Abstract
The oxadiazoles are a class of antibacterials discovered by in silico docking and scoring of compounds against the X-ray structure of a penicillin-binding protein. These antibacterials exhibit activity against Gram-positive bacteria, including against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE). They show in vivo efficacy in murine models of peritonitis/sepsis and neutropenic thigh MRSA infection. They are bactericidal and orally bioavailable. The oxadiazoles show promise in treatment of MRSA infection.
Introduction
The modern pharmaceutical industry had its beginnings in the development of the first penicillins, β-lactam antibiotics isolated from the mold Penicillium notatum that inhibit penicillin-binding proteins (PBPs) involved in bacterial cell-wall synthesis. This success created the impetus in exploration of natural products for discoveries of the early classes of antibiotics [1]. The heyday of this period came to be known as the Golden Age of Antibiotics, spanning the 1950s through 1970s. The success was so profound that it created a perception of glut in the field in the subsequent years. Many pharmaceutical companies abandoned research on antibiotics, for reasons that have been elaborated elsewhere and will not be repeated here [2, 3]. The departure of Big Pharma created a void that smaller companies and universities tried to fill. Notwithstanding the return from this exodus of a handful of companies in the past few years, the field moved in new directions, largely away from natural products. A number of new strategies have been applied to antibiotic discovery, including continued screening of natural products [4], high-throughput screening of synthetic compound libraries [5], target-directed rational design [6] and in silico docking and scoring [7, 8] of critical targets. The discovery of the oxadiazole class of antibacterials, the subject of this report, came out of the in silico search with a penicillin-binding protein (PBP) as the target [9].
We docked and scored a 1.2-million ZINC library of drug-like compounds with the X-ray structure of PBP2a of methicillin-resistant Staphylococcus aureus (MRSA). The mecA gene of the mec operon encodes PBP2a. When the first strains of S. aureus that came to be known as MRSA were identified in the early 1960s [10], it was noted that they became broadly resistant to the entire class of β-lactam antibiotics [11]. The resistance profile for MRSA often has an inducible phenotype, it involves detection of the β-lactam antibiotic in the growth medium, an information that is transduced to the cytoplasm [12–14]. The ensuing transcriptional derepression of a set of genes leads to the expression of PBP2a. This enables the organism to survive in the face of the challenge by β-lactam antibiotics, as PBP2a can perform the critical cell-wall crosslinking reaction that other PBPs are incapable of, as they would be inhibited by the antibiotic [15, 16]. This selection pressure has been important for effective global dissemination of MRSA over the previous half a century. The organisms that are collectively referred to as MRSA remain a major clinical problem to the present day [16].
We reasoned that the clinical success of β-lactam antibiotics over the previous several decades [17–19] warranted exploration of other inhibitor classes for PBPs. The argument was that the cell wall, and specifically PBPs as biosynthetic enzymes for it, remain important targets for antibiotics. The point is underscored as the cell wall is a critically important macromolecular entity in bacteria, which is absent in mammalian organisms. Inhibition of the crosslinking event, performed by certain PBPs (transpeptidases), could not be sustained by the bacterium and would lead to lethal consequences. It was in this light that the list of the docked and scored compounds for binding to PBP2a was scrutinized to identify molecules that had the potential as antibacterial candidates.
