Abstract
How chaperones, insertases and translocases facilitate insertion and folding of complex cytoplasmic proteins into cellular membranes is not fully understood. Here, we utilize single-molecule force spectroscopy to observe YidC, a transmembrane chaperone/insertase, sculpting the folding trajectory of the polytopic α-helical membrane protein lactose permease (LacY). In the absence of YidC, unfolded LacY inserts individual structural segments into the membrane; however, misfolding dominates the process so that folding cannot be completed. YidC prevents LacY from misfolding by stabilizing the unfolded state from which LacY inserts structural segments stepwise into the membrane until folding is completed. During stepwise insertion, YidC and membrane together stabilize the transient folds. Remarkably, the order of insertion of structural segments is stochastic, thereby indicating that LacY can fold along variable pathways towards the native structure. Since YidC is essential in membrane protein biogenesis and LacY a paradigm for the major facilitator superfamily, our observations have general relevance.
INTRODUCTION
The biogenesis of membrane proteins in bacteria requires insertion into the cytoplasmic membrane by two evolutionarily highly conserved SecYEG and YidC pathways1–3. The translocase SecYEG is critical for the insertion of proteins into the plasma membrane in bacteria and archaea, the endoplasmic reticulum in eukaryotes, and the chloroplast thylakoid membrane in plants4. The membrane protein insertase YidC can either couple with the SecYEG pathway to facilitate folding and assembly of membrane proteins inserted by the Sec translocase or function as an independent pathway5. In the YidC pathway, insertase activity catalyzes insertion, folding and assembly of proteins into the bacterial cytoplasmic membrane2. Thus, YidC can function both as an insertase and as a chaperone. This YidC pathway and those of its family members--Oxa1 in mitochondria and Alb3 in chloroplasts--are highly conserved6,7. While chaperones suppress misfolding and aggregation of soluble proteins to increase their folding8, it is not understood how insertion and folding of integral membrane proteins in general and cytoplasmic membrane proteins in particular are assisted by chaperones and insertases. Furthermore, since basic elements of cytoplasmic membrane protein biogenesis are conserved in pro- and eukaryotes, insight into how YidC facilitates insertion and folding of membrane proteins is highly relevant.
Single-molecule force spectroscopy (SMFS) is the method of choice for the in vitro characterization of mechanical unfolding and refolding of single proteins9. In particular, atomic force microscopy (AFM)-based SMFS can be applied to observe the unfolding and folding pathways of individual proteins in native or synthetic membranes at the level of secondary structural elements10–13. In the cellular context, unfolded proteins are exposed to various specific and non-specific interactions with macromolecules. Among these, molecular chaperones are crucial for preventing unfolded polypeptides from misfolding and aggregating8. A few SMFS studies describe the effect of chaperones on the folding trajectories of soluble14–16 and β-barrel outer membrane proteins17. However, it has not been possible so far to monitor how chaperone and translocon together guide folding of cytoplasmic integral membrane proteins.
Here we investigated the effect of YidC on the insertion and folding of single lactose permeases from Escherichia coli (LacY) under physiologically relevant conditions. LacY, a paradigm for the major facilitator superfamily (MFS)18, has 12 mostly irregular transmembrane α-helices organized into two pseudo-symmetrical six-helix bundles connected by a relatively long cytoplasmic loop19,20. It has been shown that LacY requires YidC to fold correctly in the membrane after insertion via the SecYEG complex21,22. In our previous work we applied SMFS to fully unfold native, functional LacY from phospholipid membranes23. Exposed to mechanical stress LacY stepwise unfolds structural segments giving rise to a highly reproducible unfolding pattern which is sensitive to the functional state23, as well as misfolding24 of the transporter. Here, we used the SMFS-based assay to unfold LacY and then to follow the refolding of the unfolded substrate into the phospholipid membrane in the absence or presence of YidC. We found that LacY alone can insert structural segments into the membrane, but that misfolding dominated this process and prevented the substrate from completing folding. YidC stabilized the unfolded LacY substrate from which structural segments could insert one after the other until folding was completed. Surprisingly, the sequence at which this insertion/folding process occurred was stochastic.
RESULTS
LacY unfolds stepwise on exposure to mechanical stress
To set up the insertion and folding experiments, we reconstituted LacY into phospholipid membranes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (PG) at a ratio of 3:1 (PE:PG)25 (Supplementary Results, Supplementary Fig. 1). At this phospholipid composition, which is close to the composition of the E. coli membrane (70–80% zwitterionic PE and 20–25% anionic PG plus cardiolipin), LacY is fully functional and adopts the native topology with the N- and C-termini on the cytoplasmic surface of the membrane19,26. For AFM imaging the LacY proteoliposomes were adsorbed to mica in buffer solution were they appear as single-layered membranes (Supplementary Fig. 1b). Within the membranes, high-resolution AFM topographs showed evenly distributed LacY particles.
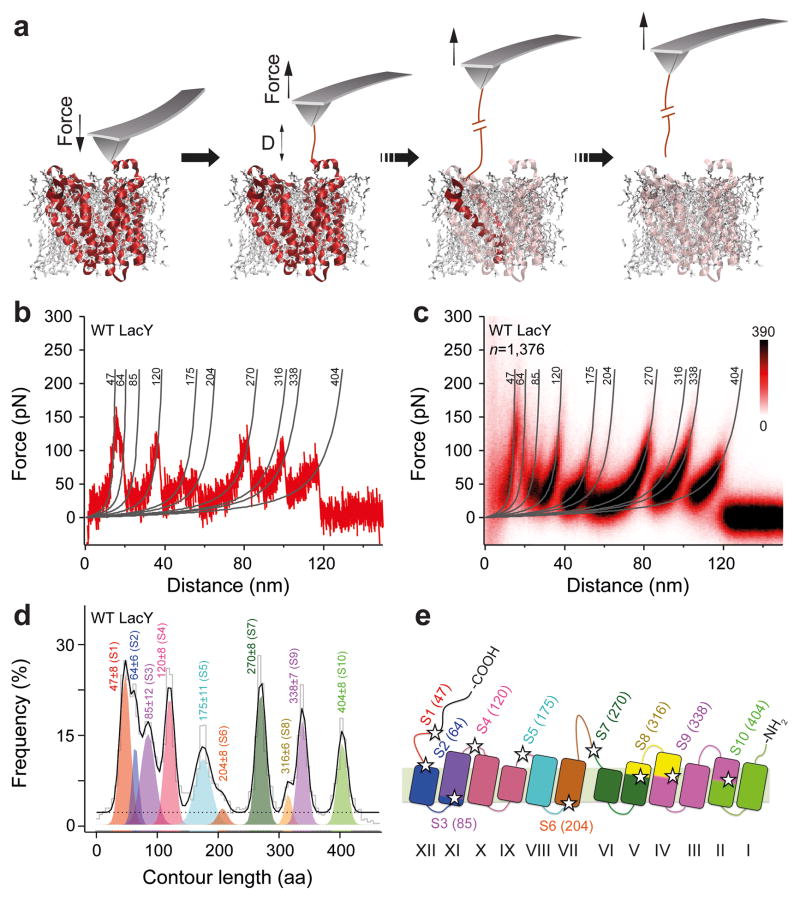
For SMFS, we proceeded as recently introduced for LacY23. Briefly, the AFM stylus was pushed gently (700 pN for 500 ms) onto LacY to non-specifically attach individual LacY polypeptides to the stylus (Fig. 1a)27. To enhance the probability of attaching the C-terminus, we extended the C-terminus of LacY with a 36 amino acid (aa) long unstructured ‘polyGly’ polypeptide [GSM(G11)EAVEEAVEEA(G11)S] followed by an 8-aa-long His-tag (His8-tag) (Supplementary Fig. 2a). The extension has little or no effect on LacY transport activity nor did it change the folding of LacY (Supplementary Fig. 2b,c)23. The stylus was then withdrawn from the membrane while recording the deflection of the AFM cantilever as a force-distance curve (Fig. 1a,b). These force-distance curves typically exhibited a saw-tooth-like pattern with 10 force peaks each indicating an unfolding step of the transporter. Repeating the experiment multiple times revealed a highly reproducible saw-tooth-like pattern (Fig. 1c). By fitting each of the 10 force peaks with the worm-like-chain (WLC) model27 we determined the contour lengths of the peptide segments unfolded in each step (Fig. 1b,c). Since we applied mechanical pulling force to the elongated C-terminus of LacY, the C-terminus was stretched until the first C-terminal bundle unfolded (Fig. 1a). This unfolding step elongated the polypeptide tethering the AFM stylus and the partially unfolded LacY molecule. Subsequent withdrawal of the stylus unfolded the transmembrane bundles stepwise in nine distinct structural segments formed by single or grouped α–helices and their connecting polypeptide loops (Fig. 1d). Thus, the force-distance curves recorded a characteristic ‘fingerprint’ spectrum for native LacY, with single force peaks corresponding to unfolding of secondary structure segments (Fig. 1d,e). The summed length of the force-distance curves corresponds to the contour length of the fully unfolded and stretched LacY polypeptide23. It has been previously shown for LacY, and for other membrane transporters and receptors that the force peaks in the native fingerprint spectrum are sensitive to the fold, functional state, ligand-binding, lipid composition, mutations and/or supramolecular assembly of the membrane protein9,23,24,28–31. It is of particular interest for this work that the characteristic fingerprint spectrum observed for native LacY is sensitive to misfolding of the transporter24. As LacY inverts topology upon depletion of PE and becomes inactive32, it was observed that the force peaks detected for the structural segments of native LacY change positions if LacY inverts topology and can be used to assign the segments changing their fold24. Thus, the comparison of the native fingerprint spectrum of LacY with that recorded of single LacY molecules can identify misfolded transporters and structural segments.
