 transgenic mice, showing a 1-kb portion of the Neurod2 (ND2) promoter (green arrow) driving expression of WIP1 (orange arrow; 1.8-kb). (B) DNA gel showing RT-PCR results from tail snips of ND2: WIP1 transgenic founder mice using primers spanning exons 4 and 5 of WIP1: expected 2485 base pair (bp) PCR product from endogenous Wip1; 180bp PCR product from WIP1 cDNA from expression construct in (A). C, empty template control; 1–15, founder mice; D1, PCR product using ND2: WIP1 construct as template. DNA ladders shown on either side of gel, with notations for 200–2000bp. (C) WIP1 expression by real-time RT-PCR in the cerebellum of P7 ND2: WIP1 transgenic, relative to Gapdh, compared to expression in the cerebellum of P7 wild-type (WT) C57Bl/6 mice, *p<0.005. (D) Hematoxylin and eosin stained P5 cerebella of WT and ND2: WIP1 transgenic mice (left-hand panel). Dotted line, cerebellar lobules (I–X). Quantitation of cerebellar lobules in P5 WT and ND2: WIP1 transgenic mice (right-hand panel) (n=3 cerebella per genotype), *p<0.05. (E) Immunohistochemical staining (left-hand panels) and quantitation (right-hand panels) of Ki-67 and the downstream marker of hedgehog pathway activation, Cyclin D1 (Ccnd1), in the external granule layer (EGL) of lobule X of the cerebellum of P5 wild-type, ND2: WIP1, and SmoA1/SmoA1 mice, *p<0.05. Bar, EGL width. (F) Immunofluorescent staining (left-hand panels) and quantitation (right-hand panels) of Ki-67 and p27Kip1 (p27), in the external granule layer (EGL) of the cerebellum of P5 wild-type, ND2: WIP1, and SmoA1/SmoA1 mice (n=3 cerebella per genotype), *p<0.005. Ki67, red; p27, green; DAPI, blue. Bar, EGL width. ML, molecular layer; IGL, internal granule layer; HPF, high-power field. Error bars, standard deviation (SD) among replicates of at least three per group. Scale bars, 100µm. All experiments were repeated at least three times or in at least three distinct cerebella.
ND2:WIP1 transgenic mice are viable and do not exhibit any morphologic abnormalities, other than fewer cerebellar lobules at P5, versus controls (Fig. 4D). Immunohistochemistry (IHC) for Ki-67 demonstrated increased proliferation in the external granule layer (EGL) of the P5 cerebellum of ND2:WIP1 and SmoA1/SmoA1 mice, compared to controls. IHC for Shh downstream targets also showed increased expression of Cyclin D1 (Ccnd1) in the P5 EGL of ND2:WIP1 and SmoA1/SmoA1 mice (Fig. 4E). Consistent with published findings, which showed that aberrant Shh signaling in the P7 cerebellum of SmoA1 mice leads to p27Kip1 (p27) mis-localization and increased cGNP proliferation throughout the EGL,20 double labeling with Ki-67 and p27 showed that, at P5, the Ki-67+ EGL in ND2:WIP1 and SmoA1/SmoA1 mice is thicker than wild-type mice. Conversely, the p27+ cell layer at the base of the EGL is thicker in wild-type, compared to ND2:WIP1 and SmoA1/SmoA1 mice (Fig. 4F). Thus, mis-localization of p27 in the developing cerebellum of ND2:WIP1 transgenic mice may alter cell cycle progression and promote MB formation.
WIP1 promotes MB tumorigenesis in a hedgehog-activated model
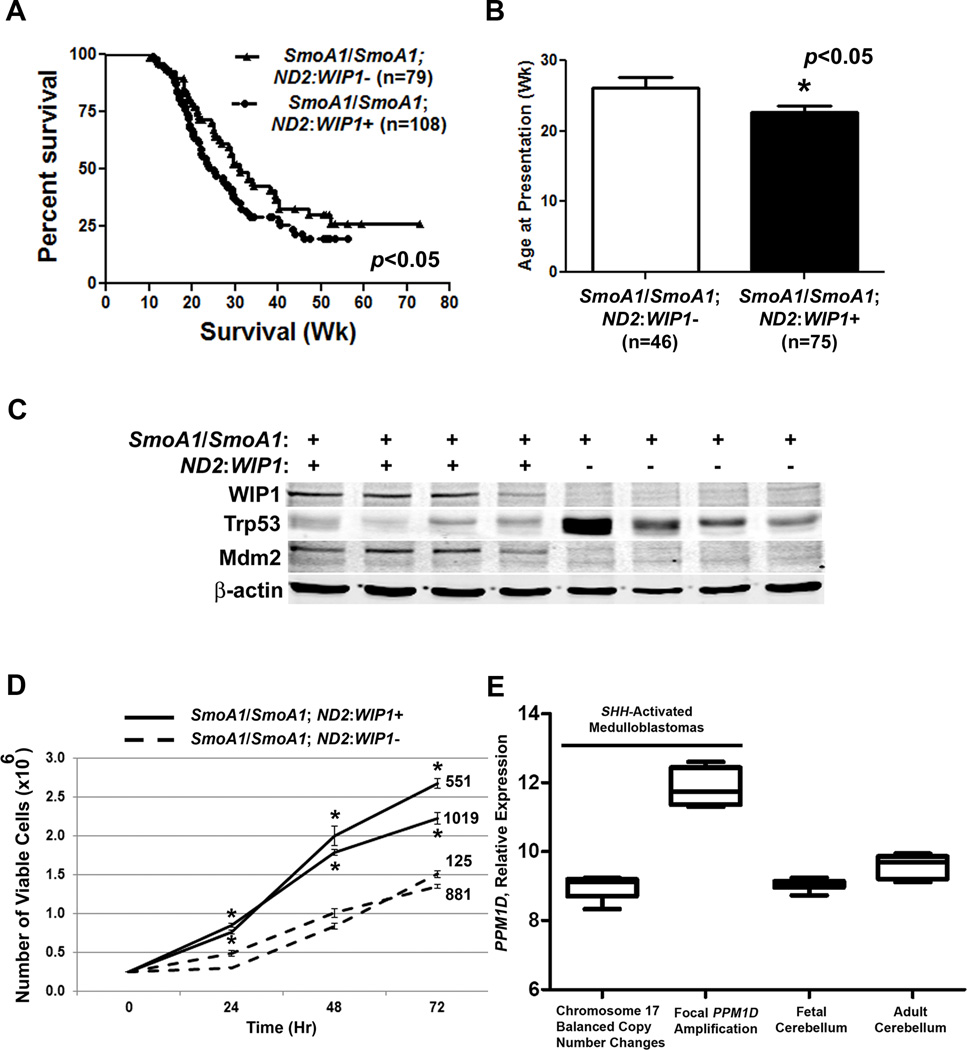
Previously published reports suggest that increased WIP1 expression alone does not promote formation of de novo tumors.14, 21 Similarly, we did not detect de novo MBs in ND2:WIP1 mice. However, when we mated ND2:WIP1 mice with SmoA1/SmoA1 mice, the incidence of MBs increased, mice presented with MB symptoms at a younger age, and SmoA1/SmoA1; ND2:WIP1+ mice exhibited reduced overall MB-related survival (Fig. 5A–B). Western blotting showed increased expression of Mdm2 and reduced expression of Trp53 in MBs derived from SmoA1/SmoA1; ND2:WIP1+ mice (Fig. 5C). And, MB cells from symptomatic SmoA1/SmoA1; ND2:WIP1+ mice exhibited increased viability compared to MB cells derived from symptomatic SmoA1/SmoA1; ND2:WIP1-mice (Fig. 5D). In human SHH-activated MBs with focal WIP1 amplification, we validated increased WIP1 expression (Fig. 5G).
