Abstract
Cellular functions in Bacteria, such as chromosome segregation and cytokinesis, result from cascades of molecular events operating largely as self-contained modules. Regulated timing of these cellular modules stems from global genetic circuits that allow precise temporal activation with respect to cell cycle progression and cell differentiation. Critically, many of these functions occur at defined locations within the cell, and therefore regulators of each module must communicate to remain coordinated in space. In this perspective, we highlight recent discoveries in Caulobacter crescentus asymmetric cell division to illuminate diverse mechanisms by which a cellular compass, composed of scaffolding and signaling proteins, directs cell cycle modules to their exact cellular addresses.
Introduction
Every cell cycle, Caulobacter crescentus divides asymmetrically to produce two different progeny, a swarmer cell and a stalked cell, each with distinct morphological features and regulatory programs (Figure 1a). The swarmer (G1-phase) cell is motile and unable to replicate its genome or to divide. The swarmer cell differentiates into a stalked (S-phase) cell by shedding its flagellum, replacing it with a stalk, and initiating DNA replication. The elongating stalked cell synthesizes a flagellum at the cell pole opposite its stalk. Cytokinesis of the inner membrane of the predivisional (G2-phase) cell compartmentalizes the replicated chromosomes into the nascent swarmer and stalked progeny, asymmetrically partitioning cell fate factors to reset their developmental programs before the conclusion of cell division [1].
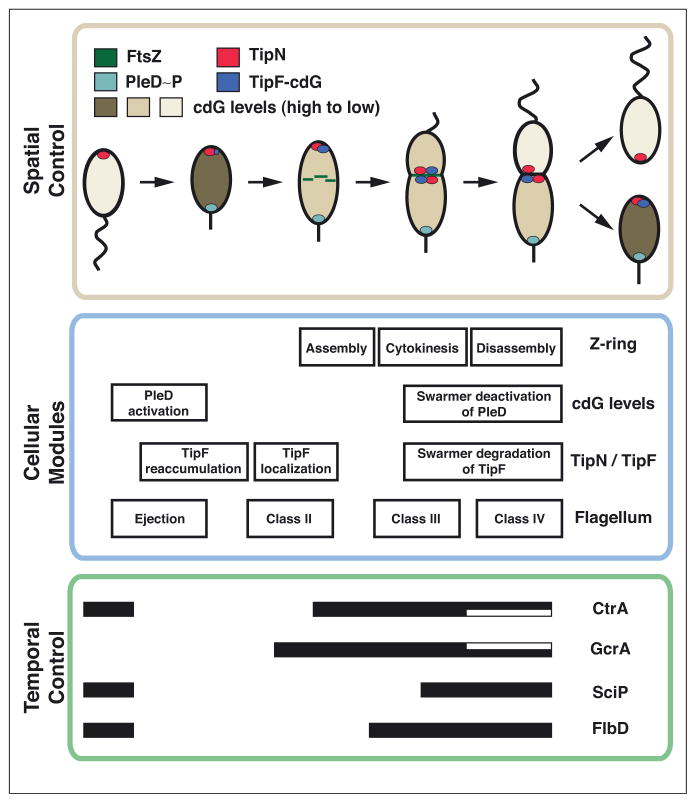
Figure 1.