Figure 1. Unfolding steps and intermediates of native WT LacY.
(a) Principle of SMFS to mechanically unfold single LacY. After AFM-imaging the proteoliposome, the stylus of the cantilever is gently pressed onto LacY (red, PDB 1PV7) to non-specifically attach the elongated C-terminus. Then the cantilever is withdrawn to stepwise unfold LacY until it has been completely unfolded and extracted from the phospholipid membrane23. (b) Force-distance curve recorded upon mechanically unfolding a single LacY from the membrane. (c) Superimposition of 1,376 force-distance curves each recorded upon unfolding of a single LacY. The resulting fingerprint spectrum shows 10 force peaks each detecting the unfolding of a stable structural segment of LacY. Fitting of each force peak with the worm-like-chain (WLC) model27 provides the contour length of the unfolded and stretched polypeptide (given in amino acids (aa) at the top of each WLC curve). (d) Contour length histogram of force peaks detected upon unfolding of LacY. Fitting the histogram with a Gaussian mixture model (black line) reveals the mean contour length of every force peak class (aa ± s.d.). The dashed line shows the uniform baseline noise. Each mean contour length of a force peak class assigns the ending of the previous structural segment and the beginning of the forthcoming structural segment. (e) The 10 stable structural segments S1–S10 color coded and mapped to the secondary structure of LacY (PDB 1PV7). Numbers give the mean contour lengths of a force peak class (in aa), which are located by stars.
Partially unfolded LacY remains stably in the membrane
The highly reproducible fingerprint spectrum characterizing the unfolding steps of LacY (Fig. 1c) indicated that the unfolding intermediates of LacY remained stably embedded in the membrane. Similarly, stable unfolding intermediates formed by individual or grouped α–helices and polypeptide loops are observed for other transmembrane proteins including members of the MFS family9,10,30,31,33. Molecular dynamics simulations also indicate that transmembrane α–helical proteins exposed to mechanical force unfold stepwise and that the partially unfolded protein remains stably folded in the membrane34,35. To test more rigorously whether partially unfolded LacY behaves similarly, we conducted control experiments in which we unfolded LacY to different extents and left the partially unfolded protein in the membrane for 2 or 5 s (Supplementary Fig. 3). We then re-applied the pulling force to the partially unfolded α-helical bundle, which exhibited the same unfolding pattern as observed for native LacY (Fig. 1c). Thus, partially unfolded LacY remained stable in the membrane and did not rearrange over a time course of seconds. The findings were consistent with in vivo experiments demonstrating that contiguous, non-overlapping LacY fragments expressed together can self-assemble to functional LacY36,37. Having shown that the transmembrane α-helical bundles of LacY unfold stepwise and leave stable unfolding intermediates in the membrane, we tested whether LacY pulled partially out of the membrane can reinsert and refold back into the membrane.
Outside the membrane LacY misfolds at high probability
Insertion and folding of proteins into the cytoplasmic membrane starts from the N-terminus 1,3,38. Thus, in our in vitro refolding experiments we wanted to characterize the insertion and folding of LacY into the membrane proceeding from the N-terminal end. As described above, we used our SMFS assay to pick up the elongated C-terminal. However, now we only unfolded and extracted a large segment of LacY consisting of the C-terminus, 10 transmembrane α–helices and the intervening loops from the membrane, thus leaving the first two N-terminal transmembrane α–helices in the membrane (Fig. 2a). We then lowered the AFM stylus into close proximity with the membrane (≈ 10–15 nm) where the unfolded portion of LacY was allowed to insert and fold for 2 s. Finally, the refolded polypeptide was probed by pulling it from the membrane again (Fig. 2b). To interpret the force-distance pattern, the native fingerprint spectrum of LacY was used as a template (Fig. 1c) to quantify whether or not the polypeptide refolded native structural segments (Supplementary Fig. 4). Each measurement was classified into one of three categories: (i) If we detected no unfolding force peaks, the LacY polypeptide did not adopt a stable fold and was classified as ‘unfolded’ (Fig. 2b). (ii) If we observed one or more unfolding force peaks at contour lengths differing from the native fingerprint spectrum, we interpreted the polypeptide having adopted a non-native fold. It was thus classified as ‘misfolding’ (Fig. 2c). (iii) If we recorded unfolding force peaks only at positions covered by the native fingerprint spectrum (Fig. 1c), the substrate was classified as refolding individual structural segments into the membrane (Fig. 2d). We then carried out 114 independent refolding experiments (Fig. 3a and Supplementary Fig. 5). Within a refolding time of 2 s, 6% of the unfolded LacY substrates remained unfolded while 41% exhibited force peaks not corresponding to the native fingerprint spectrum of LacY and thus representing misfolding. 53% of the refolding experiments exhibited force peaks corresponding to the native fingerprint spectrum and were classified as having folded native structural segments. Close inspection of the force peaks in the single refolding experiments indicated that the folding of single native structural segments occurred as a stochastic process (Supplementary Fig. 5). However, in none of the experiments was a refolding spectrum with all the force peaks of native LacY observed. Thus, although LacY refolded individual structural segments into the membrane, the polypeptide did not complete folding.
Figure 2. Refolding single lactose permeases.
