Abstract
The rise of multidrug resistant (MDR) Gram-negative pathogens complicates our ability to treat bacterial infections with antibiotics. MDR efflux pumps play a major role in the acquisition and expression of the MDR phenotype. The major MDR efflux pumps in the Gram-negative pathogens are the resistance-nodulation-division (RND) superfamily pumps. Efflux pump inhibitors (EPIs) that target RND superfamily pumps could play an important role in the clinic as an adjunctive therapy to increase antibiotic efficacy, decrease resistance, and attenuate virulence in Gram-negative pathogens. Here, we review recent advances in the discovery and structurally enabled optimization of a novel series of RND-targeting pyranopyridine EPIs currently in the early stages of lead optimization.
Introduction
The increased prevalence of multidrug resistant (MDR) Gram-negative pathogens poses a serious threat to our ability to effectively treat infections. Overexpression of multidrug efflux pumps, such as resistance-nodulation-division (RND) superfamily pumps, play a major role in the acquisition and expression of the MDR phenotype. In addition, RND pumps are required for virulence and biofilm formation in Gram-negative pathogens [1]. The prototypical RND family pump system is the AcrAB-TolC efflux pump of Escherichia coli (3), orthologs of which are found in all clinically-relevant Gram-negative pathogens, including the highly MDR organisms Pseudomonas aeruginosa (MexAB-OprM and MexXY-OprM) and Acinetobacter baumanii (AdeABC) [1]. A potent, drug-like efflux pump inhibitor (EPI) that targets the RND family pumps would be valuable as an adjunctive therapy to increase the efficacy of an appropriate antibiotic, decrease antibiotic resistance, and attenuate virulence in Gram-negative pathogens.
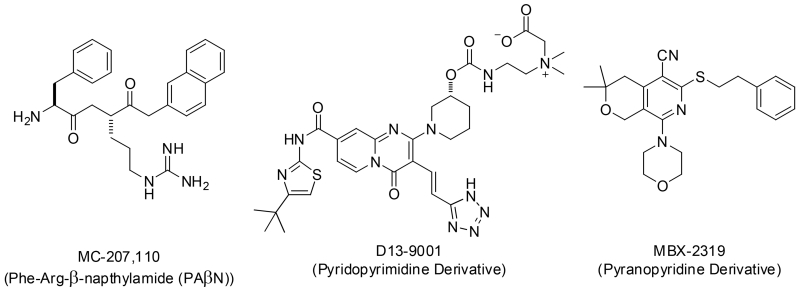
Over the last 16 years a number of attempts to develop RND EPIs for clinical use have been reported, however, to date none have been successful. The first RND EPIs to be reported were a family of peptidomimetic EPIs, including the widely used research compound PAβN (MC-207 110). Compounds in this series are competitive inhibitors of the RND efflux pumps in P. aeruginosa and other Gram-negative organisms, and they were developed for use in combination with levofloxacin as an adjunctive therapy to treat P. aeruginosa infections [2-7]. Although compounds in this series were validated using in vivo infection models [4,5,7], nephrotoxicity across the series prevented further development [8]. A second example of EPI drug development involves a series of pyridopyrimidine EPIs that culminated in D13-9001, a lead compound which was advanced to preclinical development [9-15]. The pyridopyrimidine EPIs are specific for the MexAB efflux pump of P. aeruginosa, but are not active against the MexXY pump of P. aeruginosa, which was likely a factor in the apparent cessation of development in 2007 [16]. Recently, 3D crystal structures of D13-9001 bound to AcrB and MexB were reported that show D13-9001 bound to a unique site (termed the “hydrophobic trap”) close to the deep substrate binding pocket of the pumps [17]. This discovery represents a significant advance in the field, potentially enabling the design of EPIs with increased potency and drug-like properties, and sparking a renewed interest in this series [18].
In this manuscript, we review recent advances in the discovery and optimization of a novel series of pyranopyridine EPIs. This series is in the early stages of lead optimization and efficacy in animal models of infection has yet to be established. The difficulties encountered during the development of previous classes of EPIs underscore some of the most significant challenges for their clinical development: potency, spectrum of activity, pharmacokinetics, and toxicity. Our recent advances in understanding the mode of inhibition and the binding site of this new series of EPIs could facilitate the rational design of analogs with properties that address the challenges mentioned above.