Figure 5. WIP1 promotes increased tumor incidence and reduced survival in a hedgehog-activated MB model.
(A) Kaplan-Meier curves showing overall, MB-related survival of SmoA1/SmoA1; ND2: WIP1-(WIP1 transgene absent by RT-PCR) versus SmoA1/SmoA1; ND2: WIP1+ (WIP1 transgene present) mice, *p<0.05. n, mice per group. (B) Age of mice at time of presentation with symptoms of medulloblastoma (MB), *p<0.05. (C) Representative western blots for WIP1, Trp53, and Mdm2 from lysates of MBs harvested from symptomatic SmoA1/SmoA1; ND2: WIP1- and SmoA1/SmoA1; ND2: WIP1+ mice. β-actin, loading control. (D) 2.5×105 MB cells from symptomatic mice were plated in triplicate in serum-containing media and assayed for viability by trypan blue exclusion, *p<0.05. (E) Relative WIP1 expression in human MBs with balanced copy number changes of chromosome 17 (n=5) or focal amplification of the WIP1 gene locus (n=4) by Affymetrix SNPs. Both MB groups exhibit a gene signature consistent with activation of hedgehog signaling. Shown for comparison is WIP1 expression in human fetal and adult cerebellum (n=6, each). Error bars, standard deviation (SD) among replicates of at least three per group. All experiments were repeated at least three times.
Wip1 knock-out suppresses MB formation
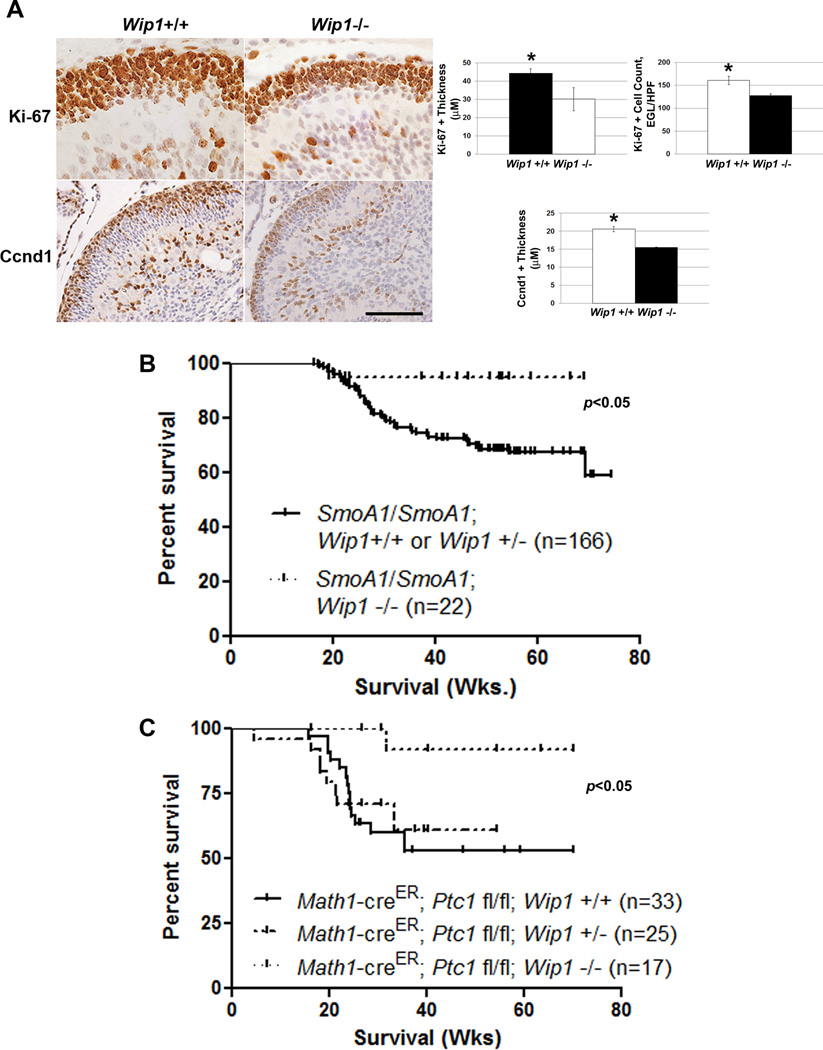
Studies published to date show that Wip1 knock-out of impairs spermatogenesis. However, Wip1−/− mice are viable and resistant to tumor formation.22 No studies to date have described abnormalities in the developing nervous system of Wip1−/− mice. Using IHC, we found that the portion of the EGL in P5 Wip1−/− mice that stained positive for Ki-67 and the downstream marker of Shh pathway activation, Ccnd1, was thinner than in P5 Wip1+/+ mice (Fig. 6A). This suggests that the P5 EGL of Wip1−/− mice contains fewer proliferating, Shh-activated cells compared to the EGL of Wip1+/+ mice. Given this finding, and since other groups have shown that Wip1 loss impairs tumor formation,15, 23 we mated Wip1−/− mice with constitutive (SmoA1/SmoA1) or conditional (Math1-creER; Ptc1 fl/fl) mouse models of Shh-activated MB. In both models, Wip1 knockout significantly suppressed de novo MB formation (Fig. 6B–C).