C. crescentus cell cycle control architecture. (a) The C. crescentus cell cycle can be viewed as a set of cellular modules (blue box), coordinated spatially (brown box) and temporally (green box). Top panel, Spatial Control. The swarmer cell has a single polar flagellum. The phosphatase PleC (orange) is localized at the flagellar pole. The swarmer cell begins to differentiate into a stalked cell. PleC becomes diffuse and is replaced at the differentiating pole with the kinase DivJ (blue). Activation of DivJ leads to the initiation of DNA replication. During this differentiation period, ejection of the flagellum permits construction of a stalk at the same cell pole. Chromosome replication and segregation then proceed simultaneously as the cell grows into a predivisional state. During this time, the cell begins assembling the cytokinesis machinery, whose core is the polymerizing GTPase FtsZ, at midcell (green). At the new cell pole, the de novo construction of a single flagellum occurs concurrently with the assembly of a set of scaffolding and signaling proteins, including PleC. As chromosome replication completes, the FtsZ ‘Z-ring’ constricts and disassembles, closing the inner membrane and separating the cytoplasm into two compartments. Cytoplasmic compartmentalization sequesters multiple signaling factors, including PleC at the new pole and DivJ at the old pole, enabling each chamber to initiate divergent genetic programs before full separation of the daughter cells. The cell type-specific presence of the master regulator CtrA, a downstream target of DivJ and PleC activity, is shown in grayscale to represent its abundance over the course of the cell cycle. Middle panel, Cellular Modules. Specific cell cycle events are shown as ‘Cellular Modules.’ Seven cellular modules are highlighted here; many more exist and have been omitted for clarity. Bottom panel, Temporal Control. Black bars indicate the abundance time period across the cell cycle for the master regulators DnaA, CcrM, GcrA, CtrA, SciP and MucR1/MucR2. Following cell division, the black top half of the bar reflects levels in the swarmer progeny and the black lower half of the bar reflects levels in the stalked progeny. Note that all master regulators except for MucR1/MucR2 are under cell cycle transcriptional control. (b) A temporally controlled transcription circuit (green) interfaces with spatially resolved signaling mechanisms (brown) to coordinate modular cellular processes (blue). Arrows indicate the connection between the modules and the proteins at the interface. Five cellular modules are highlighted here; many more exist and have been omitted for clarity. The genetic circuit guides the timing of expression of genes in all five modules. The activation and inhibition relationships between DnaA, CcrM, GcrA, CtrA, SciP, and MucR1/2 are illustrated. Spatial control results from the differential localization of distinct scaffolding proteins, which localize signaling proteins to the cell poles to generate cellular compartment-specific signaling states. Members of the compartment-sensing component (single domain response regulator DivK and diguanylate cyclase PleD) are modulated by two membrane histidine kinases PleC and DivJ, that act as swarmer and stalked cell determinants, respectively, each residing in a different pole. The compartment-sensing component modulates the localization of the flagellum and the stalk and also provides inputs for the asymmetry determination module. Proteins involved in the asymmetry determination module are shown in Figure 2a.
Asymmetric division requires mechanisms for directing each daughter cell to initiate differential expression of genes as a function of cell cycle progression. Transcript levels of at least 400 genes (~10% of the annotated open reading frames in the C. crescentus genome) vary over the course of the cell cycle, including those encoding factors required for the initiation of DNA replication, chromosome segregation, cytokinesis, and biogenesis of the stalk, flagellum, pili, and chemotaxis machinery [2,3•]. A cyclical genetic circuit involving seven master regulator proteins (CtrA, GcrA, DnaA, SciP, MucR1, MucR2, and CcrM) drives cell cycle regulated transcription [4], accounting for 60% of all cell cycle regulated genes [3•] (Figure 1a,b). These regulators peak in abundance out of phase with each other, promoting distinct events at specific times during the cell cycle stages (Figure 1a). However, temporal control of gene expression alone is not sufficient to specify the subcellular locations of cellular machinery. The core genetic circuit operates in concert with a cellular compass, composed of signaling and scaffolding proteins, to direct cellular processes in both time and space (Figure 1b).
Specific cell cycle events are organized into functional cellular modules, such as defining a division plane or assembling a flagellum. These modules are self-contained such that, once initiated, the following steps operate as a cascade without the need for external signals (i.e., flagellum assembly, compartment identification). A module communicates with other modules, first to resolve when and where to execute its function, and second to communicate the completion of its function (e.g. completing DNA segregation). Two design principles emerge from the cell cycle control diagram shown in Figure 1b. First, the system is highly integrated, with every module communicating with at least two other modules. Second, every cellular module integrates both temporal and spatial inputs. Below, we provide three examples illustrating different aspects of how temporally controlled genetic modules interface with spatial signals: (i) the regulation of Z-ring polymerization via combinatorial transcriptional control and a gradient of an inhibitory protein, (ii) the control of the levels and activity of the CtrA master regulator via spatially resolved signaling and proteolysis, and (iii) the regulation of flagellum assembly and ejection via differential concentration of a second messenger.