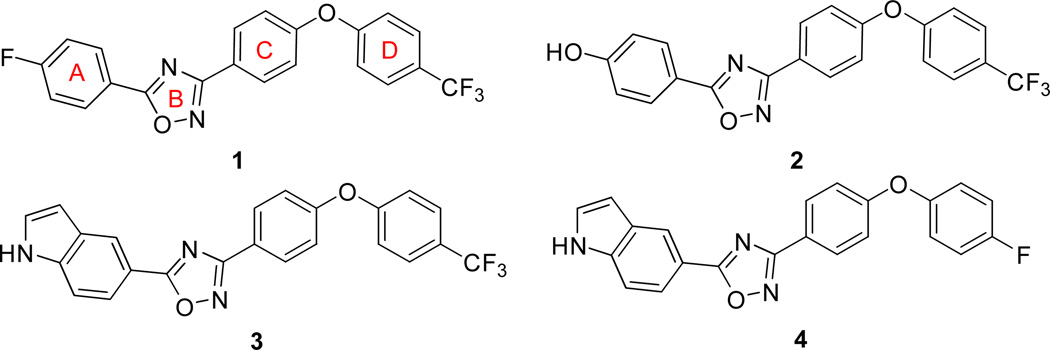
The docking and scoring procedure ranked the library compounds and we selected over 100 of the top-ranking molecules based on the pragmatic consideration of ease of synthesis for further study. These compounds were examined for their antibacterial activity at this stage with living bacteria, as opposed to their inhibitory properties with the recombinant purified PBP2a. If a compound did not exhibit antibacterial activity with living bacteria, we abandoned it. The bar was set high to weed out molecules early in discovery. We performed the antibacterial testing with the ESKAPE panel of bacteria, comprised of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacteriaceae species (the underlined first letters for the names of the genera makes the acronym), which account for the majority of the nosocomial resistant organisms [20, 21]. These studies identified oxadiazole 1 as a hit (Figure 1), with modest minimal-inhibitory concentrations (MICs) of ≥64 μg/mL against the Gram-positive organisms (E. faecium and S. aureus). Whereas the poor activity could have been the basis for abandoning this hit, the activity was reproducible and was of interest since no antibacterial activity had been attributed to this molecular scaffolding previously and the compound class was amenable to facile synthesis. These considerations prompted us to explore the oxadiazoles further. It was fortunate that merely the third oxadiazole that was synthesized in our labs improved the MIC to 2 µg/mL (compound 2, Figure 1) against the tested S. aureus strain, which justified additional efforts on this molecular template [9].
Figure 1.
Oxadiazole compounds with antibacterial activity synthesized based on an in silico search with a penicillin-binding protein (PBP) as the target.
Structure-activity relationship and the mode of action
We showed that antibacterial 2 inhibited cell-wall biosynthesis and not replication, transcription or translation using macromolecular synthesis assays. In this assay, the incorporation of radiolabeled precursors – [methyl- 3H]-thymidine, [5–3H]-uridine, L-[4,5-3H]-leucine or D-[2,3-3H]-alanine into the DNA, RNA, protein or peptidoglycan, respectively, of S.aureus strains in the log phase of growth, was monitored in the presence of sub-MIC concentrations of the antibacterial. Antibiotics such as ciprofloxacin, rifampicin, tetracycline and fosfomycin, known to inhibit the respective pathway, were used as positive controls [9]. Antibacterial 2 inhibited PBP2a, which was consistent with the search paradigm. The compound was tested against a broader panel of Gram-positive bacteria and displayed MIC values ranging from 1 to 2 μg/mL against an extended panel, including against MRSA and VRE, with the exception of Streptococci (Table 1) [9].
Table 1.
Minimal-inhibitory concentrations of oxadiazoles against Gram-positive bacteria, as determined by the microdilution MIC method.
| Bacterial strains | 2h | 3 | 4 | vancomycin | linezolid |
|---|---|---|---|---|---|
| S. aureus ATCC 29213a | 2 | 4 | 2 | 1 | 2 |
| S. aureus ATCC 27660 b | 2 | 4 | 2 | 2 | 1 |
| S.aureus N315 (NRS70)b | 2 | 2 | 1 | 1 | 1 |
| S. aureus NRS100 (COL)b | 2 | 2 | 2 | 2 | 1 |
| S. aureus NRS119c | 2 | 2 | 1 | 1 | 32 |
| S. aureus NRS120c | 2 | 2 | 2 | 1 | 32 |
| S. aureus VRS1d | 2 | 2 | 1 | 64 | 1 |
| S. aureus VRS2e | 2 | 2 | 1 | 32 | 1 |
| S. epidermidis ATCC 35547 | 2 | 2 | 1 | 4 | 0.5 |
| S. haemolyticus ATCC 29970 | 2 | 4 | 2 | 4 | 1 |
| B. licheniformis ATCC 12759 | 2 | 8 | 2 | 0.5 | 0.5 |
| E. faecalis ATCC 29212a | 2 | 8 | 4 | 2 | 1 |
| E. faecalis 201 (Van S)f | 2 | 4 | 4 | 1 | 1 |
| E. faecalis 99 (Van R)g | 2 | 4 | 4 | 128 | 1 |
| E. faecium 119-39A (Van S)f | 1 | 1 | 1 | 0.5 | 1 |
| E. faecium 106 (Van R)g | 2 | 2 | 1 | 128 | 1 |
| E. faecium NCTC 7171 | 2 | 2 | 1 | 0.5 | 1 |
A quality-control strain to monitor accuracy of MIC testing;
mecA positive, resistant to methicillin, oxacillin, and tetracycline; susceptible to vancomycin and linezolid;
mecA positive, resistant to ciprofloxacin, gentamicin, oxacillin, penicillin, and linezolid;
vancomycin-resistant MRSA (vanA) clinical isolate from Michigan;
vancomycin-resistant MRSA (vanA) clinical isolate from Pennsylvania;
vancomycin-susceptible clinical isolate;
vancomycin-resistant clinical isolate.
