Abstract
A phylogenetic analysis of 14 complete simian virus 40 (SV40) genomes was conducted in order to determine strain relatedness and the extent of genetic variation. This analysis included infectious isolates recovered between 1960 and 1999 from primary cultures of monkey kidney cells, from contaminated poliovaccines and an adenovirus seed stock, from human malignancies, and from transformed human cells. Maximum-parsimony and distance methods revealed distinct SV40 clades. However, no clear patterns of association between genotype and viral source were apparent. One clade (clade A) is derived from strain 776, the reference strain of SV40. Clade B contains isolates from poliovaccines (strains 777 and Baylor), from monkeys (strains N128, Rh911, and K661), and from human tumors (strains SVCPC and SVMEN). Thus, adaptation is not essential for SV40 survival in humans. The C terminus of the T-antigen (T-ag-C) gene contains the highest proportion of variable sites in the SV40 genome. An analysis based on just the T-ag-C region was highly congruent with the whole-genome analysis; hence, sequencing of just this one region is useful in strain identification. Analysis of an additional 16 strains for which only the T-ag-C gene was sequenced indicated that further SV40 genetic diversity is likely, resulting in a provisional clade (clade C) that currently contains strains associated with human tumors and human strain PML-1. Four other polymorphic regions in the genome were also identified. If these regions were analyzed in conjunction with the T-ag-C region, most of the phylogenetic signal could be captured without complete genome sequencing. This report represents the first whole-genome approach to establishing phylogenetic relatedness among different strains of SV40. It will be important in the future to develop a more complete catalog of SV40 variation in its natural monkey host, to determine if SV40 strains from different clades vary in biological or pathogenic properties, and to identify which SV40 strains are transmissible among humans.
It has recently been recognized that naturally occurring genetic variants of simian virus 40 (SV40) exist (18, 23, 26, 27, 31, 34, 37-40, 46). As more genomic sequences became available, it was apparent that isolates differed from the reference strain SV40-776 and from each other. Major genetic variation is localized in two regions of the viral genome: the noncoding regulatory region and the C-terminal nucleotides (approximately 300) of the carboxy-terminal region of the T-antigen gene (T-ag-C), referred to as the variable domain (39, 40).
Variations in the T-ag-C gene region were frequently detected in human tumor-associated sequences, ruling out the possibility that positive findings were the result of laboratory contamination of specimens (4, 7, 26, 27, 40, 44-46). The T-ag-C sequence was shown to be stable during tissue culture passage of SV40 isolates (24). In contrast, the SV40 regulatory region may contain large insertions, deletions, or duplications (25), and rearrangements have been observed to occur within individual infected monkeys (23, 31) and during passage of SV40 in certain cultured cells (32).
Comparative study of entire genomic sequences is one of the best methods for determining the evolutionary relationships among organisms (11, 48). In the detailed analysis reported here, we examined the known complete SV40 genomic sequences, as well as described partial sequences from the SV40 T-ag-C region.
The specific goals of this study were (i) to examine the patterns of genetic variation in the complete genomes of SV40 isolated from human, nonhuman, and vaccine sources, (ii) to determine if phylogenies based on the T-ag-C gene are congruent with phylogenies based on whole-genomic sequences, and (iii) to examine the genetic variability of T-ag-C genes across available samples for which the entire genomic sequences are unknown.
MATERIALS AND METHODS
SV40 sequences analyzed.
The viral strains and associated sequences that were analyzed are listed in Table 1. The origin of each sequence and its GenBank accession number are shown. There are two entries for strains 776 and 777, because each was sequenced twice by using different source materials and varied slightly in sequence. SV40 reference strain 776 (SV40-776) was a BamHI clone in pBR322 (pSVB-3) obtained from G. Khoury. That sequence was previously reported. SV40-776* was an EcoRI clone (pWTSV40) prepared from a laboratory stock of SV40-776 (22). Strain 777 was obtained from A. M. Lewis, Jr. (see below). SV40-777* was a BamHI clone of SV40-777 in pBR322 made in 1983, which was obtained from M. Bastin. Because SVCPC and SVMEN are identical in sequence, they were treated as a single entry for this study and are sometimes designated SVCPC(SVMEN).
TABLE 1.
SV40 sequences analyzed in this studya
| SV40 strain or sequence designation | GenBank accession no. | Origin | Complete sequence known | Infectious virus isolate | Date isolated or detected | Reference(s) |
|---|---|---|---|---|---|---|
| 776 | J02400 | Adenovirus type 1 vaccine seed stock | + | + | 1960 | 6, 42 |
| 776* | AF316139 | From 776 | + | + | 22 | |
| VA45-54 | AF156107 | Monkey kidney cells (rhesus) | + | + | 1960 | 14, 24, 39, 42 |
| Baylor | AF155359 | Type 2 Sabin poliovaccine from 1956 | + | + | 1961 | 24, 28, 30, 39, 41 |
| PA-57 | M99362 | Monkey kidney cells (patas) | 0 | + | 1961 | 16 |
| 777 | AF332562 | Inactivated poliovaccine | + | + | 1962 | 12; this report |
| 777* | AY271817 | Rodent cells transformed by 777 | + | Before 1979 | This report | |
| Rh911 | AF316140 | Monkey kidney cells (rhesus) | + | + | 1962 | 13 |
| N128 | AY120890 | Monkey kidney cells (rhesus) | + | + | 1965 | 10; this report |
| PML-1(EK) | AY271816 | Human brain | + | + | 1970 | 18, 29, 39, 47 |
| SVCPC(SVMEN) | AF156108 | Choroid plexus carcinoma and a | + | + | 1995 | 26, 39 |
| human meningioma | + | + | 1984 | 21, 39 | ||
| K661 | AF038616 | SIV-infected monkey | + | + | 1998 | 23 |
| H328 | AF316141 | SIV-infected monkey | + | + | 1998 | 23 |
| H388 | AF168993 | SIV-infected monkey | 0 | + | 1998 | 23 |
| T302 | AF168994 | SIV-infected monkey | 0 | + | 1998 | 23 |
| 6593 | AF168995 | SIV-infected monkey | 0 | + | 1998 | 23 |
| I508 | AF169001 | SIV-infected monkey | 0 | + | 1998 | 23 |
| GM00637H | AF345344 | Transformed human fibroblasts | + | + | 1999 | This report |
| MC028846B | AF180737 | Salk vaccine from 1955 | + | 0 | 1999 | 34 |
| CPP15 | AF136000 | Human brain tumor | 0 | 0 | 1995 | 26 |
| GN13 | AF136001 | Human brain tumor | 0 | 0 | 1995 | 26 |
| EP14 | AF136003 | Human brain tumor | 0 | 0 | 1995 | 26 |
| OST-2 | AF168996 | Human bone tumor | 0 | 0 | 1997 | 27 |
| OST-3 | AF168997 | Human bone tumor | 0 | 0 | 1997 | 27 |
| OST-9 | AF168998 | Human bone tumor | 0 | 0 | 1997 | 27 |
| NHL-8 | - | Human lymphoma (HIV positive) | 0 | 0 | 2002 | 45 |
| NHL-28 | - | Human lymphoma (HIV positive) | 0 | 0 | 2002 | 46 |
| NHL-170 | - | Human lymphoma | 0 | 0 | 2002 | 46 |
| NHL-416 | - | Human lymphoma | 0 | 0 | 2002 | 46 |
| Men-99 | - | Human meningioma | 0 | 0 | 2003 | 5 |
Abbreviations and symbols: SIV, simian immunodeficiency virus; HIV, human immunodeficiency virus; +, yes; 0, no; blank, not known.