Z-ring dynamics: coupling DNA replication and segregation, cell cycle progression, and cell division
Cytokinesis in C. crescentus is driven by the tubulin homolog FtsZ, which polymerizes while bound to GTP [5]. Polymerized FtsZ filaments encircle the division site as a loosely knit suprastructure (Z-ring), tethered to the cytoplasmic membrane via FzlC and FtsA [6]. Subsequently, the Z-ring recruits at least 20 different proteins to the ‘divisome’ complex, which can then execute cytokinesis [5]. Hydrolysis of GTP destabilizes the FtsZ filaments, leading to constriction.
The timing of divisome assembly and disassembly is controlled transcriptionally and post-translationally to ensure that cell division occurs only upon completion of chromosome replication and segregation. Transcription of ftsZ largely occurs in S-phase, concurrent with the initiation of DNA replication (Figure 1a), as a result of combinatorial transcriptional control by the global regulators DnaA, GcrA, and CtrA [3•,4]. Additionally, deletion of the methyltransferase ccrM dramatically reduces ftsZ expression [7], likely by decreasing GcrA-dependent transcription [8••]. Notably, a ccrM deletion can be rescued by mutations in the phosphoenolpyruvate-carbohydrate phosphotransfer system (PTS) that upregulates ftsZ transcription, suggestive of additional layers of nutrient-sensing transcriptional regulation [9,10]. Indeed, several studies have linked expression levels of GcrA-regulated transcription to levels of the alarmone (p)ppGpp [8••,10,11].
At the protein level, the NAD+/NADH dependent factors KidO and GdhZ inhibit FtsZ polymerization. These factors are thought to tune cytokinesis timing by preventing premature cytokinesis, promoting Z-ring disassembly upon completion of cytokinesis, and providing a mechanism for the cell to inhibit cytokinesis during periods of starvation [12–14]. ClpXP-mediated proteolysis of KidO, GdhZ, and FtsZ, further regulate the timing of Z-ring constriction.
Z-ring assembly must occur at midcell following completion of DNA replication to prevent chromosome breakage and thus must respond to a compass cue reporting on DNA replication and segregation. In Escherichia coli, Z-ring positioning is achieved via the MinCDE system [15]. In C. crescentus, the essential ATPase MipZ, which is closely related to MinD, not only regulates Z-ring positioning, but also spatially connects cell division with DNA replication and segregation [16] (Figure 1b). MipZ directly stimulates the GTPase activity of FtsZ to inhibit its polymerization [16]. The FtsZ-inhibitory activity of MipZ is spatially restricted such that it is lowest near the midcell before cell division. This localization is achieved by direct physical interaction of MipZ with the ParB centromere-binding protein. ParB, in turn, tightly localizes to the self-organizing multimeric polar matrix PopZ at both cell poles upon the segregation of the origin region of the chromosome [17–19]. The MipZ concentration gradient that descends from both poles reaches low levels only in the middle of the cell, permitting FtsZ polymerization at that location [20]. Thus, MipZ localization reports on chromosome segregation, thereby intimately linking replication and segregation of the chromosome to placement of the division site.
Spatially, the Z-ring also directs two other critical aspects of C. crescentus asymmetry, described below. Critically, constriction of the Z-ring and closure of the inner membrane compartmentalizes the predivisional cell [21], which asymmetrically sequesters cell fate factors and enables completion of S->G2 transition (Figure 1a). Further, before constriction, the Z-ring recruits the ‘birthmark’ protein TipN, which marks the new cell pole for flagellum construction in both daughter cells (Figure 3), coordinating cytokinesis of the mother cell with the polarity of daughter cells [22,23].
Figure 3.
