Abstract
Poliovirus-encoded nonstructural polypeptide 2C is a multifunctional protein that plays an important role in viral RNA replication. 2C interacts with both intracellular membranes and virus-specific RNAs and has ATPase and GTPase activities. Extensive computer analysis of the 2C sequence revealed that in addition to the known ATPase-, GTPase-, membrane-, and RNA-binding domains it also contains several “serpin” (serine protease inhibitor) motifs. We provide experimental evidence suggesting that 2C is indeed capable of regulating virus-encoded proteases. The purified 2C protein inhibits 3Cpro-catalyzed cleavage of cellular transcription factors at Q-G sites in vitro. It also inhibits cleavage of a viral precursor by the other viral protease, 2Apro. However, at least three cellular proteases appear not to be inhibited by 2C in vitro. The 2C-associated protease inhibitory activity can be depleted by anti-2C antibody. A physical interaction between 2C and His-tagged 3Cpro can be demonstrated in vitro by coimmunoprecipitation of 2C with anti-His antibody. Deletion analysis suggests that the 2C central and C-terminal domains that include several serpin motifs are important for 3Cpro-inhibitory activity. To examine the 2C protease inhibitory activity in vivo, stable HeLa cell lines were made that express 2C in an inducible fashion. Infection of 2C-expressing cells with poliovirus led to incomplete (or inefficient) processing of viral precursor polypeptides compared to control cell lines containing the vector alone. These results suggest that 2C can negatively regulate the viral protease 3Cpro. The possible role of the 2C protease inhibitory activity in viral RNA replication is discussed.
Poliovirus is the prototype member of the picornavirus family with a plus-sense RNA genome of 7,440 nucleotides, which is covalently linked to a small viral protein (VPg) at its 5′ end and polyadenylated at its 3′ end (34, 56). The positive-strand mRNA, which lacks VPg, codes for a single large polyprotein, which is processed into mature proteins by viral proteases 2Apro, 3Cpro and 3CDpro (reviewed in references 39 and 71). Biochemical and genetic evidence suggests that most of the poliovirus nonstructural proteins are involved in viral RNA replication in the form of precursors, mature polypeptides, or both (71). The viral RNA polymerase precursors 3CDpro, the VPg precursor 3AB and a cellular polypeptide, poly(rC)-binding protein, have been shown to interact with the 5′-cloverleaf structure of viral RNA, leading to the formation of a functional complex important for viral RNA replication (3, 4, 31, 47, 72). The precursor proteins 3AB and 3CDpro also interact with the 3′-untranslated region of viral RNA in the absence of other proteins (31). The initiation of poliovirus RNA synthesis appears to be primed by a protein-nucleotidyl covalent complex (VPg-pU or VPg-pUpU) (10, 50, 52).
Although it was initially thought to be a member of the cysteine protease family, mutational analyses, amino acid sequence comparison, and three-dimensional modeling have suggested that 3Cpro adopts a fold similar to that found in serine proteases such as chymotrypsin (12, 28, 30, 32, 33, 37, 45). The other viral protease, 2Apro, also possesses a chymotrypsin-like fold that is related to smaller serine proteases such as α-lytic proteinase (12). Although the major function of these proteases is to process viral precursor polypeptides, both proteases are also involved in the shutoff of host cell metabolism. Although 2Apro is involved in the shutoff of host cell translation (13, 29, 41), 3Cpro has been shown to cleave and inactivate a number of host cell transcription factors leading to the inhibition of cellular transcription (19, 61, 70, 75).
The 2C protein of poliovirus is 329 amino acids long (37.5 kDa) and is highly conserved among picornaviruses (5). Genetic analyses have implicated the 2C polypeptide in a number of functions during viral replication such as uncoating (40), host cell membrane rearrangement (18), RNA replication (reviewed in reference 71), and encapsidation (69). The exact role of 2C in these processes, however, is not known. Many mutations in the 2C region have been found to be lethal. Studies with nonlethal 2C mutants suggest that 2C has at least two functions in RNA replication: a cis-acting guanidine-sensitive function required for initiation and a trans-acting function required for elongation (71). More recent results utilizing an in vitro translation/replication system (10, 43), which produces viable poliovirus, have shown that 2C is required prior to or during initiation of minus-strand RNA synthesis (11). During poliovirus infection, 2C and its precursor 2BC migrate to the rough endoplasmic reticulum, where they induce the formation of smooth membrane vesicles that bud off and become the site of viral RNA synthesis: the replication complex (2, 14-16, 18, 62, 65). Although the 2C protein lacks a defined membrane-binding domain, the N-terminal region encompassing amino acids 21 to 54 appears to be important for membrane binding (24). The N-terminal fragment of 2C containing an amphipathic helix (49) is sufficient to target a soluble protein, chloramphenicol acetyltransferase (CAT), to cellular membrane (23). A second amphipathic α-helix located at the C terminus has also been implicated in 2C membrane interaction (66). Apart from its membrane-binding ability, 2C also binds to viral RNA in vitro (57) and appears to be attached to viral RNA in the replication complex that forms on virus-induced smooth vesicles (14). Purified 2C interacts specifically with the 3′-terminal cloverleaf of the minus-strand RNA through the sequence UGUUUU when present in the context of a double-stranded structure (7). This interaction requires an intact stem-loop B within the minus-strand cloverleaf (8). In addition to 2C, two host cell proteins bind to the 3′-terminal cloverleaf of the minus-strand RNA (59, 60). The 2C protein also has ATPase and GTPase activities (42, 58). Point mutations targeted to the conserved amino acids of the nucleoside triphosphate (NTP)-binding motif result in nonviable viruses that do not replicate their RNA, although such point mutants display normal proteolytic processing patterns (42, 67). The 2C protein also contains a zinc-binding cysteine-rich motif (53). The 2C ATPase activity is inhibited by low concentrations of guanidine hydrochloride (54), a potent inhibitor of picornavirus RNA replication in vivo.
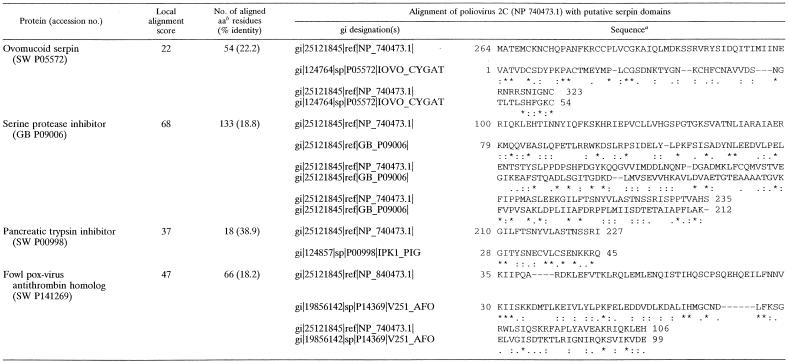
In order to gain further understanding into the functions of this multifunctional viral protein (2C), its protein sequence was analyzed extensively. The main objectives were to find distantly related proteins of known functions and align related sequences to the 2C protein to identify possible motifs or domains of interest. As expected, the first 76 matches corresponded to related single-stranded, plus-strand RNA virus sequences. A second group of homology included several known ATPases, ATP- and GTP-binding proteins, and viral helicases. A more intriguing finding was a third group of proteins that showed homology to 2C. This group included protease inhibitors, particularly serine protease inhibitors or “serpins” (26, 76). Serpins are believed to inhibit protease activity by binding to the catalytic pocket of proteases and blocking their ability to associate with their substrates. The inhibitors may be cleaved by the protease, but at much slower rates than normal substrates. Many of these proteins are characterized by having several repeats of short inhibitor domains of ca. 50 to 60 amino acids. Pairwise local alignments to the protease inhibitors showed that there were several possible serpin motifs scattered throughout the 2C sequence (Table 1 and Fig. 1). In addition, 2C was found to be homologous to the proteosome regulatory subunits from various organisms (data not shown). These observations suggest the interesting possibility that 2C may regulate viral or cellular protease activity.
TABLE 1.
Serpin domain alignment of 2C sequences
Symbols: *, identical in sequence and alignment; :, conserved substitutions observed; ., semiconserved substitutions.
aa, amino acid.
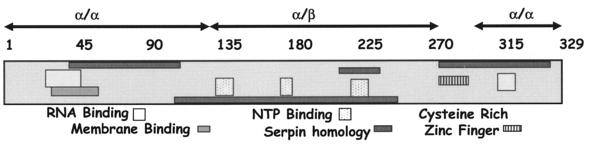
FIG. 1.
Schematic diagram of domain structures of 2C. A linear diagram of 2C shows the locations of putative domains as determined by computer analyses and prior studies from various laboratories. The numbers at the top indicate the amino acids from N (1) to C (329) termini of the 2C polypeptide.
Here we demonstrate that the purified 2C protein is indeed capable of inhibiting the activity of both the viral proteases 3Cpro and 2Apro in vitro. The regions in the center of the protein and at the C terminus that include three predicted serpin motifs appear to be important for its protease inhibitory activity. A physical interaction between 3C and 2C can be demonstrated by coimmunoprecipitation of 2C by an antibody that recognizes 3Cpro. Infection by poliovirus of HeLa cell lines that express 2C in an inducible fashion results in a processing pattern consistent with slower processing of a number of poliovirus precursor polypeptides. These results suggest that the poliovirus 2C protein is capable of regulating 3Cpro activity both in vitro and in poliovirus-infected cells.