Viruses used for molecular cloning.
Several viral isolates were cloned and sequenced as part of this study. SV40 strains Rh911 and N128 were independent isolates recovered from uninoculated rhesus monkey kidney cultures in the early 1960s and were from samples held in storage at Baylor College of Medicine for over 25 years. SV40-Rh911 was an American isolate from 1964 (13). The virus used for cloning was from passage 2 in CV-1 cells made in 1970 at The Wistar Institute (Philadelphia, Pa.); it was obtained from A. Girardi in 1971 and remained archived at −70°C at Baylor College of Medicine until April 1996, when an aliquot was seeded into CV-1 cells to prepare a virus stock for cloning. The passage history of SV40-N128 is unclear; it was isolated in Russia in 1965 (10) and was obtained from M. Nachtigal in 1971. SV40-777, recovered originally from an inactivated poliovaccine, was from an archived stock that was obtained from A. M. Lewis, Jr., in July 1998. SV40-GM00637H cells (virus-producing SV40-transformed human fibroblast cells containing integrated and episomal SV40 genomes) were obtained in November 1999 from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository through Coriell Cell Repositories (Camden, N.J.). These cells were chosen to determine whether SV40 developed adaptive genetic changes for growth in human fibroblast cells (17).
Cloning of virus DNA and DNA sequence analysis.
Cloning of SV40 strains Rh911, N128, 777, and GM00637H was performed as described in detail previously (23). Briefly, when the cytopathic effect was advanced in SV40-infected CV-1 cells, a Hirt extraction (15) was performed; the resulting cleared lysate was digested with proteinase K and extracted with phenol, and the DNA was precipitated with isopropanol. After being washed once with 70% ethanol, the DNA was suspended in Tris-EDTA buffer (pH 8), cut with restriction enzyme EcoRI, and cloned into EcoRI-digested pUC-19 plasmid that had been pretreated with shrimp alkaline phosphatase (USB Corp., Cleveland, Ohio). DNA sequences were determined from plasmid clones that were purified by using a Qiagen (Valencia, Calif.) Plasmid Midi-prep kit. Double-stranded DNA sequencing using automated ABI Prism primer extension technology was performed commercially (SeqWright, Inc., or Lone Star, Inc., both in Houston, Tex.); both DNA strands were sequenced. The primers used for SV40 genomic DNA sequencing were purchased from Invitrogen Corp. (Carlsbad, Calif.), and their sequences are shown in Table 2.
TABLE 2.
DNA sequencing primers used for determination of complete SV40 genomic sequence
| Primera | Sequence (5′-3′) |
|---|---|
| JL 14-5236 | GCAGAGGCCGAGGCCGCCTCGG |
| RS 244-268 | CCACACCCTAACTGACACACATTCC |
| RS 598-622 | GCTACTGTGTCTGAAGCTGCTGCTG |
| RS 671-649 | GCAGCAGCGGCCTCTCCAGCAGC |
| RS 842-818 | GCAAAGCGCTCACACCAGTCACAG |
| RS 987-1012 | CAGTGTTCAGTATCTTGACCCCAGAC |
| RS 1095-1072 | CTGTGAGGTGAGCCTAGGAATGTC |
| RS 1358-1382 | GCCAAAGTCCTAATGTGCAGTCAGG |
| RS 1512-1487 | GGGGCCATCTTCATAAGCTTTTAGAG |
| RS 1664-1685 | CCTCAAATGGGCAATCCTGATG |
| RS 2068-2090 | GCAGATGAACACTGACCACAAGG |
| RS 2142-2121 | GGATCAGGAACCCAGCACTCC |
| RS 2249-2223 | GCTCATCAAGAAACACTGTGGTTGCTG |
| RS 2437-2462 | CAGGAGGACACAGAGGGTGGATGGGC |
| RS 2723-2699 | CCTCCCACACCTCCCCCTGAACCTG |
| RS 2933-2959 | GCCAATCTAAAACTCCAATTCCCATAG |
| RS 3325-3305 | GGGATTATTTGGATGGCAGTG |
| RS 3520-3497 | CTACATTAGCAGCTGCTTTGCTTG |
| RS 3610-3635 | GAATCCATTTTGGGCAACAAACAGTG |
| RS 3682-3657 | CAGGCTCTGCTGACATAGAAGAATGG |
| RS 3890-3866 | CAGCCCAGCCACTATAAGTACCATG |
| RS 3927-3953 | GTACTGAAATTCCAAGTACATCCCAAG |
| RS 4085-4058 | GAGGAAAGTTTGCCAGGTGGGTTAAAG |
| JL 4129-4150 | TATTCCTTATTAACCCCTTTAC |
| RS 4277-4254 | GTAACCTTTATAAGTAGGCATAAC |
| RS 4391-4413 | GCAATTCTGAAGGAAAGTCCTTG |
| RS 4415-4391 | CCCAAGGACTTTCCTTCAGAATTGC |
| JL 4527-4550 | GGCATTCCACCACTGCTCCCATTC |
| RS 4822-4800 | GCTGTGCTTACTGAGGATGAAGC |
| JL 5079-5056 | CTGATGAGAAAGGCATATTTAAAA |
Numbers represent nucleotide positions in SV40 reference strain 776 (SV40-776). From Stewart et al. (40).