Regulation of flagellum assembly and ejection. Top panel, Spatial Control. A diagram of the C. crescentus cell cycle highlights proteins that spatially regulate flagellum assembly and ejection. The distributions of FtsZ (green), cdG (brown), PleD (light blue), TipF (dark blue), and TipN (red) are shown throughout the cell cycle. The swarmer cell, with low cdG, has no TipF, as TipF is degraded when not bound to cdG. Upon differentiation and rising levels of cdG, TipF begins to accumulate with TipN at the new pole. TipF accumulation at the new pole permits recruitment of the initial flagellar base components. Z-ring assembly in early predivisional cells eventually leads to a relocalization of TipN and TipF at the division plane, defining the future ‘new poles’ for the incipient daughter cells; concurrently, the cascade leading to assembly of a new flagellum continues. Constriction of the Z-ring generates two separate cellular compartments. In the swarmer compartment, cdG levels decrease again, leading to degradation of TipF. FtsZ is also degraded as the cells prepare to separate. PleD does not localize to the cell poles when unphosphorylated and inactive. Middle panel, Cellular Modules. Events leading to flagellum assembly and ejection are highlighted. Lower panel, Temporal Control. Black bars indicate the abundance time period across the cell cycle for the flagellar regulators CtrA, GcrA, SciP and FlbD, with split white bars representing the lack of a factor in the nascent swarmer (top) or stalked (bottom) cell compartment.
CtrA levels and activity: regulating asymmetry
The two-component signaling protein CtrA is a critical regulator of C. crescentus asymmetry through its dual rules as an inhibitor of the initiation of DNA replication and as a transcription factor that controls the expression of least 90 cell cycle regulated genes [24,25]. Unlike any other master regulator in C. crescentus, CtrA must be phosphorylated (CtrA~P) to be active as a transcription factor. CtrA levels and its phosphorylation state are regulated by the asymmetry determination module (Figures 1b and 2a), which implements either a CtrA activation or CtrA degradation cascade depending on inputs from the compartment-sensing and the polar scaffolding spatial regulators (Figures 1b and 2a) [26]. Thus, upon compartmentalization of the predivisional cell, CtrA drives asymmetry by remaining phosphorylated and active in the swarmer compartment while being dephosphorylated and degraded in the stalked compartment [27] (Figure 2b).
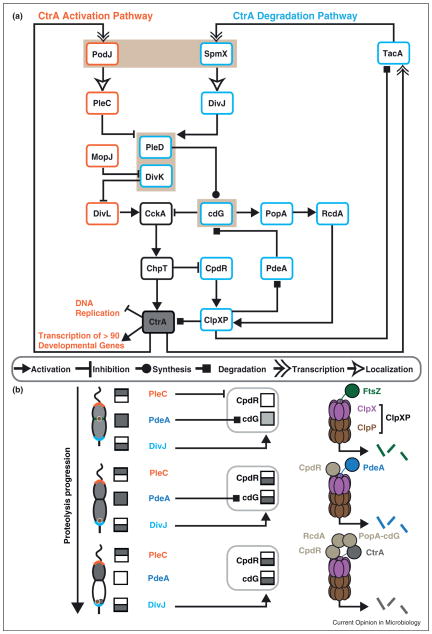
Figure 2.
Regulation of CtrA levels and activity. (a) A more detailed pathway diagram of the regulatory network driving CtrA activation (orange) and CtrA degradation (blue), which was simplified in Figure 1a. Highlighted in brown: polar scaffolds PodJ and SpmX and the compartment-sensors PleD and DivK. The members of the pathways communicate via activation, inhibition, synthesis (of cdG), localization, transcriptional control (by CtrA and TacA), and degradation (by ClpXP and PdeA). Different arrowheads represent these distinct communication modes. (b) Snapshots of ClpXP dependent proteolysis as a function of cell cycle progression. Dark gray boxes represent active signaling function; white boxes represent a lack of activity. Divided boxes represent compartment-specific activity. Top: approximately 90 minutes into the cell cycle, ClpXP (purple and brown) degrades FtsZ (green) at the division plane. PleC phosphatase activity maintains low cdG levels and high CckA activity. High CckA activity maintains high CpdR~P levels, the inactive form. Middle: once compartmentalization completes, PleC is no longer present in the stalked compartment, deactivating CckA and promoting unphosphorylated CpdR (light brown) specifically in the stalked compartment. Unphosphorylated CpdR localizes ClpXP to the stalked pole and promotes degradation of the cdG phosphodiesterase, PdeA (dark blue). Bottom: once PdeA is degraded, cdG levels can rise again in the stalked compartment, allowing PopA to bind cdG and, with RcdA (light brown), direct CtrA (gray) to ClpXP for degradation.