Figure 6. Wip1 knock-out suppresses MB formation in hedgehog-activated MB models.
(A) Immunohistochemical staining (left-hand panels) and quantitation (right-hand panels) of Ki-67+ and Cyclin D1 (Ccnd1)+ cells in the external granule layer of the cerebellum of P5 Wip1+/+ and Wip1−/− mice (n=3 cerebella per genotype), *p<0.005. Scale bar, 100µm. (B) Kaplan-Meier curves showing overall, MB-related survival of SmoA1/SmoA1; Wip1+/+, SmoA1/SmoA1; Wip1+/−, and SmoA1/SmoA1; Wip1−/− mice. (C) Kaplan-Meier curves showing overall, MB-related survival of Math1-creER; Ptc1 fl/fl; Wip1+/+, Math1-creER; Ptc1 fl/fl; Wip1+/−, and Math1-creER; Ptc1 fl/fl; Wip1−/− mice following treatment with 1mg tamoxifen by oral gavage per mouse at P7.
WIP1 inhibition suppresses hedgehog signaling and proliferation
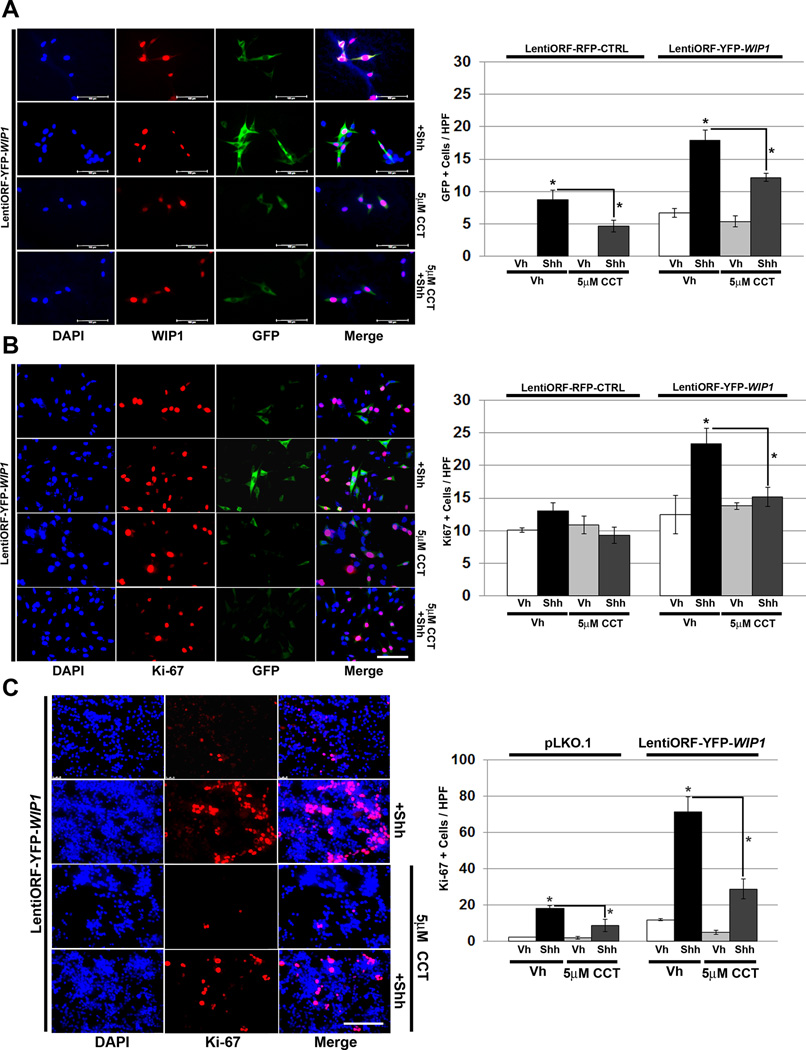
To examine the effects of pharmacologic WIP1 inhibition, we treated shh-EGFP cells with the WIP1 small molecule inhibitor, CCT007093. Shh stimulation of shh-EGFP cells transduced with YFP-WIP1 resulted in increased activation of the Gli promoter, with increased GFP, and increased Ki-67, relative to controls. Treatment of YFP-WIP1-transduced shh-EGFP cells with CCT007093 significantly inhibited both GFP and staining for Ki-67, following stimulation with Shh (Fig. 7A–B). Similarly, treatment of YFP-WIP1-transduced GNPs with CCT007093 significantly inhibited proliferation following Shh stimulation (Fig. 7C).
Figure 7. WIP1 inhibition suppresses hedgehog-mediated cell growth.
(A–B)Twenty-four hours after plating 1×105 shh-EGFP cells, media was changed to serum-free media containing vehicle (Vh), DMSO, CCT007093 (CCT, 5µM), and/or Shh (3µg/mL). Cells were also transduced with LentiORF-RFP-CTRL or LentiORF-YFP-WIP1 lentivirus. 48 hours later, cells were fixed in 4% paraformaldehyde, permeabilized, incubated with α-WIP1 or α-Ki-67 antibody, and mounted using media containing DAPI (left-hand panels). Quantification of cells, per high power field (HPF), that express green fluorescent protein (GFP) or Ki-67 (right-hand panels). Bars, mean of cell counts from 10 representative fields for each experimental condition. (C) Twenty-four hours after plating 1×106 P5 GNPs, media was changed to serum-free media containing vehicle, 5µM CCT, and/or Shh (3µg/mL). Cells were also transduced with lentivirus containing pLKO.1 or LentiORF-YFP-WIP1. 48 hours later, cells were fixed in 4% paraformaldehyde, permeabilized, incubated with α-Ki-67 antibody, and mounted using media containing DAPI (left-hand panel). Quantification of cells from (C), per high power field (HPF), that express Ki-67 (right-hand panel). Error bars, standard deviation (SD) among replicates of at least three per treatment. Scale bars, 100µm. All experiments were repeated at least three times. *p<0.005.