Whole-genomic phylogenetic analysis.
A multiple sequence alignment of 14 whole-genomic SV40 sequences was performed with ClustalX V1.81 (43) by using the default parameters. The noncoding regulatory region of SV40 is subject to insertions, deletions, and/or duplications (INDELS) (23, 25, 31, 32). Alignment gaps were treated as missing data in the whole-genome analysis; therefore, part of the regulatory region (nucleotides 29 to 246 [according to SV40-776 numbering]) was excluded from the analysis. These nucleotides (29 to 246) encompass part of the early promoter and part of the enhancer (24, 25); the core ori was essentially intact (24, 25). Under these conditions, there were few alignment gaps and no ambiguous positions in the alignment. Phylogenetic trees were constructed by the maximum-parsimony and neighbor-joining methods. Maximum parsimony was implemented by use of PAUP* 4.0b (version 4.0b; Illinois Natural History Survey, Champaign, Ill.), and gaps were treated as missing data; however, treating gaps as a fifth character did not significantly alter the outcome. The data were bootstrapped with 1,000 replicates by using the full heuristic search option.
The neighbor-joining method (36) was implemented by using MEGA-2 (Molecular Evolutionary Genetics Analysis, version 2.1; Arizona State University, Tempe). Gaps were excluded from the analysis; however, including gaps under the “pairwise deletion” option did not alter the tree topology. Distances were calculated by using the two-parameter model of Kimura (20). The same analyses were performed for a 315-position alignment of T-ag-C sequences on the same taxa.
Congruence between data sets.
Congruence between the whole-genome and the T-ag-C data sets was assessed by the partition metric (8) and the quartets method (9) implemented in COMPONENT (version 2.00a; University of Auckland, Auckland, New Zealand). The partition metric indicates the level of disagreement between sets of trees; specifically, the number of clusters found in one or the other tree, but not in both. In contrast, the quartets measure is an indication of the number of quartets (smallest unrooted sets of four taxa) that are clearly resolved and explicitly agree. One hundred unrooted maximum-parsimony trees were generated by nonparametric bootstrap replication for the following data sets: the whole-genomic data set (G); an alternate whole-genomic data set (G+); the T-ag-C data set for the same taxa (TAG); the T-ag-C data set for the same taxa, treating gaps as a fifth character (TAG5th); and two sets of randomly generated trees (RND and RND+). The following sets of 100 trees were compared in a pair-wise manner (100 × 100 = 10,000 total comparisons) by using COMPONENT (version 2.00a) for both the partition metric and quartets: (G) × (G+), (G) × (TAG), (G) × (TAG5th), and (RND) × (RND+). These comparisons provide an indication of the level of congruence between data sets, relative to the internal congruence in the whole genomic data set as opposed to sets of randomly generated trees.
Preliminary survey of all T-ag-C sequences.
The maximum-parsimony and neighbor-joining methods were implemented as in the whole-genome analysis, except that gaps were treated as fifth characters in the preliminary survey of the genetic variation in all available T-ag-C sequences (14 sequences were acquired by entire genome sequencing, and an additional 16 were obtained by PCR from a variety of sources). In contrast to the regulatory region, gaps in the T-ag-C region consist of small INDELs and are less likely to be phylogenetically problematic; therefore, they were included in the analysis. Treating gaps as missing data resulted in similar, yet less-resolved, tree topologies.
RESULTS
Whole-genomic phylogenetic analysis.
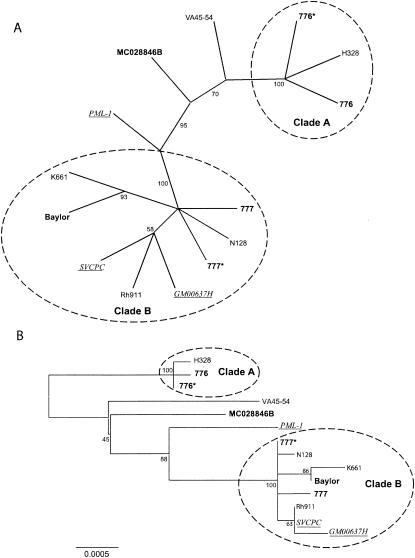
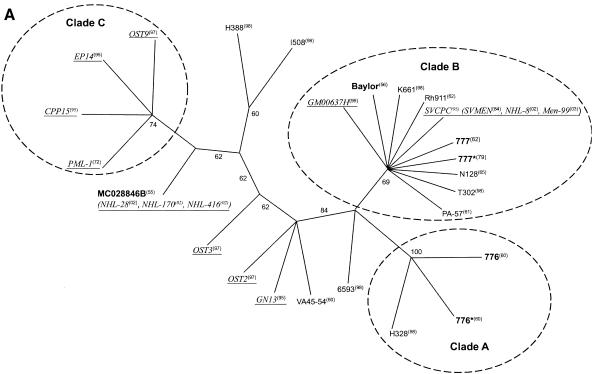
The whole-genome alignment consisted of 5,303 positions, 98.9% of which were invariant. When gaps were excluded from the analysis, 26 positions were parsimony informative and 32 were uninformative. The strict-consensus maximum-parsimony tree was highly consistent (consistency index [Ci], 0.967; rescaled consistency index [Rc], 0.942) and had an overall length of 60 steps. Two clades were consistently resolved, by both parsimony and distance methods (Fig. 1): clade A (containing 776, 776*, and H328) and clade B [containing 777, 777*, N128, GM00637H, Rh911, SVCPC(SVMEN), Baylor, and K661]. Isolates that grouped to clade B were derived from all three source populations (monkeys, contaminated vaccines, and humans). The polymorphisms detected in viruses within clades A and B and their locations are itemized in Table 3. Several isolates were ungrouped (PML-1, MC028846B, and VA45-54).