MATERIALS AND METHODS
Plasmids.
pGEM-CREB was a gift from R. Gaynor (University of Texas). Poliovirus 2A was cloned into pET-21b between BamHI and XhoI enzyme sites after PCR amplification by using the following forward and reverse primers: 5′-GTG TAA GGA TCC GGG ATT CGG ACA CCA AAA CAA AGCG-3′ and 5′-AAT TTA CTC GAG TTG TTC CAT GGC TTC TTC TTC GTAG-3′ incorporating appropriate restriction sites (underlined). 3CD was cloned into pET-15b as described earlier (7), and wild-type (wt) 2C and the 2C deletion mutants were cloned into pGEM, pET-15b, and pET-21b as previously described (7, 24). In addition, 2C and the deletion mutants were subcloned into the in vitro expression vector pET21b (Novagen) with a carboxy-terminal V5 epitope tag. The 2C coding sequence was PCR amplified by using a forward 5′-terminal primer, which annealed to the first 13 to 15 nucleotides of the coding sequence. Immediately upstream of this, an NdeI endonuclease restriction site was inserted flanked by nucleotides complementary to the parental vector. Adjacent to the 3′-terminal end of the coding sequence, a six-nucleotide spacer is located, followed by the EcoRV, BstXI, NotI, XhoI, XbaI, ApaI, and SacII restriction sites. These are immediately followed by the V5 epitope. The reverse 3′-terminal primer was complementary to the last 15 nucleotides of the V5 epitope, followed by a HindIII restriction site and 9 nucleotides of the parental vector. The 5′-NdeI and 3′-HindIII restriction sites were used to subclone the sequence into the pET21b plasmid. A similar scheme was used for each of the 2C deletion mutants.
Cell lines.
2C expressing cell lines and controls were made by using the original ClonTech Tet On/Off system vectors, pTet-On, pTRE2, and pTK-Hyg. HeLa cells were transfected with the pTet-On plasmid and selected with G418 (900 μg/ml). Selection was continued for 2 months to ensure stable integration of the plasmid. HeLa-pTet-On cell lines were clonally selected. These cell lines were then used to construct the 2C-expressing cell line and the vector-alone control cell line. The 2C gene was PCR amplified with the primers 5′-GGTCGGCCGCATATGGGTGACAGTTGGTTG and 3′-CTGATATCAGAATAGCAGTAGCAGCATGTCTAGTGGTCCTTGAAACAAAGCCTCC. The resulting PCR product was gel purified and digested with EagI and EcoRV. The pTRE2 vector was digested with EagI and EcoRV prior to ligation. In each case, the vector pTRE2 (with or without 2C) was transfected into the HeLa-pTet-On cells along with the pTk-Hyg plasmid, which carries the hygromycin resistance gene according to the ClonTech protocol. Stable cell lines were selected by using G418 (900 μg/ml) and hygromycin B (400 μg/ml). 2C is expressed in these cell lines in the presence of doxycyclin (1-3 μg/ml). The presence of the 2C gene inside the cells was confirmed by PCR (Amersham Ready-To-Go PCR beads) of the genomic DNA purified with Qiagen's genomic DNA kit. Expression of 2C RNA in the cell line was confirmed by reverse transcription-PCR (RT-PCR) (Amersham First-Strand-Ready-To-Go beads) of the total cellular RNA purified by using the Qiagen RNA purification kit. The cell lines containing 2C or vector alone were grown in medium lacking tetracycline but containing fetal bovine serum (ClonTech). The presence of the 2C sequence in the cell line was checked by PCR before each experiment with the cell line. This was necessary because the selection marker and the 2C gene were present on different plasmids in these cells, and the possibility existed that the cell line could lose the gene upon repeated passages.
Poliovirus infection and plaque assay.
The 2C cell line (E2) and vector control cell line (9E) were grown overnight (10 h) in the presence of doxycycline (2 μg/ml). The following morning the cells were infected with poliovirus at a multiplicity of infection of 10 in methionine-deficient medium. The infection was allowed to continue for 3 to 4 h. At the end of this time, 5 μg of actinomycin D/ml was added to the cells. The cells were incubated for 10 to 15 min and then pulsed with 10 μCi of [35S]methionine (specific activity, 1,000 Ci/mmol; ICN Biochemicals) for 10 to 15 min, followed by a chase with 100 mM cold methionine. Samples were taken every 20 min after the chase. Extracts were made by repeated freeze-thawing of the cells, and viral proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography. Labeled viral precursor polypeptides were quantified by using the Image J program provided by the NIH. For plaque assay, HeLa cells (∼3.2 × 106) were grown in 60-mm culture plates to near confluency in Dulbecco modified Eagle medium with 10% fetal bovine serum and newborn calf serum. 2C and vector HeLa-pTet-On cell lines were incubated with 2 μg of doxycycline/ml for 10 h before the assay. Incubation of doxycycline with the cell lines for longer periods (24 h) resulted in fewer observed plaques. For the assay, cells were aspirated and washed, incubated with 200 μl of various dilutions of wt poliovirus (Mahoney) for 30 min at 37°C in a CO2 incubator. The plates were rocked every 5 min during this time. The virus was then aspirated off, and an agarose overlay consisting of 51% of 1.8% Noble agar, 47% of 2× Dulbecco modified Eagle medium without serum, and 2% fetal bovine serum was added to the cells, and the mixture was allowed to harden at room temperature for about 15 min. The plates were incubated at 37°C for 24 h. Then, 2.5 ml of a 0.03% neutral red solution was added to the plates. The plates were then incubated in the dark at 37°C for 2 h and aspirated. After 24 h of further incubation at 37°C in the dark, plaques were counted.
Purification of 2C, 2Apro, and 3Cpro proteins.
Poliovirus 3Cpro was cloned into pQE30 (Qiagen) as described earlier and purified from Escherichia coli M15 pREP4 strain after IPTG (isopropyl-β-d-thiogalactopyranoside) induction (63). Poliovirus 2Apro was purified from E. coli strain BL21(DE3) harboring pET21b recombinant plasmid, and the protein was purified 3.5 h after IPTG induction. The protein was purified by using Talon resin under denaturing conditions by using a protocol similar to that described earlier (7). Poliovirus-encoded 2C protein was isolated from BL21(DE3) cells 3.5 h after IPTG induction from the supernatant fraction under nondenaturing conditions. The final eluate from the Talon resin (ClonTech) was dialyzed against buffer A (50 mM Tris [pH 7.4], 10% glycerol, 1 mM dithiothreitol [DTT], 50 mM KCl). The presence of 2C was monitored by checking RNA-binding activity by using a mobility shift assay and Western analysis with polyclonal antibody to 2C as described previously (7).
Protease reactions.
The indicated amounts of purified viral 3C and 2A proteases (3C at 0.5 and 2.0 μg and 2A at ∼20 ng [unless stated otherwise]) were added to in vitro-translated [35S]methionine-labeled CREB or viral 3CD proteins. Reactions were incubated at 30°C for 4 h, followed by the addition of 2× SDS sample buffer to terminate the reaction. In some reactions, 3Cpro or 2Apro was preincubated for at least 30 min at 30°C with the indicated amounts of 2C before the addition of labeled substrate to the reaction. Thrombin and enterokinase were obtained commercially (Novagen, Inc.), and cleavage reaction protocols were followed according to the manufacturer's instructions. Enterokinase was diluted to 0.06 U with dilution buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 2 mM CaCl2, 50% glycerol) before use, whereas thrombin was diluted to 0.005 U with cleavage buffer (20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2). The reactions were incubated overnight at room temperature, followed by the addition of 2× SDS-PAGE sample buffer. The samples were analyzed on 4 to 20% gradient gels.
Western blot analysis.
Purified protein or reactions containing cleaved proteins were mixed with SDS sample buffer, resolved on an SDS-PAGE gel, and transferred onto nitrocellulose membrane as described earlier (7). The polyclonal antibody used for detection of 2C was a third bleed from rabbits injected with purified full-length 2C used at a 1:100 dilution.
The 48-kDa cleavage control protein used in the thrombin and enterokinase protease assays was blocked according to the manufacturer's instructions. The nitrocellulose was incubated with an S-protein alkaline phosphatase conjugate at room temperature for 30 min, and the complex was visualized by using a chromogenic substrate BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium kit (Kirkegaard and Perry Laboratories).
Immunoprecipitation.
In vitro-translated, [35S]methionine-labeled CAT or poliovirus 2C protein was incubated with the appropriate antibody in 1× radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.5], 0.5% deoxycholate, 1% NP-40, 150 mM NaCl, 2.5 mg of bovine serum albumin [BSA]/ml) with aprotinin. Proteins were incubated for 3 h on ice; the specific polyclonal or preimmune sera appropriate for the reaction was then added, and incubation continued for an additional 2 h on ice. The anti-His monoclonal antibody was obtained from ClonTech. The immune complexes were precipitated by using preswollen protein A-Sepharose. The Sepharose-immune complex was washed extensively (five to six times) with 1× radioimmunoprecipitation assay, followed by two washes with phosphate-buffered saline and recovery of the immunoprecipitated protein complex by elution with 30 μl of 2× SDS sample buffer.
Immunodepletion assay.