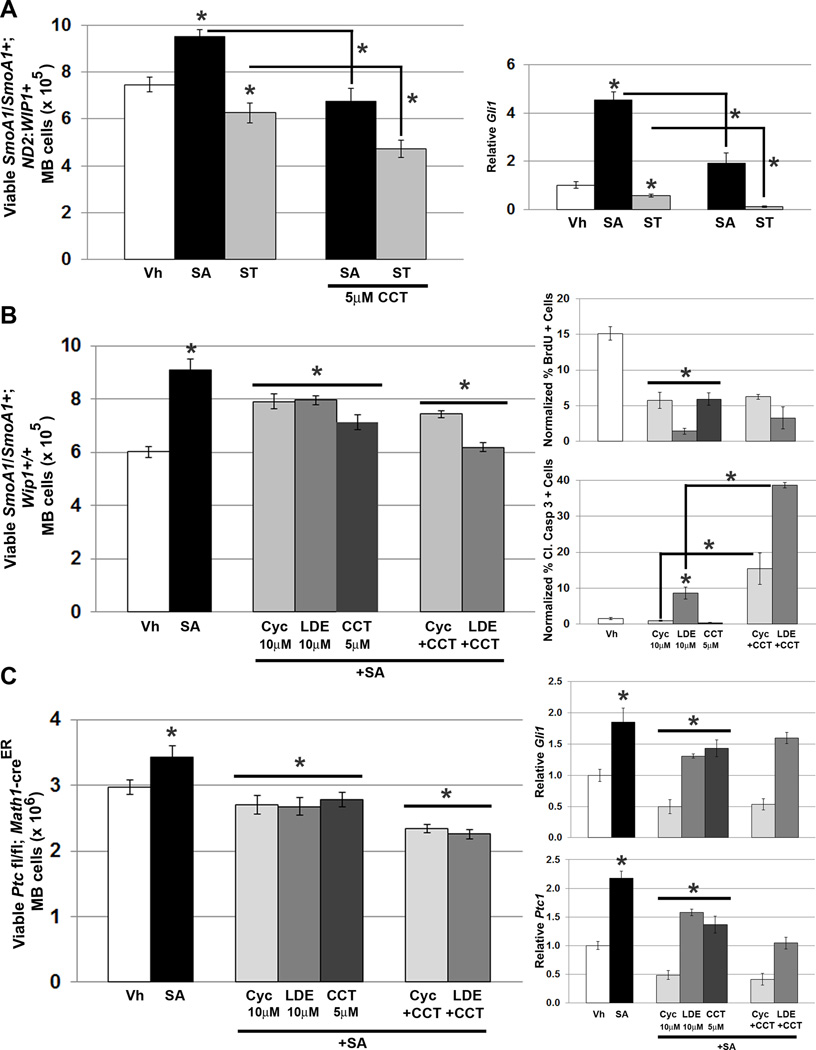
We also examined pharmacologic WIP1 inhibition in MB cells cultured from SmoA1/SmoA1 or tamoxifen-induced, Math1-creER+; Ptc1 fl/fl mice. The small molecules SAG and SANT-1 stimulate or inhibit Shh signaling, respectively. SAG stimulated, while SANT-1 inhibited the growth of MB cells derived from SmoA1/SmoA1; ND2:WIP1+ mice in short-term cultures. Treatment of MB cells from a SmoA1/SmoA1; ND2:WIP1+ mouse with lentivirus that knocked down Wip1 (not shown) or with CCT007093 inhibited the growth-promoting effects of SAG and potentiated the anti-proliferative effects of SANT-1 (Fig. 8A, left). Effects of WIP1 inhibition on cell viability correlated with reduced expression of the downstream Shh target Gli1 in CCT007093-treated SmoA1/SmoA1; ND2:WIP1+ MB cells (Fig. 8A, right).
Figure 8. WIP1 inhibition augments inhibition of hedgehog signaling in hedgehog-activated MB cells.
(A). Twenty-four hours after plating 1×106 MB cells from symptomatic SmoA1/SmoA1; ND2: WIP1+ mice, media was changed to serum-free media containing vehicle (Vh), DMSO, 200nM SAG, or 100nM SANT-1, with or without CCT007093 (5µM, CCT). Viable cells were determined by trypan blue exclusion (left-hand panel), *p<0.05. Real-time, RT-PCR for Gli1, relative to Gapdh and normalized to vehicle-treated controls, using cells from (A) (right-hand panel), *p<0.05. (B) Twenty-four hours after plating 1×106 MB cells derived from symptomatic SmoA1/SmoA1; Wip1+/+ or (C) tamoxifen-induced Math1-creER; Ptc1 fl/fl mice (left-hand panels), media was changed to serum-free media containing vehicle (Vh), 200nM SAG, 10µM cyclopamine (Cyc), 10µM LDE225 (LDE), or 5µM CCT, with or without Cyc or LDE. Cells were incubated under tissue culture conditions for another 24 hours. Viable cells were determined by trypan blue exclusion (left-hand panel), *p<0.05. Real-time, RT-PCR for Gli1and Ptc1, relative to Gapdh and normalized to vehicle-treated controls, in cells from (C) (right-hand panels), *p<0.05. Medulloblastoma cells from symptomatic SmoA1/SmoA1; Wip1+/+ were also treated with Vh, 10µM Cyc, 10µM LDE, or 5µM CCT, with or without Cyc or LDE, followed by incubation with 3µg/mL BrdU for four hours, fixation in 4% PFA, permeabilization, and incubation with α-BrdU antibody, followed by incubation with Alexa Fluor 594-conjugated secondary antibody. Shown is quantitation of BrdU (B, top-right panel) or Cleaved Caspase 3 (B, bottom-right panel) immunofluorescence, relative to DAPI using CellProfiler software, *p<0.005. Error bars, standard deviation (SD) among replicates of at least three per treatment. All experiments were repeated at least three times.
We also studied the effects of small molecules that inhibit Shh and WIP1 signaling alone and together in the SmoA1/SmoA1; Wip1+/+ and tamoxifen-induced Math1-creER; Ptc1 fl/fl MB models, which do not have high Wip1 expression. Treatment of MB cells with CCT007093 alone resulted in a similar suppression of cell viability, compared to treatment with either Shh small molecule inhibitor: cyclopamine or the clinically-relevant drug LDE22524 (Fig. 8B–C). Reduction in cell viability correlated with reduced expression of Gli1 and Ptc1 (Fig. 8C, right-hand panels). Combined treatment with CCT007093 and a Shh-inhibiting drug resulted in a further reduction in MB cell viability, although this did not appear to be associated with further reductions in expression of Gli1 and Ptc1. We also employed BrdU incorporation and IF for Cleaved Caspase 3 to assay for proliferation and cell death, respectively, in MB cells isolated from symptomatic SmoA1/SmoA1+; Wip1+/+ mice. Cyclopamine and LDE225, as well as CCT007093, significantly inhibited MB cell proliferation, alone or in combination, under cell culture conditions. However, LDE225, alone or in combination with CCT007093, also significantly increased MB cell death (Fig. 8B, right-hand panels).