FIG. 1.
Phylogenetic tree for SV40 based on complete genome sequences. (A) Unrooted consensus tree of 1,000 bootstrap replicates of the whole-genome data, generated by the maximum-parsimony method. Conventions for labels are as follows: monkey isolates are in roman type; vaccine isolates are in boldface type; human isolates are in underlined italic type. (B) Consensus tree of 1,000 bootstrap replicates of the whole-genome data, generated by the neighbor-joining method. Distances are proportional to the number of mutational changes, after the Kimura (20) correction has been applied; the scale is in proportion of changes relative to the whole genome. The scale bar (0.0005) represents the number of substitutions per site. Numbers proximal to nodes in the tree indicate the proportion of 1,000 bootstrap replications; only bootstrap values >50% are shown. Two clades (A and B) are resolved, with several outlier sequences.
TABLE 3.
Polymorphisms in SV40 strains within clades A and Ba
| Clade | Nucleotide position in:
|
Base identified in virus strain (GenBank no.):
|
Gene or region | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SV40-776 | SVCPC | SV40-776 (J02400) | 776* (AF316139) | H328 (AF316141) | 777 (AF332562) | 777* (AY271817) | SVCPC (AF156108) | Baylor (AF155359) | Rh911 (AF316140) | N128 (AY120890) | GM00637H (AF345344) | K661 (AF038616) | ||
| A | 948 (C) | 876 (T) | C | T | T | VP2/VP3 | ||||||||
| 4579 (A) | 4516 (A) | A | A | C | T-ag intron | |||||||||
| B | 749 (C) | 677 | G | C | C | C | C | C | C | C | VP2 | |||
| 849 (G) | 777 | G | G | A | G | A | G | A | G | VP2 | ||||
| 945 (T) | 873 | C | T | T | T | T | T | T | T | VP2/VP3 | ||||
| 1727 (C) | 1655 | C | C | C | C | C | C | A | C | VP1 | ||||
| 2057 (G) | 1985 | G | G | G | G | G | G | T | G | VP1 | ||||
| 2751 (A) | 2679 | G | G | G | G | G | G | G | G | T-ag | ||||
| 2757 (G) | 2686 | A | A | A | A | A | A | A | A | T-ag | ||||
| 2796 (A) | 2733 (A) | ins 9 nt | ins 9 nt | ins 9 nt | ins 9 nt | ins 9 nt | ins 9 nt | ins 9 nt | ins 9 nt | T-ag | ||||
| 2801 (C) | 2738 | C | C | C | C | C | C | C | del 3 nt | T-ag | ||||
| 2817 (G) | 2754 | A | A | A | A | A | A | A | A | T-ag | ||||
| 2874 (A) | 2811 | A | A | A | A | A | A | A | del 9 nt | T-ag | ||||
| 2907 (T) | 2844 | A | A | A | A | A | A | A | A | T-ag | ||||
| 2912 (C) | 2849 | C | C | C | T | C | C | C | C | T-ag | ||||
| 2951 (T) | 2888 | C | C | C | C | C | C | C | C | T-ag | ||||
| 3117 (T) | 3054 | C | C | C | C | C | C | C | C | T-ag | ||||
| 3755 (A) | 3692 | G | G | G | G | G | G | G | G | T-ag | ||||
| 4045 (T) | 3982 | T | T | T | T | T | G | T | T | T-ag | ||||
| 4071 (T) | 4008 | A | A | A | A | A | A | A | A | T-ag | ||||
| 4110 (C) | 4047 | T | T | T | T | T | T | T | T | T-ag | ||||
| 4299 (C) | 4236 | T | T | T | T | T | T | T | T | T-ag | ||||
| 4642 (G) | 4579 | T | T | T | T | T | T | T | T | T-ag | ||||
| 4839 (C) | 4776 | T | T | T | T | T | T | T | T | t-ag | ||||
| 4879 (C) | 4816 | T | T | T | T | T | T | T | T | t-ag | ||||
| 5164 (C) | 5101 | C | C | C | A | C | C | C | A | Regulatory | ||||
| 5209 (C) | 5146 | T | T | T | T | T | T | T | T | Regulatory | ||||
Sequences given are for the noncoding strand; ins, insertion; del, deletion; nt, nucleotides; T-ag, large tumor antigen; t-ag, small tumor antigen.
T-ag-C phylogenetic analysis.
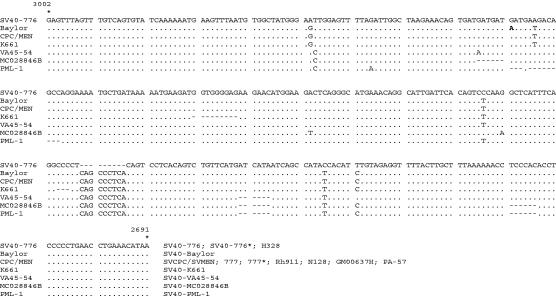
Analyses of the SV40 C-terminal T-ag variable domain sequences were performed to determine if the T-ag-C sequences are capable of resolving relationships that are congruent with the whole-genome analysis. If the T-ag-C sequence phylogeny is highly congruent with the whole-genome analysis, then rapid and cost-effective strain identification becomes possible, as does a preliminary analysis of a larger number of samples. A partial listing of T-ag-C sequences is shown in Fig. 2. The T-ag-C alignment consisted of 315 positions; when gaps were treated as missing data, 93% of the positions were invariant, with 9 parsimony-informative and 10 uninformative sites (Ci = 0.558; Rc = 0.400) and an overall length of 34 steps (data not shown). When gaps were included as a fifth character, then only 68% of the positions were invariant, with 51 parsimony-informative and 49 uninformative sites (Ci = 0.966; Rc = 0.93) and an overall length of 59 steps.
FIG. 2.
Nucleotide sequences of the T-ag-C domain from several SV40 isolates. The sequence given is for the coding strand, and the numbering is according to the system for SV40-776. Alignments are compared to the sequence of SV40-776; a dot represents identity, and a dash represents a deleted nucleotide.