Immunoglobulin G (IgG) was isolated from hyperimmune rabbit sera injected with bacterially expressed glutathione S-transferase-2C fusion protein by using a standard protocol (Pierce Chemicals). Purified 2C (∼1.5 μg of protein) was incubated with either immune IgG or preimmune sera for 3 h on ice, after which preswollen protein A-Sepharose beads (in 10 mM Tris [pH 7.4]) were added, followed by incubation overnight on ice. The protein A-Sepharose complex was pelleted by centrifugation, and the resulting supernatant were designated depleted 2C extracts. These extracts were used in the 3Cpro cleavage assay. For heat denaturation experiments, 2C was denatured by placement in a heating block preset at the indicated temperature for the required time. The heated samples were spun at room temperature for 5 min in a microfuge, and the supernatant was saved and used in the appropriate cleavage assay.
In vitro transcription and translation.
The clones for in vitro transcription were linearized with the appropriate restriction enzymes. pCREB was linearized with HindIII, gel purified, and in vitro transcribed by using SP6 RNA polymerase. The 2C clone was linearized with AvaII before transcription by the T7 RNA polymerase. The mRNA obtained was alcohol precipitated and subsequently translated in rabbit reticulocyte lysate according to the manufacturer's instructions (Promega). 3CD and CAT constructs were translated by using the Promega coupled transcription-translation system. In vitro-synthesized proteins were labeled with 40 μCi of [35S]methionine (specific activity, >1,000 Ci/mmol; Amersham) during translation.
In vitro RNA polymerase II transcription assays.
RNA polymerase II transcription reactions were carried out by using a plasmid, pA326 (75), containing the TATA initiator promoter as a template for in vitro reactions. Transcription reaction mixtures containing 700 ng of template DNA, 100 μg of HeLa nuclear extracts, 0.5 mM each of four ribonucleoside triphosphates, 40 U of RNasin, 5 mM MgCl2, and 1 mM DTT in a reaction volume of 25 μl. These reaction mixtures were incubated at 30°C for 90 min, and the reactions were terminated by the addition of 200 μl of stop solution (300 mM Tris-HCl [pH 7.4], 2 mM EDTA, 300 mM sodium acetate, 250 μg of tRNA/ml). The RNA was extracted with phenol and phenol-chloroform, followed by ethanol precipitation. The RNA was annealed with 50,000 cpm of Sp6 primer, 25 mM Tris-HCl [pH 8.3], 25 mM KCl, 5 mM MgCl2, 0.25 mM spermidine, and 5 mM DTT in a reaction volume of 11 μl. The mixture was incubated for 20 min at 55°C, followed by 10 min at room temperature. This annealed mixture was analyzed by the primer extension method. The primer extension reaction contained the annealed mixture described above plus 10 U of avian myeloblastosis virus reverse transcriptase (Life Science, Inc.), 1 μl of 5× avian myeloblastosis virus reverse transcriptase buffer (250 mM Tris-HCl [pH 8.3], 50 mM MgCl2, 250 mM KCl, 50 mM DTT, 2.5 mM spermidine, 1.5 mM deoxynucleoside triphosphates), and 2.8 mM sodium pyrophosphate in a 20-μl reaction volume. The reaction mixture was incubated at 42°C for 1 h. The reactions were stopped by the addition of 20 μl of stop dye (80% formamide, 0.01% xylene cyanol, 0.01% bromophenol blue), loaded onto an 8% acrylamide-8 M urea gel, and subjected to electrophoresis.
RESULTS
2C inhibits 3Cpro-mediated cleavage of transcription factor CREB in vitro.
In order to gain further understanding into the function of 2C, its amino acid sequence was analyzed extensively. The main goals were (i) to find distantly related proteins of known functions and (ii) to align related sequences to the 2C protein in order to identify possible motifs or domains of interest. Initial profile search was performed by using a nonredundant protein database containing entries from the SWISS-PROT sequence data bank at the European Molecular Biology Laboratory (EMBL) (6), the protein identification resource of the National Biomedical Research Foundation, and the Gert peptide database (6, 64). Protein sequences with a Z score higher than 6.0 were then characterized further by reading their annotations in the database by using local alignment programs. Table 1 shows pairwise local alignments between poliovirus 2C and various protease inhibitors. Four potential protease inhibitory domains were scattered throughout the entire 2C sequence. The longest serpin motif identified by the computer program was present in the 2C central domain (amino acids 100 to 235), which also included a shorter motif (amino acids 210 to 227) with a high degree of homology (Table 1 and Fig. 1). Two other serpin motifs were located at the N- and C-terminal regions of 2C spanning amino acid residues 35 to 106 and 264 to 323, respectively (Table 1).
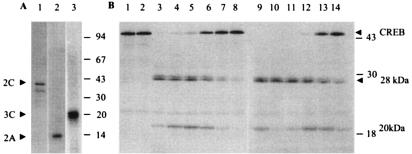
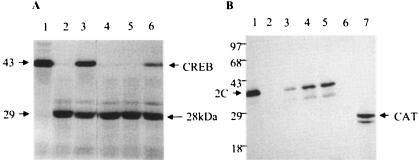
To verify experimentally whether the 2C polypeptide indeed contains protease inhibitory activity, the 2C protein was expressed in bacteria and purified under nondenaturing conditions. Bacterially expressed, poliovirus 3Cpro and 2Apro were also purified to near homogeneity (63, 73). Figure 2A shows analysis of purified 3Cpro, 2C and 2Apro by SDS-PAGE, followed by silver staining. Purified preparations of both 2Apro and 3Cpro showed single polypeptides with the appropriate molecular weights. The highly purified 2C protein consisted of mainly two polypeptides: the full-length 2C protein and a truncated form of 2C. Both polypeptides were found previously to specifically react with anti-2C (7). The transcription factor cyclic AMP-responsive element binding protein (CREB), shown to be directly cleaved by 3Cpro (74), was used as a 3Cpro substrate in an in vitro cleavage assay. In vitro-translated [35S]methionine-labeled CREB was incubated with buffer (lane 1), 2C (lane 2), 3Cpro (lane 3), or increasing concentrations of 2C in the presence of a constant amount of 3Cpro (lanes 4 to 8) (Fig. 2B). The following lanes in Fig. 2 (lanes 9 to 14) show a similar experiment except that a fourfold-higher amount of 3Cpro was used in this experiment compared to those shown in lanes 1 to 8. Incubation of the labeled CREB with 2C alone had no significant effect on the CREB protein (lane 2) compared to the control (lane 1). Proteolytic cleavage of CREB by 3Cpro into two major fragments (28 and 20 kDa) was apparent at both concentrations of 3Cpro (lanes 3 and 9, Fig. 2B). We have shown previously that the two fragments of CREB are generated by 3Cpro-mediated cleavage of the Q-G pair at position 172 to 173 of CREB (74). The 28-kDa proteolytic fragment of CREB migrated as a doublet. The precise reason for this is not known. However, 3Cpro-mediated cleavage of an additional Q-G bond between amino acids 187 and 188 of CREB could be responsible for the production of the doublet. Increasing the concentration of 2C in the reaction clearly inhibited the cleavage of CREB by 3Cpro, as evidenced by the reduction of both the proteolytic fragments concomitant with the appearance of full-length CREB (lanes 4 to 8 and lanes 10 to 14). Quantification of the full-length CREB polypeptide showed >90% inhibition of 3Cpro-mediated cleavage of CREB at the highest concentration of 2C (lane 8). The inhibition of CREB cleavage at higher concentrations of 3Cpro was significantly lower than that observed at the lower concentration of the protease (compare lanes 6 and 12). We then tested a few other viral nonstructural proteins for their ability to block 3Cpro-mediated cleavage of CREB. As presented in Fig. 3A, both 2C and 2BC were able to almost totally block the proteolytic cleavage of CREB by 3Cpro (lanes 5, 8, and 9). The viral RNA polymerase, 3Dpol, however, had no detectable effect in blocking 3Cpro activity (lane 7). The viral protein 2B showed low but detectable activity compared to the control (lanes 5 and 6). Thus, 2C and 2BC were the major polypeptides that efficiently blocked 3Cpro activity among the viral proteins tested. An extract prepared from bacteria that harbored the plasmid lacking the 2C insert did not inhibit 3Cpro activity significantly (lane 10). To rule out the possibility that 2C-mediated inhibition of 3Cpro activity was specific to cellular transcription factor CREB, two other transcription factors, TBP (TATA-binding protein) and an RNA polymerase I factor, SL-1, were tested in this assay. The results were very similar to that observed with CREB; the 3Cpro-mediated cleavage of both TBP and SL-1 was almost completely blocked by 2C, suggesting the protease inhibitory effect of 2C was not limited to CREB (data not shown). These results imply that 2C's protease inhibitory effect is most probably mediated through 3Cpro rather than through the protease substrates.
FIG. 2.
Poliovirus 3Cpro activity is inhibited by the 2C polypeptide. (A) Totals of 300 ng of 2C (lane 1) and 2A (lane 2) and 800 ng of 3C (lane 3) were analyzed by SDS-PAGE, followed by silver staining as described in Materials and Methods. (B) Totals of 500 ng (lanes 3 to 8) and 2 μg (lanes 9 to 14) of 3Cpro was preincubated with buffer (lane 1), 500 ng of 2C (lane 2), or various amounts of purified 2C prior to labeled substrate addition (1 μl of in vitro-translated, [35S]methionine-labeled CREB) as described in Materials and Methods. Reactions were terminated by the addition of 2× SDS sample buffer (1:1 [vol/vol]), and proteins were resolved by SDS-14% PAGE, followed by autoradiography. The numbers on the left show the position and approximate molecular mass (in kilodaltons) of the marker proteins. The arrowheads on the right indicate the migration of full-length and the proteolyzed CREB products (∼28 and ∼20 kDa). Lanes 4 through 8 contain 50, 125, 250, 375, or 500 ng of 2C, respectively. Lanes 10 through 14 contain 50, 125, 250, 375, or 500 ng of 2C, respectively.
