Abstract
It is well established that lactate secreted by fermenting cells can be oxidized or used as a gluconeogenic substrate by other cells and tissues. Within the fermenting cell itself, however, it is generally assumed that lactate is produced to replenish NAD+ and then is secreted. Here we explored the possibility that cytosolic lactate is metabolized by the mitochondria of fermenting mammalian cells. We found that fermenting HeLa and H460 cells utilize exogenous lactate carbon to synthesize a large percentage of their lipids. With high-resolution mass spectrometry, we found that both 13C and 2-2H labels from enriched lactate enter the mitochondria. The lactate dehydrogenase (LDH) inhibitor oxamate decreased respiration of isolated mitochondria incubated in lactate, but not isolated mitochondria incubated in pyruvate. Additionally, transmission electron microscopy (TEM) showed that LDHB localizes to the mitochondria. Taken together, our results demonstrate a link between lactate metabolism and the mitochondria of fermenting mammalian cells.
For every mole of glucose metabolized through glycolysis, the enzyme glyceraldehyde 3-phosphate (G3P) dehydrogenase reduces two moles of NAD+ to NADH. In most non-proliferating cells, these NADH are ultimately oxidized back to NAD+ via the mitochondrial electron transport chain when oxygen levels are sufficient. If dehydrogenation of G3P outstrips the rate at which aerobic respiration and/or electron shuttles can regenerate NAD+, however, cells must use an alternative route to oxidize NADH. In mammalian cells, this route is provided by lactic acid fermentation, in which lactate dehydrogenase (LDH) oxidizes NADH back to NAD+ while simultaneously reducing the glycolytic end-product pyruvate to lactate1.
Although the lactate produced during fermentation is sometimes referred to as a “metabolic waste product”, productive utilization of lactate is well established. In the 1920’s, it was demonstrated that lactate produced by active muscle is transported to the liver for conversion back to glucose via gluconeogenesis Cori cycling1,2. Later, it was shown that skeletal muscle-derived lactate is also transported to the heart for oxidation as a fuel source3. These processes of shuttling lactate integrate metabolic pathways at the tissue level, but similar phenomena have also recently been posited to take place at a smaller scale. The two best known examples of the latter are the astrocyte-neuron lactate shuttle and the reverse-Warburg effect4,5. Both of these models, which remain controversial, hypothesize that lactate secreted by fermenting cells is oxidized by neighboring cells for energy.
By applying high-resolution metabolomic technologies and stable isotope labeling, here we investigated whether lactate can be used for energy in the fermenting cell itself. When lactate is used as an energy source, lactate carbon is ultimately inserted into the tricarboxylic acid (TCA) cycle in the mitochondria. It is generally assumed that lactate is first oxidized to pyruvate in the cytosol prior to entering the mitochondria1,6. However, cytosolic oxidation of lactate to pyruvate does not support the main objective of fermentation (i.e., to recycle NADH to NAD+ in the cytosol). Here we consider the alternative possibility that lactate is oxidized to pyruvate in the mitochondria7,8. Specifically, we consider oxidation of lactate in the intermembrane space of the mitochondria and/or in the mitochondrial matrix.
Results
Solid-State NMR Shows Lactate is Metabolized to Lipids
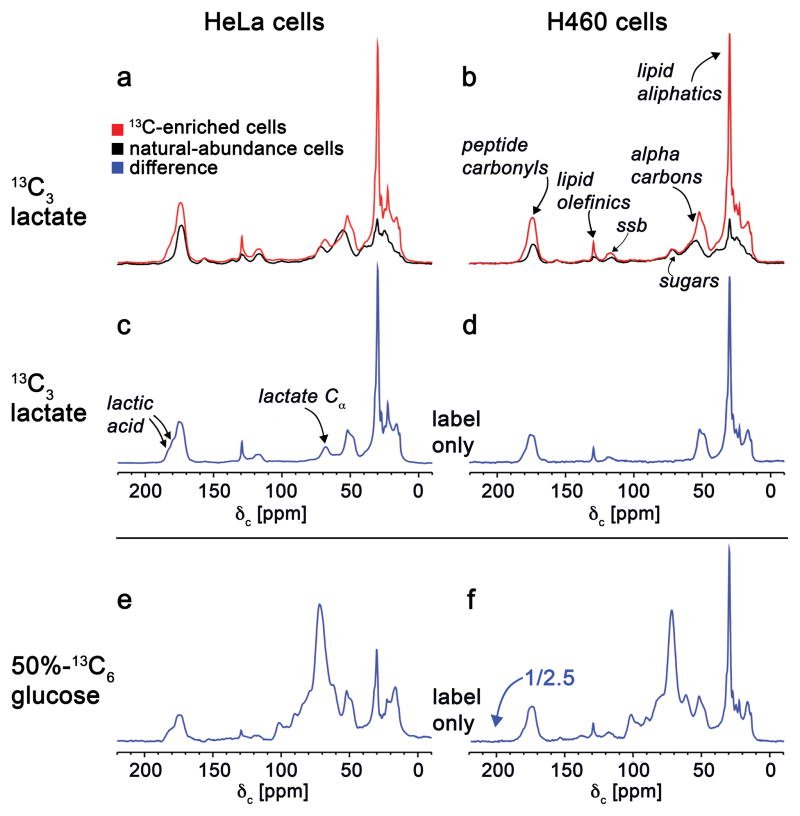
To examine lactate utilization in rapidly proliferating and fermenting cells, we cultured HeLa cells and H460 lung cancer cells in media supplemented with 3 mM 13C3-lactate for 48 hours. Afterwards, we lyophilized the cells and analyzed the whole-cell pellets by solid-state NMR. A strength of this approach is its ability to simultaneously track 13C labels in small molecules as well as macromolecules (e.g., proteins and DNA). To account for natural-abundance contributions to the NMR spectra, we prepared parallel samples under identical conditions but with natural-abundance lactate. The cross-polarization magic angle spinning (CPMAS) spectra from each experiment are shown in Fig. 1a–b. The difference between the natural-abundance and 13C-labeled spectra represents the fate of 13C3-lactate carbons (Fig. 1c–d). These data show that a major fate of lactate carbon is lipids.
Figure 1.
125-MHz cross-polarization magic-angle spinning 13C NMR spectra of intact HeLa cells (left column) and H460 lung-cancer cells (right column). The full spectra obtained from cells labeled for 48 hours with 3 mM 99%-13C3-lactate (red, panels a and b) are compared to the corresponding spectra of unlabeled cells (black, panels a and b). The label-only difference spectra are in blue. The label-only difference spectrum shown in panel c resulted from subtracting the black and red spectra shown in panel A. Similarly, the label-only difference spectrum shown in panel d resulted from subtracting the black and red spectra shown in panel b. Label-only difference spectra were also obtained from cells labeled with 24 mM 50%-13C6-glucose (panels e and f, the latter scaled down by a factor of 2.5). The glucose labeling resulted in secreted lactate with four times the 13C concentration of the exogenous lactate labeling of panels c and d (see Supplementary Fig. 1).
Given that the major source of lactate in vivo is glucose, we cultured cells in uniformly 13C-labeled glucose (13C6-glucose) to compare the fates of glucose and lactate carbons. We determined that the extracellular concentration of 13C3-lactate reaches ~10 mM after 48 hours from cells cultured in 12 mM 13C6-glucose (Supplementary Results, Supplementary Fig. 1). Strikingly, on a per mole of 13C basis, labeled lactate produces more 13C-enriched lipids than labeled glucose (Fig. 1e–f). This result is due in part to the number of competing pathways that utilize glucose1. We have shown previously that 24% of glucose carbon has fates other than lactate in HeLa cells9. A liquid chromatography/mass spectrometry (LC/MS)-based comparison of 13C6-glucose fates to 13C3-lactate fates reveals that, as expected, glucose carbon is routed to pathways that lactate carbon is not, such as the pentose phosphate pathway (Supplementary Fig. 2, Supplementary Tables 1–2). These data suggest that no gluconeogenesis (i.e., conversion of lactate to glucose) occurs in HeLa and H460 cells under the experimental conditions here.