reproduced from O Daniel et al [9].
Meanwhile, we synthesized a collection of oxadiazoles in efforts to define the structure-activity relationship for this class. Modifications of ring A (see the chemical structure 1, Figure 1) generated a series of active antibacterials. Loss of activity was observed when the hydrogen-bond donor at the 4-position of ring A was replaced with halogens or other hydrogen-bond accepting moieties, as did several hetero-aromatic systems and aliphatic heterocycles. Although compounds with pyrazoles at ring A showed very good in vitro antibacterial activity, they exhibited some toxicity to mammalian cells. Antibacterial 3 (also designated as ND-421 in some publications), is a lead molecule of this series.
Isomeric arrangements of heteroatoms in ring B retained activity. Other five-membered ring species, such as pyrazoles and isoxazoles produced antibacterial activity. Several structural variations in ring C were well tolerated. Replacing the bridging oxygen with a sulfide or an amine did not affect the antibacterial activity. Fused C and D rings were mostly inactive or had a higher MIC values of 8 μg/mL [22]. Replacing the trifluoromethyl at the 4-position of the diphenyl ether moiety with a fluorine (antibacterial 4) did not affect activity. A number of substitutions in ring D were tolerated [22].
It is known that β-lactam antibiotics often target more than one PBP for inhibition, as each bacterium has several closely related PBPs [23]. This multiplicity of targeting is actually an aspect of the success of β-lactams [18]. Hence, it is likely that oxadiazoles would target more than a single PBP in various organisms. As such the MIC that is measured is a composite of the inhibition profile for the collection of PBPs that are inhibited. To gain insight into the structural attributes that impart antibacterial activity, a three-dimensional quantitative structure-activity relationship (3D-QSAR) model was developed based on the experimental structure-activity analysis [24]. In this model, the partial least square regression method was applied to a training set of 77 synthetic oxadiazole compounds (represented by both active and inactive compounds), to build a 3D map of the geometric and electrostatic properties. In order to validate the reliability of this in silico model, it was used to predict the activities of another 25 test set compounds, which had not influenced the model. The model revealed certain important antibacterial structure-activity features of oxadiazole compounds. Large substitutions at the para-position of the ring A were found to result in loss of activity. At the same position, electropositive functional groups (such as amines) were favored, while placement of electronegative groups (for example, a carboxylate) at this position resulted in loss of activity. Steric map predicted that presence of bulky group at the position of ring D was important for activity, but variations were possible. For example, a cyclopentyl or trifluoromethyl group in place of the aromatic ring in the D position produced active compounds. Certain aliphatic groups at this position are also tolerated.
Molecular attributes of oxadiazoles
Microdilution MIC values for antibacterial 3 were mostly similar to those of 2, ranging within 1–4 μg/mL. As with β-lactam antibiotics, the oxadiazoles are bactericidal, consistent with inhibition of PBPs as the target. The minimal-bactericidal concentrations (MBCs) for oxadiazoles were either the same as the MIC or two-fold higher. The inoculum effect is a clinically relevant factor in assessing antibiotic efficacy, assessing whether the extent of the growth of bacteria could influence antibacterial activity. The oxadiazoles do not display an inoculum effect. XTT cell-viability assays with HepG2 cells were performed to assess potential toxicity of the oxadiazoles. Compared to compound 2, the indole variant 3 was found to be three-fold less toxic (IC50 values of 25.8 μg/mL vs. 75.7 μg/mL) [9, 25]. Antibacterials 2 and 3 were metabolically stable.