The major groupings were the same regardless of whether gaps were treated as missing data or as fifth characters, but treating gaps as missing data resulted in very little resolution. The parsimony consensus trees resulting from treating gaps as fifth characters are presented because of higher resolution, higher Ci index, and slightly higher congruence to the whole-genome data as measured by quartet(s) (see “Congruence between data sets” below [Fig. 3]). Both distance and parsimony methods resolved the same groupings as the whole-genome data, although the topology is slightly different for intermediate taxa.
FIG. 3.
Phylogenetic tree for SV40 based on T-ag-C sequences from completely sequenced genomes. (A) Unrooted consensus tree of 1,000 bootstrap replicates of T-ag-C data (for the same taxa as the whole-genome analysis), generated by the maximum-parsimony method. Labeling conventions are the same as for Fig. 1. (B) Consensus tree of 1,000 bootstrap replicates of T-ag-C data, generated by the neighbor-joining method. Distances are proportional to the number of mutational changes, after the Kimura (20) correction has been applied; the scale is in proportion of changes relative to the T-ag-C gene. Only bootstrap values >50% are shown. The same two clades (A and B) as those based on complete genome analyses (Fig. 1) were resolved, with the same isolates remaining ungrouped.
Congruence between data sets.
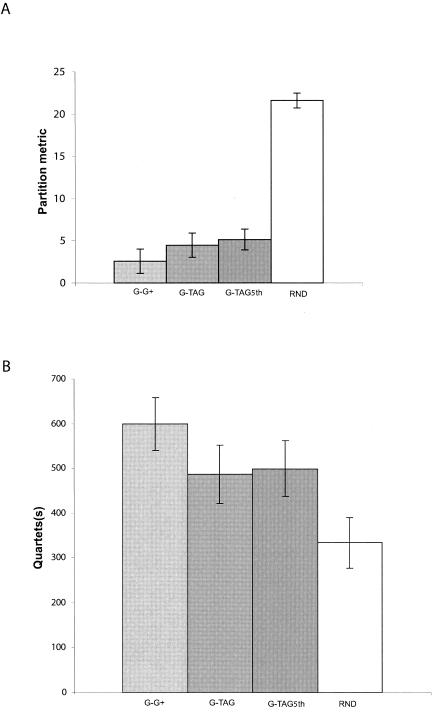
The partition metric is a measure of dissimilarity (the number of clusters in one tree or the other tree but not in both). Penny and Hendy (33) computed exact probabilities for comparing two trees up to 16 taxa by using the partition metric. The probability that two trees with 14 taxa have a partition metric <6 is very small (P < 0.01). The whole-genomic and T-ag-C data sets have few partitions that disagree, in contrast to comparisons between randomly generated trees (Fig. 4A). The comparison between the T-ag-C data set (TAG) and the whole-genome data set (G) showed a level of congruence similar to that observed between two alternate whole-genomic data sets (G × G+). The quartets metric(s) indicates strict agreement between resolved quartets between trees. The whole-genomic and T-ag-C data sets have a large number of quartets that agree (Fig. 4B). Both metrics indicate that the T-ag-C data set is highly congruent with the whole-genome data set. The levels of congruence relative to randomly generated trees and to internal congruence of the whole-genomic data set are also indicated (Fig. 4A and B).
FIG. 4.
Illustration of congruence between phylogenetic trees based on SV40 complete-genome sequences and sequences of T-ag-C only. (A) Averages and standard deviations of the partition metric for a comparison of 10,000 pair-wise comparisons between sets of trees (100 each) generated by nonparametric bootstrap replicates using maximum-parsimony analysis. (B) Averages and standard deviations of quartets for a comparison of 10,000 pair-wise comparisons between sets of 100 trees generated by nonparametric bootstrap replicates using maximum-parsimony. Abbreviations: G, the whole genomic data set; G+, 100 additional bootstrap replicate trees of the genomic data set; TAG, the T-ag-C data set for the same taxa; TAG5th, the T-ag-C data set for the same taxa, treating gaps as a fifth character; RND, a comparison of two sets of randomly generated trees.
Preliminary survey of all T-ag-C sequences.
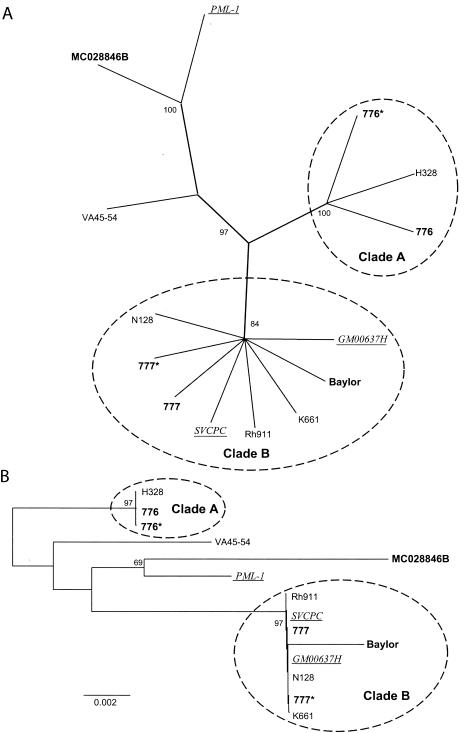
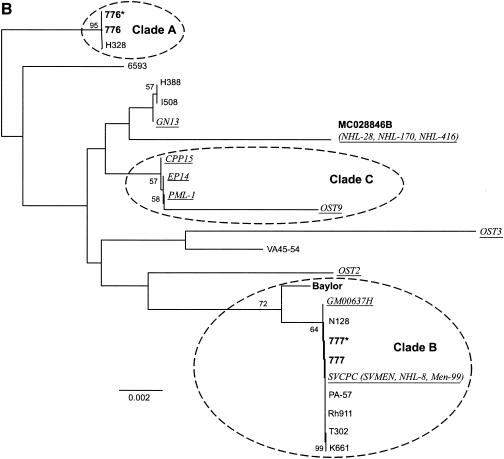
The consensus of maximum-parsimony trees of all available T-ag-C sequences (Ci = 0.718; Rc = 0.587; total length = 142) is indicated in Fig. 5A. The previously identified clusters (clades A and B) were clearly resolved. In addition, a third group (clade C) was resolved by the majority of trees (Fig. 5B). All three clades were resolved by both distance and parsimony methods. Clade C consists of human isolate PML-1 and sequences that were associated with human tumors.