We next turned our attention to quantitating the distribution of 13C fates in biomass by solid-state NMR. First, we quantified the percentage of 13C-labeled lactate that produces 13C-labeled lipids. This can be determined by comparing integrated areas from the difference spectra shown in Fig. 1c–d. The calculation shows that most of the labeled lactate produces labeled protein and labeled lipids, with labeled lipids representing ~ 50% of the total label. Next, we examined the percentage of the total lipid pool that is derived from lactate (via any metabolic route). This quantitation proved challenging because it relies on accurately measuring the intracellular isotopic enrichment of lactate, which changes as a function of culture time. Nonetheless, considering that the amount of labeled lipids shown in Fig. 1c–d is approximately 10% of the total lipid pool (based on the natural-abundance peak) and that at 10 minutes after introduction of 13C3-lactate the intracellular isotopic enrichment of lactate has decreased to 24% (Supplementary Fig. 3), we estimate that a substantial fraction of the total lipids are produced from lactate.
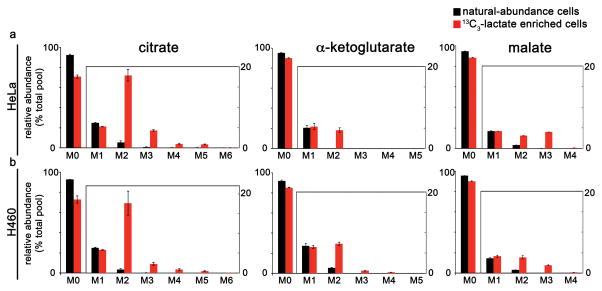
These solid-state NMR results show that exogenous labeled lactate can be used efficiently as a nutrient source of 13C by HeLa and H460 cells. At media concentrations of 3 mM, exogenous lactate provides an important carbon contribution to the total pool of lipids. Isotopologue patterns from LC/MS analysis of whole-cell lysates were consistent with metabolism of exogenous lactate via LDH to produce pyruvate for entry into the TCA cycle (Fig. 2). The M+2 isotopologues in Fig. 2 result from the insertion of pyruvate carbon into the TCA cycle by the pyruvate dehydrogenase complex followed by citrate synthase. The M+2 isotopologue of α-ketoglutarate and malate is substantially smaller than that of citrate. We also repeated the experiment in glutamine-free media and observed a similar trend (Supplementary Fig. 4). Together, these results are consistent with citrate carbon being used to synthesize lipids. After establishing exogenous lactate utilization, we sought to determine the route of lactate entry into the TCA cycle, specifically, the location of LDH-catalyzed conversion to pyruvate.
Figure 2.
HeLa and H460 cells cultured in 13C3-lactate have labeled TCA cycle intermediates. The patterns shown represent the isotopologue distribution of citrate, α-ketoglutarate, and malate as measured by LC/MS. Data are from (a) HeLa cells or (b) H460 lung cancer cells cultured for 3 hours in standard media (containing natural-abundance glucose) that was supplemented with either 3 mM 13C3-lactate (red) or natural-abundance lactate (black). Data shown are mean values ± s.d. (n = 3).
Isolated Mitochondria Metabolize 13C-Labeled Lactate
We considered two potential pathways of lactate metabolism. In the first pathway, lactate is metabolized to pyruvate by cytosolic LDH. In the alternative pathway, lactate is imported into the mitochondria and metabolized by mitochondrial LDH in the intermembrane space and/or the matrix. Analysis of whole cells treated with 13C3-lactate cannot distinguish between these two routes of lactate metabolism because the product is mitochondrial pyruvate in both cases. Instead, we isolated mitochondria from HeLa cells grown in unlabeled media and incubated them in respiration buffer containing 13C3-lactate (as well as unlabeled glutamine and unlabeled malate) for 30 minutes. After rinsing the samples with unlabeled buffer, we extracted metabolites and analyzed them by LC/MS. We detected 13C3-lactate, indicating its uptake into the mitochondria. We also measured TCA cycle intermediates that had incorporated 13C label, supporting the finding that lactate carbons enter the isolated mitochondria (Supplementary Fig. 5). The M+2 isotopologue of citrate was enriched at 12%, while the M+3 isotopologue of citrate was enriched at 3%. In contrast, the M+2 and the M+3 isotopologues of malate had much lower enrichments (~ 1%). When we repeated the experiment without adding glutamine to the mitochondrial buffer, we saw similar labeling distributions of TCA cycle intermediates (Supplementary Fig. 5b). These data are consistent with lactate being metabolized to pyruvate by mitochondrial LDH in the intermembrane space or the matrix, and this pyruvate then entering the TCA cycle by either the pyruvate dehydrogenase complex and citrate synthase or by pyruvate carboxylase. The enrichment of the M+2 isotopologue in citrate together with the decreased enrichment of the M+2 isotopologue in malate is consistent with a truncated TCA cycle.
Interpretation of the above data is complicated by the possibility that the integrity of the mitochondrial membrane was damaged during the mitochondrial isolation process, allowing indiscriminate influx of lactate into the mitochondria. A second complicating factor is the possibility that cytosolic LDH adhered to the outer mitochondrial membrane during sample preparation. Each complication is evaluated below.
Integrity of Isolated Mitochondria
To assess the integrity and metabolic function of the isolated mitochondria, we measured their oxygen consumption rates (OCRs) in the presence of various substrates and inhibitors (Supplementary Fig 6). The basal OCR of isolated HeLa mitochondria was measured as 1.35 nmol/min/mg. The inner and outer mitochondrial membrane integrities were assessed by the respiratory ratio (State 3/State 4) and the OCR after introduction of cytochrome c, respectively. The respiratory ratio was greater than 8, and the addition of cytochrome c did not change the OCR, indicating that the respiratory capacity of the isolated mitochondria was intact. Further evidence of mitochondrial integrity was obtained through analysis of the respiration buffer in a separate experiment. Isolated mitochondria were incubated in buffer containing labeled pyruvate for 30 minutes. Subsequent LC/MS analysis of the buffer revealed no labeled lactate (Supplementary Fig 7). A comparable analysis was performed with labeled lactate, and no labeled pyruvate was detected in the respiration buffer (Supplementary Fig 7). These data indicate no extra-mitochondrial LDH activity and no unregulated transport of small organic acids, which might be expected if the mitochondrial membrane integrity had been compromised, allowing leakage of metabolites and enzymes.
2H Labels Enter the Mitochondria of Intact Cells
The second complication to the discernment of which pathway lactate takes as it moves into the mitochondria is the possibility of LDH bound to the outer mitochondrial membrane. It has been shown previously that cytosolic LDH adheres to the outer mitochondrial membrane in some preparations10. Although the above results indicate that this is not the case for our samples, we wanted to test if cytosolic oxidation of lactate is coupled to transport. The 13C labeling that would result from active LDH adhered to the outer mitochondrial membrane during mitochondrial isolation cannot be easily distinguished from the 13C labeling that would result from mitochondrial LDH activity. To resolve the ambiguity, we employed a different isotope labeling scheme using 2-2H-lactate.
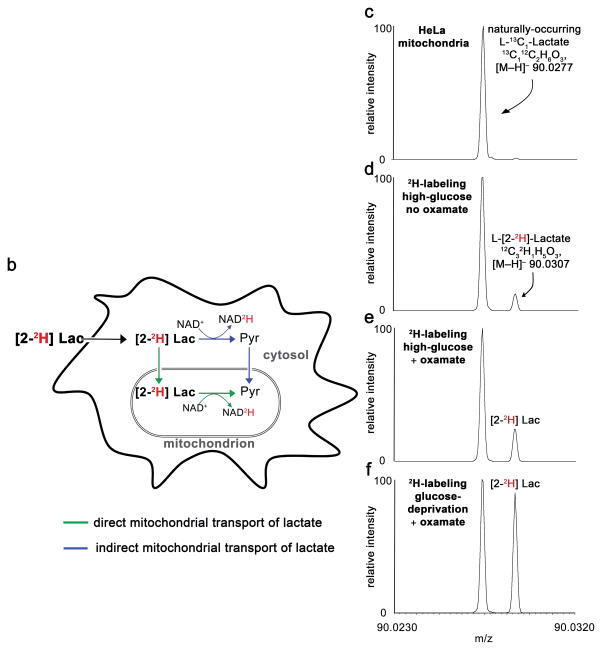
To test for mitochondrial LDH activity, we cultured HeLa cells in 3 mM 2-2H-lactate. When 2-2H-lactate is oxidized to pyruvate, the 2H label is transferred to NAD+, thereby forming deuterated NADH and unlabeled pyruvate (Fig 3a). If this pyruvate molecule is transported into the mitochondria and reduced to lactate by mitochondrial LDH, we would detect only unlabeled mitochondrial lactate (Fig 3b). In contrast, transport of 2-2H-lactate into the mitochondria would result in a pool of labeled mitochondrial lactate (Fig. 3b). This simplified assessment ignores the potential contribution of the malate-aspartate shuttle, which we consider below.