(a) Schematics of the refolding experiment. After attaching the AFM stylus to the C-terminus, the cantilever is withdrawn to sequentially unfold the LacY (red) until leaving the last structural segment S10 in the membrane (blue). The force-distance curve recording this partial unfolding process shows force peaks of the fingerprint spectrum of native LacY (Fig. 1c). Approaching the AFM stylus close to the membrane surface (≈10–15 nm) relaxes the unfolded polypeptide. After a refolding time of 2 s the stylus is completely withdrawn to characterize whether the LacY substrate remained unfolded, misfolded or folded structural segments. (b–d) Typical force-distance curves showing the before unfolded LacY polypeptide unfolded (b), misfolded (c) and having folded some structural segments (d). Unfolding (U) is indicated when the force-distance curve of the before ahead unfolded portion of LacY showed no force peak and thus no structures refolded; misfolding (M) is indicated when the force-distance curve of the before ahead unfolded portion of LacY showed at least one force peak at a position not corresponding to the native unfolding force peak pattern of WT LacY; folding (F) is indicated when the force-distance curve of the before ahead unfolded portion of LacY showed only force peaks corresponding to the native unfolding force peak pattern of LacY in terms of mean ± s.d. (Fig. 1c and Supplementary Fig. 4). Experiments were conducted in 50 mM KPi at pH 7.2 and 25°C.
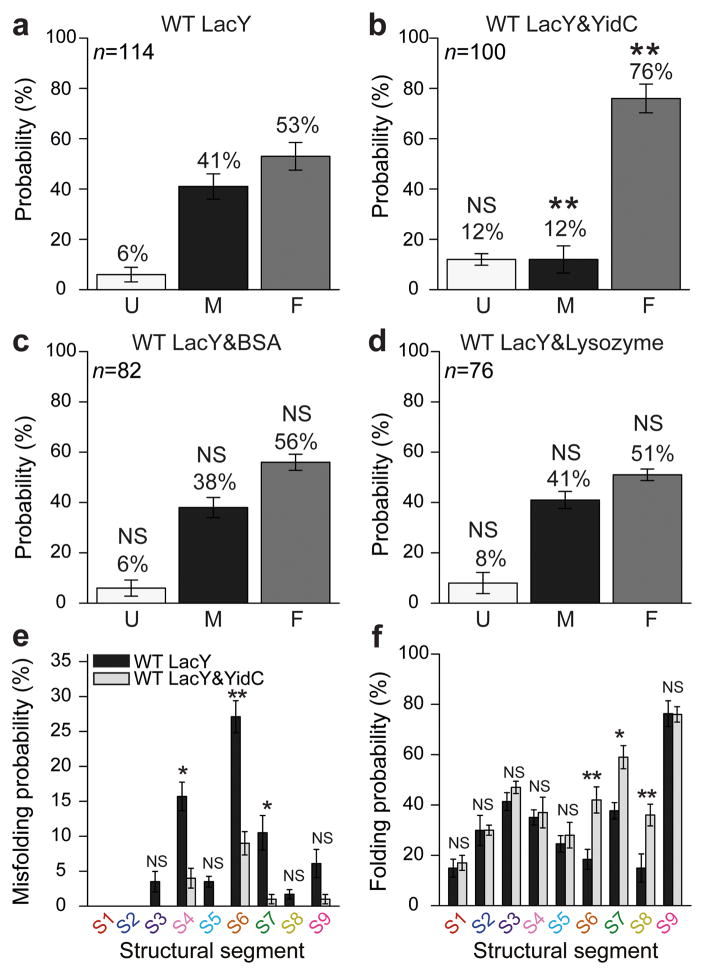
Figure 3. YidC suppresses misfolding and assists folding of LacY.
Probability of the unfolded LacY polypeptide to remain unfolded (U) or to show misfolding (M) and refolding (F) states (Fig. 2) in the absence (a) and presence (b) of YidC. (c,d) Control experiments refolding unfolded LacY in the presence of either 1 μM BSA or 1 μM lysozyme. (e) Probability of structural segments S1–S9 to misfold in the absence and presence of YidC. (f) Probability of structural segments to fold in the absence and presence of YidC. Refolding experiments of WT LacY alone (a) or of WT LacY co-reconstituted with YidC (b) were conducted as described in Fig. 2 after a refolding time of 2 s. Refolding experiments analyzed in e and f are those from a and b. n gives the number of single LacY refolding experiments analyzed. Error bars indicate the s.e.m. as estimated by resampling using 4,000 iterations. Differences of refolding experiments conducted in the absence and presence of YidC are considered significant. Significance tests with respect to WT LacY data are indicated: NS, P ≥ 0.01, * P < 0.01 and ** P < 0.001 as determined by two-tailed Z-test. Statistical tests obtained from Chi-square and two-tailed Fischer’s exact tests are additionally provided (Supplementary Tables 1–4). For single refolding experiments see Supplementary Figs. 5 and 9.
YidC supports native folding intermediates
To test whether YidC has an effect on insertion and folding of LacY, we purified and co-reconstituted both proteins into PE:PG (3:1) membranes. Co-reconstitution was verified by SDS/PAGE and AFM imaging (Supplementary Fig. 1) and by SMFS (Supplementary Fig. 6). Mechanical unfolding of single membrane proteins of the co-reconstituted sample by SMFS revealed force-distance curves with two different patterns in equal frequency. One pattern was identical to that of native LacY with 10 force peaks (Supplementary Fig. 6a)23. The second pattern exhibited only eight force peaks that extended towards much longer contour length than observed for LacY (Supplementary Fig. 6b). The YidC polypeptide with a C-terminal His10-tag is much longer than LacY (561 versus 461 aa), which argued that the longer second pattern corresponded to the unfolding of YidC into a fully stretched conformation. Indeed, reconstitution of YidC alone showed that the second pattern of force-distance curves (Supplementary Fig. 7). We questioned how LacY and YidC in the co-reconstituted sample distributed within the membrane. To answer this, we conducted spatially resolved SMFS of co-reconstituted LacY and YidC (Supplementary Fig. 8). The experiments revealed that LacY and YidC were in close proximity (< 10 nm). Such close proximity can be the result of a kinetically stable association of LacY and YidC. Alternatively, an unstable association may have been facilitated by lateral diffusion, which is reduced but not impaired by adsorption to mica39. Thus, co-reconstitution of LacY and YidC indicated that both proteins were present in the phospholipid membrane and in close proximity.
To test YidC-assisted insertion and folding, we repeated the LacY refolding experiments in the presence of YidC (Supplementary Fig. 9). Typically, we observed LacY that refolded a few but not all structural segments in 2 s time. Following the criteria described above, we classified each refolding experiment as ‘Unfolded’, ‘Misfolding’ or ‘Refolding’ (Fig. 3a,b). The refolding trajectories of LacY detecting the folding of native structural elements increased from 53 to 76% in the presence of YidC, trajectories detecting the unfolded state increased slightly from 6 to 12%, and trajectories detecting the misfolded state decreased from 41 to 12%. Interestingly, the force required to unfold structural segments of native LacY (94.3 ± 27.7 pN; mean ± s.d.; n = 614) was similar to that required to unfold segments refolded in the absence (93.5 ± 30.1 pN; n = 408) or presence (84.3 ± 25.3 pN; n = 493) of YidC. Thus, structural segments of native and refolded LacY had similar mechanical stability. Control experiments conducted in the presence of lysozyme or BSA did not increase the probability of the unfolded LacY polypeptide to insert and fold (Fig. 3c,d). The observations rationalized that YidC protected LacY from misfolding and supported the folding of individual structural segments such as observed for native LacY. However, the refolding experiments also showed that the LacY substrate did not establish all of the force peaks observed with native LacY.