The asymmetry determination module is composed of two protein signaling pathways differentially localized to the two cell poles in predivisional cells (Figure 2a). This differential localization is mediated by a distinct set of scaffolds at each cell pole (Figures 1b and 2a). The CtrA activation pathway is localized to the new pole primarily through interactions with the polar scaffolding protein PodJ [28,29] and to a lesser extent with TipN [22,23]. Meanwhile, the lysozyme-like factor SpmX localizes the histidine kinase DivJ at the old pole, activating the CtrA degradation pathway [30]. In addition, the self-assembling pole-organizing protein PopZ, that is localized at one pole in the swarmer cell and both poles following the differentiation of the swarmer cell into the stalked cell, is critical for the differential localization members of both pathways to their corresponding poles [31].
A linchpin of the asymmetry determination module is the bifunctional hybrid histidine kinase CckA [32]. When stimulated as a kinase, CckA autophosphorylates and transfers its phosphate to the phosphotransfer protein ChpT [32–34], which in turn passes the phosphate to either CtrA, resulting in its activation, or to CpdR, resulting in inhibition of CtrA degradation (Figure 2a). When CckA is stimulated as a phosphatase, phosphoryl groups are siphoned back from CtrA through ChpT to CckA, where they are hydrolyzed. This reverse process inhibits the activation of CtrA and promotes CtrA degradation by shutting off the flow of phosphate to CpdR. Thus, the dual functions of CckA act as a switch between either executing CtrA activation or promoting CtrA degradation.
The function of CckA is regulated both in space and time via the composition of the pole it resides in, as well as via an interface with members of the compartment-sensing proteins DivK and PleD. First, CckA is primarily delocalized in swarmer cells, is localized to the stalked pole in some stalked cells, and subsequently localized either to both poles or only at the swarmer pole in predivisional cells [35,36]. Accumulation of CckA at the new cell pole, mediated by the pseudokinase DivL, promotes kinase activity in the swarmer compartment [37•,38] (Figure 2a). Second, DivK, when phosphorylated, binds to DivL, and this DivL–DivK~P complex can inhibit CckA kinase activity [39,40•]. PleD, when phosphorylated, synthesizes cyclic-di-GMP (cdG), which can bind CckA and inhibit its kinase activity [37•,41•]. The levels of DivK~P and PleD~P are modulated by the swarmer fate determinant PleC and the stalked fate determinant DivJ (Figure 2a). In predivisional cells, DivJ is activated, perhaps indirectly, by KidO (in addition to KidO’s previously described role in regulating FtsZ polymerization) [12], phosphorylates DivK and PleD, and elevates cdG levels specifically in the stalked compartment. PleC dephosphorylates DivK~P and PleD~P, permitting the activity of CckA in the swarmer compartment [42–45]. Notably, PleD is one of eight diguanylate cyclases in C. crescentus [45], but it has been shown to be a critical contributor to cdG levels at the stage of predivisional compartmentalization [44,45]. Upon swarmer to stalked differentiation, DivJ replaces PleC at the stalked pole, allowing for a surge in DivK~P and cdG that triggers the clearing of CtrA. Thus, CckA integrates information about the progression of assembly of the CtrA activation pathway and of cellular compartmentalization to regulate the phosphorylation state and stability of CtrA.
