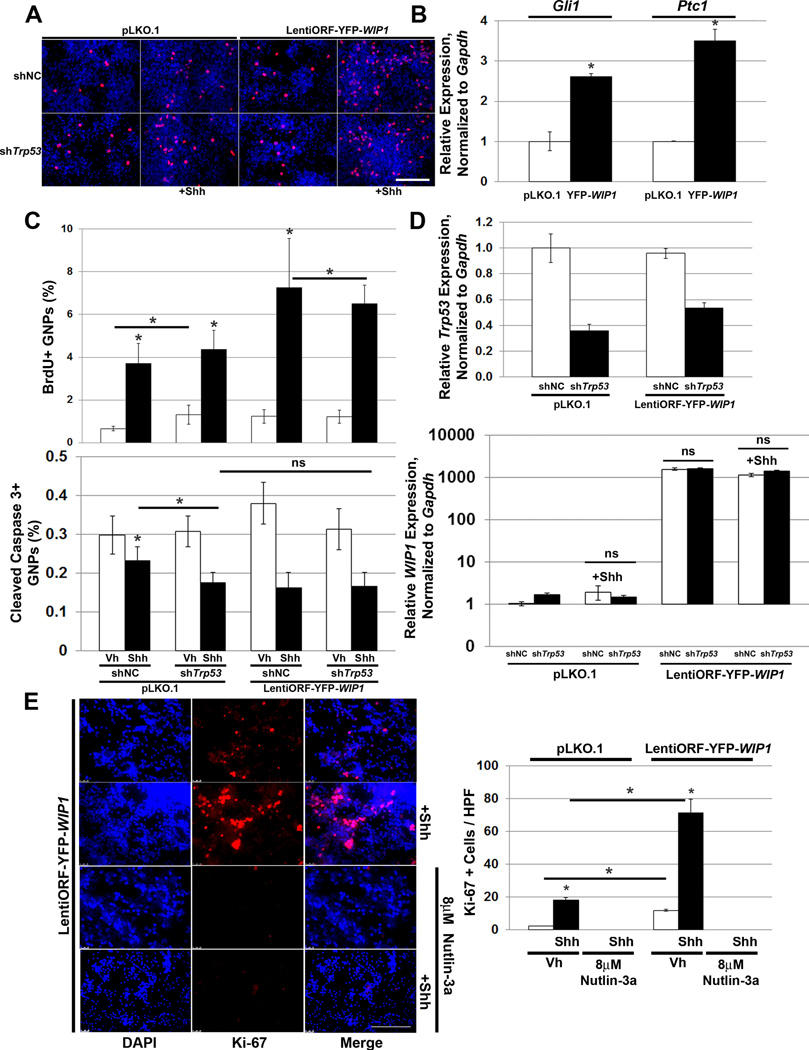
Figure 3. WIP1 enhances hedgehog signaling and proliferation of cerebellar granule neuron precursor cells.
(A) Three hours after plating 1×106 post-natal day five (P5) cerebellar granule neuron precursor cells (GNPs), media was changed to serum-free media containing vehicle or Shh (3µg/mL) and cells were transduced with lentivirus containing shNC or shTrp53 in combination with lentivirus containing empty vector or LentiORF-YFP-WIP1. 48 hours later, cells were incubated with 3µg/mL BrdU for four hours, fixed in 4% paraformaldehyde (PFA), permeabilized, and incubated with α-BrdU antibody, followed by Alexa Fluor 594-conjugated secondary antibody. Shown are representative photomicrographs 48 hours after viral transduction. BrdU, red; DAPI, blue. Scale bar, 100µm. (B) Lentivirus-transduced GNPs from (A) were examined by real-time, RT-PCR for Gli1 and Ptc1, relative to Gapdh and normalized to empty vector-transduced controls, *p<0.005. (C) Quantitation of GNPs, from (A), that fluoresce positive for BrdU (top panel) or Cleaved Caspase 3 (bottom panel), relative to DAPI, using CellProfiler software, *p<0.005. (D) Real-time, RT-PCR for Trp53 (top panel) or WIP1 (bottom panel), relative to Gapdh and normalized to vehicle-treated, shNC-transduced controls, *p<0.005. (E) Twenty-four hours after plating 1×106 P5 GNPs, media was changed to serum-free media containing vehicle, 8µM Nutlin-3a, and/or Shh (3µg/mL). Cells were also transduced with lentivirus containing pLKO.1 or LentiORF-YFP-WIP1. 48 hours later, cells were fixed in 4% PFA, permeabilized, incubated with α-Ki-67 antibody, and mounted using media containing DAPI (left-hand panel). Scale bar, 100µm. Quantitation of cells from (C) that express Ki-67, per high power field (HPF) (right-hand panel), *p<0.005. Bars, mean of cell counts from 10 representative fields for each experimental condition. Error bars, standard deviation (SD) among replicates of at least three per treatment. All experiments were repeated at least three times.