FIG. 3.
2BC inhibits 3Cpro activity. (A) In vitro-translated, [35S]methionine-labeled CREB was incubated with buffer (lane 1) or 500 ng each of bacterially expressed, affinity-purified 2B (lanes 2 and 6), 3Dpol (lanes 3 and 7), 2BC (lanes 4 and 9), and 2C (lane 8) in the absence (lanes 1 to 4) or presence (lanes 5 to 9) of 500 ng of 3Cpro. Lane 10 is the same as lane 5 except that 500 ng of an extract from bacteria harboring the plasmid without the 2C insert was used. (B) 2Apro activity is inhibited by 2C. Various amounts of 2C were preincubated with 20 ng of purified 2Apro before addition to the reactions containing [35S]methionine-labeled 3CD. Reactions were terminated by the addition of 2x SDS sample buffer and proteins resolved on a 14% SDS polyacrylamide gel. All reactions contain labeled [35S] methionine-labeled 3CD. Lane 1, control reaction with labeled 3CD as substrate plus buffer; lane 2, 500 ng of 2C; lane 3, 20 ng of 2Apro; lanes 4 to 7, reactions containing 2Apro (20 ng) preincubated with 125 ng (lane 4), 250 ng (lane 5), 375 ng (lane 6), 500 ng (lane 7) of 2C, or 500 ng of heat-inactivated 2C (lane 8) before the addition of labeled 3CD substrate. The arrowheads indicate full-length 3CD and proteolyzed products 3C′ and 3D′, respectively. Positions of the migration of molecular markers are indicated on the right (in kilodaltons).
2C inhibits 2A pro-catalyzed cleavage of poliovirus 3CD precursor polypeptide.
To examine whether 2C is also capable of inhibiting the other poliovirus-encoded protease 2Apro, in vitro-translated [35S]methionine-labeled 3CD precursor was used as a substrate. Viral protease 2Apro cleaves a Tyr-Gly bond within the polymerase sequence of 3CD to generate 3C′ (∼37 kDa) and 3D′ (∼34 kDa) (43, 67). The 3C′ contains the entire 3C, as well as part of the 3D sequence. As can be seen in Fig. 3B, incubation of 3CD with bacterially expressed, purified 2Apro resulted in the cleavage of 3CD to two polypeptides that migrated at ca. 33 and 39 kDa (compare lanes 1 and 3). The molecular masses of these polypeptides are consistent with those of 3C′ and 3D′ observed in poliovirus-infected cells (38). Addition of 2C inhibited cleavage of 3CD in a dose-dependent manner (lanes 4 to 7), and the generation of both 3C′and 3D′ was almost totally blocked at the highest concentration of 2C (lane 7). As a negative control, we used heat-inactivated 2C, which had lost its ability to block 2Apro activity (lane 8). Also, affinity-purified proteins from bacteria harboring the plasmid without the 2C insert was unable to block both 3Cpro and 2Apro activities (data not shown). Moreover, 2Apro-mediated cleavage of the TATA-binding protein, TBP at the 34th Y-G bond (73) was also inhibited by 2C (data not shown). These results suggest that 2C is capable of inhibiting 2Apro activity in vitro.
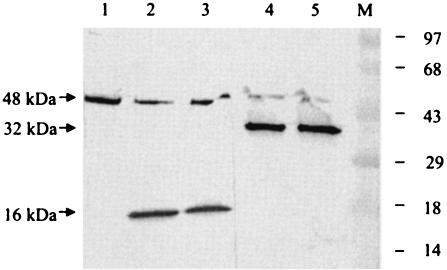
2C does not inhibit cellular enterokinase and thrombin activities.
To determine whether 2C inhibits cellular protease activity, we initially examined nonspecific cleavage of bovine serum albumin by trypsin. We found no inhibitory activity of 2C against trypsin (data not shown). We also examined the effect of 2C on two other cellular proteases: a nonserine protease (enterokinase) and a serine protease (thrombin). In the protease assay, the control peptide (∼48 kDa), provided by the manufacturer (Novagen), is cleaved by enterokinase and thrombin to 16- and 32-kDa fragments, respectively. As can be seen in Fig. 4, neither the enterokinase nor thrombin catalyzed cleavage of the control peptide can be inhibited by the amount of 2C (Fig. 4, lanes 2 and 3 and lanes 4 and 5) sufficient to almost totally inhibit cleavage of CREB (Fig. 2). It should be noted that the same 2C preparation was used for both experiments (shown in Fig. 2 and 4) and that these experiments were performed concurrently. Thus, 2C does not appear to inhibit at least three cellular proteases.
FIG. 4.
Effect of 2C protein on cellular proteases. Two eukaryotic proteases, thrombin, and enterokinase were used in this experiment. The activity of the proteases was monitored by Western blot analysis after cleavage of a control protein of 48 kDa supplied by the manufacturer (Novagen, Inc.) in the presence or absence of 2C polypeptide as detailed in Materials and Methods. Lane 1, control protein plus buffer; lane 2, control protein in the presence of 0.06 U of enterokinase; lane 3, same as lane 2 but also containing 500 ng of purified 2C; lane 4, control protein and 0.005 U of thrombin; lane 5, same as lane 4 but also containing 500 ng of purified 2C. The numbers on the right indicate the positions of the prestained molecular weight markers (lane M). The migrations of the 32- and 16-kDa cleaved products and the full-length protein are marked by arrows.
The protease inhibitory activity is depleted by anti-2C antibody.
The ability of 2C to inhibit 3Cpro-mediated cleavage of CREB was completely blocked by heating the protein at 80°C, suggesting that the protease inhibitory activity associated with 2C was not due to a contaminating small molecule in the preparation (Fig. 3B). To determine whether the 2C polypeptide was indeed responsible for the protease inhibitory activity, the purified 2C preparation was incubated with anti-2C IgG or preimmune IgG. After incubation, protein A-Sepharose was added to the reaction, and the protein A-bound antigen-antibody complex was removed by centrifugation. The remaining supernatants were then assayed for inhibition of 3Cprocatalyzed cleavage of CREB. As can be seen in Fig. 5A, depletion of 2C by anti-2C leads to almost total inhibition of 2C protease inhibitory activity compared to the reaction treated with preimmune IgG (compare lanes 4 and 5 with lane 6). Some nonspecific inhibition of 2C protease inhibitory activity was observed with the preimmune IgG (lane 6) compared to the untreated sample (lane 3). This is due to nonspecific binding of 2C to protein A-Sepharose (data not shown). In this experiment both the 20-kDa proteolyzed product of CREB and the 18-kDa marker were not detected because they ran off the gel.
FIG. 5.
The protease inhibitory activity of 2C is depleted specifically by 2C antibody. (A) A 1.5-μg portion of 2C was incubated with 3 (lane 4) or 4.5 (lane 5) μg of anti-2C IgG or 4.5 μg (lane 6) of preimmune IgG for 2 h at 4°C. The antigen-antibody complex was removed after incubation with protein A-Sepharose, and the supernatants were assayed for 2C-mediated inhibition of 3Cpro activity as described in Materials and Methods. [35S]methionine-labeled CREB was incubated with 2C (lane 1), 3C (lane 2), or both 2C and 3C (lane 3) and analyzed directly. Anti-2C depleted supernatants from reactions containing 3 μg of anti-2C IgG (lane 4), 4.5 μg of anti-2C IgG (lane 5), and 4.5 μg of preimmune IgG (lane 7) were assayed for inhibition of 3Cpro activity. (B) Analysis of physical interaction between 3Cpro and 2C. A fixed amount (200,000 cpm) of in vitro-translated [35S]methionine-labeled 2C was incubated with various amounts of unlabeled, purified His-tagged 3Cpro as described in Materials and Methods. An antibody directed against the His epitope was used to immunoprecipitate the 2C-3C complex. Lane 1, [35S]methionine-labeled 2C protein after immunoprecipitation with anti-2C IgG; lanes 2 to 5, [35S]methionine-labeled 2C was preincubated in the presence of 5 μg of BSA (lane 2) or 1.25 (lane 3), 2.5 (lane 4), or 5 (lane 5) μg of unlabeled His-3Cpro, and the 2C-3C complex was immunoprecipitated with anti-His. [35S]methionine-labeled CAT was preincubated with 5 μg of unlabeled His-3Cpro and immunoprecipitated with anti-His (lane 6) or anti-CAT (lane 7) antibodies. The arrows indicate positions of full-length 2C and CAT, and the numbers to the left correspond to the position and approximate molecular masses (in kilodaltons) of marker proteins.
2C can form a complex with 3Cpro in vitro.