The AAA+ protease ClpXP degrades CtrA in a cell cycle dependent manner. In all bacteria, ClpXP uses adaptor accessory factors to modulate its substrate specificity [46]. In C. crescentus, ClpXP targets more than 30 proteins for degradation [47], including KidO, GdhZ, and FtsZ, as described above. In the case of CtrA, it has been shown that CpdR is a required adaptor for CtrA degradation in vivo [48]. Notably, in vitro, the ClpXP recognition degradation tag at the C-terminus of free CtrA molecules is sufficient for its efficient degradation. However, two additional adaptors, RcdA and the cdG receptor PopA, are required for degradation of CtrA when bound to the chromosome [49,50], the relevant in vivo state of CtrA in G1→S and S→G2 transitions. The compartment-sensing module coordinates the availability of these three adaptors, in a hierarchical manner, to bind ClpXP at the right time and place [51••]. In late predivisional cells, ClpXP localizes to midcell, where it degrades FtsZ, leading to closure of the inner membrane [52] (Figure 2b). Closing of the inner membrane stimulates deactivation of CckA as a kinase in the stalked compartment, shutting off the flow of phosphate to CpdR. When unphosphorylated, CpdR is localized to the stalked pole, together with ClpXP, by an unknown factor (Figure 2b). The localized CpdR/ClpXP complex leads to the degradation of the phosphodiesterase PdeA [53,54]. Degradation of PdeA in turn promotes the accumulation of cdG, allowing for binding of RcdA and PopA-cdG to the degradation complex, finally allowing for the degradation of CtrA (Figure 2b).
Flagellum dynamics: coupling cell division and cell cycle state
C. crescentus builds a single flagellum at the new cell pole of predivisional cells. This assembly process remarkably coordinates the construction of an approximately 50-protein machine following two principles: (i) just-in-time synthesis, in which proteins are made only when needed, and (ii) an inside-out assembly in which the envelope components are inserted ahead of the external parts [55]. Despite the apparent complexity of this assembly, its spatiotemporal coordination with cell cycle progression is regulated by a small number of factors. Temporal coordination is achieved by an interface to the genetic circuit (via the global regulator GcrA and its reactivation of CtrA in late S-phase), and its spatial coordination results from an interface with the compartment-sensing component (via cdG levels) and an interface to the cell division machinery (via FtsZ and TipN) (Figure 3a).
Temporally, activation of CtrA~P during late S-phase initiates flagellum assembly by inducing transcription of genes that compose the flagellum base (Class II genes, Figure 3). Expression of Class II genes is repressed by SciP [56,57], ensuring that the flagellar base is made only once. Class II genes include, in addition to structural genes, three transcriptional regulators: the RNA polymerase sigma subunit σ54, the σ54-dependent transcriptional activator FlbD, and its regulator FliX [55]. These three proteins work together to regulate the ordered expression of class III and class IV genes that code for the basal body, the hook, and the filament. The system is tuned to prevent the expression of later flagellar parts until completion of assembly of the earlier substructures [55,58,59]. Some class IV flagellin genes are under the direct control of CtrA~P and are not expressed together with class II genes because they are repressed by MucR1/2 (Figure 1) [60••]. Thus, the activation of CtrA~P in the swarmer compartment sets off a self-propagating cascade of transcriptional events to direct ordered flagellum assembly.
Spatially, the localization of TipN to the Z-ring during cell division ultimately marks the flagellum assembly site at the new cell pole (Figure 3). At the new pole, TipN recruits the cdG binding protein TipF, which in turn seeds the assembly of several factors involved in flagellar positioning and basal body building [22,61•]. Deletion of tipF results in non-flagellated cells, while deletion of tipN results in multiple, misplaced flagella [22]. TipF protein levels rise concurrently with CtrA, in early predivisional cells, immediately preceding production of the basal flagellar components (Figure 3).