MBX2319, a pyranopyridine EPI
In 2014, we reported the discovery and initial characterization of the pyranopyridine (Pypy) MBX2319 (Fig. 1) as an inhibitor of the AcrAB efflux pump of E. coli [19] that is structurally distinct from the previously reported EPIs. MBX2319 was found through a high-throughput screen for small molecules that potentiate the antibacterial activity of ciprofloxacin (CIP) against E. coli (manuscript in preparation). MBX2319 does not exhibit membrane-disruptive or intrinsic antibacterial activity (MIC ≤ 100 μg/mL), but it potentiates antibiotics that are substrates of AcrB, including fluoroquinolones, β-lactams, chloramphenicol, minocycline, erythromycin, and linezolid. Concentrations of MBX2319 ranging between 3.1 – 12.5 μM induce a 4-fold shift in the MICs of these antibiotics in a standard checkerboard assay, activity consistent with MICs observed with the isogenic ΔacrB strain. The spectrum of EPI activity of MBX2319 covers Shigella flexneri, Klebsiella pneumoniae, Salmonella enterica serovar Typhimuriam, and Enterobacter cloacae, but does not include Pseudomonas aeruginosa. Notably, MBX2319 is a very potent EPI against P. aeruginosa in the presence of Polymyxin B nonapeptide (PMBN), which selectively permeabilizes that outer membrane [20], indicating that MBX2319 is active against the RND-type pumps of P. aeruginosa, but cannot penetrate the highly selective outer membrane of this organism (manuscript in preparation). Based on these observations, the major obstacle to broad spectrum Gram-negative activity of the Pypy class of EPIs is penetration of the outer membrane of P. aeruginosa.
Figure 1.
Select RND efflux pump inhibitors (EPIs).
Initial SAR
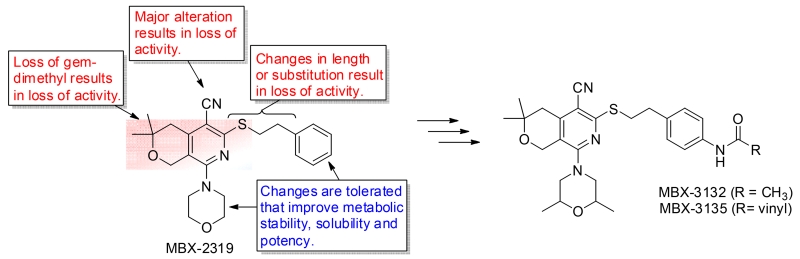
To assess the structure activity relatationships (SAR) of MBX2319, we systematically varied the substituents around the pyranopyridine core and measured their effect on EPI activity, cytotoxicity, solubility, and stability in the presence of liver microsomes [21]. The results of these studies produced a molecular activity map of the scaffold (Fig. 2). This study identified regions on the scaffold that are critical for EPI activity and those that can be modified to improve potency, metabolic stability and solubility. In terms of activity, features necessary for the maintenance of activity among analogs surveyed include the gem dimethyl moiety of the tetrahydropyran, the length of the dimethylenesulfide linker, and the nitrile group. In contrast, the pendant morpholine moiety and aryl group were both broadly amenable to substitution, permitting alterations that improve both overall activity and metabolic stability (as measured with liver microsomes). Introduction of 2,6-dimethyl functionality to the morpholinylyl group improved microsomal stability and EPI activity, and replacement of the morpholinyl group with 2-methoxyethylpiperazinyl improved aqueous solubility with a slight reduction of EPI activity [21]. Combination of 2,6-dimethylmorpholinyl and an acetamide (MBX3132) or acrylamide (MBX-3135) on the phenyl group resulted in a 10- to 20-fold improvement in EPI activity against E. coli as compared to MBX2319 in checkerboard MIC and time-kill assays. MBX3132 and MBX3135 significantly alter the Michealis-Menton kinetics of the AcrAB-TolC pump in E. coli for nitrocefin efflux (increase Km and Vmax) at concentrations as low as 10 nM, suggesting these compounds interfere specifically with the activity (binding or extrusion) of the pump [22].
Figure 2.
Structure activity relationships (SAR) of the pyranopyridines.