Figure 3.
Deuterium labels from 2H-lactate enter the mitochondria. (a) Schematic showing location of 2H label as a function of LDH activity. (b) Schematic showing two pathways (blue and green) that result in mitochondrial lactate. Only the pathway shown in green results in deuterated mitochondrial lactate. (c–f) Mass spectra from isolated mitochondria showing that 13C1-lactate (from natural abundance) and 2-2H-lactate (from exogenous label) can be resolved. The 2-2H-lactate peak is only detected at levels above natural abundance in mitochondria from cells cultured in 2-2H-lactate. Data are from mitochondria isolated from HeLa cells cultured for 45 minutes in high-glucose media supplemented with (c) 3 mM natural-abundance lactate or (d) 2-2H-lactate. The bottom spectra show increased 2-2H-lactate labeling from HeLa mitochondria. These data result from (e) culturing HeLa cells for 45 minutes in high-glucose media and 100 mM oxamate during mitochondrial isolation or (f) culturing HeLa cells for 45 minutes in glucose-free media and 100 mM oxamate during mitochondrial isolation.
We first demonstrated that the deuterium label of 2-2H-lactate did not undergo non-enzymatic, free exchange with water over the time scale of our experiment (Supplementary Fig. 8). We then cultured HeLa cells in 2-2H-lactate for 45 minutes, isolated the mitochondria, rinsed them with isolation buffer, extracted their metabolites, and measured the labeling of lactate with high-resolution mass spectrometry. Because we used a mass spectrometer with sufficient resolving power to experimentally distinguish the natural-abundance 13C peak of lactate from the 2H peak, we did not have to rely on computational methods for deconvolution. Under these conditions, we detected 2-2H-lactate from the mitochondria. In contrast, we detected only the expected natural-abundance level of deuterated lactate from control samples in which no 2-2H-lactate was added to the culture medium (Fig. 3c–d).
Increasing 2H Enrichment in Mitochondria of Intact Cells
It is important to note that our protocol to isolate mitochondria from intact cells required ~45 minutes (see Online Methods). Although the cells are maintained at 4 °C during the isolation procedure, metabolism remains active and the mitochondria are still respiring and consuming unlabeled substrate from the buffer. We suspected that this could result in significant dilution of metabolite labeling and low isotopic enrichment.
When we cultured HeLa cells in 2-2H-lactate for 45 minutes and then isolated their mitochondria for LC/MS analysis, we measured an isotopic enrichment for 2-2H-lactate of 0.4% from the mitochondria (Fig. 3d). We speculated that this low isotopic enrichment was because (i) very little lactate was transported into the mitochondria, (ii) equilibration of deuterium label with the large pool of redox-active hydrides during dynamic equilibrium of lactate and pyruvate, and/or (iii) the label was metabolically diluted during the mitochondrial isolation by production of unlabeled lactate. To assess metabolic dilution, we incubated HeLa cells in 13C3-lactate for 45 minutes. We then transferred the cells to non-labeled buffer and kept them on ice for 45 minutes prior to quenching metabolism with liquid nitrogen and organic solvent. This preparation mimicked that of the mitochondrial isolation. When we measured the isotopic enrichment of 13C3-lactate from these whole-cell lysates, we found that it was 0.3%. This result shows that, after the 45-minute preparation time required to isolate mitochondria, there is minimal labeled lactate remaining in the cell. Thus, we concluded that the low level of 2-2H-lactate measured from isolated mitochondria (Fig. 3d) was not indicative of low mitochondrial transport.
We next aimed to show that we could manipulate 2-2H lactate labeling from isolated mitochondria by modifying the culture conditions. To reduce metabolic dilution of lactate label, we treated cells with oxamate (an inhibitor of LDH). We also sought to increase the isotopic enrichment of lactate by removing glucose from the media. To test the effects of oxamate and glucose deprivation on lactate labeling, we incubated HeLa cells in media containing 13C3-lactate without glucose. After incubating the cells for 45 minutes in non-labeled buffer containing oxamate, we then measured the isotopic enrichment of 13C3-lactate from whole cells and found that we could increase it substantially (Supplementary Fig. 9).
When we cultured HeLa cells in 2-2H-lactate without glucose for 45 minutes, isolated their mitochondria while incubating them in oxamate, and then measured the isotopic enrichment of 2-2H-lactate from the mitochondria by LC/MS, we found that 2-2H-lactate label increased by ~7.5 fold from 0.4% to 3% (Fig. 3e–f). This condition-dependent increase in the isotopic enrichment of lactate supports our interpretation that the observed deuterium labeling from isolated mitochondria results from mitochondrial lactate and is not from non-physiological contamination.
2H Labels Enter Isolated Mitochondria
The glycerol phosphate shuttle cannot transfer deuterium label from the cytosol to the mitochondria, however, the malate-aspartate shuttle can. We did observe 2H-malate in mitochondria isolated from cells labeled with 2-2H-lactate. In some cells we examined (e.g., 3T3 fibroblasts), the enrichment of 2H-malate was an order of magnitude smaller than the enrichment of 2-2H-lactate, and the enrichment of 2H-malate approached its natural-abundance level (Supplementary Fig 10). In all cells we studied, however, 2H-malate had a measurable, non-zero value. This raises the question of the potential contribution of the malate-aspartate shuttle to the enrichment of 2-2H-lactate in isolated mitochondria. To evaluate the possibility that lactate labeling is a result of only the malate-aspartate shuttle, we repeated the 2-2H-lactate labeling experiment in isolated mitochondria and omitted malate and glutamate from the mitochondrial buffer (we note that the mitochondria have limited respiration under these conditions, see later).
Mitochondria from HeLa cells were isolated, incubated in respiration buffer containing 2-2H-lactate for 30 min, rinsed, and extracted for LC/MS analysis. The integrity and functionality of the isolated mitochondria were confirmed as described above by measuring mitochondrial OCRs in the presence of various substrates and inhibitors (Supplementary Fig. 6). We determined that 70% of the mitochondrial lactate pool was enriched for deuterium under these conditions (Supplementary Fig. 11a). We did not observe deuterium label on malate (Supplementary Fig. 11b). These results support a pool of transported lactate in the intermembrane space or mitochondrial matrix. Here, because metabolism in the isolated mitochondria was quenched immediately after 2-2H-lactate labeling, the enrichment percentage is much higher than that shown in Fig. 3, where 45 min was needed to isolate the mitochondria from whole cells after labeling. Notably, not all detected lactate is deuterated in Supplementary Fig. 11a. This result is consistent with rapid pyruvate-lactate exchange (i.e., some of the unlabeled pyruvate produced from 2-2H-lactate reforms lactate).