Oxadiazoles 2 and 3 exhibit high oral bioavailability. Among the antibiotics currently used for treatment of MRSA infections, only the oxazolidinones (linezolid and tedizolid) are orally bioavailable. Antibacterials 2 and 3 exhibit 100% and 97% oral bioavailability, respectively. Pharmacokinetic studies of 2 and 3 after oral administration showed long terminal half-lives of 18.6 h and 40 h, respectively. Oxadiazole 2 had a modest clearance of 18.9 mL/min/kg and modest volume of distribution (Vd) of 0.37 L/kg, while 3 had a three-fold lower clearance yet (5.68 mL/min/kg) and 13-fold higher Vd (4.73 L/kg) (Table 2).
Table 2.
Pharmakokinetic parameters of oxadiazoles compared to linezolida
| Drugsb | F (%) |
AUC (μg.min/m L) |
t½β (h) | CL (mL/min/ kg) |
Vd (L/kg) |
Tmax (h) |
Cmax (μg/mL) |
|
|---|---|---|---|---|---|---|---|---|
| 2 | iv | 2,650 | 22 | 18.9 | 0.37 | |||
| po | 100 | 2,620 | 40 | 2 | 2.5 | |||
| 3 | iv | 3,520 | 9.6 | 5.68 | 4.73 | |||
| po | 97 | 4,060 | 18.6 | 6 | 2.34 | |||
| Linezo lidc |
iv | 606 | 0.55 | 16.3 | 0.63 | |||
| po | 72.5 | 440 | 0.65 | 0.33 | 6.5 |
F - oral bioavailablity; AUC - area under the curve; t½β- terminal half-life; t½elim - elimination half-life; CL – clearance; Vd – Volume of distribution; Tmax – Time at which peak serum concentration was achieved; Cmax – peak serum concentration.
Compound 2 was administered iv and po at 50 mg/kg, 3 at 20 mg/kg and linezolid at 10 mg/kg.
Data reproduced from Slatter et al. 2002 [29] for comparison.
Two animal models, the murine peritonitis/sepsis and the neutropenic thigh infection models, have been used to document the in vivo efficacy of the oxadiazoles. In both models, outbred female ICR mice were used, to provide a heterogeneous population akin to humans. The choice of these mice set the bar high for attrition of compounds early in the discovery phase. The peritonitis model was used first in exploring in vivo efficacy due to its simple end points (death or survival) and availability of results within 48 h. In this model, the mice were infected intraperitoneally with MRSA containing mucin as adjuvant to inhibit the immunological response for 2–3 h [26]. The animals were dosed with the oxadiazoles, vancomycin (positive control) or vehicle (negative control) given iv by tail-vein injection at 30 min and 8 h after infection. The efficacy of the compound after oral administration was also evaluated in a separate study with a single dose of the oxadiazoles, linezolid (comparator positive control) or vehicle given by oral gavage one hour after infection. In both cases the mice were monitored for 2 days with survival as the end point. The peritonitis model was also used to determine the dose-response curves for the oxadiazoles by the iv or po routes of administration. The median effective dose (ED50) for 2 after was found to be 40 mg/kg after iv administration and 44 mg/kg after oral administration. Antibacterial 3 had a six-fold lower ED50 at 7.6 mg/kg after iv administration. A single oral dose of 3 had an excellent ED50 of 3.1 mg/kg, which was comparable to that of linezolid at 2.8 mg/kg [9, 25].
The neutropenic thigh-infection model was also used as a second model to test the efficacy of the oxadiazoles. This model is more clinically relevant, resembling the MRSA complicated skin and tissue infections commonly seen in the clinic. The end point was the reduction of MRSA colony counts in the treated groups compared to the untreated ones. The mice were rendered neutropenic by cyclophosphamide injections prior to infection by either linezolid-resistant or linezolid-sensitive clinical MRSA strains. One hour after the infection, mice were treated with a single oral dose of an oxadiazole, linezolid or vehicle. After 48 hours the infected thighs were harvested and bacterial colonies were counted. Oxadiazole 3 showed equivalence in efficacy to linezolid in treating infection by the linezolid-susceptible strain, but showed superiority in the infection by the linezolid-resistant strain (unpublished data).