YidC protects α-helices V–VII and IX–X from misfolding
Which structural regions of LacY were prone to misfolding in the absence of YidC and which were preferentially inserted and folded in the presence of YidC? To answer this query, we analyzed the LacY refolding experiments carried out in the absence of YidC to obtain the positions of every misfolded force peak. These positions were compared with those in the native fingerprint spectrum of LacY. To approximate the position of a misfolded structural segment, we assigned the force peak indicating misfolding to the closest force peak detected in the native fingerprint spectrum. In the absence of YidC, structural segments S4, S6 and S7 showed the highest probability for misfolding (Fig. 3e). Segments S6 and S7 contain transmembrane α-helices V, VI, VII and the long cytoplasmic loop19. Within these secondary structures helix VII is weakly hydrophobic and considered to be the ‘weakest link’ with respect to LacY folding, stability and topogenesis32,40. Although Asp237 and Asp240 in helix VII form salt bridges with Lys358 (helix XI) and Lys319 (helix X), respectively41, in the LacY structure they become negatively charged as the salt bridge is broken when the partner helix is pulled from the membrane. Such mutual stabilization of secondary structures from S4, S6 and S7, which is crucial for LacY folding and function, may explain the high misfolding probability observed. The situation changed markedly in the presence of YidC, which suppressed misfolding of these structural segments and enhanced the probability of folding properly into the membrane (Fig. 3e,f).
In the absence of YidC, structural segment S6, which encompasses transmembrane α-helix VII, shows the highest probability to misfold (27%) among all structural segments of LacY (Fig. 3e). As shown previously, α-helix VII tends to misfold upon depletion of PE24. Misfolding of α-helix VII may thus disturb the folding of α-helix X, which interacts with α-helix VII via the 240/319 salt bridge41. Interestingly, structural segment S4, which contains α-helices IX and X, showed the second highest misfolding probability (16%). Furthermore, 61% of the refolding experiments in which misfolding of segment S4 was detected also showed that segment S6 was misfolded, while in only 6% of all cases segment S4 misfolded and segment S6 folded correctly. The data thus suggested that misfolding of the structural segment with α-helices IX and X and of the segment with α-helix VII was coupled. Possibly, misfolding of α-helix VII destabilized α-helices IX and X since the stabilizing salt bridges and hydrogen bond network between residues Tyr236 and Asp240 in α-helix VII, Arg302 in α-helix IX, and Lys319, His322, and Glu325 in α-helix X cannot be formed19. The chaperoning function of YidC thus appeared to preferentially protect misfolding of the structural segments encompassing α-helices VII, IX and X.
YidC supports insertion and folding steps to completion
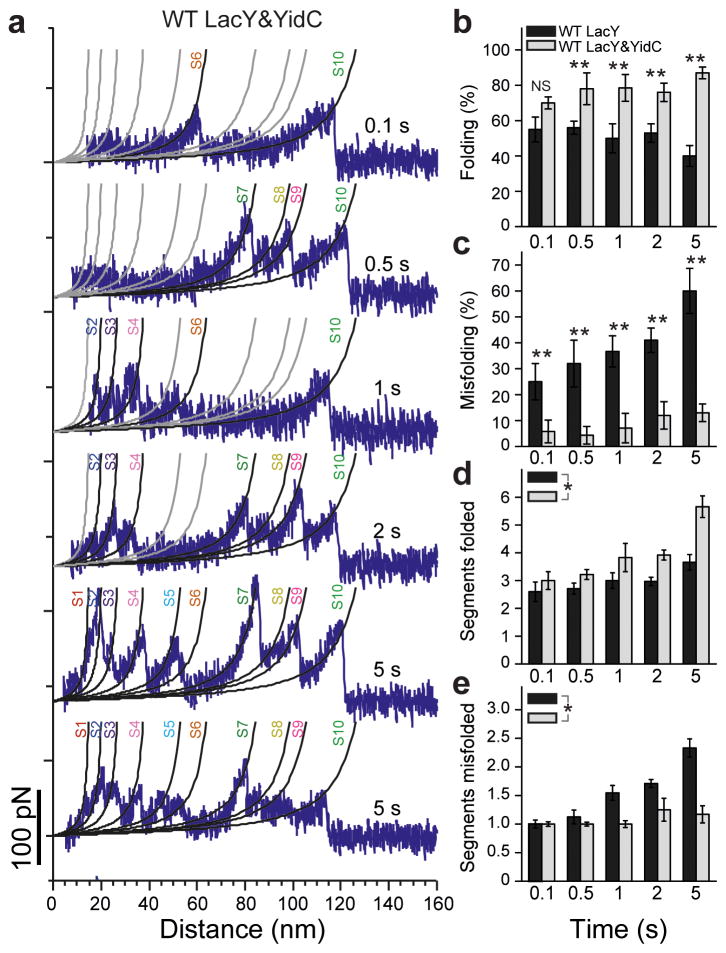
In the absence or presence of YidC, unfolded LacY could insert and fold structural segments in the membrane. Our single-molecule refolding experiments indicated that in both cases, the individual structural segments inserted and folded in a stochastic process (Supplementary Figs. 5 and 9). This result is surprising, as we expected a priori that YidC would determine the sequence of insertion and folding. Within the refolding time of 2 s, our refolding experiments exhibited the correct folding of a few but not of all structural segments. Refolding of all structural segments as observed with native LacY was not observed. Therefore, we questioned whether increasing the refolding time would increase the number of structural segments that refold properly. Unfolded LacY was thus allowed to refold for times ranging from 0.1 to 5 s in the absence or presence of YidC and the folding intermediates were characterized (Fig. 4 and Supplementary Fig. 10). In the absence of YidC, increasing the refolding time clearly increased the number of force peaks and thus the number of refolding steps (Supplementary Fig. 10). However, with increasing refolding time the fraction of correctly folded curves decreased (Fig. 4b), while the probability of detecting misfolding events increased strongly (Fig. 4c). Consequently, at 5 s refolding time, misfolding of the LacY substrate dominated (≈ 60%). No LacY remained unfolded and the number of correctly folded structural segments remained constant for all refolding times (Fig. 4d), while with increasing refolding time the number of misfolded segments increased by ≈ 130% to ≈ 2.3 (Fig. 4e). Thus, with increasing refolding time, force peaks corresponding to misfolded segments did not transform into force peaks of the fingerprint spectrum of native LacY. Moreover, in none of the refolding experiments conducted at extended times a complete set of force peaks like that of native LacY was observed.
Figure 4. LacY shows variable folding intermediates towards full refolding.
(a) Force-distance curves recorded of unfolded LacY refolding in the presence of YidC at refolding time varying from 0.1 to 5 s (experiments conducted as described in Fig. 2). WLC curves with the structural segment indicated are taken from the native fingerprint spectrum of LacY (Fig. 1c). WLC curves matching the refolding force peaks in terms of mean ± s.d. (Supplementary Fig. 4) are colored black, those not matching any peak are colored grey. (b,c) Probability of the unfolded LacY substrate towards folding structural segments and misfolding over the refolding time and in the absence (black) and presence (grey) of YidC. In the absence of YidC 204 refolding experiments were analyzed and 206 in the presence of YidC. For single refolding experiments see (a) and Supplementary Fig. 10. (d,e) Number of folded segments per folding substrate and misfolded segments per misfolding substrate over the refolding time and in the absence (black) and presence (grey) of YidC. Error bars indicate the s.e.m. as estimated by resampling using 4,000 iterations. Significance with respect to WT LacY data indicates: NS, P ≥ 0.01, * P < 0.01 and ** P < 0.001 as determined by two-tailed Z-test (b,c) (Supplementary Tables 1,2). The increase (slope) of the number of segments folded (d) and misfolded (e) over the refolding time was significantly different in the presence vs absence of YidC (* P < 0.01 as determined by Analysis of covariance (ANCOVA)).