Discussion
WIP1 is a serine/threonine protein phosphatase that is important in the DNA damage response. Through its phosphatase activity on proteins crucial for cell cycle regulation or DNA damage response, including Atm, p53, Chk1, Chk2, Mdm2, Mdm4, p38MAPK, and γ-H2AX, WIP1 functions in a negative feedback control loop to maintain homeostasis following exposure to DNA-damaging agents.25 However, gene amplification, high-level expression, or specific C-terminal mutations can result in WIP1 functioning as an oncoprotein.
On its own, Wip1 overexpression does not promote spontaneous tumor formation. But, Wip1 cooperates with validated oncogenes, including c-myc, Erbb2, Ras, and E1A, to transform mouse embryonic fibroblasts.22, 26 Double transgenic mice that express both Erbb2 and Wip1 under control of the MMTV promoter exhibit a shorter onset to the development of breast tumors.21 And, amplification of the chromosomal locus of WIP1, 17q22-q23, has been described in numerous human malignancies that are wild-type for TP53, including MB.9, 26, 27
By fluorescence in situ hybridization and comparative genomic hybridization we previously identified 64% of MBs with amplification of the WIP1 locus.28 We have described increased WIP1 expression in non-Wingless (WNT) activated MBs,29 a signature that is often associated with inferior patient survival.30 And, we have shown that increased WIP1 expression in MB is associated with poor overall and event-free survival. Recently, we have shown increased WIP1 expression in Group 3&4 MBs, which have a high incidence of metastasis at diagnosis. And, we have described an important interaction between WIP1 and CXCR4 signaling that may promote MB metastasis.31
Following up on the work of Northcott et al. in which they showed that a subset of MBs that have a gene signature of active SHH signaling also have high-level amplification of the chromosomal locus of WIP1,9 we found that WIP1 overexpression in NIH/3T3 cells and cGNPs enhanced proliferation and expression of Shh target genes in response to Shh stimulation. Overall, our findings were in agreement with Pandolfi et al. in which they show that WIP1 increased the transcriptional activity of GLI1 in the colon cancer cell line HCT116.15 Interestingly, neither wild-type nor phosphatase-deficient, mutant (WIP1 D314A) WIP1 activated a GLI-dependent promoter in HCT116 cells. But, in patient-derived SSM2c melanoma and MCF7 breast cancer cells, which express high levels of WIP1 at baseline, transfection with exogenous WIP1 increased activation of a GLI-dependent promoter and GLI1 mRNA expression. This suggests that cross-talk between WIP1and SHH signaling may be modulated by tumor or cell line-specific factors that are still unknown.
Investigators have previously shown that WIP1 inactivates p5332 and that p53 and GLI1 function in a negative feedback loop.33 We investigated whether the interaction between WIP1 and SHH signaling in MB is p53-dependent. Using expression plasmids that contain a luciferase reporter, investigators have shown that p53 negatively regulates the activity, localization, and expression of GLI1.34 We found that WIP1 overexpression enhanced proliferation, while stimulation with Shh inhibited apoptosis. Furthermore, knock-down of Trp53 did not significantly affect Shh-stimulated proliferation in the presence or absence of overexpressed WIP1. This suggests that WIP1 augments Gli1 expression through p53-independent pathways. That Nutlin-3a significantly suppressed Shh-stimulated proliferation of cerebellar GNPs transduced with empty vector or YFP-WIP1, suggests that, in cGNPs, the effects of high levels of p53 override whatever growth support is provided by Shh or transduction with WIP1 lentivirus.
To further understand WIP1 mechanisms in vivo, we developed a mouse in which WIP1 is expressed under control of the Neurod2 promoter (ND2:WIP1). Compared to wild-type controls, the EGL of P5 ND2:WIP1 mice exhibited increased proliferation and expression of the Shh downstream target Ccnd1. Although a few >12-month-old ND2:WIP1 mice exhibited lethargy or preferential turning in one direction, we were unable to find histologic evidence of MB in these mice. This is consistent with the findings of other groups who have been unable to promote tumor formation in vivo by overexpressing WIP1 alone, but suggests that activation of growth promoting signaling pathways by WIP1 may predispose cells in the early post-natal EGL of ND2:WIP1 mice to malignant transformation in the presence of other oncogenes.14, 21, 26
Investigators have previously shown that co-injected of RCAS-WIP1 and -Shh expression constructs into neonatal mice increased the incidence of MB.35 And, knock-down of PTCH1 accelerated the growth of flank xenografts of WIP1 high-expressing patient-derived melanoma cells in athymic-nude mice.15 We validated increased WIP1 expression in SHH-activated MB tissues with focal amplification of the WIP1 locus, compared to cerebellar controls and compared to MBs with balanced alterations in chromosome 17. And, when we mated ND2:WIP1 transgenic with SmoA1/SmoA1 mice, MB incidence increased and survival decreased. This confirmed an important interaction between WIP1 and SHH signaling that promotes MB tumorigenesis.
Since other groups have shown that Wip1 loss inhibits tumor formation23 and that WIP1 is a viable target for cancer therapy,15, 25, 36–38 we examined the effects of Wip1 loss or inhibition in Shh-activated models of MB. Wip1 knock out reduced the thickness of the proliferating Ki-67+ or Ccnd1+ P5 EGL. Wip1 knock out also significantly suppressed MB formation in two independent Shh-activated mouse models of MB. Treatment with a lentivirus that knocks down Wip1 or with the WIP1 inhibitor CCT007093 blocked the effects of Shh stimulation and potentiated the effects of the SHH pathway-inhibiting drugs SANT-1, cyclopamine, or the clinically-relevant SHH inhibitor, LDE225, on the growth of either SmoA1/SmoA1 or tamoxifen-induced Math1-creER+; Ptc1 fl/fl MB cells in vitro. These findings suggest that WIP1 is an important, druggable molecular target for MB treatment.