Electron Microscopy Localizes LDHB to the Mitochondria
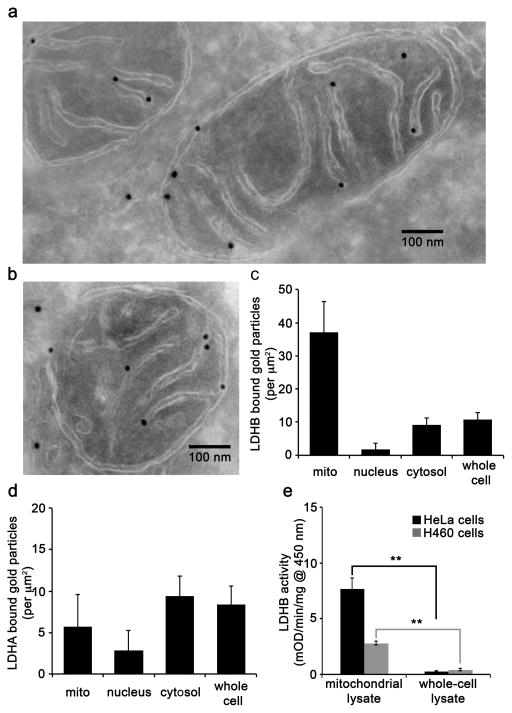
Functional LDH is a homo or hetero tetramer composed of LDHA and LDHB subunits11. Although LDHA and LDHB are expressed ubiquitously, LDHA is the predominant isoform found in skeletal muscle and therefore is commonly referred to as the M subunit12,13. In contrast, LDHB is the predominant isoform found in heart muscle and is often referred to as the H subunit14. Interestingly, LDHA has been found to have a higher affinity for pyruvate and a higher Vmax for pyruvate reduction compared to LDHB15. Consequently, it has been suggested that LDHA preferentially reduces pyruvate to lactate, while LDHB supports conversion of lactate to pyruvate in cells that utilize lactate as a nutrient source for oxidative metabolism or gluconeogenesis15,16. Since we were interested in the conversion of lactate to pyruvate, we first assessed LDHB in HeLa cells. We used transmission electron microscopy (TEM) with immunogold labeling to localize LDHB to subcellular compartments. Our results support that gold-labeled LDHB localizes to the inner mitochondrial membrane, although we cannot determine if it is on the intermembrane or the matrix side (Fig. 4a–b). A semi-quantitative analysis of LDHB localization to the mitochondria, nucleus, and cytosol, revealed that there was a much greater density of gold-labeled LDHB associated with the mitochondria relative to other locations, including the nucleus and cytosol (Fig. 4c). In contrast to LDHB, TEM experiments with gold-labeled LDHA did not show localization to the mitochondria (Fig. 4d, Supplementary Fig. 12a). Data from enzyme-linked immunosorbent assay (ELISA) experiments with LDHA performed on HeLa mitochondrial and HeLa whole-cell lysates provided similar distributions (Supplementary Fig. 12b).
Figure 4.
LDHB is present in HeLa mitochondria. (a–b) Electron micrographs of cryosectioned HeLa cells immunolabeled with primary anti-LDHB and secondary antibody conjugated to 18 nm gold nanoparticles. Scale bar, 100 nm. (c) Semi-quantitative analysis of LDHB localization in the mitochondria (mito), nucleus, cytosol, and whole cell. The data were obtained from images of 10 random cells taken at 20,000X. The area of the mitochondria, nucleus, and cytosol were calculated by using ImageJ. Error bars represent s.d. from n = 10 images. (d) Semi-quantiative analysis of LDHA localization in the mitochondria (mito), nucleus, cytosol, and whole cell from HeLa cells immunolabeled with primary anti-LDHA and secondary antibody conjugated to 18 nm gold nanoparticles. The data were obtained from images of 10 random cells taken at 20,000X. The area of the mitochondria, nucleus, and cytosol were calculated by using ImageJ. Error bars represent s.d. from n = 10 images. (e) LDHB activity was detected in both whole-cell and isolated mitochondrial lysates from HeLa and H460 cells. LDHB activities were measured by an immunocapture enzymne activity assay. Data shown are the averages ± s.d. from n = 3 replicates (**p < 0.01, Student’s t-test).
Mitochondrial Lysates Show LDHB Activity
We also evaluated LDHB activity in lysates from HeLa and H460 cells. We detected LDHB activity in both whole-cell and isolated mitochondrial lysates from HeLa and H460 cells (Fig. 4e). On a per mg of protein basis, the mitochondria had higher LDHB activity, further suggesting that LDHB activity is concentrated in mitochondria. With only these results shown in Fig. 4e and the TEM data above, however, we note that we cannot determine the location of LDHB with respect to the intermembrane space or the matrix.
Mitochondria use Lactate as a Carbon Source to Respire
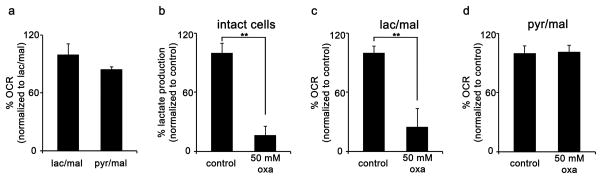
By using isotope tracing, we have shown that isolated mitochondria can metabolize lactate. These data suggest that isolated mitochondria can use lactate as a carbon source to respire. To test this, we incubated HeLa mitochondria in buffer containing either malate and lactate or malate and pyruvate. We then measured OCR. We note that our HeLa mitochondria do not respire with only lactate or pyruvate as a carbon source in the buffer. Similarly, our HeLa mitochondria did not respire well with only malate as a carbon source in the buffer. We found that HeLa mitochondria respire when incubated in buffer containing malate and pyruvate in addition to buffer containing malate and lactate (Fig. 5a). We next examined if we could reduce the OCR of HeLa mitochondria incubated in malate and lactate by inhibiting lactate utilization with oxamate. We first demonstrated that oxamate inhibits LDH in HeLa cells by measuring decreased lactate production from whole cells (Fig. 5b). We then treated isolated HeLa mitochondria incubated in malate and lactate with oxamate. We measured a significantly decreased oxygen respiration rate (Fig. 5c). In contrast, control mitochondria incubated in malate and pyruvate were not affected by oxamate (Fig. 5d). These data show that isolated HeLa mitochondria can use lactate carbon to respire.
Figure 5.
HeLa mitochondria use lactate as a carbon source to respire. (a) OCR of isolated HeLa mitochondria incubated with the following substrates: 10 mM lactate, 5 mM malate, and 5 mM ADP (lac/mal, n = 3); or 10 mM pyruvate, 5 mM malate, and 5 mM ADP (pyr/mal, n = 3). (b) Lactate-production assay from intact HeLa cells treated with 50 mM of oxamate shows a ~70% decrease in lactate production relative to control cultures after 6 hours. (c) OCR of isolated HeLa mitochondria incubated with 10 mM lactate, 1 mM malate, and 5 mM ADP (control, n = 3); or 10 mM lactate, 1 mM malate, 5 mM ADP, and 50 mM oxamate (oxa, n = 3). (d) OCR of isolated HeLa mitochondria incubated with the following substrates: 10 mM pyruvate, 1 mM malate, and 5 mM ADP (control, n = 3); or 10 mM pyruvate, 1 mM malate, 5 mM ADP, and 50 mM oxamate (oxa, n = 3). Data shown are averages ± s.d. from n = 3 replicates. (**p < 0.01, Student’s t-test).
Discussion
Although lactate was once recognized as a “metabolic waste product”, productive utilization of lactate as a source of energy is now well established17–20. Consistent with this paradigm, we have shown here that lactate is used by HeLa and H460 cells as an important source of labeled carbon to synthesize lipids during proliferation (Fig. 1). We describe two models by which lactate can be utilized. In the first model, lactate is oxidized to pyruvate in the cytosol with the exchange of carbon labels from lactate to pyruvate21. In the second model, lactate is oxidized to pyruvate by LDH associated with the mitochondria (Fig. 3b). Our data support the existence of the second model, but we note that we have not quantitated its contribution relative to the first. A benefit to having mitochondrial LDH is that a fermenting cell can use its own lactate carbon as a source of energy. The model may be particularly relevant to cancer cells, most of which have highly elevated levels of glycolysis and lactate production.