The situation changed when we refolded the LacY substrate in the presence of YidC (Fig. 4a,b). Again, the force peaks detected for refolded LacY increased with the refolding time. Again, the force peaks indicating the folding steps after certain refolding times occurred stochastically (Fig. 4a and Supplementary Fig. 10b), which showed that LacY inserted and folded structural segments in variable sequences. However, in the presence of YidC the force peaks of the refolded structural segments were at the same positions as in native LacY. The probability of detecting LacY with correctly folded segments increased with refolding time from ≈ 70% to ≈ 87%, while the probability of misfolding increased from ≈ 6% to ≈ 13% (Fig. 4b,c). No unfolding was detected at 5 s, and with longer refolding times, the number of insertion and folding steps increased until only force peaks typical of native LacY were observed (Fig. 4d). Thus, refolding of structural segments dominated over misfolding. Finally, ≈ 9% of these refolding experiments showed all force peaks observed for the fingerprint spectrum of native LacY. In summary, the refolding experiments recorded in the presence of YidC showed that the stepwise insertion of structural segments proceeds until LacY fully refolds into the native structure.
DISCUSSION
We introduced an SMFS-based assay to study how YidC assists the insertion and folding of LacY into a phospholipid membrane under physiological relevant conditions. The assay, which allows characterization of insertion and folding of individual structural segments with single membrane proteins, contrasts most biophysical experiments carried out in bulk that refold polypeptides from a chaotrope-denatured state into detergent micelles or phospholipid bilayers42,43. Importantly, the method can be readily applied to study the folding of a wide range of membrane proteins.
With our SMFS-based assay, we observe that partially unfolded LacY can self-insert and fold structural segments into a phospholipid membrane (Fig. 5). The process is prone to misfolding, and with increasing refolding time misfolding dominates over the many folding steps required to complete the operation. Consequently, we do not observe the complete refolding of LacY in the absence of YidC. With increased refolding time, the number of LacY substrates showing correctly folded structural segments decreases while misfolding increases. Therefore, it is unlikely that misfolded LacY substrates can transform into natively folded proteins. YidC thus protects LacY from misfolding by stabilizing the unfolded substrate and promoting stepwise insertion and folding of α-helix-containing segments into the membrane. These insertion and folding steps proceed until the fingerprint spectrum of native LacY is obtained. Surprisingly, the folding steps assisted by YidC do not occur in a specific order. The results highlight the unique chaperoning function of YidC, which stabilizes unfolded LacY by suppressing misfolding, as well as an insertase activity that facilitates stepwise insertion and folding of structural segments into the membrane until LacY completed folding.
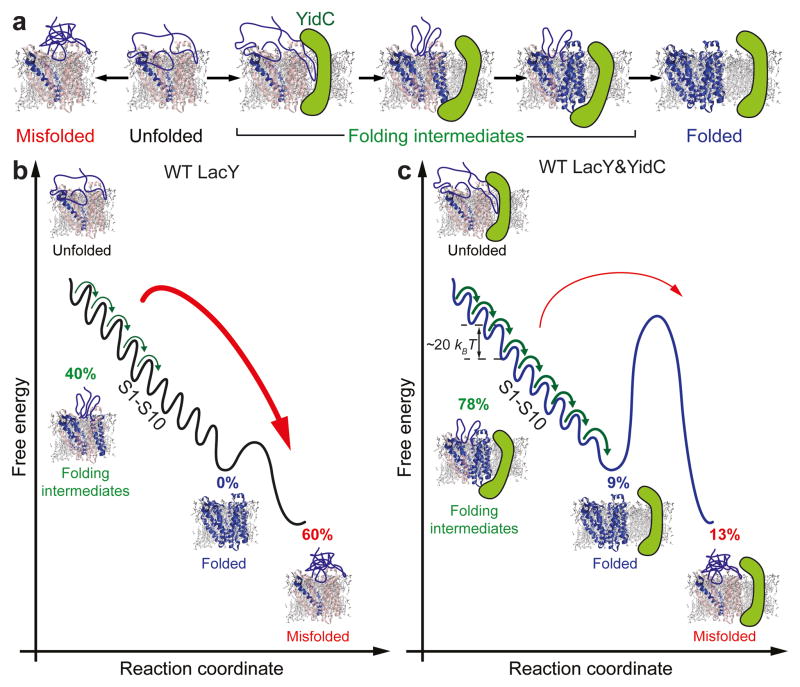
Figure 5. Schematic model of the folding free-energy landscape of LacY sculpted by YidC.
(a) LacY partially unfolded and extracted from the membrane misfolds at high probability. YidC prevents LacY from misfolding and funnels the unfolded substrate to the native folded state via numerous folding intermediates. (b) In the absence of YidC the unfolded LacY substrate inserts single structural segments into the membrane. After 5 s none of the LacY substrates remained unfolded and because the structural segments show a higher misfolding probability the substrate predominantly misfolds (≈ 60%). (c) YidC stabilizes the unfolded LacY substrate and reduces misfolding from ≈ 60% to ≈ 13%. From this stabilized unfolded state ≈ 87% (≈ 78% folding intermediates and ≈ 9% completing folding) of the LacY substrates insert and fold structural segments into the membrane. The unfolded LacY substrate inserts and folds one structural segment after the other until all structural segments S1–S10 folded thereby completing the folding process towards the natively folded state. However, the sequence at which the individual segments insert appears stochastic. After 5 s, ≈ 9% of all substrates completed folding into the native LacY structure. Probabilities were taken from refolding experiments conducted at 5 s refolding time (Fig. 4). As previously revealed from dynamic SMFS the average free-energy stabilizing a folded structural segment of LacY is ≈ 20 kBT[23], which sums up to ≈ 200 kBT for natively folded LacY.
The phospholipid bilayer is equivalent to a free-energy sink that determines how unfolded LacY substrates prevented from misfolding by YidC insert and fold (Fig. 5). Once a structural segment is inserted into the lipid bilayer, it is thermodynamically stable and remains folded (Supplementary Fig. 3). Such long-term stability has been demonstrated for fragments of LacY and of other membrane proteins10,11,17,36,37,44. Our refolding experiments further show that after the first structural segment has been inserted the polypeptide stretches remaining outside of the membrane are maintained in an unfolded state by YidC until the entire LacY substrate completed folding. By this unique coordination of insertion and folding steps YidC and membrane prevent misfolded species and promote folding (Fig. 5). Energetically, perhaps YidC protects weakly interacting structural regions of LacY (e.g., α-helices VII, IX and X) by increasing the free-energy barrier to misfolding and lowering the free-energy barrier to folding. The increase in correct refolding with time is a direct reflection of altering free-energy barriers.
It is not precisely clear how YidC prevents LacY from misfolding and supports correct folding. The X-ray structure of YidC suggests that folding of a polypeptide is facilitated by transient interactions with a hydrophilic groove in YidC22,45. This groove, which is opened towards the lipid bilayer and the cytoplasmic side45,46, contains several conserved hydrophilic residues and the highly conserved Arg366, which generates a positively charged surface47. We observed that YidC was particularly important for the correct folding of structural segment S6 with α-helix VII. This result suggests that interaction of the positively charged hydrophilic groove of YidC with negatively charged, weakly hydrophobic α-helix VII reduced its misfolding propensity considerably. Subsequently, once properly inserted and folded α-helix VII could establish the salt bridges and hydrogen network needed to stabilize the structural segment S4 encompassing α-helices IX and X. Notably, a weakly hydrophobic α-helix VII appears to be common among sugar permeases in the MFS32,40. This common property suggests that the YidC-assisted insertion and folding of α-helix VII is of general importance.