Materials and Methods
Lentiviral Particle Production and Infection
RFP control and YFP-WIP1 Precision LentiORF expression constructs (Thermo Scientific) were modified in the Custom Cloning Core Facility (CCCF). EGFP-negative control and shTrp53 short hairpin RNA lentiviral expression constructs have been previously described.31 psPAX2 and pVSVG plasmids (gifts, Dr. H. Trent Spencer, Emory University). pLKO.1 (gift, Dr. Rita Nahta, Emory University). Production and transduction with lentiviral particles were previously described.29
Cell Culture
shh-EGFP and shh-LIGHT2 cells (gifts, James Chen, Stanford University) have been described previously.16 Cells were maintained in DMEM medium with high glucose, 1x PenStrep (100 U/mL Penicillin and 100µg/mL Streptomycin), 10% (vol/vol) heat-inactivated FBS at 37°C in 5% CO2 under selection with 400µg/mL Zeocin (Life Technologies). Cells were authenticated by STR profiling within the last 6 months and are screened for mycoplasma contamination every 6 months (MycoAlert Detection Kit; Lonza Group).
Cerebellar GNPs were generated, as described.39 A Percoll gradient was used to purify re-suspended GNPs, as previously described.13 Cells were plated onto poly-dl-ornithine-coated 24-well plates with glass coverslips for histology experiments or plated onto poly-dl-ornithine-coated 6-well plates for RT-PCR experiments. Shh (3µg/mL) was added to samples within the Shh treatment groups. Media containing serum was replaced with serum-free media with and without Shh after a 3 hour incubation period. At this time, 20µl of respective viruses were added to each well. Immunofluorescence or RT-PCR was performed on samples 48 and 72 hours post-treatment.
Proliferation Assays
1×105-2.5×105 shh-EGFP cells were plated on 6-well plates or 1×106 GNPs on poly-L-ornithine-coated 24-well plates in triplicate and under conditions of serum starvation. Twenty-four hours later, cells were transduced with RFP-tagged control or YFP-tagged WIP1 lentivirus with or without control (pLKO.1) or shTrp53 lentivirus, and treated with vehicle or 3µg/mL Shh-N. Alternatively, cells transduced with control or YFP-tagged WIP1 lentivirus were treated for 24 hours with vehicle, 8µM Nutlin-3a (Selleck, Houston, TX) , or 5µM CCT007093 (Selleck). After Shh-N stimulation and/or drug treatment, cells were harvested with 0.25% trypsin EDTA (Mediatech) and counted by trypan blue exclusion using a Cellometer Vision CBA (Nexcelom).
Luminescence
24-well tissue culture plates were coated with 1X Poly-L-Ornithine (Corning Inc., Lowell, MA, 25–055-CV). 2.5×105 Shh-LIGHT2 cells17 were plated per well in 500µl media containing DMEM with 10%FBS and 1% P/S. 24 hours later, cells were changed to DMEM with 0.5% FBS and transduced with red fluorescent protein (RFP)-tagged control (RFP-CTRL) or yellow fluorescent protein (YFP)-tagged WIP1 (YFP-WIP1) lentivirus. 24 hours later, cells were stimulated with vehicle or Shh (3µg/mL) for 24 hours. Luciferase activity was assayed using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, # E1910), according to manufacturer’s instructions.
Cell Cycle Analysis
Forty-eight hours after transducing shh-EGFP cells with control or YFP-tagged WIP1 lentivirus and stimulating with vehicle or 3µg/mL Shh-N, 10µM EdU was added to media and incubated for 5 hours. Cells were harvested by scraping, fixed, permeabilized, and incubated with Click-iT® EdU reagents, including FxCycle™ Violet, per the manufacturer’s protocol (Life Technologies). Samples were analyzed using a BD FACS Canto II cytometer (BD Biosciences, San Jose, CA, USA) with BD FACS Diva software.
Quantitative Real-Time, RT-PCR
Total cellular RNA was extracted, as previously described.29 qRT-RT-PCR reactions containing cDNA, Sybr Green PCR Master Mix (Life Technologies) and primers for WIP1, Gli1, Ptc1, Trp53, and/or Glyceraldehyde-3 Phosphate Dehydrogenase (Gapdh) were run on an ABI 7500 Real-Time PCR Cycler (Life Technologies) using absolute quantification with a standard curve. Primer sequences available upon request. Absolute gene expression was determined based on standard curves. Target gene expression was normalized to Gapdh expression.
Western Blotting
Protein was extracted, quantified, and probed with antibodies, as described.40 Whole cell lysates were extracted using RIPA buffer (Cell Signaling). Antibodies used included p53 (Cell Signaling, Danvers, MA), Mdm2 (EMD Millipore, Billerica, MA), WIP1 (Bethyl, Montgomery, TX), and β-actin (Sigma-Aldrich, St. Louis, MO). Secondary antibodies Alexa Fluor 680 goat anti-mouse IgG (Life Technologies) or IRDye 800 goat anti-rabbit IgG (Rockland, Gilbertsville, PA) were used at a dilution of 1:5,000. Immunoblots were imaged as previously described.29
Tissue Handling and Immunohistochemistry
Mouse brains were sectioned sagittally down the midline and fixed in formalin. Tissue blocks were paraffin-embedded and cut into 5µm sections. Following antigen retrieval, tissues were blocked and incubated with α-Ki67 (1:500; Vector, VP-RMO4) or Ccnd1 (1:100; Thermo Scientific, SP4) overnight at 4°C, then incubated with biotinylated goat anti-rabbit IgG (1:300; Vector, BA-1000). Slides were stained with hematoxylin and mounted using VectaMount (Vector). Images were captured with an Olympus DP70 digital camera, and were analyzed using the DP Controller software package (Olympus). Images were processed for publication using Adobe Photoshop Elements 5.0 (Adobe Systems).
BrdU Labeling
BrdU (Fisher Scientific, # BP 2508250) was added to medium (3µg/ml) for 4 hrs, and fixed in 4% paraformaldehyde (PFA). Cells were incubated with α-BrdU (BD Biosciences, San Jose, CA, #347580) (1:1000), and then Alexa Fluor 594-conjuated Affinipure F (ab’)2 fragment goat anti-mouse IgG (H+L) secondary antibody (Jackson ImmunoResearch, West Grove, PA, code:115–586-003). Coverslips were mounted with Vectashield with DAPI (Vector, Burlingame, CA, H-1500). Images were obtained from 10 representative areas on each coverslip using a Leica DM2500 microscope with a DFC 365FX digital camera and the Leica Application Suite-Advanced Fluorescence software package (Leica Microsystems).