Mechanism of Action
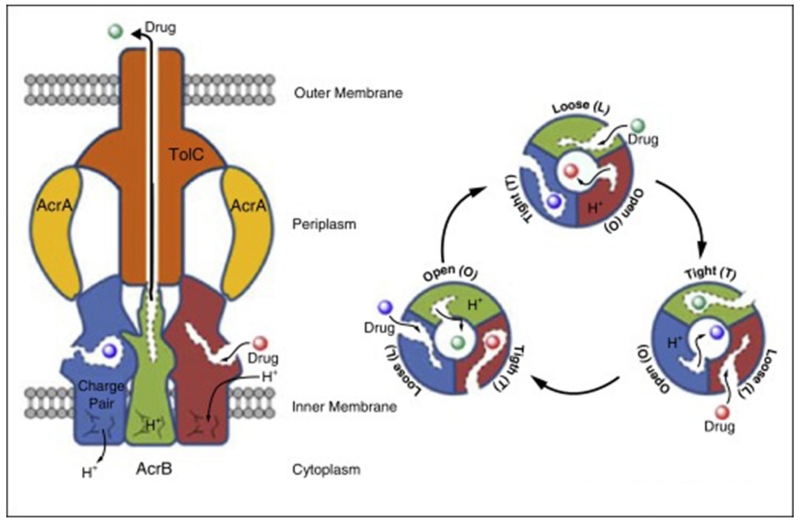
Our initial mechanism of action studies indicated that the primary target of MBX2319 in E. coli is the integral membrane transporter AcrB [19]. AcrB is part of a tripartite pump that includes the outer membrane channel TolC and the periplasmic protein adaptor AcrA that stabilizes the interaction between AcrB and TolC (4) (Fig. 3). Crystal structures of AcrB show that the pump is an asymmetrical homotrimer [23-25], in which each protomer adopts a different conformation representing a distinct step in the translocation pathway [26-28]. The conformations of the individual protomers are described as loose (L), tight (T), and open (O), corresponding to the initial substrate interaction, poly-specific binding, and extrusion of substrates to the TolC channel, respectively [29]. The AcrB transporter extrudes substrates from the periplasmic space into the TolC channel similarly to that of a peristaltic pump that is driven by proton motive force [26,30]. Substrates first interact with a binding cleft near the inner membrane in the L protomer. A conformational change to that of the T protomer forces the substrate into the deep binding pocket, where it interacts with the polyspecific binding site. During conversion to O protomer, the deep binding pocket collapses, forcing the substrate into central water filled channel that communicates with TolC.
Figure 3.
The structure of the tripartite RND family efflux pump AcrAB-TolC (left) and a cartoon illustrating the peristaltic mechanism of the AcrB pump (right).
A mechanistic understanding of EPIs has only recently become detailed, although they have long been thought to inhibit substrate binding to the deep pocket.
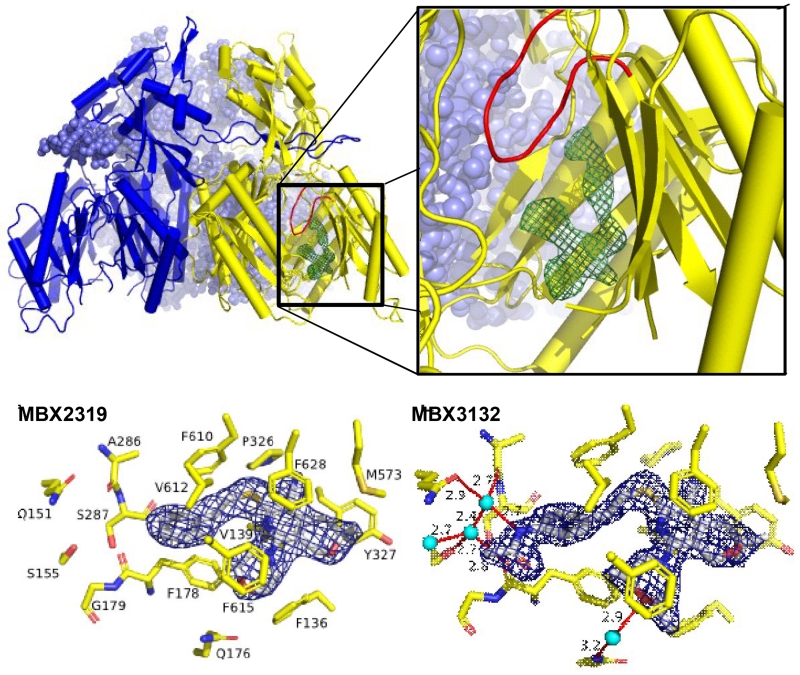
To gain insights into the molecular mechanism of inhibition, Attilio Vargiu and Hiroshi Nikaido utilized Molecular Dynamic (MD) simulations to determine the binding site of MBX2319 in the substrate binding pocket of AcrB [31]. These studies predicted that MBX2319 binds to the same “hydrophobic trap” that was identified in the x-ray crystal structure of D13-9001 bound to AcrB [17]. Shortly after this work was published, Hanno Sjuts and Martin Pos (Goethe University Frankfurt am Main) solved the co-crystal structure of MBX2319 bound to AcrBper, an engineered protein consisting of a soluble form of the periplasmic “porter” domain of AcrB (AcBper) [22]. Based on this structure, we found that MBX2319 was bound to the hydrophobic trap of the T protomer, where it interacts the hydrophobic residues lining the deep binding pocket and hydrophobic trap (Fig. 4). The pyridine ring of MBX2319 is engaged an extensive π-π stacking interaction with the aromatic side chain of F628, and the phenyl and morpholinyl groups interact with F178 and F615. Van der Waals interactions are apparent between the side chain of F610 and the dimethylenesulfide linker, and between the gem dimethyl group and the side chains of Y327 and M573. The results of MD