Resistance to oxadiazoles
An in vitro study for selection of resistance for oxadiazole 2 was performed [27]. Seven serial passages of S. aureus COL strain with increasing levels of oxadiazole antibiotic were made, starting from the half-MIC concentration of the antibacterial 2 (1 μg/mL) with subsequent two-fold increases in concentration. Additionally four passages were carried out at 8 μg/mL, which was the solubility limit of this specific antibacterial, hence the highest concentration tested. Single colonies isolated from these passages were tested for MIC and for homogeneity. Two strains were selected with MIC values of 4 μg/mL and >8 μg/mL designated as COLI and COLR, respectively. Compared to the wild-type COL strain, neither of these resistant strains displayed any changes in susceptibility patterns to other antibiotics such as ampicillin, imipenem, linezolid or vancomycin, indicating that the resistance mechanism to oxadiazole might be unique. Whole-genome sequencing of the COLR strain revealed 31 mutations, which were confirmed by PCR-based sequencing. The majority of the mutations in the coding region involved structural genes such as those involved in cell-envelope biogenesis, transport and binding or cell-wall stress stimulon promoter genes. When compared to the COLI strains, the unique mutations found in COLR were P70Q substitution in the putative thioredoxin TRX and T172I substitution in MmpL. The MmpL showed sequence similarity to the corresponding protein in Mycobacterium tuberculosis [28] and deletion of this gene from our COLR strain resulted in a reproducible two-fold decrease in MIC of the strain to the oxadiazole, but not against other antibiotics tested. Annotated as a putative member of the resistance nodulation and cell-division family of proteins, the family of genes that mmpL belongs to encodes large membrane proteins, which are known to function as scaffolds for biosynthesis of cell-wall associated lipids. However, deletion of the putative trx gene showed no change in susceptibility patterns for the oxadiazole or for other antibiotics It is known that the thioredoxin systems are involved in protection of cells against oxidative stress. But the actual function of the putative trx gene is not fully understood. However, it is conceivable that this gene might play a role in fitness adaptation. These mutations were the result of a stepwise accumulation of mutations, which suggest that development of resistance to oxadiazoles would require the collective effect of mutations at multiple points, rather than alterations at a single site. Even though the exact mechanism of development of resistance to oxadiazoles in not known, we assume that simultaneous occurrences of mutations in the mmpL and trx genes by themselves or in combination with other mutations would be required to attain high-level resistance to oxadiazoles.
Conclusions
The oxadiazoles were discovered recently by in silico docking and scoring of compounds of known three-dimensional chemical structures. The compounds target cell-wall synthesis, are orally bioavailable, are bactericidal against a range of Gram-positive bacteria, share a number of desirable pharmacokinetic attributes and exhibit good in vivo activity in murine models of MRSA infection. A key attractive feature of this class of antibacterials is the simplicity of the structure(s) that lends itself to facile and efficient total synthetic preparation. The oxadiazoles hold significant promise in treatment of infections by Gram-positive bacteria.
Acknowledgments
This work was supported by grants AI90818 and AI104987 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Dias DA, Urban S, Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2:303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Shlaes DM. The abandonment of antibacterials: why and wherefore? Curr Opin Pharmacol. 2003;3:470–473. doi: 10.1016/j.coph.2003.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 6.Fabbretti A, Gualerzi CO, Brandi L. How to cope with the quest for new antibiotics. FEBS Lett. 2011;585:1673–1681. doi: 10.1016/j.febslet.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 7.Kapetanovic IM. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem Biol Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S. Computer-aided drug discovery and development. Methods Mol Biol. 2011;716:23–38. doi: 10.1007/978-1-61779-012-6_2. [DOI] [PubMed] [Google Scholar]
- 9. O'Daniel PI, Peng Z, Pi H, Testero SA, Ding D, Spink E, Leemans E, Boudreau MA, Yamaguchi T, Schroeder VA, et al. Discovery of a new class of non-beta-lactam inhibitors of penicillin-binding proteins with Gram-positive antibacterial activity. J Am Chem Soc. 2014;136:3664–3672. doi: 10.1021/ja500053x. ••This article outlines our discovery of oxadizoles as an effective antibacterial against Gram-positive bacteria, its mode of action and in vivo efficacy.