FIG. 5.
Phylogenetic tree for SV40 based on all available T-ag-C sequences. (A) Unrooted consensus tree of 1,000 bootstrap replicates of all available T-ag-C sequences, generated by maximum-parsimony analysis. Labeling conventions are the same as for Fig. 1. Sequences in parentheses are from independent sources (Table 1) but are the same as the sequence preceding the parentheses. The year of sample origin, isolation, or detection is indicated in superscript in parentheses. (B) Consensus tree of 1,000 bootstrap replicates of all available T-ag-C data, generated by the neighbor-joining method. Distances are proportional to the number of mutational changes, after the Kimura (20) correction has been applied; the scale is in proportion to changes relative to the T-ag-C gene. Labeling conventions are the same as for Fig. 1 and Fig. 5A. Clades A and B are supported by the whole-genome analysis (Fig. 1). Clade C is of interest because it consists of a human isolate and sequences associated with tumors in humans. Only bootstrap values >50% are shown.
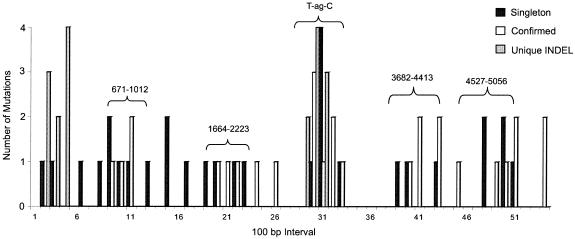
Polymorphic regions in SV40 genome.
The whole-genome sequences were further analyzed to identify other polymorphic regions in addition to the T-ag-C terminal domain. The entire viral genome was divided into 100-bp intervals and scored for different types of mutations (Fig. 6). “Singletons” occur only once and might include PCR errors or base identification errors, “confirmed” mutations occur in more than one sequence at that nucleotide position, and “unique INDEL” events represent unique gaps. In addition to T-ag-C, other polymorphic regions exist, as indicated in Fig. 6. The use of existing sequencing primers (Table 2) covering positions 671 to 1012, 1664 to 2223, 3682 to 4413, and 4527 to 5056 (SV40-776 numbering), in addition to T-ag-C, would capture 89% of the confirmed known polymorphisms and 63% of the singletons.
FIG. 6.
Polymorphisms in the sequence alignment plotted against position in the alignment for the entire genome of SV40 (divided into 100-bp intervals). Singletons have been detected only once, confirmed changes have occurred in more than one sequence, and unique INDEL changes represent gaps. The T-ag-C gene region has the highest level of polymorphism. Four other polymorphic regions that can be analyzed by using existing DNA-sequencing primers were identified (Table 2); nucleotide positions (SV40-776 numbering) are shown over those regions.
DISCUSSION
This study is the first to examine evolutionary relationships in polyomavirus SV40. Although the overall level of genetic variation in SV40 is minimal, distinct clades were found, and these are strongly supported by whole-genome analysis using neighbor-joining and maximum-parsimony methods. The same groups were resolved when T-ag-C data alone were used, and the two data sets did not appear to be significantly incongruous. The survey of T-ag-C sequences from all available sources indicated that additional genetic variation is likely to exist; the same clades were resolved as with the whole-genome analysis, as well as a group that exclusively contains human isolates (PML-1 and sequences associated with human cancers).
A portion of the regulatory region was excluded from this survey of SV40 genetic variation, because it is unclear whether the large insertions and deletions that are a characteristic of the region should be treated as single or multiple mutation events. The regulatory region contains enhancer and promoter sequences as well as the viral origin of DNA replication (ori). Genetic changes can occur within the SV40 regulatory region during viral growth in vivo (23, 31) and possibly in vitro under certain conditions (32) through mechanisms that are not understood. Genetic changes at the regulatory region typically consist of duplication or deletion (or both) of enhancer and/or promoter sequences. These changes occur relative to the structure of a viral species-specific basic regulatory region termed archetypal or protoarchetypal (3, 25, 35). In contrast, the T-ag-C region of SV40 strains can vary in sequence and in length but does not appear to change in response to growth in vitro or in vivo (18, 23, 24, 31). We and others have previously proposed the identification of SV40 strains based on T-ag-C sequences (18, 23, 24, 31, 40). The analyses described here confirmed the validity of the use of the T-ag-C domain for genotyping SV40 isolates and tissue-associated sequences.
The data suggest that several SV40 genotypes are common to different population sources (monkeys, contaminated vaccines, and humans). A possible explanation for this pattern is that the variation present in the simian population of SV40 was sampled during the manufacture of the poliovaccines and was transferred to the human population. Data from the present study based on known isolates and genomic fragments indicate that monkey and vaccine populations contain A and B clade viruses, whereas the human population contains representatives in the B and C clades. It is of interest that vaccine-derived viruses appear to overlap with the monkey and human populations, supporting the hypothesis that contaminated vaccines played a role in the introduction of SV40 into humans (Fig. 5B). It should be noted that clade B isolates from humans have not diverged from monkey isolates of clade B genotype, showing that adaptation is not essential for viral survival in humans. It is possible that clade C is representative of viruses that have undergone genetic change during adaptation to human hosts. However, since DNA viruses evolve slowly, it is more likely that clade C occurs at some frequency in monkey populations but remains undiscovered. It will be necessary to have a more comprehensive knowledge of SV40 genetic variation in simian hosts to be able to interpret the significance of the relative distribution of strains found in humans. If a particular genotype has a higher frequency in humans than in monkeys, it could be the result of selection, creating a founder effect in the new host environment. It is also possible that some viral sequences detected in human tumors represent dead-end infections unable to be transmitted from human to human. Additional sampling is necessary to determine if any particular genetic variant is found exclusively in human populations.
Prior to this analysis, phylogenetic studies of whole genomes of polyomaviruses had been performed only for JC virus (1-3, 19). The results obtained for SV40 are similar in that genotypes can be distinguished by analyzing a genomic region with sequence variability outside the viral regulatory region. This includes the T-ag-C region for SV40 and the V-T intergenic region between the T-antigen and VP1 coding regions for JC virus.