simulations indicate that the major contributors to the tight binding between MBX2319 and AcrBper are the interactions with the side chains of F178 and F628. The binding site of MBX2319 on the T protomer of AcrB appears to be similar to that of the pyridopyrimidine inhibitor D13-9001 [17], suggesting they have similar mechanisms of inhibition. Two hypotheses for the mechanism of inhibition of the pyranopyridines, which are not mutually exclusive, are as follows: 1) binding to the hydrophobic trap inhibits conformational changes required for pump function, and 2) the portion of the inhibitor that protrudes into the substrate binding pocket sterically hinders substrate binding.
Figure 4.
Three dimensional structures of MBX2319 and MBX3132 bound to AcrBper. Top. The three dimensional co-crystal structure MBX2319 bound to AcrBper. Bottom. Detailed views of MBX2319 and MBX3132 bound to the hydrophobic trap. Adapted from Sjuts et al. [22].
Co-crystal structures of the analogs MBX3132 and MBX3135 provided a molecular basis for the increased EPI activity of these compounds. The acetamide and acrylamide groups of MBX3132 and MBX3132, respectively, engage in a highly ordered and complex network of hydrogen bonds centered on a highly coordinated solvent water molecule (Fig. 4), which serves as a hydrogen bond donor to the carbonyl backbone oxygen of A286 and the Q151 side chain of AcrB. In addition, the morpholinyl group of these compounds forms a bridging hydrogen bond through a water molecule to the side chain of Q176. Thus, both structural and MD simulation data provide a consistent molecular explanation for the observed increased potency of MBX3132 and MBX3135. Significantly, the combination of the structural and MD simulation data provides a powerful platform for the design and evaluation of potent EPIs with improved drug-like properties.
Newer Compounds
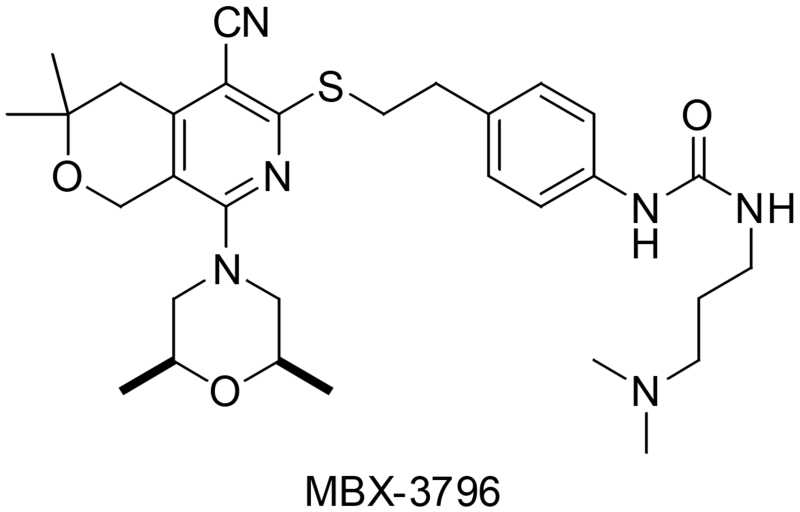
Current optimization efforts are leveraging existing SAR and structural analyses of pyranopyridines bound to AcrB to improve ADME properties and maintain potency, improving the solubility, stability and minimizing the plasma protein binding of lead compounds. This work has identified “hot spots” of metabolic activity such as the C4’ amide of MBX3132 that can be modified to improve stability. Additionally, the placement of polar functionality in water-filled pockets (identified in the crystal structure) allowed for improved solubility profiles and lessened plasma protein binding. Further co-crystal structures of new analogs bound to ACBper have allowed us to use an iterative strategy. Based on this work, compounds such as MBX3796 (Fig. 5) have been identified as exemplar of the next-generation EPIs, well tolerated at 10 mg/kg IV and exhibiting a promising PK profile with an AUC ~10,000 and a CL < 1000 mL/hr/kg (manuscript in preparation). Further efforts are identifying compounds with improved solubility and slower clearance mechanisms to enable in vivo proof-of-concept experiments.