Immunofluorescence Quantitation
Quantitation of BrdU or Cleaved Caspase 3 positive cells was preformed using CellProfiler.41 Two analysis pipelines were used to identify nuclear labeling (DAPI or BrdU) or cytoplasm labeling (Cleaved Caspase 3) in .TIFF images: (1) Color to Gray module converted images to greyscale, and (2) Identify Primary Objects module was used to optimize program recognition of individual positive cells. Positive cells were identified based on diameter and fluorescence intensity. BrdU or Cleaved Caspase 3-positive counts were divided by DAPI-positive nuclei counts. Cell counting was single blinded: the experimenter was conscious of experimental details, but the unbiased observer, who counted the cells, was blinded from sample details.
Immunofluorescence
Coverslips were placed into 24-well plates, coated with poly-L-ornithine, and seeded with 5×104 shh-EGFP cells or 1×106 GNPs and treated as described for Proliferation. After 48 hours, cells were fixed with 4% PFA and blocked, then incubated with α-WIP1 (1:100; Bethyl, A300–664A) or Ki67 (1:500; Vector, VP-Rmo4) overnight at 4°C, then incubated with secondary antibody (1:500; Alexa Fluor® 594 goat anti-rabbit IgG, A-11012, Life Technologies). Coverslips were mounted on slides with Vectashield with DAP1 (Vector, H-1500). High-magnification images were obtained from 10 representative areas on each coverslip using a Leica DM2500 microscope with a DFC 365FX digital camera and Leica Application Suite-Advanced Fluorescence (Leica Microsystems).
P5 brains were sectioned sagittally down the midline, and fixed in 10% formalin. Following antigen retrieval, tissues were blocked, and incubated with α-Ki67 (SP6) (1:250 dilution, Abcam, ab16667) overnight at 4° C, followed by incubation with secondary antibody (Alexa Fluor 549 goat anti-rabbit IgG. Life Technologies, A11012). P27 (Kip1) (1:100 dilution, BD Transduction laboratories, 610241) was then added, followed by a second secondary antibody (Alexa Fluor 488 goat anti-mouse IgG. Life Technologies, A11029). Tissues were mounted on slides with Vectashield mounting medium with DAPI (Vector, H-1500), and imaged at 40X, as above.
Mice
Full-length WIP1 was isolated from a Tre-tight WIP1 expression plasmid (gift, Lawrence A. Donehower, Baylor College of Medicine, Houston, TX) by restriction digest using KpnI-HindIII and ligated between XbaI-ApaI into a pcDNA3.1/V5-His B vector (gift, Dr. James Olson, Fred Hutchinson Cancer Center, Seattle, WA) that already contained the NeuroD2 promoter between EcoRI-XhoI. WIP1 orientation was confirmed and sequenced. A fragment containing the NeuroD2 promoter, WIP1, and a polyadenylic acid sequence was isolated by restriction digest (HindIII-AvrII, 4036 bp), purified using Qiagen columns, and provided to the Emory Transgenic Mouse & Gene Targeting Core for zygote (E0.5 fertilized eggs) injection. Transgene expression was quantified on genomic DNA derived from tail snips using reverse transcription-PCR (RT-PCR) with the following primers: TCCTGGGAAGTGATGGACTTTGGA (forward) and ACGATGGCACTTGTGTTATCTGCC (reverse).
SmoA1 transgenic mice, on a C57Bl/6 background (gift, James Olson, Fred Hutchinson Cancer Center, Seattle, WA), were mated for at least six generations and then test crossed with wild-type C57Bl/6 mice to ensure a genotype of SmoA1/SmoA1.19 Similarly, ND2:WIP1 transgenic mice were mated with SmoA1/SmoA1 mice for at least six generations and then test crossed with wild-type C57Bl/6 mice to ensure the desired genotypes. Mice used for the MB study represent at least six generations, beginning with the F2 generation.
Wip1+/− mice (gift, Dr. Xiongbin Lu, MD Anderson Cancer Center, Houston, TX), on a 129Sv background, were bred to obtain desired genotypes.42 Wip1+/− mice were mated with SmoA1/SmoA1 mice for at least six generations and test crossed with wild-type C57Bl/6 mice to ensure desired genotypes. Math1-creER; Ptc1 fl/fl mice43 were mated with Wip1+/− mice for at least six generations to obtain desired genotypes. All mice were maintained on a mixed C57Bl/6 × 129Sv background. Mice were administered 1mg tamoxifen by gavage on P7. All mice used for MB studies represent at least six generations, beginning with the F2 generation.
All mice were housed in an AALAC–accredited facility and maintained in accordance with NIH guidelines. All animal care and experiments were approved by the Emory University IACUC (Protocol # DAR-2002073).
Statistical Analysis
Sample size was chosen based on a previous report.44 Simple randomization was used in experiments where randomization was based on a single sequence of random assignments (control versus treated).45 Mice bearing tumors were selected based on symptoms, described previously.46 Gene expression in human MB samples has been described in the Vanner dataset;47 copy number was determined using Affymetrix SNP 6.09. The Vanner dataset was run on the Affymetrix 1.0T platform. All results were analyzed using a two-tailed Student’s t-test or one-way ANOVA in Microsoft Excel or Graphpad Prism 4 software to assess statistical significance. Normality of data was determined by Chi-square testing and by reviewing graphical (i.e. boxplot) distribution of data. Values of p < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We acknowledge Drs. James Olson and Xiongbin Lu for providing mice and expression constructs, and Dr. Oskar Laur in the Emory University CCCF for plasmid construction. Work was supported by the NIH (1R01CA172392, R.C.C.; CA159859, M.D. Taylor), St. Baldrick’s Foundation (R.C.C.), CURE Childhood Cancer Foundation (R.C.C.), and the Dr. Mildred-Scheel Foundation (M.R.).