The terminology used for describing SV40 isolates is currently inconsistent and becomes more difficult with the realization that SV40 clades exist. Based on this study, we recommend the following terminology. The term “strain” should describe particular isolates. The word “genotype” should be utilized when distinguishing among SV40 isolates based on T-ag-C sequences. Viruses with the same genotype can vary at the regulatory region or the viral coat protein coding regions and should be referred to as “variants.” A “clade” (or “genogroup”) is a phylogenetic group of viruses with similar genotypes. In general, viruses within a clade differ very slightly. For example, SVCPC and SV40-777 differ by 3 of 5,180 nucleotides (in the VP2 gene) and are 99.94% similar. The T-ag-C region appears to be useful as a rapid means of classifying genotypes of SV40, as the whole-genome and T-ag-C data sets are highly congruent (disregarding rearrangements in the viral regulatory region).
This analysis, establishing SV40 genetic diversity, raises new questions. It emphasizes the need to learn more about the natural distribution of transmissible SV40 strains in monkeys. The biological significance of these groupings is unclear but warrants further investigation. It is of interest that preliminary findings from hamster studies indicate that isolate SVCPC (clade B) is more tumorigenic in vivo than VA45-54 (ungrouped) (R. A. Vilchez, C. Brayton, C. Wong, P. Zanwar, D. E. Killen, J. L. Jorgensen, and J. S. Butel, unpublished data). It will be important to determine the relative pathogenicity and tissue tropisms of representative viruses of clades A, B, and C. It would be informative, also, to obtain sequence data from additional polymorphic regions in the viral genome (Fig. 6) to increase phylogenetic resolution of known sequences. The ability to incorporate additional full-length genomes from humans, as well as monkeys, would also contribute to higher phylogenetic resolution. Greater numbers of T-ag-C sequences amplified from monkey and human tissues would help determine whether any human-specific viral clades do, in fact, exist. Finally, direct evidence of human-to-human transmission of SV40 would indicate if the sequences associated with human tumors represent transmissible variants or if they are at an evolutionary dead end.
Acknowledgments
This work was supported in part by a grant from the Center for Biologics Evaluation and Research, Food and Drug Administration, to J.S.B.; by grant CA096951 from the National Cancer Institute to J.S.B.; and by grants from the National Space Biomedical Research Institute (NASA Cooperative Agreement NCC-9-58) to G.E.F., R.C.W., and J.S.B.
We thank X. M. Dai for excellent technical assistance.
REFERENCES
- 1.Agostini, H. T., A. Deckhut, D. V. Jobes, R. Girones, G. Schlunck, M. G. Prost, C. Frias, E. Pérez-Trallero, C. F. Ryschkewitsch, and G. L. Stoner. 2001. Genotypes of JC virus in East, Central and Southwest Europe. J. Gen. Virol. 82:1221-1231. [DOI] [PubMed] [Google Scholar]
- 2.Agostini, H. T., D. V. Jobes, and G. L. Stoner. 2001. Molecular evolution and epidemiology of JC virus, p. 491-526. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss, Inc., New York, N.Y.
- 3.Agostini, H. T., C. F. Ryschkewitsch, E. J. Singer, and G. L. Stoner. 1997. JC virus regulatory region rearrangements and genotypes in progressive multifocal leukoencephalopathy: two independent aspects of virus variation. J. Gen. Virol. 78:659-664. [DOI] [PubMed] [Google Scholar]
- 4.Arrington, A. S., and J. S. Butel. 2001. SV40 and human tumors, p. 461-489. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. John Wiley & Sons, New York, N.Y.
- 5.Arrington, A. S., M. S. Moore, and J. S. Butel. 2004. SV40-positive brain tumor in scientist with risk of laboratory exposure to the virus. Oncogene 23:2231-2235. [DOI] [PubMed] [Google Scholar]
- 6.Buckler, C. E., and N. P. Salzman. 1986. Annotated nucleotide sequence and restriction site lists for selected papovavirus strains, p. 379-447. In N. P. Salzman (ed.), The Papovaviridae, vol. 1. The polyomaviruses. Plenum Press, New York, N.Y. [Google Scholar]
- 7.Butel, J. S. 2001. Increasing evidence for involvement of SV40 in human cancer. Dis. Markers 17:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, W. H. E. 1985. Optimal algorithms for comparing trees with labeled leaves. J. Classification 2:7-28. [Google Scholar]
- 9.Day, W. H. E. 1986. Analysis of quartet dissimilarity measures between undirected phylogenetic trees. Syst. Zool. 35:325-333. [Google Scholar]
- 10.Deichman, G. I., and T. E. Kluchareva. 1966. Loss of transplantation antigen in primary simian virus 40-induced tumors and their metastases. J. Natl. Cancer Inst. 36:647-655. [DOI] [PubMed] [Google Scholar]
- 11.Field, K. G., G. J. Olsen, D. J. Lane, S. J. Giovannoni, M. T. Ghiselin, E. C. Raff, N. R. Pace, and R. A. Raff. 1988. Molecular phylogeny of the animal kingdom. Science 239:748-753. [DOI] [PubMed] [Google Scholar]
- 12.Gerber, P. 1962. An infectious deoxyribonucleic acid derived from vacuolating virus (SV40). Virology 16:96-97. [DOI] [PubMed] [Google Scholar]
- 13.Girardi, A. J. 1965. Prevention of SV40 virus oncogenesis in hamsters. I. Tumor resistance induced by human cells transformed by SV40. Proc. Natl. Acad. Sci. USA 54:445-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardi, A. J., B. H. Sweet, V. B. Slotnick, and M. R. Hilleman. 1962. Development of tumors in hamsters inoculated in the neonatal period with vacuolating virus, SV40. Proc. Soc. Exp. Biol. Med. 109:649-660. [DOI] [PubMed] [Google Scholar]
- 15.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Hsiung, G. D., and W. H. Gaylord, Jr. 1961. The vacuolating virus of monkeys. I. Isolation, growth characteristics, and inclusion body formation. J. Exp. Med. 114:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, K. C., E. F. Yamasaki, and R. M. Snapka. 1999. Maintenance of episomal SV40 genomes in GM637 human fibroblasts. Virology 262:457-469. [DOI] [PubMed] [Google Scholar]