Figure 5.
Structure of a newer analog of MBX3132,
Challenges
An insightful description of the unique challenges generally facing the clinical development of EPIs has been published by Lomovskaya and Bostian [8], and these apply fully to the pyranopyridines. One key issue is that developing an effective antibacterial therapy requires the EPI to be combined with an antibiotic. A combination therapy approach requires that the pharmacokinetics and mode of action of both compounds are compatible to maximize synergistic activity and minimize toxicity. This will likely involve choosing an antibiotic with a mode of action that is similar to the EPI (e.g. AUC/MIC or time over MIC dependent) and adjusting the PK of parameters of the EPI to match the antibiotic. Adjusting the dosing regimen to achieve maximal efficacy while minimizing toxicity will also be a major challenge. The choice of the antibiotic partner is also complicated by the rise of MDR organisms that carry acquired resistance mechanism for several antibiotics. EPIs can increase susceptibility of organisms in which efflux overexpression is the major mode of resistance to a given antibiotic, or one that carries low level resistance mutations in the gene(s) that encode the antibiotic target (e.g. gyrA mutations that confer resistance to FQs). EPIs will not increase susceptibility of organisms carrying extended spectrum beta-lactamases to beta lactams.
Highlights.
Efflux pumps play a major role in multidrug resistance (MDR) in bacteria.
RND family efflux pumps are the major MDR pumps in the Gram-negative pathogens.
Efflux pump inhibitors (EPIs) increase antibiotic efficacy and decrease resistance.
Recent advances in optimizing novel RND-targeting pyranopyridine EPIs are reviewed.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases (Grant R44 AI100332).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. *This is a comprehensive review of recent research on the role that efflux pumps play in MDR in several important human pathogens.
- 2.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renau TE, Leger R, Filonova L, Flamme EM, Wang M, Yen R, Madsen D, Griffith D, Chamberland S, Dudley MN, et al. Conformationally-restricted analogues of efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2003;13:2755–2758. doi: 10.1016/s0960-894x(03)00556-0. [DOI] [PubMed] [Google Scholar]
- 4.Renau TE, Leger R, Flamme EM, Sangalang J, She MW, Yen R, Gannon CL, Griffith D, Chamberland S, Lomovskaya O, et al. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem. 1999;42:4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 5.Renau TE, Leger R, Flamme EM, She MW, Gannon CL, Mathias KM, Lomovskaya O, Chamberland S, Lee VJ, Ohta T, et al. Addressing the stability of C-capped dipeptide efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2001;11:663–667. doi: 10.1016/s0960-894x(01)00033-6. [DOI] [PubMed] [Google Scholar]
- 6.Renau TE, Leger R, Yen R, She MW, Flamme EM, Sangalang J, Gannon CL, Chamberland S, Lomovskaya O, Lee VJ. Peptidomimetics of efflux pump inhibitors potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2002;12:763–766. doi: 10.1016/s0960-894x(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 7.Watkins WJ, Landaverry Y, Leger R, Litman R, Renau TE, Williams N, Yen R, Zhang JZ, Chamberland S, Madsen D, et al. The relationship between physicochemical properties, in vitro activity and pharmacokinetic profiles of analogues of diamine-containing efflux pump inhibitors. Bioorg Med Chem Lett. 2003;13:4241–4244. doi: 10.1016/j.bmcl.2003.07.030. [DOI] [PubMed] [Google Scholar]
- 8.Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic--a vision for applied use. Biochem Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama K, Ishida Y, Ohtsuka M, Kawato H, Yoshida K, Yokomizo Y, Hosono S, Ohta T, Hoshino K, Ishida H, et al. MexAB-OprM-specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 1: discovery and early strategies for lead optimization. Bioorg Med Chem Lett. 2003;13:4201–4204. doi: 10.1016/j.bmcl.2003.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama K, Ishida Y, Ohtsuka M, Kawato H, Yoshida K, Yokomizo Y, Ohta T, Hoshino K, Otani T, Kurosaka Y, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 2: achieving activity in vivo through the use of alternative scaffolds. Bioorg Med Chem Lett. 2003;13:4205–4208. doi: 10.1016/j.bmcl.2003.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama K, Kawato H, Watanabe J, Ohtsuka M, Yoshida K, Yokomizo Y, Sakamoto A, Kuru N, Ohta T, Hoshino K, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 3: Optimization of potency in the pyridopyrimidine series through the application of a pharmacophore model. Bioorg Med Chem Lett. 2004;14:475–479. doi: 10.1016/j.bmcl.