While our results show that more LDHB is localized to HeLa mitochondria relative to other cellular locations, we cannot resolve where in the mitochondria lactate oxidation is occurring. One possibility is that lactate is oxidized by LDHB located in the intermembrane space. Such compartmentalization of LDHB may facilitate substrate channeling. If LDHB is positioned near a mitochondrial pyruvate transporter for example, this could decrease local concentrations of pyruvate and therefore help promote lactate oxidation. Indeed, this type of arrangement has been proposed to constitute a “lactate-malate-aspartate shuttle” in which lactate oxidation is coupled to the malate-aspartate shuttle in the intermembrane space to translocate reducing power to the mitochondria6.
An alternative possibility is that lactate is directly transported across the mitochondrial inner membrane and oxidized by LDHB in the matrix. Even though intermembrane LDH and matrix LDH would achieve the same function of oxidizing lactate to pyruvate while simultaneously reducing NAD+ to NADH, the location of the enzyme determines where NADH is produced. NADH cannot directly cross the inner mitochondrial membrane and the ratio of NAD+ to NADH in the cytosol and the mitochondria is known to regulate reactions in each respective compartment1. Thus, a matrix LDH could have important regulatory roles. Moreover, unlike intermembrane LDH, the existence of matrix LDH and a lactate transporter would enable the hydride of lactate to carry its reducing power into the mitochondria independent of the malate-aspartate shuttle. This could represent a mechanism for carrying reducing equivalents produced by glycolysis into the mitochondria that compensates for limitations in the glycerol phosphate and malate-aspartate shuttles when glycolytic flux is increased. It has been suggested that lactate production in cells with elevated glycolysis compensates for the inability of the glycerol phosphate and malate-aspartate shuttles to keep pace in recycling NADH to NAD+22–24. For example, it has been shown that the malate-aspartate shuttle’s transfer of NADH in myocardium is outpaced by glycolysis under high cardiac workloads, despite oxygen availability and adequate blood flow25. In ascites tumor cells, it was determined that when the malate-aspartate shuttle operates at maximum capacity it can only oxidize 13% of the NADH produced by glycolysis23. Thus, increased production of lactate in cancer cells may be partially due to limitations of the malate-aspartate shuttle. Instead of secreting lactate from the cancer cell as a “waste product”, direct mitochondrial transport of lactate and its subsequent metabolism could provide a route for capturing both carbon and reducing equivalents produced by glycolysis. This may be an important mechanism to support growth in rapidly dividing cells, such as cancer. Notably, direct transport of lactate into the mitochondrial matrix would require a mitochondrial transport protein. It has been suggested previously that import of lactate into the mitochondrial matrix may be accomplished by a monocarboxylate transporter26.
Independent of whether lactate oxidation occurs in the intermembrane space or the mitochondrial matrix, our results indicate that selective inhibition of LDHB may be an attractive cancer therapeutic that warrants further investigation.
Online Methods
Materials
Solvents and chemicals for liquid chromatography were obtained from Sigma-Aldrich. Stable isotopes were obtained from Cambridge Isotope Laboratories. Cell-culture media and reagents were purchased from Life Technologies, unless otherwise specified. HeLa cells were obtained from the Tissue Culture Support Center of Washington University in St Louis. The human lung carcinoma cell line (H460) was a gift from the laboratory of Dr. Leah Shriver (University of Akron). The mouse fibroblast 3T3-L1 cell line was obtained from the American Tissue Culture Collection (ATTC CL-173).
Cell culture
All cells were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM, 4.5 g/L D-glucose) containing 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin at 37 °C with 5% CO2 unless otherwise noted. Isotope labeling was achieved by culturing cells in media supplemented with either 13C- or 2H-labeled nutrients as specified in each experimental section. Both HeLa and H460 cells were tested for mycoplasma contamination. Cells were examined by using a mycoplasma detection kit (Lonza, MycoAlert). Both cell lines were determined to be free of mycoplasma contamination. Test readings were: positive control, 26.06; negative control, 0.15; HeLa cells, 0.45; and H460 cells, 0.62.
Statistical analysis
All isotope-labeling experiments were performed in pairs, with parallel cultures being identical other than replacement of natural-abundance nutrients (D-glucose or L-lactate) with uniformly 13C-labeled nutrients (D-13C6-glucose, 99% or L-13C3-lactate, 98%) or L-2-2H-lactate (98%). Experiments were performed with n = 3 samples per group. All p-values were calculated by using a two-tailed Student’s t-test.
Metabolite extraction
Cell or mitochondrial pellets were extracted by using methanol/acetonitrile/water (2:2:1), with the solvent volumes adjusted to maintain a ratio of 1 mL solvent per 1 mg of dried material. Following the previously described extraction protocol27, samples were vortexed for 30 seconds and incubated 1 minute in liquid nitrogen (LN2). The samples were then thawed at room temperature, vortexed and LN2-incubated two more times prior to a 10-minute sonication. After 1 h at −20 °C, the samples were centrifuged at 14,000 rpm for 10 min. The resulting supernatant was dried on a speedVac, and then reconstituted in acetonitrile/water (1:1). The final volume of reconstitution solvent was adjusted to 100 μL per 1 mg of the starting dried material. For extraction of liquid samples, 200 μL of media or buffer was extracted with 800 μL methanol/acetonitrile (1:1), vortexed for 30 s, sonicated at 25 °C for 15 minutes, and incubated at −20 °C for an hour. The samples were centrifuged and the supernatant was then collected, dried, and reconstituted in 100 μL acetonitrile/water (1:1).
Mitochondrial isolation
Mitochondria were isolated from cells according to the previously described protocol28, 29 with minor modifications, as detailed below. Cells were harvested, pelleted, and re-suspended in cold mitochondrial isolation media [MIM, 300 mM sucrose, 10 mM sodium 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 0.2 mM ethylenediaminetetraacetic acid (EDTA), and 1mg/mL Bovine Serum Albumin (BSA), pH 7.4] at a ratio of 1 × 108 cells per 1 mL of MIM. Cells were then homogenized with a glass-teflon potter for 20 strokes. After homogenization, samples were centrifuged at 700 × g and 4 °C for 7 minutes to separate mitochondria from the remaining cellular material. The supernatant was decanted and the remaining pellet was homogenized a second time in MIM to recover more mitochondria. The supernatants were combined and centrifuged at 10,000 × g (4 °C) for 10 minutes to obtain a mitochondrial pellet. Mitochondrial pellets were washed with cold BSA-free MIM before determining their total protein content by Bradford protein assay (Bio-Rad). Isolated mitochondria were studied with various buffer conditions and substrates as specified in each experimental section. Two tables summarizing mitochondrial experiments, the various conditions used, and associated figures are provided in Supplementary Table 3.
Assessment of mitochondrial integrity
The function and integrity of isolated mitochondria was assessed by using the high-resolution OROBOROS oxygraph-2k respirometry (Oroboros Instruments)30. Each chamber was filled with 2.3 mL of mitochondrial respiration media [MiR05, 110 mM sucrose, 60 mM K-lactobionate, 20 mM HEPES, 20 mM taurine, 10 mM potassium phosphate monobasic (KH2PO4), 3 mM magnesium chloride (MgCl2), 0.5 mM ethylene glycol tetraacetic acid (EGTA), 0.1% BSA, pH 7.1] and air saturated. HeLa mitochondria (300 μg mitochondrial protein) were added to each chamber and equilibrated for 4 minutes. Subsequently, the metabolic substrates (5 μL of 2 M pyruvate and 12.5 μL of 0.8 M malate) were added and then other substrates (ADP and succinate), inhibitors (rotenone, Rot; oligomycin, Olig; antimycin A, AntA), or uncouplers (carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazone, FCCP) to induce state 2, 3, and 4 respiration or to examine mitochondrial function31. Each addition of substrate, inhibitors, or uncoupler was allowed 2–4 minutes to stabilize. Finally, cytochrome c (0.05 mg/mL) was added to test the outer mitochondrial membrane integrity. The inner membrane integrity was assessed by measuring the respiratory control ratios (state 3/state 4)30.