To determine whether 2C physically interacts with 3Cpro to bring about inhibition of 3Cpro activity, in vitro-translated [35S]methionine-labeled 2C was incubated with purified, unlabeled His-tagged 3Cpro, and the resulting complex was immunoprecipitated with anti-His antibody. The ability of anti-His antibody to specifically coimmunoprecipitate labeled 2C would indicate physical association between His-3C and 2C. Lane 1 of Fig. 5B shows analysis of in vitro-translated [35S]methionine-labeled 2C after immunoprecipitation with anti-2C. Immunoprecipitation with anti-His monoclonal antibody of reactions where a constant amount of labeled 2C was incubated with increasing amounts of unlabeled, His-tagged 3Cpro showed coprecipitation of 2C (lanes 3 to 5). In fact, the intensity of the 2C protein immunoprecipitated with anti-His antibody increased almost linearly with increasing concentrations of unlabeled His-3Cpro present during preincubation with 2C (lanes 3, 4, and 5). However, the [35S]methionine-labeled 2C protein preincubated with the equivalent amount of BSA comparable to that of His-3Cpro used in lane 5 could not be immunoprecipitated by anti-His antibody (lane 2). An unrelated protein (35S-labeled CAT), when preincubated with 3Cpro, was not coimmunoprecipitated by anti-His antibody (lane 6), although the labeled CAT protein from the same reaction can be immunoprecipitated by anti-CAT antibody (lane 7). Immunoprecipitation of [35S]methionine-labeled 2C by anti-2C antibody did not reveal any significant difference in the amount of 2C between samples analyzed in lanes 2 to 5 (data not shown). In this experiment, ca. 20% of the labeled 2C was immunoprecipitated by the anti-His antibody at the highest concentration of His-3Cpro (data not shown). These results suggest that the 2C polypeptide is capable of forming a complex with the viral protease 3Cpro in vitro.
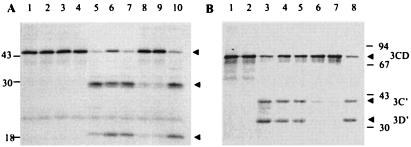
2C deletion mutations affect its ability to inhibit 3Cpro.
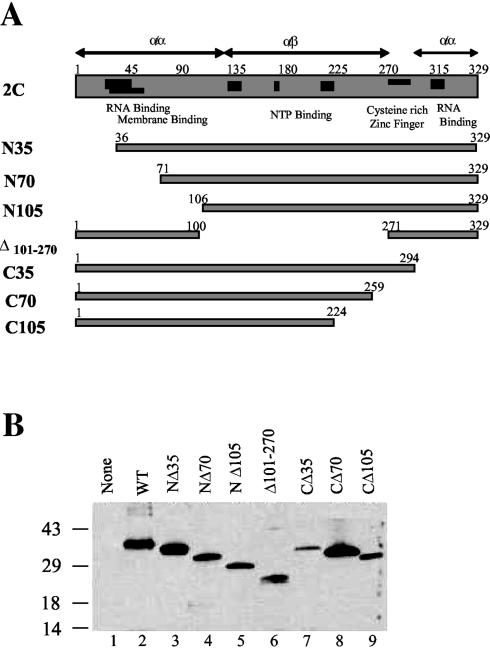
In order to determine which region(s) of 2C might be involved in the inhibition of 3Cpro activity, various deletion mutants of 2C were constructed (Fig. 6A). Both the wt and various mutant 2C polypeptides (V5-tagged) were expressed in bacteria and purified by affinity chromatography (Fig. 6B).
FIG. 6.
Expression and purification of recombinant wt 2C and deletion mutants. (A) Diagram of wt and various 2C deletion mutants; (B) V5-tagged wt 2C or various 2C mutants were expressed in E. coli and purified by affinity chromatography. The figure shows a Western blot of wt 2C (lane 2) and deletion mutants NΔ35 (lane 3), NΔ70 (lane 4), NΔ105 (lane 5), Δ101-270 (lane 6), CΔ35 (lane 7), CΔ70 (lane 8), or CΔ105 (lane 9) with anti-V5 tag antibody. In lane 1, proteins purified thorough an affinity column from bacterial extracts without the 2C plasmid were analyzed.
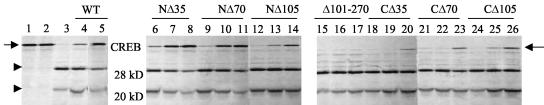
When CREB was incubated with 3Cpro in the presence of increasing amounts of 2C protein, various mutants exhibited distinctively different levels of inhibitory capability (Fig. 7). The mutants NΔ35 (lanes 6 to 8) and NΔ70 (lanes 9 to 11) showed no significant difference in the protease inhibitory ability compared to the wt 2C (lanes 4 and 5). The remaining amino-terminal deletion mutant NΔ105 (lanes 12 to 14) showed ca. 55% inhibitory activity compared to the wt protein (lanes 4 to 5). In contrast, the carboxyl-terminal deletions demonstrated only 25% (CΔ35, lanes 18 to 20), 27% (CΔ70, lanes 21 to 23), and 39% (CΔ105, lanes 24 to 26) of the inhibitory activity of the wt protein. The most dramatic decrease in 3Cpro-inhibitory activity was observed in the mutant containing the central deletion (mutant Δ101-270; lanes 15 to 17). The inhibitory activity for this mutant compared to wt 2C was only 3%. These results suggest that the central domain of 2C, which includes two separate but overlapping serpin motifs (Table 1) is important for 3Cpro-inhibitory activity, although the C-terminal domain also plays a role in protease inhibition.
FIG. 7.
Effect of 2C mutations on its 3C inhibitory activity. First, 2 μg of 3Cpro was preincubated with various amounts of purified wt or mutant 2C polypeptides as indicated. The preincubated proteins were then added to reactions containing [35S]methionine-labeled CREB. Reactions were terminated by the addition of 2× SDS sample buffer (1:1 [vol/vol]), and proteins were resolved on 14% SDS-acrylamide gels. The arrow and arrowheads on the left show the migration of full-length CREB and the proteolyzed CREB products (28 and 20 kDa), respectively. All lanes contain [35S]methionine-labeled CREB, and all lanes except lanes 1 and 2 contain 3Cpro. Lanes 1 and 2, CREB alone and CREB incubated with buffer, respectively; lane 3, same as lane 2 except 3Cpro was added instead of buffer; lanes 4 and 5, same as lane 3 except that 0.25 and 1.0 μg of wt 2C was included; lanes 6, 9, 12, 15, 18, 21, and 24, same as lane 3 except 0.25 μg of various mutant 2C proteins (as indicated at the top) was added; lanes 7, 10, 13, 16, 19, 22, and 25, same as lane 3 except 0.5 μg of various 2C mutants were added; lanes 8, 11, 14, 17, 20, 23, and 26, same as lane 3 except 1.0 μg of various 2C mutants was added.
2C reverses 3Cpro-mediated inhibition of RNA polymerase II-mediated transcription in vitro.
Poliovirus shuts off the host cell transcription catalyzed by all three cellular RNA polymerases (20). We have shown previously that transcription shutoff by poliovirus can be recapitulated in vitro by 3Cpro treatment of uninfected cell extracts and is due to 3Cpro-mediated cleavage of TBP (20, 75). To verify the 3Cpro-inhibitory activity of 2C in an independent assay that does not involve the use of reticulocyte lysates, we tested whether purified 2C would inactivate 3C's ability to shutoff pol II transcription in HeLa cell extracts. Purified 3Cpro was preincubated with purified 2C or equivalent amount of the Δ101-270 2C mutant. These mixtures were then added to HeLa cell extracts, and polymerase II transcription was carried out. As can be seen in Fig. 8, 3Cpro inhibited transcription from the TATA-initiator promoter significantly (compare lane 3 with lane 1). The 3Cpro-mediated inhibition of transcription can be completely restored by wt 2C (lane 4) but not by the Δ101-270 2C mutant (lane 5), which is defective in blocking 3Cpro activity. The mutant protein alone did not alter transcription significantly compared to the control (lanes 1 and 6). These results are consistent with our previous finding that TBP cleavage by 3Cpro is inhibited by 2C in reticulocyte lysates (data not shown).
FIG. 8.
Reversal of 3Cpro-mediated transcription inhibition by 2C in vitro in HeLa cell extract. In vitro RNA polymerase II-catalyzed transcription was performed in HeLa nuclear extracts as described in Materials and Methods. RNA polymerase II transcription was carried out in the presence of 100 μg of HeLa nuclear extract plus 2C buffer (lane 1), 2.5 μg of 2C (lane 2), 1 μg of 3Cpro (lane 3), 2.5 μg of wt 2C and 1 μg of 3Cpro (lane 4), 2.5 μg of mutant (Δ101-270) 2C and 1 μg of 3Cpro (lane 5), or 2.5 μg of mutant (Δ101-270) 2C (lane 6). The arrow indicates the appropriate transcript.
2C expression in HeLa cells affects proteolytic processing of poliovirus precursor polypeptides.
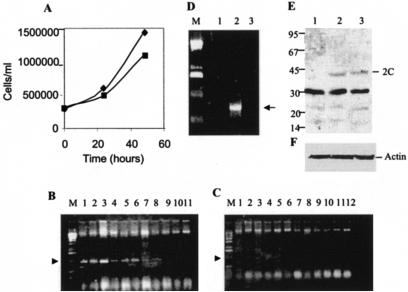
To determine whether the 2C polypeptide is capable of regulating proteolytic processing of poliovirus precursor polypeptides in vivo, we wished to make a cell line that expresses the 2C protein. After repeated attempts we were unable to obtain any cells that constitutively express 2C. We, therefore, used the Tet On/Off system to make a cell line, which expresses 2C in an inducible manner. Stable cell lines were selected by using G418 and hygromycin B. 2C is expressed in these cell lines in the presence of doxycycline (1 to 3 μg/ml). Genomic DNA PCR assay confirmed the presence of the 2C DNA sequence in the 2C cell lines but not in the control cell line (Fig. 9B and C). Clonally selected cell lines that contain 2C DNA (Fig. 9B, lane 2) and vector alone (Fig. 9C, lane 9) were selected for further studies. RT-PCR of total RNA from doxycycline-treated cells confirmed the presence of 2C RNA only in the 2C expressing cell line but not in the vector alone cells or in normal HeLa cells (Fig. 9D, lanes 1 to 3). The 2C protein could be detected in the cell line (called E2) but not in the vector control line (called 9E) by Western analysis with a polyclonal antibody to 2C (Fig. 9E, lanes 1 to 3). The level of the 2C protein in E2 cells was relatively low. The cells that expressed higher levels of 2C after induction grew poorly, suggesting that higher levels of 2C expression was detrimental to cell growth (data not shown). However, the E2 line expressing relatively low levels of the 2C protein grew efficiently, and the growth rate of these cells was comparable to that of the control cell line (Fig. 9A). We also did not find any significant difference in the overall protein synthesis between the 2C and vector control cells (data not shown). A nonspecific protein, which migrated slightly above the 30-kDa marker, was found to cross-react with the 2C antibody and was present in both cell lines with or without the 2C insert. In addition, small amounts of two other cellular polypeptides of 23 and 18 kDa were also found to cross-react with the 2C antibody nonspecifically.