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K, Kuru N, Ohtsuka M, Yokomizo Y, Sakamoto A, Kawato H, Yoshida K, Ohta T, Hoshino K, Akimoto K, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 4: Addressing the problem of poor stability due to photoisomerization of an acrylic acid moiety. Bioorg Med Chem Lett. 2004;14:2493–2497. doi: 10.1016/j.bmcl.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K, Nakayama K, Kuru N, Kobayashi S, Ohtsuka M, Takemura M, Hoshino K, Kanda H, Zhang JZ, Lee VJ, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 5: Carbon-substituted analogues at the C-2 position. Bioorg Med Chem. 2006;14:1993–2004. doi: 10.1016/j.bmc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida K, Nakayama K, Ohtsuka M, Kuru N, Yokomizo Y, Sakamoto A, Takemura M, Hoshino K, Kanda H, Nitanai H, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem. 2007;15:7087–7097. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida K, Nakayama K, Yokomizo Y, Ohtsuka M, Takemura M, Hoshino K, Kanda H, Namba K, Nitanai H, Zhang JZ, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 6: exploration of aromatic substituents. Bioorg Med Chem. 2006;14:8506–8518. doi: 10.1016/j.bmc.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. *An excellent review of the relationship between the structure, function, and inhibition of RND-type multi-drug efflux pumps in Gram-negative pathogens.
- 17.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. **This is an important paper that describes the first 3D structure of an efflux pump inhibitor bound to the RND-type pumps AcrB and MexB. The structures revealed the pyridopyrimidine inhibitor D13-9001 binds to a unique site, called the “hydrophobic trap”, near the deep substrate binding pocket. The structure suggested a mechanism of efflux pump inhibition for D13-9001, which was explored in several supporting experiments that are reported in this paper.
- 18.Inoue Y. Pacifichem 2015. Honolulu, HI: 2015. SBDD approach of the novel inhibitor of bacterial multidrug efflux transporter. [Google Scholar]
- 19.Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D’Souza S, Walker GC, Peet NP, Nikaido H, Bowlin TL. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014;58:722–733. doi: 10.1128/AAC.01866-13. *This paper describes the initial characterization of MBX2319, a novel pyranopyridine efflux pump inhibitor that exhibits potent activity against the major RND-type pumps of the Enterobacteriaceae.
- 20.Vaara M, Vaara T. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature. 1983;303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen ST, Kwasny SM, Ding X, Cardinale SC, McCarthy CT, Kim HS, Nikaido H, Peet NP, Williams JD, Bowlin TL, et al. Structure-activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg Med Chem. 2015;23:2024–2034. doi: 10.1016/j.bmc.2015.03.016. *This work describes the first exploration of the structure activity relationships of the pyranopyridine efflux pump inhibitors. By systematically varying the various substituents that are attached to the pyrano[yridine core, a detailed “activity map” for efflux pump inhibition, solubility, and first pass metabolism was generated.
- 22.Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim HS, Ding X, Ornik AR, Ruggerone P, Bowlin TL, Nikaido H, et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci U S A. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. **This work decribes two major advances. The first advance is the design and construction of a soluble version of the periplasmic, substrate-binding domain of AcrB (AcrBper), which is structurally identical to the periplasmic domain of full-length AcrB, an integral membrane protein. The second advance is the application of the AcrBper construct to rapidly generate 3D structures of MBX2319 and three analogs bound to the “hydrophobic trap”, which will enable structure guided drug design.
- 23.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 24.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 25.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eicher T, Seeger MA, Anselmi C, Zhou W, Brandstatter L, Verrey F, Diederichs K, Faraldo-Gomez JD, Pos KM. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. Elife. 2014;3 doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 28.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 29.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Su CC, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother. 2014;58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]