Our finding that the stepwise insertion and folding appears to be a stochastic process is surprising, as current models expect translocases like YidC to act processively. A sequential insertion model scheme seems logical in spite of the structural details known of translocons38,48. This model is currently challenged by models assuming the stochastic insertion of structural segments, which after insertion anneal to the functional structure of the membrane protein38,49,50. Our experiments in which we observe the stochastic insertion of structural segments until finally the polypeptide assembled the native fold of LacY, supports the latter model. Insertion of cytoplasmic membrane proteins in vivo involves SecYEG and/or YidC2. Interestingly, recent structures of YidC reveal a structural arrangement45 lacking a polypeptide- conducting channel as observed for SecYEG3. Presumably, this structural particularity allows YidC to guide the insertion of individual structural segments like α-helices and loops via various co-existing folding pathways towards the native LacY structure. Since the YidC insertase is an essential component of the biogenesis pathway of many cytoplasmic membrane proteins, our mechanistic findings likely have general relevance. In this regard our experiments may be seen as a first milestone towards studying the mechanisms by which YidC, SecYEG and the SecYEG/YidC complex insert and fold membrane proteins.
ONLINE METHODS
LacY and YidC engineering, expression and purification
LacY
A 36 amino acids (aa) long “polyGly” peptide followed by a His8-tag extension [GSM(G11)EAVEEAVEEA(Gly11)S(His8)] was engineered to the LacY C-terminus using QuikChange® II PCR and plasmid pT7-5/LacY as template. Both polyGly LacY and WT LacY were purified from E. coli XL1-Blue (Stratagene) transformed with pT7-5 plasmids harboring given mutant genes by using Co(II) affinity chromatography as prescribed51. LacY eluted from the Co(II)-Talon column was concentrated and washed with 50 mM sodium phosphate (NaPi), pH 7.5 and 0.01% (w/w) dodecyl-β-D-maltopyranoside (DDM, Maumee) on an Amicon Ultra-15 concentrator with a 30 kDa cut-off (Millipore).
YidC
Expression and purification of WT YidC with a His10-tag cloned in pT7-7 plasmid was performed similar to LacY except that the expression strain used was E. coli BL21 (DE3) and the DDM concentration used was 0.03%. YidC eluted from the Co(II)-Talon column was concentrated and washed with 150 mM NaCl, 50 mM sodium phosphate (NaPi), pH 7.5 and 0.03% (w/w) DDM on an Amicon Ultra-15 concentrator with a 30 kDa cut-off. All protein preparations were at least 95% pure as judged by staining after sodium dodecyl sulfate polyacrylamide (SDS) gel electrophoresis (Supplementary Fig. 1).
For the reconstitution of LacY and YidC into proteoliposomes containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE, Avanti Polar Lipids) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (PG, Avanti Polar Lipids) (ratio 3:1) was accomplished by using the dilution method25. Briefly, LacY and YidC were mixed at equimolar (1:1) concentration and incubated for 30 min at room temperature before reconstitution. Protein concentrations of LacY, YidC (for the reconstitution of LacY or YidC) or the protein mixture of LacY and YidC (for the co-reconstitution of LacY and YidC) were dissolved in 0.01% DDM solution was then mixed with phospholipids dissolved in 1.2% octyl glucoside (OG, Maumee) to yield a lipid-to-protein ratio of 5 (w/w). The mixture was kept on ice for 20 min and then quickly diluted 50-fold in 50 mM NaPi, pH 7.5. Proteoliposomes were harvested by centrifugation for 1 h at 100,000g, suspended in the same buffer and flash frozen in liquid nitrogen.
Single-molecule unfolding and refolding experiments
Proteoliposomes containing WT LacY reconstituted in PE:PG (ratio 3:1), YidC reconstituted in PE:PG (3:1), or WT LacY co-reconstituted with YidC in PE:PG (3:1) were adsorbed to freshly cleaved mica in 50 mM potassium phosphate (KPi), pH 7.2 for 20 min. The samples were then gently rinsed with the same buffer to remove weakly attached proteoliposomes. For all AFM-based experiments we used the same AFM (Nanowizard Ultra, JPK Instruments AG) at room temperature and in buffer solution. Si3N4 cantilevers (OMCL RC800PSA, Olympus) having a nominal spring constant of 0.05 N·m−1 were used for all measurements. Cantilevers were calibrated in SMFS buffer applying the equipartition theorem at the beginning and at the end of each experiment. Before SMFS, the reconstituted membrane proteins were imaged by AFM (Supplementary Fig. 1)27. These AFM images showed the proteins distributed evenly over the surface of the membrane. On average, the reconstitutions of LacY or/and YidC showed approximately the same low density of membrane proteins. To enhance the probability of attaching LacY molecules to the AFM stylus, we defined a grid (similar to that shown in Supplementary Fig. 8) of thousands of pixels distributed over the protein membrane. For each topographic pixel one or several force-distance curves were recorded. To record a force-distance curve, the AFM stylus was pushed to the membrane applying ≈ 700 pN for 500 ms. To unfold LacY or YidC completely the cantilever was then withdrawn 0.7 μm·s−1 while recording a force-distance curve. The distance travelled by the AFM stylus was chosen sufficiently to fully unfold and extract the polypeptide from the membrane. For refolding, the cantilever was retracted for 100 nm at 0.7 μm·s−1 to partially unfold LacY while recording a force-distance curve. The cantilever was then re-approached towards the membrane at 1 μm·s−1 and kept 10–15 nm above the membrane surface to relax the unfolded polypeptide chain. After controlled refolding times of 0.1–5 s, which allowed the unfolded polypeptide to reinsert into the membrane, the cantilever was retracted for 250 nm at 0.7 μm·s−1 to completely unfold the protein.
The probability of non-specifically attaching the AFM stylus to the “polyGly” elongated C-terminal end of LacY was ≈ 0.1% (n = 2,974), which was ≈ 10-fold higher than the probability of attaching the non-elongated C-terminal end23. This attachment is temporary and the polypeptide slipped from the AFM stylus after several seconds27,33. Thus, within a refolding time of 2 s the C-terminal end slipped from the AFM stylus in ≈ 91% of all cases (n = 1,160). Together the low attachment rate and the high slip off rate considerably lowered the throughput of the single-molecule refolding experiments. Therefore, in a normal experimental day we could record only from one to four successful refolding experiments. In average recording more than 100 refolding experiments required more than 47 experimental days. For each experimental day we had to prepare a new LacY sample and AFM cantilever. Taken together several hundred fresh samples and AFM cantilevers were prepared to acquire the high number of force-distance curves and refolding experiments presented in the manuscript.
SMFS data selection and analysis
The data recorded in several hundred experiments (details see above) were pooled to obtain statistically sufficient amounts of data for each experimental condition. Every force-distance curve of the pooled data was corrected for deflection sensitivity and force offset by applying the standard AFM data processing software (JPK Instruments, software version 5.0.12). A fully unfolded and stretched LacY polypeptide [417 aa extended by a 36 aa long polyGly tail and a His8-tag] has a contour length 461 aa and exhibits force peak patterns extending to a pulling distance of ≈ 120 nm. To ensure that only force-distance curves recording the full unfolding of LacY from mechanically pulling the terminal end were analyzed, we selected force-distance curves showing force peak patterns extending to > 110 nm.
A fully unfolded and stretched YidC polypeptide has a contour length 561 aa (551 aa extended by an His10-tag) and exhibits the force peak patterns extending to ≈ 150 nm. To ensure that only force-distance curves recording the full unfolding of YidC from mechanically pulling one terminal end were analyzed, we selected curves showing force peak patterns extending to > 130 nm.