Preparing HeLa and H460 cells for solid-state NMR
Samples labeled with D-13C6-glucose were prepared by culturing cells in standard media containing 50% D-13C6-glucose and 50% natural-abundance D-glucose for 48 hours. Samples labeled with L-13C3-lactate were prepared by growing cells in high-glucose media supplemented with 3 mM L-13C3-lactate for 48 hours. For the control cultures, 3 mM natural-abundance L-lactate was supplied to the culture media. Cells were then washed with PBS three times, scraped from the plate, pelleted, and snap-frozen in LN2. Frozen cell pellets and spent media were subsequently lyophilized and analyzed by solid-state NMR.
Preparing 13C3-lactate labeled HeLa and H460 cells for LC/MS analysis
Cells were grown to ~70% confluency in a 100-mm culture dish. The culture media was then exchanged for fresh media supplemented with 3 mM L-13C3-lactate. Parallel samples were prepared by culturing cells under the same conditions with 3 mM natural-abundance L-lactate. Cells were washed with phosphate-buffered saline (PBS) and HPLC-grade water, quenched with 1 mL cold HPLC-grade methanol, scraped from the plate, and pelleted. Pellets were dried on a SpeedVac and subsequently lyophilized. Dried samples were weighed and extracted by using the protocol described above. Experiments were performed with n = 3 cultures per sample group.
Isotopic labeling of isolated mitochondria
Mitochondria were isolated from cells by using the protocol described above. Next, mitochondria (0.45 mg mitochondrial protein) were incubated for 30 minutes in respiration buffer [buffer C, 125 mM potassium chloride (KCl), 20 mM HEPES, 3 mM magnesium acetate, 0.4 mM EGTA, 0.3 mM dithiothreitol (DTT), 5 mM KH2PO4, 0.2% BSA, pH 7.1] containing metabolic substrates. Two conditions were used: (i) 5 mM malate, 4 mM glutamine, and 3 mM lactate; or (ii) 5 mM malate and 10 mM lactate. Labeling was achieved by incubating mitochondria in buffer and replacing the natural-abundance L-lactate with L-13C3-lactate. Experiments were performed with n = 3 replicates of isolated mitochondria per sample group. Mitochondria were then pelleted, washed with PBS and HPLC-grade water, snap-frozen in LN2, and extracted as described above.
Direct mitochondrial transport of 2-2H-lactate
Mitochondrial samples were prepared in two ways for LC/MS analysis to examine direct mitochondrial transport of L-2-2H-lactate. One experiment involved labeling isolated mitochondria (A) and the other involved labeling whole cells (B).
A. Labeling isolated mitochondria with 2-2H-lactate
Isolated mitochondria from unlabeled HeLa cells were prepared according to the method described above (Online Methods section “Mitochondrial isolation”). Isolated mitochondria were then incubated in buffer C containing 3 mM sodium L-2-2H-lactate or natural-abundance L-lactate for 30 minutes. Each condition included three independent mitochondrial incubations (n = 3). Mitochondrial samples were then washed, pelleted, and extracted for targeted LC/MS analysis (see Online Methods section “LC/MS/MS analysis”).
B. Labeling whole cells with 2-2H-lactate (prior to mitochondrial isolation)
HeLa and 3T3-L1 fibroblast cells were cultured in high-glucose media containing 3 mM L-2-2H-lactate for 45 minutes. Mitochondria were then isolated, washed, pelleted, and extracted for targeted LC/MS analysis of lactate (see Online Methods section “LC/MS/MS analysis”). This procedure led to relatively low levels of isotopic enrichment for 2-2H-lactate (0.4%) due to metabolic dilution occurring while mitochondria were being isolated. To minimize metabolic dilution and maximize isotopic enrichment of 2-2H-lactate, we repeated the experiment with various concentrations of glucose and the LDH inhibitor oxamate. We optimized experimental conditions by labeling cells for 45 minutes with 10 mM L-13C3-lactate. We then washed the cells, spun them down, and suspended them in a minimal amount of cold MIM. The cell pellets were stored on ice for 45 minutes to mimic the mitochondrial isolation procedure. The cell pellets were extracted for LC/MS analysis of lactate after 45 minutes. The conditions providing the highest isotopic enrichment of 13C3-lactate (1.3%) were selected for a 2-2H-lactate labeling experiment as described below.
HeLa cells were grown to confluence in a T-150 flask, refreshed with glucose-free media, and cultured for 4 hours. After 4-hours, cells were supplemented with L-2-2H-lactate at a final concentration of 10 mM and cultured for another 45 minutes. Each flask of labeled cells were washed, harvested, and pelleted. Each cell pellet was then re-suspended in 500 μL of cold MIM with 100 mM oxamate, and the mitochondria were isolated according to the procedure detailed above. The isolated mitochondrial pellets were then extracted and analyzed by targeted LC/MS analysis to determine 2-2H-lactate labeling. Two independent experiments were performed and provided consistent results.
Transmission Electron Microscopy
For immunolocalization of LDHB (or LDHA) in HeLa cells, cells were fixed in 1% paraformaldehyde/0.01% glutaraldehyde (Polysciences Inc.) in 100 mM PIPES/0.5 mM MgCl2, pH 7.2 for 1 hour at 4 °C and then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose/20% polyvinyl pyrrolidone in piperazine-N,N′-bis(2-ethanesulfonic acid) [PIPES]/MgCl2 at 4 C°. Embedded cells were trimmed, frozen in LN2, and sectioned with a Leica Ultracut UCT7 cryo-ultramicrotome (Leica Microsystems Inc.). Sections of 50-nm were blocked with 5% FBS/5% Normal Goat Serum (NGS) for 30 minutes and subsequently incubated with rabbit anti-LDHB antibody (Cat. No. PA5-27505, Thermo Scientific; dilution 1:100) or rabbit anti-LDHA antibody (Cat. No. PA5-17183, Thermo Scientific; dilution 1:100) for 1 hour, followed by secondary goat anti-rabbit IgG (H+L) antibody conjugated to 18 nm colloidal gold (Cat. No. 111-215-144, Jackson ImmunoResearch; dilution 1:30) for 1 hour. Sections were washed in PIPES buffer followed by a water rinse, and stained with 0.3% uranyl acetate/2% methylcellulose. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques). All gold labeling experiments were conducted in parallel with controls omitting the primary antibody. These controls were consistently negative at the concentration of colloidal gold conjugated secondary antibodies used.
Lactate dehydrogenase B activity assay
A LDHB activity assay kit was purchased from abcam® and used according to the manufacturer’s protocol. The assay determines LDHB activity by measuring changes of NADH, which is the product of the LDHB catalyzed reaction: L-lactate + NAD+ → pyruvate + NADH. The NADH generated is coupled to the reduction of a reporter dye to yield a colored product at a 1:1 ratio. The change of NADH is measured by monitoring increases in the absorbance of the dye at 450 nm. LDHB was immunocaptured from lysates, and substrates (lactate and NAD+) were then added to measure LDHB activity. Whole-cell lysates were obtained from incubating HeLa cells or H460 cells in the extraction buffer provided from the assay kit at a ratio of 2 × 107 cells per 1 mL of buffer. Mitochondrial lysates were prepared from mitochondria isolated from 5 × 107 cells and then incubated in 100 μL of extraction buffer. The total protein content of the cell or mitochondrial lysate was determined by a bicinchoninic acid (BCA) protein assay (Life Technologies). Lysate samples (100 μL each) were incubated in the antiLDHB-coated microplate for 2 hours, aspirated, and then washed. Activity solution provided by the kit (100 μL per well) was added to each well and the absorbance of each sample was measured at a wavelength of 450 nm every minute. Both whole-cell and mitochondrial lysates included n = 3 replicates. Each lysate sample was obtained from a different culture dish and processed in parallel.
Lactate dehydrogenase A ELISA
A LDHA ELISA assay kit was purchased from U.S. Biological. HeLa cells were harvested, washed, and then re-suspended in cold PBS at a concentration of 1 × 108 cells/mL. After 3 freeze-thaw cycles, the cell suspension was centrifuged at 1,500 × g for 10 minutes to remove cellular debris. HeLa mitochondria were isolated from 4 × 107 cells, washed and pelleted, and re-suspended in 100 μL cold PBS. The protein content of each lysate sample was determined by using a Bradford protein assay (Bio-Rad). Sample lysates and standard (100 μL per well) was added to an antiLDHA-coated microplate and incubated for 2 hours at 37 °C, aspirated, and washed. After incubating in detection reagents and washing with buffer, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added and incubated for 15 minutes at 37 °C. The assay was terminated with stop solution and read immediately at 450 nm. The lysate samples from whole cells and mitochondria included n = 3 replicates. Each replicate was obtained from a different culture dish and processed in parallel.