Footnotes
Conflict of Interest
Drs. Castellino’s, Kenney’s, and Taylor’s work has been funded by the NIH. Remaining authors declare no conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.von Hoff K, Hinkes B, Gerber NU, Deinlein F, Mittler U, Urban C, et al. Long-term outcome and clinical prognostic factors in children with medulloblastoma treated in the prospective randomised multicentre trial HIT’91. Eur J Cancer. 2009;45:1209–1217. doi: 10.1016/j.ejca.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Torres CF, Rebsamen S, Silber JH, Sutton LN, Bilaniuk LT, Zimmerman RA, et al. Surveillance scanning of children with medulloblastoma. N Engl J Med. 1994;330:892–895. doi: 10.1056/NEJM199403313301303. [DOI] [PubMed] [Google Scholar]
- 3.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samkari A, White J, Packer R. SHH inhibitors for the treatment of medulloblastoma. Expert Rev Neurother. 2015:1–8. doi: 10.1586/14737175.2015.1052796. [DOI] [PubMed] [Google Scholar]
- 5.Ellison D. Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol Appl Neurobiol. 2002;28:257–282. doi: 10.1046/j.1365-2990.2002.00419.x. [DOI] [PubMed] [Google Scholar]
- 6.Burnett ME, White EC, Sih S, von Haken MS, Cogen PH. Chromosome arm 17p deletion analysis reveals molecular genetic heterogeneity in supratentorial and infratentorial primitive neuroectodermal tumors of the central nervous system. Cancer Genet Cytogenet. 1997;97:25–31. doi: 10.1016/s0165-4608(96)00319-6. [DOI] [PubMed] [Google Scholar]
- 7.Tabori U, Baskin B, Shago M, Alon N, Taylor MD, Ray PN, et al. Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J Clin Oncol. 2010;28:1345–1350. doi: 10.1200/JCO.2009.23.5952. [DOI] [PubMed] [Google Scholar]
- 8.Frank AJ, Hernan R, Hollander A, Lindsey JC, Lusher ME, Fuller CE, et al. The TP53-ARF tumor suppressor pathway is frequently disrupted in large/cell anaplastic medulloblastoma. Brain research Molecular brain research. 2004;121:137–140. doi: 10.1016/j.molbrainres.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 11.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 13.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 14.Doucette TA, Yang Y, Pedone C, Kim JY, Dubuc A, Northcott PD, et al. WIP1 enhances tumor formation in a sonic hedgehog-dependent model of medulloblastoma. Neurosurgery. 2012;70:1003–1010. doi: 10.1227/NEU.0b013e31823e5332. discussion 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandolfi S, Montagnani V, Penachioni JY, Vinci MC, Olivito B, Borgognoni L, et al. WIP1 phosphatase modulates the Hedgehog signaling by enhancing GLI1 function. Oncogene. 2012 doi: 10.1038/onc.2012.502. [DOI] [PubMed] [Google Scholar]
- 16.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 19.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 20.Bhatia B, Malik A, Fernandez LA, Kenney AM. p27(Kip1), a double-edged sword in Shh-mediated medulloblastoma: Tumor accelerator and suppressor. Cell Cycle. 2010;9:4307–4314. doi: 10.4161/cc.9.21.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demidov ON, Kek C, Shreeram S, Timofeev O, Fornace AJ, Appella E, et al. The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene. 2007;26:2502–2506. doi: 10.1038/sj.onc.1210032. [DOI] [PubMed] [Google Scholar]
- 22.Nannenga B, Lu X, Dumble M, Van Maanen M, Nguyen TA, Sutton R, et al. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Mol Carcinog. 2006;45:594–604. doi: 10.1002/mc.20195. [DOI] [PubMed] [Google Scholar]
- 23.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, et al. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- 24.Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20:1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- 25.Emelyanov A, Bulavin DV. Wip1 phosphatase in breast cancer. Oncogene. 2014 doi: 10.1038/onc.2014.375. [DOI] [PubMed] [Google Scholar]
- 26.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 27.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, et al. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63:1876–1883. [PubMed] [Google Scholar]
- 28.Castellino RC, De Bortoli M, Lu X, Moon SH, Nguyen TA, Shepard MA, et al. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J Neurooncol. 2008;86:245–256. doi: 10.1007/s11060-007-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buss MC, Read TA, Schniederjan MJ, Gandhi K, Castellino RC. HDM2 promotes WIP1-mediated medulloblastoma growth. Neuro Oncol. 2012;14:440–458. doi: 10.1093/neuonc/nos001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buss MC, Remke M, Lee J, Gandhi K, Schniederjan MJ, Kool M, et al. The WIP1 oncogene promotes progression and invasion of aggressive medulloblastoma variants. Oncogene. 2015;34:1126–1140. doi: 10.1038/onc.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Nguyen TA, Zhang X, Donehower LA. The Wip1 phosphatase and Mdm2: cracking the “Wip” on p53 stability. Cell Cycle. 2008;7:164–168. doi: 10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- 33.Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. Embo J. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, et al. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci U S A. 2008;105:4838–4843. doi: 10.1073/pnas.0712216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doucette TA, Yang Y, Pedone C, Kim JY, Dubuc A, Northcott PD, et al. WIP1 Enhances Tumor Formation in a Sonic Hedgehog-Dependent Model of Medulloblastoma. Neurosurgery. 2012 doi: 10.1227/NEU.0b013e31823e5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–1044. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 37.Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nature chemical biology. 2014;10:181–187. doi: 10.1038/nchembio.1427. [DOI] [PubMed] [Google Scholar]
- 38.Yoda A, Toyoshima K, Watanabe Y, Onishi N, Hazaka Y, Tsukuda Y, et al. Arsenic trioxide augments Chk2/p53-mediated apoptosis by inhibiting oncogenic Wip1 phosphatase. J Biol Chem. 2008;283:18969–18979. doi: 10.1074/jbc.M800560200. [DOI] [PubMed] [Google Scholar]
- 39.Parathath SR, Mainwaring LA, Fernandez LA, Campbell DO, Kenney AM. Insulin receptor substrate 1 is an effector of sonic hedgehog mitogenic signaling in cerebellar neural precursors. Development. 2008;135:3291–3300. doi: 10.1242/dev.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castellino RC, De Bortoli M, Lin LL, Skapura DG, Rajan JA, Adesina AM, et al. Overexpressed TP73 induces apoptosis in medulloblastoma. BMC Cancer. 2007;7:127. doi: 10.1186/1471-2407-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 42.Choi J, Nannenga B, Demidov ON, Bulavin DV, Cooney A, Brayton C, et al. Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control. Mol Cell Biol. 2002;22:1094–1105. doi: 10.1128/MCB.22.4.1094-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 45.Suresh K. An overview of randomization techniques: An unbiased assessment of outcome in clinical research. J Hum Reprod Sci. 2011;4:8–11. doi: 10.4103/0974-1208.82352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, et al. The Smo/Smo model: hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer research. 2008;68:1768–1776. doi: 10.1158/0008-5472.CAN-07-5092. [DOI] [PubMed] [Google Scholar]
- 47.Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.