- 18.Ilyinskii, P. O., M. D. Daniel, C. J. Horvath, and R. C. Desrosiers. 1992. Genetic analysis of simian virus 40 from brains and kidneys of macaque monkeys. J. Virol. 66:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jobes, D. V., S. C. Chima, C. F. Ryschkewitsch, and G. L. Stoner. 1998. Phylogenetic analysis of 22 complete genomes of the human polyomavirus JC virus. J. Gen. Virol. 79:2491-2498. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Krieg, P., and G. Scherer. 1984. Cloning of SV40 genomes from human brain tumors. Virology 138:336-340. [DOI] [PubMed] [Google Scholar]
- 22.Lednicky, J., and W. R. Folk. 1992. Two synthetic Sp1-binding sites functionally substitute for the 21-base-pair repeat region to activate simian virus 40 growth in CV-1 cells. J. Virol. 66:6379-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lednicky, J. A., A. S. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lednicky, J. A., and J. S. Butel. 1997. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J. Gen. Virol. 78:1697-1705. [DOI] [PubMed] [Google Scholar]
- 25.Lednicky, J. A., and J. S. Butel. 2001. Simian virus 40 regulatory region structural diversity and the association of viral archetypal regulatory regions with human brain tumors. Semin. Cancer Biol. 11:39-47. [DOI] [PubMed] [Google Scholar]
- 26.Lednicky, J. A., R. L. Garcea, D. J. Bergsagel, and J. S. Butel. 1995. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology 212:710-717. [DOI] [PubMed] [Google Scholar]
- 27.Lednicky, J. A., A. R. Stewart, J. J. Jenkins III, M. J. Finegold, and J. S. Butel. 1997. SV40 DNA in human osteosarcomas shows sequence variation among T-antigen genes. Int. J. Cancer 72:791-800. [DOI] [PubMed] [Google Scholar]
- 28.Lednicky, J. A., C. Wong, and J. S. Butel. 1995. Artificial modification of the viral regulatory region improves tissue culture growth of SV40 strain 776. Virus Res. 35:143-153. [DOI] [PubMed] [Google Scholar]
- 29.Martin, J. D. 1989. Regulatory sequences of SV40 variants isolated from patients with progressive multifocal leukoencephalopathy. Virus Res. 14:85-94. [DOI] [PubMed] [Google Scholar]
- 30.Melnick, J. L., and S. Stinebaugh. 1962. Excretion of vacuolating SV-40 virus (papova virus group) after ingestion as a contaminant of oral poliovaccine. Proc. Soc. Exp. Biol. Med. 109:965-968. [DOI] [PubMed] [Google Scholar]
- 31.Newman, J. S., G. B. Baskin, and R. J. Frisque. 1998. Identification of SV40 in brain, kidney and urine of healthy and SIV-infected rhesus monkeys. J. Neurovirol. 4:394-406. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill, F. J., J. E. Greenlee, and H. Carney. 2003. The archetype enhancer of simian virus 40 DNA is duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells. Virology 310:173-182. [DOI] [PubMed] [Google Scholar]
- 33.Penny, D., and M. D. Hendy. 1985. The use of tree comparison metrics. Syst. Zool. 34:75-82. [Google Scholar]
- 34.Rizzo, P., I. Di Resta, A. Powers, H. Ratner, and M. Carbone. 1999. Unique strains of SV40 in commercial poliovaccines from 1955 not readily identifiable with current testing for SV40 infection. Cancer Res. 59:6103-6108. [PubMed] [Google Scholar]
- 35.Rubinstein, R., B. C. Schoonakker, and E. H. Harley. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J. Virol. 65:1600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sangar, D., P. A. Pipkin, D. J. Wood, and P. D. Minor. 1999. Examination of poliovirus vaccine preparations for SV40 sequences. Biologicals 27:1-10. [DOI] [PubMed] [Google Scholar]
- 38.Simon, M. A., P. O. Ilyinskii, G. B. Baskin, H. Y. Knight, D. R. Pauley, and A. A. Lackner. 1999. Association of simian virus 40 with a central nervous system lesion distinct from progressive multifocal leukoencephalopathy in macaques with AIDS. Am. J. Pathol. 154:437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, A. R., J. A. Lednicky, U. S. Benzick, M. J. Tevethia, and J. S. Butel. 1996. Identification of a variable region at the carboxy terminus of SV40 large T-antigen. Virology 221:355-361. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, A. R., J. A. Lednicky, and J. S. Butel. 1998. Sequence analyses of human tumor-associated SV40 DNAs and SV40 viral isolates from monkeys and humans. J. Neurovirol. 4:182-193. [DOI] [PubMed] [Google Scholar]
- 41.Stinebaugh, S., and J. L. Melnick. 1962. Plaque formation by vacuolating virus, SV40. Virology 16:348-349. [DOI] [PubMed] [Google Scholar]
- 42.Sweet, B. H., and M. R. Hilleman. 1960. The vacuolating virus, S.V.40. Proc. Soc. Exp. Biol. Med. 105:420-427. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilchez, R. A., and J. S. Butel. 2003. SV40 in human brain cancers and non-Hodgkin's lymphoma. Oncogene 22:5164-5172. [DOI] [PubMed] [Google Scholar]
- 45.Vilchez, R. A., J. A. Lednicky, S. J. Halvorson, Z. S. White, C. A. Kozinetz, and J. S. Butel. 2002. Detection of polyomavirus simian virus 40 tumor antigen DNA in AIDS-related systemic non-Hodgkin lymphoma. J. Acquir. Immune Defic. Syndr. 29:109-116. [DOI] [PubMed] [Google Scholar]
- 46.Vilchez, R. A., C. R. Madden, C. A. Kozinetz, S. J. Halvorson, Z. S. White, J. L. Jorgensen, C. J. Finch, and J. S. Butel. 2002. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet 359:817-823. [DOI] [PubMed] [Google Scholar]
- 47.Weiner, L. P., R. M. Herndon, O. Narayan, R. T. Johnson, K. Shah, L. J. Rubinstein, T. J. Preziosi, and F. K. Conley. 1972. Isolation of virus related to SV40 from patients with progressive multifocal leukoencephalopathy. N. Engl. J. Med. 286:385-390. [DOI] [PubMed] [Google Scholar]
- 48.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]