Flagellum assembly also responds to the spatial and temporal fluctuations of cdG, as cdG frequently regulates changes in motility state throughout bacteria [62]. TipF directly binds to cdG in late S-phase, when cdG levels are still high, enabling the TipF-cdG complex to bind TipN [61•]. When cdG levels drop in the swarmer compartment, TipF can no longer bind the ligand, releasing TipN and leading to TipF proteolysis. Thus, the drop in cdG levels resets the flagellar polarity cascade for the next cell cycle. Upon differentiation of the swarmer cell into a stalked cell, new DivJ synthesis enables activation of PleD and an increase in cdG. The elevated cdG concentration in late G1 and S phases is required for flagellum ejection [63] and eventually for the positioning of a new complement of TipF. In the absence of pleD, the majority of cells have a flagellum at the stalked pole, indicating that the flagellum was not ejected [63], while constitutive PleD activity interferes with motility [64].
Concluding remarks
Establishment of cellular polarity requires spatial coordination of multiple cellular processes. The asymmetric division of C. crescentus requires oscillating cell-type specific gene expression programs. In addition to temporal coordination by a master genetic circuit, C. crescentus implements an efficient intracellular compass composed of mechanisms for coordinating cell polarity, sequestering cell fate factors, and establishing intracellular concentration gradients of proteins. The progress in our understanding of these phenomena highlights promising directions for future study, including how cells achieve high temporal resolution of gene expression through combinatorial transcriptional regulation and how multi-input and dual-function proteins can coordinate distinct cellular pathways. Understanding the spatial mechanisms coordinating these processes at a single-cell and detailed molecular level presents an exciting and challenging new frontier for systems biology.
Acknowledgments
We thank Michael D. Melfi, W. Seth Childers, and Alex R. S. von Diezmann for helpful comments and discussions. We acknowledge support from the Gordon and Betty Moore Foundation [GBMF 2550.03 to Life Sciences Research Foundation to K.L.] and the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science [to K.L.] and NIH [R01 GM32506 and R35-GM118071-01 to L.S.].
Footnotes
Competing interests
The authors declare no competing interests.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 2.Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 3•.Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, McAdams HH, Shapiro L. The global regulatory architecture of transcription during the caulobacter cell cycle. PLoS Genet. 2015;11:e1004831. doi: 10.1371/journal.pgen.1004831. Global analysis of Caulobacter transcription start sites reveals that many cell cycle controlled promoters respond to several of the master regulators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panis G, Murray SR, Viollier PH. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev. 2015;39:120–213. doi: 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 5.Meier EL, Goley ED. Form and function of the bacterial cytokinetic ring. Curr Opin Cell Biol. 2014;26:19–27. doi: 10.1016/j.ceb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Meier EL, Razavi S, Inoue T, Goley ED. A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol Microbiol. 2016;101:265–280. doi: 10.1111/mmi.13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez D, Kozdon JB, McAdams HH, Shapiro L, Collier J. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 2014;42:3720–3735. doi: 10.1093/nar/gkt1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Haakonsen DL, Yuan AH, Laub MT. The bacterial cell cycle regulator GcrA is a σ 70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev. 2015;29:2272–2286. doi: 10.1101/gad.270660.115. GcrA functions as a non-canonical transcription factor, directly binding RNA polymerase and catalyzing RNA polymerase open complex formation at GANTC methylation sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez D, Collier J. Genomic adaptations to the loss of a conserved bacterial DNA methyltransferase. MBio. 2015;6:1–12. doi: 10.1128/mBio.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanselicio S, Viollier PH. Convergence of alarmone and cell cycle signaling from trans-encoded sensory domains. MBio. 2015;6:1–10. doi: 10.1128/mBio.01415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanselicio S, Bergé M, Théraulaz L, Radhakrishnan SK, Viollier PH. Topological control of the Caulobacter cell cycle circuitry by a polarized single-domain PAS protein. Nat Commun. 2015;6:7005. doi: 10.1038/ncomms8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 2015;34:1–16. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaufay F, De Bolle X, Hallez R. Metabolic control of cell division in α-proteobacteria by a NAD-dependent glutamate dehydrogenase. Commun Integr Biol. 2016;9:e1125052. doi: 10.1080/19420889.2015.1125052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 16.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 17.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ptacin JL, Gahlmann A, Bowman GR, Perez AM, von Diezmann ARS, Eckart MR, Moerner WE, Shapiro L. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci U S A. 2014;111:E2046–E2055. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiekebusch D, Michie KA, Essen L-O, Löwe J, Thanbichler M, Dimerization L, Drive B, Formation G. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell. 2012;46:245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci U S A. 2003;100:8235–8240. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 24.Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:13600–13605. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matroule J-Y, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Ryan KR, Judd EM, Shapiro L. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J Mol Biol. 2002;324:443–455. doi: 10.1016/s0022-2836(02)01042-2. [DOI] [PubMed] [Google Scholar]