Every force peak of a force-distance curve corresponding to unfolding of LacY or YidC was fitted by the worm-like-chain (WLC) model using a persistence length of 0.4 nm and contour length of 0.36 nm per aa[27,52]. Each WLC gives the contour length (number of aa) of the polypeptide unfolded and stretched in an unfolding force peak. Histograms were generated showing how frequently force peaks were detected at given contour lengths. To calculate the frequency of the occurrence of force peaks at a given contour length the counts of force peaks were divided through the total number of force-distance curves analyzed. These histograms were fitted with the Gaussian mixture model53. Accordingly to this model, each i-th observed contour length li originates from one force peak class s = 1, …. , M with probability πs or originates from background noise with probability π0. We found 10 force peaks classes for native LacY and eight force peak classes for YidC (Fig. 1 and Supplementary Fig. 7). The mean contour length μs for a given force peak class s was described by a Gaussian distribution with variance σs2. The probability density f of li can be presented as a mixture of Gaussians and background noise with weights πs and π0 respectively.
| (Equation 1) |
ϕ(li, μs, σs2) is the probability density of the Gaussian distribution and g(li) the background noise. The expectation optimization algorithm was applied to find parameters of the Gaussian mixture model (π, μ, σ2). The most probable force peak class si was assigned to any given contour length li with the Bayes classifier by setting:
| (Equation 2) |
Every detected contour length was assigned to a force peak class each of which being mapped to the secondary structure of LacY23. The length of C-terminal polyGly and His8-tag extension were taken into account for mapping the force peak classes. If the force peak class located the beginning or end of a stabilizing structural segment on the mica-facing side of the membrane or within the membrane the thickness of the membrane was taken into account27.
Control of YidC reconstitution
To show the successful reconstitution of YidC in PE:PG membranes SMFS unfolding experiments were performed. Eight different membrane patches were characterized by SMFS. Recorded force-distance curves were superimposed revealing the unique force peak pattern of YidC (Supplementary Fig. 7). This force peak pattern served as the native fingerprint spectrum of YidC.
Control of LacY and YidC co-reconstitution
To show the successful co-reconstitution of LacY and YidC in PE:PG membranes SMFS unfolding experiments were performed. Twelve different membrane patches were imaged by AFM (Supplementary Fig. 1) and then characterized by SMFS. Recorded force-distance curves were classified in two classes and superimposed to show their common features. One class of force-distance curves revealed the native fingerprint spectrum recorded for WT LacY (Fig. 1 and Supplementary Fig. 6) while the other class of force-distance curves showed the native fingerprint spectrum recorded for YidC (Supplementary Figs. 6 and 7). The ratio of force-distance curves representing the unfolding of LacY or YidC approached 1:1.
Refolding experiments, data analysis and classification
Unfolding and extraction of the last stable segment of LacY occurred at pulling distances > 100 nm (force peak at 404 aa, Fig. 1), thus we stopped initial unfolding of LacY at distances of 100 nm and brought back the unfolded polypeptide into close proximity (≈ 10 nm) to the membrane surface were the polypeptide was held in the relaxed state. After a defined relaxation time left the unfolded protein to refold, the AFM stylus was fully withdrawn to unfold LacY completely. Force-distance curves recorded in these refolding experiments were analyzed as follows: The force-distance curve recording the initial partial unfolding of LacY was horizontally shifted to match the native fingerprint spectrum of WT LacY (Fig. 1b,c). If this initial unfolding force-distance curve did not match the native fingerprint spectrum the experiment was discarded. After this, the same horizontal shift applied to fit the initial force-distance curve to the native fingerprint spectrum was applied to the second unfolding force-distance curve recorded from the refolded LacY substrate. The force peaks in both unfolding force-distance curves were then fitted using the WLC model to determine their contour lengths and classified as having remained unfolded, having misfolded, or having inserted and folded structural segments. The classification criteria have been described in the Results and Supplementary Fig. 4. Briefly, a force-distance curve was classified to present a LacY substrate remaining unfolded if no interactions were recovered after the refolding time passed. A force-distance curve was classified to present a LacY substrate having misfolded if it detected at least one force peak at a position not matching the mean ± s.d. of a force peak position detected in the native unfolding force peak pattern of WT LacY (Fig. 1b,c). A force-distance curve was classified to present a LacY having folded one or more structural segments if it detected at least one force peak at a position matching the mean ± s.d. of a force peak position detected in the native unfolding force peak pattern of WT LacY (Fig. 1). Force-distance curves detecting refolding and misfolding events were classified to present a misfolded LacY. To test whether unspecific interactions play a role in determining the outcome of the refolding experiments, WT LacY has been refolded as described above but in the presence of either 1 μM BSA (Sigma-Aldrich) or 1 μM lysozyme (Fluka Analytical). To examine the role of YidC in the outcome of the refolding experiments, WT LacY co-reconstituted with YidC has been refolded as described above. All refolding experiments were analyzed following the same procedure and classified applying the same criteria described above.
Statistical data analysis
Approximately the same sample size of ≈ 100 single refolding experiments was taken for each tested group (WT LacY, WT LacY & YidC, WT LacY & BSA, WT LacY & lysozyme). A sample size of ≈ 100 provides a standard error of ≤ 0.05 (5%) for each of the three categories, unfolded (U), misfolding (M), and folding (F), of a group was classified (Figs. 3 and 4). In order to test the significance of the difference between tested groups (WT LacY, WT LacY & YidC, WT LacY & BSA, WT LacY & lysozyme) two-tailed Z-tests, two-tailed Fisher’s exact tests and Chi-square tests were performed for each refolding time and all categories (Supplementary Tables 1–4). Tested samples were independent and the data were categorical. For Fisher’s exact tests and Chi-square tests 2×3 contingency tables were analyzed (tested groups (WT LacY & YidC, WT LacY & BSA or WT LacY & lysozyme) vs control (WT LacY with possible outcomes U, M, and F). There were no interactions between row and column classifications. The differences between categories/groups were considered not significant (NS) when P ≥ 0.01, and significant when * P < 0.01 and highly significant when ** P < 0.001. Error bars (s.e.m.) shown in Figs. 3 and 4 were estimated by resampling using 4,000 iterations. Statistical analysis was performed using R.
Supplementary Material
Acknowledgments
We thank R.E. Dalbey for providing plasmid pT7-7 encoding YidC with a His10-tag at the C-terminus, R. Newton, J. Thoma, S. Hiller and R.E. Dalbey for encouraging and constructive comments, and S. Weiser for assistance. This work was supported by the Eidgenössische Technische Hochschule Zürich (to D.J.M.), the Swiss National Science Foundation (Grants 205320_160199 to D.J.M.), the National Center of Competence in Research “NCCR Molecular Systems Engineering” (to D.J.M.), the European Union Marie Curie Actions program through the ACRITAS Initial Training Network (FP7-PEOPLE-2012-ITN, Project 317348 to D.J.M.), National Institutes of Health Grant DK51131 (to H.R.K.), as well as National Science Foundation Grant MCB-1129551 (to H.R.K.).