FIG. 9.
Inducible expression of 2C in HeLa cells. A HeLa cell line that expresses 2C when induced by doxycycline was prepared as described in Materials and Methods. (A) Rate of cell growth for the 2C cell line (E2 [▪]) versus that of the control line with vector alone (9E [♦]) for up to 48 h. (B) Genomic DNA was used to detect 2C sequence from various antibiotic-resistant colonies transformed with a vector containing the 2C sequence by PCR with appropriate primers. The arrowhead indicates the position of the appropriate PCR product. (C) Genomic DNA from various colonies transformed with the vector alone was analyzed by PCR. (D) Total RNA isolated from E2 (lane 2) and 9E (lane 3) cells after 10 h of induction with doxycycline and from HeLa cells (lane 1) was analyzed to detect 2C RNA with appropriate primers. The arrowhead indicates the position of the RT-PCR product. (E) Fifty micrograms of total protein from the E2 cell line (lanes 2 and 3, duplicate) or 9E (lane 1) were used for Western blot with a polyclonal antibody to 2C.
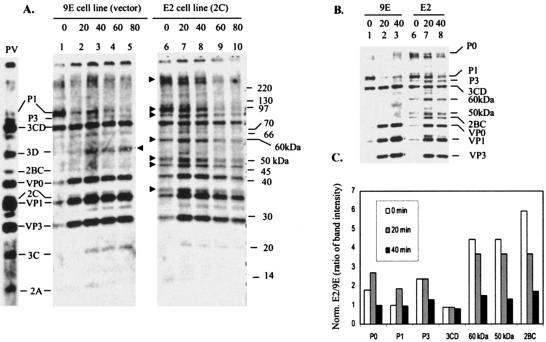
To examine whether inducible expression of 2C has any effect on proteolytic processing of viral proteins, the 2C cell line (E2) and vector control cell line (9E) were grown overnight (10 h) in the presence of doxycycline (2 μg/ml) and then infected with poliovirus. The infection was allowed to continue for 3 h. At the end of this time, 5 μg of actinomycin D/ml was added to the cells. After 15 min, the cells were pulsed with [35S]methionine for 10 min, followed by a chase with 100 mM cold methionine. Samples of approximately equal number of cells were obtained every 20 min after the chase and analyzed by SDS-PAGE. As can be seen in Fig. 10, the processing of the P1 precursor was relatively fast and essentially complete by 40 min in vector control cells (lanes 1 to 3). This processing event generated VP0, VP1, and VP3. In these cells, very little P3 precursor was detected at zero time of chase (lane 1); however, relatively high amounts of 3CD were present at zero time, indicating that the cleavage at the 3B-3C junction occurs relatively quickly. The 3C and 3D polypeptides were detectable after 20 to 40 min of chase (lanes 2 and 3), and the intensity of these bands increased slightly after 40 to 80 min of chase (lanes 3 to 5). Although P1 processing appeared to be normal with the appearance of VP0, VP1, and VP3 in the 2C-expressing cells, there was a significant difference in the processing patterns of the 2C and vector control cells (lanes 6 to 10). First, after 10 min of labeling (no chase), the intensity of viral precursor proteins, such as the P1, P3, 2BC, 60-kDa, and 50-kDa (just above 2BC) proteins and the large polyprotein (P0) migrating more slowly than the 220-kDa marker, was higher than in vector control cells (compare lane 6 with lane 1). Also, the intensity of the P3 precursor increased significantly after 20 min chase compared to that at zero time, suggesting delayed processing of the P0 precursor in the infected E2 cells (lanes 6 and 7, Fig. 10). This increase in P3 can be correlated with the decrease in the intensity of the P0 precursor. In general, the levels of VP0, VP1, VP3, 3Dpol, and 3Cpro were lower in 2C-expressing cells than in control cells lacking the 2C insert. In particular, there was a striking difference in the amount of 3Dpol between the E2 and 9E cells (compare lanes 3 to 5 with lanes 8 to 10). Significant amounts of viral precursors remained even after 40 min of chase in E2 cells, which could be seen clearly in a lower exposure of the gel (compare lanes 7 and 8 with lanes 2 and 3, Fig. 10B). The overall level of the 2C polypeptide was higher in E2 cells than in 9E cells. This was true even at the zero time of chase (compare lanes 1 and 6, Fig. 10A), which could be due to residual translation from the 2C mRNA expressed in these cells from the vector encoding the 2C sequence. This is possibly due to incomplete shutoff of existing mRNAs at 3 h postinfection. The possibility that higher amounts of 2C is made from viral precursors in E2 cells seems unlikely since a significant amount 2BC, a precursor of 2C, is found accumulated in E2 cells (Fig. 10A, lane 7). However, generation of 2C from a larger precursor, e.g., 2BC-3ABCD, cannot be ruled out.
FIG. 10.
Proteolytic processing of viral precursor polypeptides in stable HeLa cell line that express 2C in an inducible fashion. (A) Pulse-chase analysis of protein processing in poliovirus-infected HeLa cells expressing the 2C polypeptide. HeLa cells lines 9E (with vector alone, lanes 1 to 5) and E2 (2C expressing cells, lanes 6 to 10) were infected at a multiplicity of infection of 10 with type 1 poliovirus as described in Materials and Methods. Cells were labeled with [35S]methionine at 3 h postinfection. The numbers above the lanes indicate times in minutes after the chase with unlabeled methionine. The lane marked PV shows the migration of poliovirus proteins from HeLa cells infected with poliovirus for 5 h. The migration of molecular markers is indicated on the right (in kilodaltons). The arrowheads pointing right indicate precursor and mature viral proteins that are either present exclusively or in higher amounts in the E2 cell line compared to the control 9E cell line. The arrowhead pointing left indicates the position of migration of 3D. (B) A lower exposure of lanes 1 to 3 and 6 to 8 from Fig. 10A is shown. (C) Quantification of viral precursor proteins from E2 and 9E cells. The viral precursor polypeptides P0, P1, P3, 3CD, 2BC, 60 kDa, and 50 kDa were quantified by densitometric scanning as described in Materials and Methods. The numerical values were normalized with respect to total cellular proteins from virus-infected E2 and 9E cells. The ratio of the numerical values for E2 to 9E cells for each polypeptide at 0, 20, and 40 min of chase are shown.
To quantify the difference in the processing of viral precursor polypeptides between E2 and 9E cells, the intensities of individual polypeptides were measured and expressed as E2 to 9E ratio for each polypeptide after normalizing for total protein in infected extracts. After a 20-min chase, the 2BC, 50-kDa, and 60-kDa polypeptides showed the highest level of difference with values close to 3.7-fold between E2 and 9E cells (Fig. 10C). The levels of P0 and P3 were >2-fold in E2 cells compared to 9E cells, whereas the P1 level was ∼1.8-fold higher in E2 cells. The 3CD ratio did not change significantly over the period of 40-min chase, although the ratio was <1. These results suggest that viral protein processing is delayed or inhibited in the 2C-expressing cells compared to cells containing the vector alone. Total incorporation of labeled methionine into virus-specific proteins, as determined by densitometric scanning of individual viral proteins, was not significantly different between the 9E and E2 cells (data not shown). This suggests that synthesis of the viral polyprotein during the pulse was not significantly affected by 2C expression. Despite the accumulation of a number of viral precursor polypeptides and a reduced level of mature proteins, the virus titer in E2 cells was reduced only by ∼2-fold compared to the 9E cells (data not shown). This is most probably due to the fact that poliovirus proteins are synthesized in far excess than required for efficient viral RNA replication and packaging. These results suggest that processing of viral precursor proteins is significantly affected in HeLa cells that express 2C.
DISCUSSION
Both genetic and biochemical studies implicate poliovirus 2C polypeptide in a number of key processes during viral infection. These processes include the initiation and elongation of viral RNA synthesis, membrane proliferation, and vesicle induction, and possibly virion assembly. The 2C polypeptide also contains ATPase and GTPase activities and is capable of binding cellular membranes and viral RNA. Here we describe a new activity associated with the 2C polypeptide, namely, its ability to negatively regulate viral protease 3Cpro.