Oxygen consumption assays
The oxygen consumption rate (OCR) of isolated mitochondria was measured by using an XFe analyzer (Seahorse Bioscience)32. Isolated HeLa mitochondria were re-suspended in cold mitochondrial assay solution [MAS, 70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA and 0.2% (w/v) fatty acid-free BSA, pH 7.2 at 37 °C] with metabolic substrates [10 mM pyruvate/5 mM malate or 10 mM lactate/5 mM malate and 10 mM pyruvate/1 mM malate or 10 mM lactate/1 mM malate] at a concentration of 0.28 μg (mitochondrial protein) per μL. A volume of 25 μL of mitochondrial suspension was transferred to each well of the XFe cell culture microplate (resulting in 7 μg of mitochondria per well) and then spun down at 2,000 × g, 4 °C for 20 minutes. After centrifugation, 155 μL of pre-warmed MAS containing substrates was added to each well without disturbing the mitochondrial layer and then inserted into the XFe analyzer. The OCR of HeLa mitochondria was monitored and compared under four different conditions: pyr/mal (n = 3), lac/mal (n = 3), pyr/mal with 50 mM oxamate (n = 3), and lac/mal with 50 mM oxamate (n = 3). During the experiment, substrate and inhibitors were introduced to examine mitochondrial coupling. ADP (5 mM, final) and oligomycin (2.5 μg/mL, final) were added to induce state 3 and 4 respiration, respectively. FCCP (4 μM, final) was used to uncouple respiration (note that the concentration of FCCP was not optimized for maximal respiration), and antimycin A (4 μM, final) was added to inhibit respiration. All the data shown in Fig. 5 (panel a, c, and d) are the OCRs of the state 3 respiration under the various conditions described above.
Lactate Production Assay
HeLa cells were transferred to a 12-well plate at 1.4 × 106 cells/mL and allowed to attach overnight. Cells were then washed and supplemented with FBS-free and low-glucose media (1g/L D-glucose), and treated with vehicle (control, n = 3) or 50 mM oxamate (oxa, n = 3). After 6 hours, the culture media was collected and extracted for analysis by liquid chromatography triple quadrupole MS. Lactate concentrations were quantitated as described below in the Online Methods section “LC/MS/MS analysis”.
Solid-state 13C NMR analysis
Experiments were performed at 12 Tesla with a transmission-line probe having a 12-mm long, 6-mm inner-diameter analytical coil, and a Chemagnetics/Varian ceramic spinning module. Samples were spun by using a thin-wall Chemagnetics/Varian 5-mm outer diameter-zirconia rotor at 7143 Hz, with the speed under active control and maintained to within ± 2 Hz. A Tecmag Libra pulse programmer controlled the spectrometer. A 2-kW American Microwave Technology (AMT) power amplifier was used to produce radio-frequency pulses for 13C (125 MHz). The 1H (500 MHz) radio-frequency pulses were generated by a 2-kW Creative Electronics tube amplifier driven by a 50-W AMT amplifier. A 2 second recycle delay was used and all final-stage amplifiers were under active control33. The π-pulse lengths were 9 μs for both 13C and 1H. Proton-carbon-matched cross-polarization transfers were made in 2.0 ms at 56 kHz. Proton dipolar decoupling was 100 kHz during data acquisition. Typical data acquisition used 2,000 scans for labeled samples (~40 mg) and 20,000 scans for natural-abundance samples. All together, 38 samples (including both HeLa and H460) were examined.
LC/MS profiling analysis and data processing with X13CMS
An aliquot of each extracted sample (8 μL) was injected onto a Luna Aminopropyl column (3 μm, 150 mm × 1.0 mm I.D., Phenomenex) coupled to an Agilent 1200 series high performance liquid chromatography (HPLC) system. The column was used in hydrophilic interaction (HILIC) mode with the following buffers and gradients: A = 95% water, 5% acetonitrile (ACN), 10 mM ammonium hydroxide (NH4OH), 10 mM ammonium acetate (NH4Ac), pH 9.5; B = 95 % ACN, 5% water; 100% B from 0–3 min, 100% to 0% B from 3–37 min and 0% B from 37–40 min. MS detection was carried out on an Agilent 6540 Q-TOF in negative ESI (electrospray ionization) mode with a mass range of 30–1500 Da.
All raw data files were converted into mzXML files by using msconvert34. The metabolite profiling analysis was performed by using the XCMS-Warpgroup package35 implemented in R with the following parameters: centWave algorithm for features detection (peakwidth = 20–200 s and ppm = 10) and obiwarp method for retention time correction. The output object, xcmsSet(), containing labeled and unlabeled data was then submitted to X13CMS36 for the isotopic labeling analysis. The peaks corresponding to the isotoplogues of each base peak were identified according to the following inputs: i.) the mass difference between 13C and 12C = 1.003355 Da, ii.) the mass of 12C = 12.000000 Da, iii) ppm = 20, iv) RTwin = 10 s and v) baseline Noise = 10,000. Data annotations were confirmed by manual inspection. All metabolite identifications were confirmed by comparing the retention time and MS/MS patterns to model standards.
Targeted LC/MS analysis of 2H-labeled metabolites
Targeted analyses were performed on a Thermo Q Exactive Plus mass spectrometer in negative mode at 140,000 resolving power. The instrument was interfaced with a Dionex UltiMate® 3000 RSLCnano LC system. Three μL of each extracted sample were injected onto a Luna Aminopropyl column (3 μm, 150 mm × 1.0 mm I.D., Phenomenex) operated in HILIC mode with the same buffers and gradients as described above.
LC/MS/MS analysis
TCA intermediates and amino acids standards were used for MS/MS parameter optimization on the Agilent 6460 triple quadruple in negative ESI mode. After optimizing MS/MS parameters, multiple reaction monitoring (MRM) experiments were performed by using an Agilent 1290 Infinity Binary LC system with a Luna Aminopropyl column (3 μm, 150 mm × 4.6 mm I.D., Phenomenex). The column was used in HILIC mode with the following buffers and gradients: A = 95% water, 5% ACN, 20 mM NH4OH, 20 mM NH4Ac; B = 100% ACN; 85% B from 0–3 min, 85–50% B from 3–15 min, 50–20% B from 15–20 min, 20–5% B from 20–25 min and 5% B from 25–35 min. Each extracted sample was injected in 10 μL aliquots and ran at a flow rate = 0.15 mL/min. MRM transitions for citrate, α-ketoglutarate, malate, and lactate were optimized by using standards. For citrate, the quantifier ion transition m/z 191.0 → 111.0 and the qualifier 191.0 → 87.0 were used with a fragmentor voltage of 72 V, a collision energy of 12 V, and a cell accelerator voltage of 7 V. For α-ketoglutarate, the quantifier ion transition m/z 145.0 → 101.0 and the qualifier 145.0 → 57.0 were used with a fragmentor voltage of 60 V, a collision energy of 4 V, and a cell accelerator voltage of 7 V. For malate, the quantifier ion transition m/z 133.0 → 115.0 and the qualifier 133.0 → 71.0 were used with a fragmentor voltage of 74 V, a collision energy of 4 V, and a cell accelerator voltage of 7 V. For lactate, the quantifier ion transition m/z 89.0 → 43.0 and the qualifier 89.0 → 41.0 were used with a fragmentor voltage of 45 V, a collision energy of 8 V, and a cell accelerator voltage of 7 V.