Footnotes
AUTHOR CONTRIBUTIONS
T.S., D.J.M. and H.R.K. designed the experiments. J.S. and D.B. cloned, expressed, purified and reconstituted LacY and YidC. T.S. performed the SMFS experiments. S.A.M. recorded AFM images. All authors analyzed experimental data and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–67. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Dalbey RE, Wang P, Kuhn A. Assembly of bacterial inner membrane proteins. Annu Rev Biochem. 2011;80:161–87. doi: 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- 3.Luirink J, Yu Z, Wagner S, de Gier JW. Biogenesis of inner membrane proteins in Escherichia coli. Biochim Biophys Acta. 2012;1817:965–76. doi: 10.1016/j.bbabio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Stephenson K. Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway. Mol Membr Biol. 2005;22:17–28. doi: 10.1080/09687860500063308. [DOI] [PubMed] [Google Scholar]
- 5.van Bloois E, et al. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J Biol Chem. 2005;280:12996–3003. doi: 10.1074/jbc.M414094200. [DOI] [PubMed] [Google Scholar]
- 6.Wang P, Dalbey RE. Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta. 2011;1808:866–75. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Saller MJ, Wu ZC, de Keyzer J, Driessen AJ. The YidC/Oxa1/Alb3 protein family: common principles and distinct features. Biol Chem. 2012;393:1279–90. doi: 10.1515/hsz-2012-0199. [DOI] [PubMed] [Google Scholar]
- 8.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 9.Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–48. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 10.Kedrov A, Janovjak H, Ziegler C, Kuhlbrandt W, Muller DJ. Observing folding pathways and kinetics of a single sodium-proton antiporter from Escherichia coli. J Mol Biol. 2006;355:2–8. doi: 10.1016/j.jmb.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Kessler M, Gottschalk KE, Janovjak H, Muller DJ, Gaub HE. Bacteriorhodopsin folds into the membrane against an external force. J Mol Biol. 2006;357:644–54. doi: 10.1016/j.jmb.2005.12.065. [DOI] [PubMed] [Google Scholar]
- 12.Damaghi M, Koster S, Bippes CA, Yildiz O, Muller DJ. One β-hairpin follows the other: exploring refolding pathways and kinetics of the transmembrane β-barrel protein OmpG. Angew Chem Int Ed Engl. 2011;50:7422–4. doi: 10.1002/anie.201101450. [DOI] [PubMed] [Google Scholar]
- 13.Bosshart PD, et al. The transmembrane protein KpOmpA anchoring the outer membrane of Klebsiella pneumoniae unfolds and refolds in response to tensile load. Structure. 2012;20:121–7. doi: 10.1016/j.str.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Bechtluft P, et al. Direct observation of chaperone-induced changes in a protein folding pathway. Science. 2007;318:1458–61. doi: 10.1126/science.1144972. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser CM, et al. Tracking UNC-45 chaperone-myosin interaction with a titin mechanical reporter. Biophys J. 2012;102:2212–9. doi: 10.1016/j.bpj.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunes JM, Mayer-Hartl M, Hartl FU, Muller DJ. Action of the Hsp70 chaperone system observed with single proteins. Nat Commun. 2015;6:6307. doi: 10.1038/ncomms7307. [DOI] [PubMed] [Google Scholar]
- 17.Thoma J, Burmann BM, Hiller S, Muller DJ. Impact of holdase chaperones Skp and SurA on the folding of beta-barrel outer-membrane proteins. Nat Struct Mol Biol. 2015;22:795–802. doi: 10.1038/nsmb.3087. [DOI] [PubMed] [Google Scholar]
- 18.Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 19.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–5. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 20.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–8. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagamori S, Smirnova IN, Kaback HR. Role of YidC in folding of polytopic membrane proteins. J Cell Biol. 2004;165:53–62. doi: 10.1083/jcb.200402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Kaback HR, Dalbey RE. YidC protein, a molecular chaperone for LacY protein folding via the SecYEG protein machinery. J Biol Chem. 2013;288:28180–94. doi: 10.1074/jbc.M113.491613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serdiuk T, et al. Substrate-induced changes in the structural properties of LacY. Proc Natl Acad Sci U S A. 2014;111:E1571–80. doi: 10.1073/pnas.1404446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serdiuk T, Sugihara J, Mari SA, Kaback HR, Muller DJ. Observing a lipid-dependent alteration in single lactose permeases. Structure. 2015;23:754–61. doi: 10.1016/j.str.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viitanen P, Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification, reconstitution, and characterization of the lac permease of Escherichia coli. Methods Enzymol. 1986;125:429–52. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- 26.Herzlinger D, Viitanen P, Carrasco N, Kaback HR. Monoclonal antibodies against the lac carrier protein from Escherichia coli. 2 Binding studies with membrane vesicles and proteoliposomes reconstituted with purified lac carrier protein. Biochemistry. 1984;23:3688–93. doi: 10.1021/bi00311a018. [DOI] [PubMed] [Google Scholar]
- 27.Muller DJ, Engel A. Atomic force microscopy and spectroscopy of native membrane proteins. Nat Protoc. 2007;2:2191–7. doi: 10.1038/nprot.2007.309. [DOI] [PubMed] [Google Scholar]
- 28.Kedrov A, Krieg M, Ziegler C, Kuhlbrandt W, Muller DJ. Locating ligand binding and activation of a single antiporter. EMBO Rep. 2005;6:668–74. doi: 10.1038/sj.embor.7400455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sapra KT, Besir H, Oesterhelt D, Muller DJ. Characterizing molecular interactions in different bacteriorhodopsin assemblies by single-molecule force spectroscopy. J Mol Biol. 2006;355:640–50. doi: 10.1016/j.jmb.2005.10.080. [DOI] [PubMed] [Google Scholar]
- 30.Ge L, Perez C, Waclawska I, Ziegler C, Muller DJ. Locating an extracellular K+-dependent interaction site that modulates betaine-binding of the Na+-coupled betaine symporter BetP. Proc Natl Acad Sci U S A. 2011;108:E890–8. doi: 10.1073/pnas.1109597108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bippes CA, et al. Peptide transporter DtpA has two alternate conformations, one of which is promoted by inhibitor binding. Proc Natl Acad Sci U S A. 2013;110:E3978–E3986. doi: 10.1073/pnas.1312959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–40. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oesterhelt F, et al. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–6. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 34.Fanelli F, Seeber M. Structural insights into retinitis pigmentosa from unfolding simulations of rhodopsin mutants. FASEB J. 2010;24:3196–209. doi: 10.1096/fj.09-151084. [DOI] [PubMed] [Google Scholar]
- 35.Kappel C, Grubmuller H. Velocity-dependent mechanical unfolding of bacteriorhodopsin is governed by a dynamic interaction network. Biophys J. 2011;100:1109–19. doi: 10.1016/j.bpj.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bibi E, Kaback HR. In vivo expression of the lacY gene in two segments leads to functional lac permease. Proc Natl Acad Sci U S A. 1990;87:4325–9. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zen KH, McKenna E, Bibi E, Hardy D, Kaback HR. Expression of lactose permease in contiguous fragments as a probe for membrane-spanning domains. Biochemistry. 1994;33:8198–206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]
- 38.Cymer F, von Heijne G, White SH. Mechanisms of integral membrane protein insertion and folding. J Mol Biol. 2015;427:999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller DJ, et al. Observing membrane protein diffusion at subnanometer resolution. J Mol Biol. 2003;327:925–30. doi: 10.1016/s0022-2836(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 40.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–35. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan L, Kaback HR. Properties of a LacY Efflux Mutant. Biochemistry. 2009;48:9250–9255. doi: 10.1021/bi9014067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–9. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 43.Harris NJ, Booth PJ. Folding and stability of membrane transport proteins in vitro. Biochim Biophys Acta. 2012;1818:1055–66. doi: 10.1016/j.bbamem.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Engelman DM, et al. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555:122–5. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 45.Kumazaki K, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–20. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 46.Shimokawa-Chiba N, et al. Hydrophilic microenvironment required for the channel-independent insertase function of YidC protein. Proc Natl Acad Sci U S A. 2015;112:5063–8. doi: 10.1073/pnas.1423817112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumazaki K, et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep. 2014;4:7299. doi: 10.1038/srep07299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapoport TA, Goder V, Heinrich SU, Matlack KE. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–75. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Van Lehn RC, Zhang B, Miller TF., 3rd Regulation of multispanning membrane protein topology via post-translational annealing. Elife. 2015;4:e08697. doi: 10.7554/eLife.08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White SH. The messy process of guiding proteins into membranes. Elife. 2015;4:e12100. doi: 10.7554/eLife.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A. 2007;104:16504–9. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 53.Kawamura S, et al. Kinetic, energetic, and mechanical differences between dark-state rhodopsin and opsin. Structure. 2013;21:426–37. doi: 10.1016/j.str.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.