Various lines of evidence strongly suggest that 2C indeed contains protease inhibitory activity. Initial computer analysis of the 2C amino acid sequence identified domains of considerable homology to protease inhibitors with Z (local alignment) scores ranging from 22 to 68 for various serpin motifs scattered throughout the 2C protein (Table 1). Experimental evidence included the demonstration that bacterially expressed, affinity purified 2C protein was capable of inhibiting both viral proteases 3Cpro and 2Apro. An E. coli extract derived from cells harboring the plasmid lacking the 2C insert did not have the protease inhibitory activity. The protease inhibitory activity was heat labile, suggesting that it was not a small molecule or that the inhibition was due to a component of the buffer. An antibody directed against the 2C protein depleted the protease inhibitory activity associated with 2C. Deletion analyses showed that the central as well as C-terminal amino acid sequences were important for 2C's ability to inhibit 3Cpro activity. Finally, poliovirus infection of HeLa cells that express 2C in an inducible fashion resulted in accumulation of a number of precursor polypeptides compared to those containing the vector alone in a pulse-chase experiment, suggesting a role of the 2C polypeptide in the regulation of poliovirus protein processing. Although the in vivo results implicate 2C in the regulation of viral protein processing, we cannot rule out the possibility that cellular changes induced by 2C could also contribute to the incomplete protein processing observed in 2C-expressing cells. The in vitro results, on the other hand, strongly suggest a role of the 2C protein in the regulation of viral proteases.
Our choice of using transcription factors as 3Cpro substrates for the detection of 2C protease inhibitory activity in vitro was based on the availability of these substrates in our laboratory and on the fact that the 3Cpro cleavage specificity was found to be similar, if not identical, for both viral precursors and transcription factors (20). Both an accessible Q-G pair and an aliphatic amino acid at position P4 were shown to be required for the efficient cleavage of TBP and CREB by 3Cpro, which closely parallels the requirement for cleavage of viral precursors by this protease (reference 20 and data not shown). Thus, although CREB was used as a substrate in the in vitro protease assay, we anticipated similar results with viral precursor proteins. Indeed, our in vivo data of viral protein processing in 2C-expressing cells are clearly consistent with the idea that 2C is capable of regulating viral protein processing by 3Cpro in poliovirus-infected cells.
The experiment demonstrating 2C's ability to block 2Apro activity utilized 3CD as the substrate. Since our immunoprecipitation experiment suggested a physical interaction between 2C and 3Cpro (Fig. 5), the possibility that 2C-mediated inhibition of 3CD cleavage (by 2Apro) could be due to 3CD-2C interaction rather than through inhibition of 2Apro could not be ruled out. This is highly unlikely, however, since this experiment was performed under conditions that did not favor 3CD (or 3C)-2C interaction. First, 2Apro was preincubated with highly concentrated 2C for 2 h in a small volume. The [35S]methionine-labeled 3CD substrate was then added to these reactions so that 2C is diluted at least 15 to 20 times in the final reaction. Under these conditions, the concentration of 2C in the final reaction (∼13 ng/ml) was not sufficient to inhibit 3Cpro (and possibly 3CD) activity. Moreover, 2Apro-mediated cleavage of TBP at the 34th tyrosine-glycine site (19, 71) was also inhibited by 2C (data not shown).
The results presented in Fig. 10 strongly suggest a role of protein 2C in the regulation of proteolytic processing of viral precursor polypeptides. The processing of viral precursors was significantly slower in 2C-expressing cells than in those containing the vector without the 2C insert. The processing defect was evidenced by accumulation of significant amounts of P0, P1, P3, and 2BC. Increased accumulation of these precursors was accompanied by decreased production of mature polypeptides VP0, VP1, VP2, 3D, and 3C in E2 cells compared to 9E cells. We also detected two viral proteins with approximate molecular masses of 60 and 50 kDa that accumulated in significant amounts in E2 cells. The 60-kDa polypeptide appears to be a 3D-related protein, as it can be immunoprecipitated by an anti-3D antibody (data not shown). A similar protein of ∼60 kDa (referred to as protein 4a) was described earlier as an unstable precursor to 3D in the membrane-associated replication complex (25). Interestingly, accumulation of a very similar 3D-related 60-kDa viral protein was detected in cells infected with a mutant poliovirus containing a point mutation (Val to Ala) at position 54 of 3Cpro (22). The 3C mutant virus was deficient in the production of 3Dpol and showed a proteolytic defect leading to accumulation of the 60-kDa polypeptide. Despite this processing defect, however, the mutant virus grew to near-wt titers (22). The processing defect imparted either by 2C expression or mutation in the 3C protein may be explained by structural changes in the protease as a result of mutation or interaction with the 2C polypeptide. However, in E2 cells the effect on viral protein processing was more profound, as evidenced by the accumulation of number of precursors rather than just the 60-kDa polypeptide. The nature of the 50-kDa polypeptide detected in E2 cells is not known at present, and its identification will require further investigation.
The results presented in Fig. 10 also suggest that proteolytic processing of P2 and P3 precursors were more affected than that of P1 in 2C-expressing cells. Although these results may suggest differential regulation of 3C versus 3CD proteolytic activity by the 2C polypeptide, these results must be considered tentative until confirmed by direct analysis of the effect of 2C in proteolytic processing of the P1 precursor by the recombinant 3CD protein. In addition, it would be interesting to determine whether 2C is capable of interacting directly with 3CD.
The protease inhibitory activity associated with the 2C protein appears to be specific with respect to viral proteases. Although both poliovirus 3Cpro and 2Apro activities were inhibited by 2C in a dose-dependent manner in vitro, three different cellular proteases—thrombin, enterokinase, and trypsin—were not inhibited by 2C (Fig. 4 and data not shown). These observations, along with the reduced processing of viral precursors in E2 cells, raise the possibility that 2C may be involved in modulating viral protease activities in viral RNA replication The most likely target for this function would be the precursor 3CDpro (or 3Cpro). The role of 3CDpro as the major protease in the processing of both structural and nonstructural precursors is becoming increasingly apparent (48). Moreover, 3Cpro-related proteolysis could be stimulated by the genome-linked protein (VPg) and its precursor 3AB in vitro (44). The precursor polypeptides 3CDpro and 3AB bind to the 5′ positive strand cloverleaf structure along with the cellular protein, PCBP2 (4, 47, 72). We have shown that 2C (and 2BC) polypeptides interact with the 3′-terminal sequences in the minus-strand RNA, which is complementary to the plus-strand 5′ cloverleaf (7). The physical proximity of 2C to both 3CD and 3AB in the replication complex may provide 2C with the opportunity to control the proteolysis of 3AB into 3A and VPg by 3CDpro (or 3Cpro) (36). Alternatively, 2C could interact with free 3CDpro (or 3Cpro) in the cytoplasm. Recent evidence also suggests that the 3Dpol-catalyzed uridylylation of VPg is stimulated by 3CD (51). Thus, prolonging the half-lives of both 3AB and 3CD by 2C/2BC may help initiation of RNA synthesis (possibly through 3AB uridylylation).
Although prior expression of 2C in HeLa cells results in inefficient processing of viral precursors during viral infection, how 2C exerts its protease inhibitory function in polyprotein processing during a normal infection is not known. The magnitude of 2C-induced inhibition of processing observed in vivo was much lower than that seen in vitro. In fact, the virus titer in E2 cells was reduced only twofold compared to the 9E cells. The regulation of viral proteases by 2C in infected cells must be precise and presumably temporal to ensure proper processing of viral precursor polypeptides, thereby ensuring efficient viral replication. The 2C/2BC polypeptides interact with intracellular membranes, virus-specific RNAs and possibly other viral and/or cellular polypeptides. Thus, the availability of free 2C (or 2BC) in poliovirus-infected cells and when and how they interact with viral proteases or their precursors will be important factors in the regulation of viral proteases.
Studies utilizing deletion mutants of 2C revealed that several regions of the protein might play a role in the regulation of 3Cpro activity (Fig. 10). This observation is consistent with the fact that many known serine protease inhibitors are characterized by having several repeats of serpin motifs (76). A deletion of the central region of the 2C protein (mutant Δ101-270) demonstrated the most dramatic loss of 3Cpro inhibitory activity. This region includes the NTP-binding domain (42, 58). The central region also includes two separate but overlapping serpin motifs (Table 1), which could explain the almost total loss of protease inhibitory activity of the Δ101-270 mutant. Alternatively, the overall structure of the protein may have been be altered by this fairly large deletion. The deletion of the C-terminal region also results in significantly decreased protease inhibitory activity of 2C. This can be seen in all three C-terminal deletion mutants including the C-terminal Δ35 mutant. This region of 2C containing a putative alpha helix motif, as well as an arginine-rich RNA-binding motif (49, 57), also contains a 54-amino-acid serpin motif (Table 1). Future studies involving more defined mutations should shed more light on the protease inhibitory activity of the 2C serpin motifs.
How does 2C negatively regulate 3Cpro activity? We believe that the protease inhibition occurs through physical interaction of 2C with 3Cpro. Indeed, results presented here (Fig. 8) indicate physical association between the two molecules. It is interesting to point out in this context that serpins generally act by binding to the active site of the protease. This binding is characterized by having a slow kinetics. The binding of serpin results in the formation of an SDS-stable protease-serpin covalent complex that eventually leads to cleavage of the serpin itself. However, another class of serpins bind but do not form a covalent complex with the protease. There is also evidence that binding of some serpins to the target protease leads to distortion of the active site of the protease and consequent loss of its catalytic activity (55). Further studies utilizing the full-length protein, as well as individual serpin domains, will be required to delineate the mechanism by which 2C inhibits 3Cpro activity.