- 10.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. doi: 10.1016/s0966-842x(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 12.Fuda CC, Fisher JF, Mobashery S. Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol Life Sci. 2005;62:2617–2633. doi: 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llarrull LI, Fisher JF, Mobashery S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob Agents Chemother. 2009;53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thumanu KCY, Fisher JF, Perrins R, Mobashery S, Wharton C. The Discrete Steps in Sensing of beta-Lactam Antibiotics by the BlaR1 Protein of Methicillin-Resistant Staphylococcus aureus bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinho MG, de Lencastre H, Tomasz A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A. 2001;98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CDC. cfdcap: Antibiotic resistance threats in the United States, 2013. Edited by. 2013. [Google Scholar]
- 17.Llarrull LI, Testero SA, Fisher JF, Mobashery S. The future of the beta-lactams. Curr Opin Microbiol. 2010;13:551–557. doi: 10.1016/j.mib.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Testero SA, Fisher JF, Mobashery S. Beta-lactam Antibiotics. In: Abraham DJaR DP, editor. Burger's Medicinal Chemistry. Wiley and Sons; 2010. pp. 259–404. [Google Scholar]
- 19.Thakuria B, Lahon K. The Beta Lactam Antibiotics as an Empirical Therapy in a Developing Country: An Update on Their Current Status and Recommendations to Counter the Resistance against Them. J Clin Diagn Res. 2013;7:1207–1214. doi: 10.7860/JCDR/2013/5239.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 21.Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 22.Ding D, Boudreau MA, Leemans E, Spink E, Yamaguchi T, Testero SA, O'Daniel PI, Lastochkin E, Chang M, Mobashery S. Exploration of the structure-activity relationship of 1,2,4-oxadiazole antibiotics. Bioorg Med Chem Lett. 2015;25:4854–4857. doi: 10.1016/j.bmcl.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgopapadakou NH. Penicillin-binding proteins and bacterial resistance to beta-lactams. Antimicrob Agents Chemother. 1993;37:2045–2053. doi: 10.1128/aac.37.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leemans E, Mahasenan KV, Kumarasiri M, Spink E, Ding D, O'Daniel PI, Boudreau MA, Lastochkin E, Testero SA, Yamaguchi T, et al. Three-dimensional QSAR analysis and design of new 1,2,4-oxadiazole antibacterials. Bioorg Med Chem Lett. 2016;26:1011–1015. doi: 10.1016/j.bmcl.2015.12.041. •This article outlines the development of a three-dimensional QSAR model to assist the design of novel analogs with improved activity.
- 25. Spink E, Ding D, Peng Z, Boudreau MA, Leemans E, Lastochkin E, Song W, Lichtenwalter K, O'Daniel PI, Testero SA, et al. Structure-activity relationship for the oxadiazole class of antibiotics. J Med Chem. 2015;58:1380–1389. doi: 10.1021/jm501661f. ••This paper outlines the exploration of the structural space for the oxadiazole class and identified an oxadiazole antibacterial that was efficacious in a mouse model of MRSA infection, exhibiting a long half-life, a high volume of distribution, and low clearance.
- 26.Frimodt-Moller N, Knudsen JD, FE . The mouse peritonitis/sepsis model. In: Academic Press, editor. Handbook of animal models of infection: experimental models in antimicrobial chemotherapy. 1999. pp. 127–136. [Google Scholar]
- 27. Xiao Q, Vakulenko S, Chang M, Mobashery S. Mutations in mmpL and in the cell wall stress stimulon contribute to resistance to oxadiazole antibiotics in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58:5841–5847. doi: 10.1128/AAC.03501-14. •This paper outlines a possible mechanism by which resistance could develop to oxadiazoles
- 28.Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol. 2012;19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slatter JG, Adams LA, Bush EC, Chiba K, Daley-Yates PT, Feenstra KL, Koike S, Ozawa N, Peng GW, Sams JP, et al. Pharmacokinetics, toxicokinetics, distribution, metabolism and excretion of linezolid in mouse, rat and dog. Xenobiotica. 2002;32:907–924. doi: 10.1080/00498250210158249. [DOI] [PubMed] [Google Scholar]