- 28.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 29.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 33.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 34.Blair JA, Xu Q, Childers WS, Mathews II, Kern JW, Eckart M, Deacon AM, Shapiro L. Branched signal wiring of an essential bacterial cell-cycle phosphotransfer protein. Structure. 2013;21:1590–1601. doi: 10.1016/j.str.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelastro PS, Sliusarenko O, Jacobs-Wagner C. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J Bacteriol. 2010;192:539–552. doi: 10.1128/JB.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of two Phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Mann TH, Childers WS, Blair Ja, Eckart MR, Shapiro L. A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat Commun. 2016;7:11454. doi: 10.1038/ncomms11454. CckA uses PAS domain sensors to respond to subcellular accumulation and c-di-GMP as input signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iniesta AA, Hillson NJ, Shapiro L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci. 2010;107:7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsokos CG, Perchuk BS, Laub MT. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell. 2011;20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Childers WS, Xu Q, Mann TH, Mathews II, Blair Ja, Deacon AM, Shapiro L. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol. 2014;12:e1001979. doi: 10.1371/journal.pbio.1001979. DivL detects the phosphorylated form of DivK in a PAS domain dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–239. doi: 10.1038/nature14473. CckA is inhibited by c-di-GMP in vivo, reinforcing the DivK inhibition circuit. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 43.Lam H, Matroule J-Y, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 44.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel zur Wiesch P, Jenal U. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genet. 2013;9:5–11. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2015;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci. 2014;111:14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, Chien P, Laub MT. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2013;87:1277–1289. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Joshi KK, Bergé M, Radhakrishnan SK, Viollier PH, Chien P. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell. 2015;163:419–431. doi: 10.1016/j.cell.2015.09.030. ClpXP uses a hierarchical set of proteolytic adaptors to degrade regulators of the CtrA pathway in a defined order. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol Microbiol. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rood KL, Clark NE, Stoddard PR, Garman SC, Chien P. Adaptor-dependent degradation of a cell-cycle regulator uses a unique substrate architecture. Structure. 2012;20:1223–1232. doi: 10.1016/j.str.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ardissone S, Viollier PH. Interplay between flagellation and cell cycle control in Caulobacter. Curr Opin Microbiol. 2015;28:83–92. doi: 10.1016/j.mib.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. A cell-type-specific protein–protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell. 2010;39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan MH, Kozdon JB, Shen X, Shapiro L, McAdams HH. An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc Natl Acad Sci U S A. 2010;107:18985–18990. doi: 10.1073/pnas.1014395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown PJB, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol. 2009;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Fumeaux C, Radhakrishnan SK, Ardissone S, Théraulaz L, Frandi A, Martins D, Nesper J, Abel S, Jenal U, Viollier PH. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Commun. 2014;5:4081. doi: 10.1038/ncomms5081. MucR1/2, ancestral zinc-finger domain proteins, work together with CtrA to control G1-phase genes in C. crescentus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Davis NJ, Cohen Y, Sanselicio S, Fumeaux C, Ozaki S, Luciano J, Guerrero-Ferreira RC, Wright ER, Jenal U, Viollier PH. De- and repolarization mechanism of flagellar morphogenesis during a bacterial cell cycle. Genes Dev. 2013;27:2049–2062. doi: 10.1101/gad.222679.113. TipF binding to cdG enables its localization, recruitment of flagellar proteins, and stability against proteolysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 64.Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]