The best-studied viral serpin is crmA, a poxvirus protein, that inhibits both cysteine and serine proteases involved in the regulation of host inflammatory and apoptotic processes (35). Although not classified as a serpin, the antiapoptotic protein, p35, of baculovirus has been shown to inhibit cellular caspases (17). Interestingly, only specific caspases such as caspase-1, -3, -6, -7, and -8 were inhibited by p35; 12 unrelated serine or cysteine proteases were not affected (77). Although three cellular proteases tested (thrombin, enterokinase, and trypsin) were not inhibited by 2C, it remains to be seen whether 2C is able to block other cellular proteases involved in apoptosis and inflammatory response. Cells infected with poliovirus undergo many changes, including both induction and inhibition of apoptosis (1, 68). Inducible expression of 3Cpro and 2Apro leads to apoptotic cell death (9, 27). It will be interesting to determine whether 2C plays any role in the inhibition of apoptosis in poliovirus-infected cells.
In summary, we report a previously unknown activity associated with the poliovirus-encoded 2C polypeptide. We believe that the protease inhibitory activity of 2C (or 2BC) plays a key role in viral replication and pathogenesis of this important group of human pathogens. Future studies will elucidate the exact sequences or domains of the 2C protein required for the protease inhibitory activity and will include molecular genetic analyses to probe the role of these regions in the virus life cycle.
Acknowledgments
This study was supported by the NIH grant AI27451.
We thank members of the Dasgupta laboratory for helpful suggestions.
REFERENCES
- 1.Agol, V. I., G. A. Belov, K. Bienz, M. S. Egger, M. S. Kolesnikova, L. I. Romanova, L. V. Sladkova, and E. A. Tolskaya. 2000. Competing death programs in poliovirus-infected cells: commitment switch in the middle of the infectious cycle. J. Virol. 74:5534-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of the poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 4.Andino, R., G. E. Reickhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argos, P., G., M. Kamer, M. Nicklin, and E. Wimmer. 1984. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant coronavirus suggest common ancestry of these virus families. Nucleic Acids Res. 12:7251-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch, A, and R. Apweiler. 1998. The SWISS-PROT protein sequence data bank and its new supplement, TREMBL. Nucleic Acids Res. 24:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus encoded 2C polypeptide specifically binds to the 3′-terminal sequence of viral negative strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′-terminus negative-strand cloverleaf requires an intact stem-loop B. Virology 280:41-51. [DOI] [PubMed] [Google Scholar]
- 9.Barco, A., E. Feduchi, and L. Carrasco. 2000. Poliovirus protease 3Cpro kills cells by apoptosis. Virology 266:352-360. [DOI] [PubMed] [Google Scholar]
- 10.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VP-g-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazan, J. F., and R. J. Flettenick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein, H. D., N. Sonenberg, and D. Baltimore. 1985. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol. Cell. Biol. 5:2913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 15.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassens, U., G. Lewinski, A. K. Samraj, H. Von Bernuth, H. Baust, K. Khazaie, and M. Los. 2003. Viral modulation of cell death by inhibition of caspases. Arch. Immunol. Ther. Exp. 51:19-27. [PubMed] [Google Scholar]
- 18.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 19.Clark, M. E., M. Lieberman, A. J. Berk, and A. Dasgupta. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das, S., and A. Dasgupta. 1993. Identification of cleavage site and determinants required for poliovirus 3Cpro-catalyzed cleavage of human TATA-binding transcription factor TBP. J. Virol. 67:3226-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta, A., et al. 2002. Effects of picornavirus proteinases on host cell transcription, p. 321-335. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 22.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 61:2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echeverri, A., R. Banerjee, and A. Dasgupta. 1998. Amino-terminal region of poliovirus 2C is sufficient for membrane binding. Virus Res. 54:217-223. [DOI] [PubMed] [Google Scholar]
- 24.Echeverri, A., and A. Dasgupta. 1995. Amino-terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 25.Etchison, D., and E. Ehrenfeld. 1980. Viral polypeptides associated with the RNA replication complex in poliovirus-infected cells. Virology 107:135-142. [DOI] [PubMed] [Google Scholar]
- 26.Gettins, P. G. 2002. Serpin structure, mechanism, and function. Chem. Rev. 102:4751-4804. [DOI] [PubMed] [Google Scholar]
- 27.Goldstaub, D., A. Gradi, Z. Bercovitch, Z. Grossman, Y. Nophar, S. Luris, N. Sonenberg, and Kahana, C. 2000. Poliovirus 2A protease induces apoptotic cell death. Mol. Cell. Biol. 20:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbalenya, A. E., V. M. Blinov, A. P. Donchenko. 1986. Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families. FEBS Lett. 194:253-257. [DOI] [PubMed] [Google Scholar]
- 29.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human translation initiation factor eIF4GII but not eIF4GI coincides with the shut-off of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grubman, M. J., M, Zellner, G. Bablanian, P. W. Mason, and M. E. Piccone. 1995. Identification of the active site residues of the 3C proteinase of foot-and-mouth disease virus. Virology 213:581-589. [DOI] [PubMed] [Google Scholar]
- 31.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 32.Ivanoff, L. A., T. Towatari, J. Ray, B. D. Korant, and S. R. Petteway, Jr. 1986. Proc. Natl. Acad. Sci. USA 83:5392-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kean, K. M., N. L. Teterina, D. Marc, and M. Girard. 1991. Analysis of putative active site residues of the poliovirus 3C protease. Virology 181:609-619. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Domer, E. A. Emini, R. Hanecak, J. J. Lee, S. Van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization, and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 35.Komiyama, T., L. T. Quan, and G. S. Salvesen. 1996. Inhibition of cysteine and serine proteases by the cowpox virus CRMA. Adv. EXP. Med. Biol. 389:173-176. [DOI] [PubMed] [Google Scholar]
- 36.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as cofactor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 37.Lawson, M. A., and B. L. Semler. 1991. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C.-K., and E. Wimmer. 1988. Proteolytic processing of poliovirus polyprotein: elimination of 2Apro-mediated alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology 166:405-414. [DOI] [PubMed] [Google Scholar]
- 39.Leong, L. E.-C., C. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 40.Li, J. P., and D. Baltimore. 1990. An intragenic revertant of poliovirus 2C mutant has an uncoating defect. J. Virol. 64:1102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloyd, R. E., M. J. Grubman, and E. Ehrenfeld. 1988. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J. Virol. 62:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirzayan, C., and E. Wimmer. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199:176-187. [DOI] [PubMed] [Google Scholar]
- 43.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 44.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg-and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 45.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. James. 1997. Refined X-ray chrystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 46.Pallansch, M. A., O. M. Kew, B. L. Semler, D. R. Omilianowski, C. W. Anderson, E. Wimmer, and R. R. Ruckert. 1984. Protein processing map of poliovirus. J. Virol. 49:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′ terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 48.Parsley, T. B., C. T. Cornell, C. T., and B. L. Semler. 1999. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J. Biol. Chem. 274:12867-12876. [DOI] [PubMed] [Google Scholar]
- 49.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N terminus of poliovirus protein 2C. Virology 199:188-199. [DOI] [PubMed] [Google Scholar]
- 50.Paul, A. V., J. H. Van Boom, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 51.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul, A.V. 2002. Possible unifying mechanism in picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 53.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2CATPase is involved in RNA replication and binding zinc in vitro. J. Virol. 74:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 55.Plotnick, M. I., L. Mayne, N. M. Schechter, and H. Rubin. 1996. Distortion of the active site of chymotrypsin complexed with a serpin. Biochemistry 35:7586-7590. [DOI] [PubMed] [Google Scholar]
- 56.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214:916-919. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 59.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubinstein, S. J., T. Hammerle, E. Wimmer, and A. Dasgupta. 1992. Infection of HeLa cells with poliovirus results in modification of a complex that binds to the rRNA promoter. J. Virol. 66:3062-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen, Y., M. Igo, P. Yalamanchili, A. J. Berk, and A. Dasgupta. 1996. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol. 16:4163-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sidman, K. D., W. George, B. Banker, and L. Hunt. 1988. The protein identification resource (PIR). Nucleic Acids Res. 16:869-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74: 8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 8:1977-1986. [DOI] [PubMed] [Google Scholar]
- 68.Tolskaya, E. A., L. I. Romanova, M. S. Kolensikova, T. A., Ivannikova, E. A., Smirnova, N. T. Raikhlin, and V. I. Agol. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 69:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weidman, M. K., P. Yalamanchili, B. Ng, W. Tsai, and A. Dasgupta. 2001. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology 291:260-271. [DOI] [PubMed] [Google Scholar]
- 71.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 72.Xiang, W., K. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′ terminal cloverleaf and 3AB/3CD of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yalamanchili, P., R. Banerjee, and A. Dasgupta. 1997. Poliovirus-encoded protease 2Apro cleaves the TATA-binding protein but does not inhibit RNA polymerase II transcription in vitro. J. Virol. 71:6881-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:120-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yalamanchili, P., K. Harris, E. Wimmer, and A. Dasgupta. 1996. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J. Virol. 70:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye, S., and E. J. Goldsmith. 2001. Serpins and other covalent protease inhibitors. Curr. Opin. Struct. Biol. 11:740-745. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, Q., J. F. Krebs, S. J. Snipas, A. Price, E. S. Alnemri, K. J. Tomaselli, and G. S. Salvesen. 1998. Interaction of the baculovirus antiapoptotic protein p35 with caspases: specificity, kinetics, and characterization of the caspase/p35 complex. Biochemistry 37:10757-10765. [DOI] [PubMed] [Google Scholar]