For the lactate production assay, triple quadrupole MS was used for lactate quantification. An aliquot of each sample (3 μL) was injected onto a Luna Aminopropyl column with the following mobile phases and gradients: A = 95% water, 5% ACN, 20 mM NH4OH, 20 mM NH4Ac; B = 100% ACN; 85% B from 0–3 min, 85% to 50% B from 3–7 min, 50% to 5% B from 7–11 min and 5% B from 11–13 min. A serial standard dilution of lactate (1000, 500, 200, 100 and 20 ng/mL) was also analyzed and used for constructing a standard curve.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health grants R01 ES022181 (GJP), R21 CA191097-01A1 (GJP), R01 HL118639-03 (RWG), and R01 EB002058 (JS) as well as grants from the Alfred P. Sloan Foundation (GJP), the Camille & Henry Dreyfus Foundation (GJP), and the Pew Scholars Program in the Biomedical Sciences (GJP). We thank Dr. Wandy Beatty at Washington University’s Molecular Microbiology Imaging Facility for acquiring the TEM images.
Footnotes
Author contributions: YJC prepared samples, carried out biochemical assays, and performed LC/MS analyses. YJC, MS, and JS performed NMR analyses. YJC, NGM, XH, MS, PAC, SLJ, RWG, JS, and GJP contributed to experimental design and data interpretation. YJC, NGM, and GJP wrote the manuscript.
Competing financial interests: G.J.P. is a scientific advisory board member for Cambridge Isotope Laboratories. R.W.G. has financial relationships with LipoSpectrum and Platomics.
References
- 1.Lehninger A, Nelson D, Cox ME. Lehninger Principles of Biochemistry. W. H. Freeman and Company; 2008. [Google Scholar]
- 2.CORI CF, CORI GT. Carbohydrate metabolism. Annu Rev Biochem. 1946;15:193–218. doi: 10.1146/annurev.bi.15.070146.001205. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen HB, Clemmesen JO, Skak C, Ott P, Secher NH. Attenuated hepatosplanchnic uptake of lactate during intense exercise in humans. J Appl Physiol. 2002;92:1677–83. doi: 10.1152/japplphysiol.00028.2001. [DOI] [PubMed] [Google Scholar]
- 4.Bonvento G, Herard AS, Voutsinos-Porche B. The astrocyte--neuron lactate shuttle: a debated but still valuable hypothesis for brain imaging. J Cereb Blood Flow Metab. 2005;25:1394–9. doi: 10.1038/sj.jcbfm.9600127. [DOI] [PubMed] [Google Scholar]
- 5.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 6.Kane DA. Lactate oxidation at the mitochondria: a lactate-malate-aspartate shuttle at work. Front Neurosci. 2014;8:366. doi: 10.3389/fnins.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto T, Hussien R, Brooks GA. Colocalization of MCT1, CD147, and LDH in mitochondrial inner membrane of L6 muscle cells: evidence of a mitochondrial lactate oxidation complex. Am J Physiol Endocrinol Metab. 2006;290:E1237–44. doi: 10.1152/ajpendo.00594.2005. [DOI] [PubMed] [Google Scholar]
- 8.Brooks GA, Dubouchaud H, Brown M, Sicurello JP, Butz CE. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc Natl Acad Sci U S A. 1999;96:1129–34. doi: 10.1073/pnas.96.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YJ, et al. Differential incorporation of glucose into biomass during Warburg metabolism. Biochemistry. 2014;53:4755–7. doi: 10.1021/bi500763u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt RB, Laux JE, Spainhour SE, Kline ES. Lactate dehydrogenase in rat mitochondria. Arch Biochem Biophys. 1987;259:412–422. doi: 10.1016/0003-9861(87)90507-8. [DOI] [PubMed] [Google Scholar]
- 11.Markert C, Shaklee J, Whitt G. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science (80- ) 1975;189:102–114. doi: 10.1126/science.1138367. [DOI] [PubMed] [Google Scholar]
- 12.Takasu T, Hughes BP. Lactate dehydrogenase isozyme patterns in human skeletal muscle. I Variation of isozyme pattern in the adult. J Neurol Neurosurg Psychiatry. 1969;32:175–9. doi: 10.1136/jnnp.32.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjödin B. Lactate dehydrogenase in human skeletal muscle. Acta Physiol Scand Suppl. 1976;436:1–32. [PubMed] [Google Scholar]
- 14.SHAW CR, BARTO E. GENETIC EVIDENCE FOR THE SUBUNIT STRUCTURE OF LACTATE DEHYDROGENASE ISOZYMES. Proc Natl Acad Sci U S A. 1963;50:211–4. doi: 10.1073/pnas.50.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson DM, Goodfriend TL, Kaplan NO. Lactic Dehydrogenases: Functions of the Two Types. Science (80- ) 1964;143:929–933. doi: 10.1126/science.143.3609.929. [DOI] [PubMed] [Google Scholar]
- 16.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–92. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elustondo PA, et al. Physical and functional association of lactate dehydrogenase (LDH) with skeletal muscle mitochondria. J Biol Chem. 2013;288:25309–17. doi: 10.1074/jbc.M113.476648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks GA. Intra- and extra-cellular lactate shuttles. Med Sci Sports Exerc. 2000;32:790–9. doi: 10.1097/00005768-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 19.van Hall G, et al. Blood lactate is an important energy source for the human brain. J Cereb Blood Flow Metab. 2009;29:1121–9. doi: 10.1038/jcbfm.2009.35. [DOI] [PubMed] [Google Scholar]
- 20.Gladden LB. Lactate metabolism: a new paradigm for the third millennium. J Physiol. 2004;558:5–30. doi: 10.1113/jphysiol.2003.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy BWC, Kettunen MI, Hu DE, Brindle KM. Probing lactate dehydrogenase activity in tumors by measuring hydrogen/deuterium exchange in hyperpolarized l-[1-(13)C,U-(2)H]lactate. J Am Chem Soc. 2012;134:4969–77. doi: 10.1021/ja300222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenhouse WV, Lehninger AL. Magnitude of malate-aspartate reduced nicotinamide adenine dinucleotide shuttle activity in intact respiring tumor cells. Cancer Res. 1977;37:4173–81. [PubMed] [Google Scholar]
- 23.Chiaretti B, Casciaro A, Minotti G, Eboli ML, Galeotti T. Quantitative evaluation of the activity of the malate-aspartate shuttle in Ehrlich ascites tumor cells. Cancer Res. 1979;39:2195–9. [PubMed] [Google Scholar]
- 24.Bücher T, Klingenberg M. Wege des Wasserstoffs in der lebendigen Organisation. Angew Chemie. 1958;70:552–570. [Google Scholar]
- 25.O’Donnell JM, Kudej RK, LaNoue KF, Vatner SF, Lewandowski ED. Limited transfer of cytosolic NADH into mitochondria at high cardiac workload. Am J Physiol Heart Circ Physiol. 2004;286:H2237–42. doi: 10.1152/ajpheart.01113.2003. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto T, Hussien R, Cho HS, Kaufer D, Brooks GA. Evidence for the mitochondrial lactate oxidation complex in rat neurons: demonstration of an essential component of brain lactate shuttles. PLoS One. 2008;3:e2915. doi: 10.1371/journal.pone.0002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanisevic J, et al. Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal Chem. 2013;85:6876–84. doi: 10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–95. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 29.Schugar RC, Huang X, Moll AR, Brunt EM, Crawford PA. Role of choline deficiency in the Fatty liver phenotype of mice fed a low protein, very low carbohydrate ketogenic diet. PLoS One. 2013;8:e74806. doi: 10.1371/journal.pone.0074806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–72. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuznetsov AV, et al. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat Protoc. 2008;3:965–76. doi: 10.1038/nprot.2008.61. [DOI] [PubMed] [Google Scholar]
- 32.Rogers GW, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stueber D, Mehta AK, Chen Z, Wooley KL, Schaefer J. Local order in polycarbonate glasses by13C{19F} rotational-echo double-resonance NMR. J Polym Sci Part B Polym Phys. 2006;44:2760–2775. [Google Scholar]
- 34.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahieu NG, Spalding JL, Patti GJ. Warpgroup: increased precision of metabolomic data processing by consensus integration bound analysis. Bioinformatics. 2016;32:268–75. doi: 10.1093/bioinformatics/btv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal Chem. 2014;86:1632–9. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.