Abstract
The human cytomegalovirus DNA polymerase includes an accessory protein, UL44, which has been proposed to act as a processivity factor for the catalytic subunit, UL54. How UL44 interacts with UL54 has not yet been elucidated. The crystal structure of UL44 revealed the presence of a connector loop analogous to that of the processivity subunit of herpes simplex virus DNA polymerase, UL42, which is crucial for interaction with its cognate catalytic subunit, UL30. To investigate the role of the UL44 connector loop, we replaced each of its amino acids (amino acids 129 to 140) with alanine. We then tested the effect of each substitution on the UL44-UL54 interaction by glutathione S-transferase pulldown and isothermal titration calorimetry assays, on the stimulation of UL54-mediated long-chain DNA synthesis by UL44, and on the binding of UL44 to DNA-cellulose columns. Substitutions that affected residues 133 to 136 of the connector loop measurably impaired the UL44-UL54 interaction without altering the ability of UL44 to bind DNA. One substitution, I135A, completely disrupted the binding of UL44 to UL54 and inhibited the ability of UL44 to stimulate long-chain DNA synthesis by UL54. Thus, similar to the herpes simplex virus UL30-UL42 interaction, a residue of the connector loop of the accessory subunit is crucial for UL54-UL44 interaction. However, while alteration of a polar residue of the UL42 connector loop only partially reduced binding to UL30, substitution of a hydrophobic residue of UL44 completely disrupted the UL54-UL44 interaction. This information may aid the discovery of small-molecule inhibitors of the UL44-UL54 interaction.
Replicative DNA polymerases are capable of synthesizing long stretches of DNA without dissociating from the template. The high processivity of these polymerases is dependent upon accessory proteins, called processivity factors, that bind to the catalytic subunit of the polymerase. As an example, the human cytomegalovirus (HCMV) DNA polymerase is composed of a catalytic subunit, Pol, or UL54, which possesses a basal DNA polymerase activity (4, 17), and an accessory protein, UL44 (7), which has been shown to bind double-stranded DNA, to interact specifically with UL54, and to stimulate long-chain DNA synthesis, possibly by increasing the processivity of the polymerase along the DNA template (7, 28). The details of the UL44-UL54 interaction and its role in UL44 function have not been yet completely elucidated. Recent data demonstrated that, like UL30, the catalytic subunit of herpes simplex virus type 1 (HSV-1) DNA polymerase, UL54, interacts with the cognate accessory subunit through the C-terminal region (15, 16), despite the fact that the C termini of UL54 and UL30 share almost no amino acid sequence homology. In addition, UL44 has been predicted to possess a structure with a “processivity fold” similar to that reported for the processivity subunit of HSV-1 DNA polymerase, UL42 (29). Indeed, the crystal structure of UL44 revealed an overall fold strikingly similar to that of UL42 (1), even though again the HCMV accessory protein has very little sequence homology to UL42.
The structure of UL44 includes an element analogous to the so-called connector loop, a long loop which connects the two topologically similar domains of UL42 (29) and is crucial for interaction with its cognate catalytic subunit, UL30 (2). Taken together, these observations suggested that the two subunits of HCMV DNA polymerase could interact in a way which is analogous to that of the two subunits of HSV DNA polymerase and therefore the UL44 connector loop could play a role in the UL54-UL44 interaction.
However, recent studies highlighted differences between the UL54-UL44 and UL30-UL42 interactions. A mutational analysis revealed that the UL54 residues crucial for interaction with UL44 are hydrophobic (16), whereas the UL30 residues important for UL42 binding are basic (1a, 2). Moreover, UL44 can form homodimers in both its crystal structure and in solution (1), unlike UL42, which is a monomer (8, 9, 22). Therefore, despite the remarkable structural similarities between UL44 and UL42, the UL54 binding site on UL44 might differ from the UL30 binding site on UL42.
To investigate whether the UL44 connector loop is indeed involved in the interaction with UL54, we engineered a series of substitutions in the connector loop of UL44 (amino acids 129 to 140) and tested the effect of each substitution on the physical and functional interactions between UL54 and UL44. Our findings highlight important similarities and differences between the UL54-UL44 and UL30-UL42 interactions. This might aid in the discovery of new drugs for the treatment of HCMV infection based on disruption of the UL54-UL44 interaction.
MATERIALS AND METHODS
Plasmids.
The pRSET-Pol plasmid, containing the HCMV strain AD169 UL54 gene, was a gift from P. F. Ertl (GlaxoSmithKline, Stevenage, United Kingdom). The pT730 plasmid, expressing HSV-1 UL30 under a T7 promoter, and the pD15-GST and pD15-UL44ΔC290 plasmids, which express glutathione S-transferase (GST) and the N-terminal 290 residues of UL44 as a GST fusion protein, respectively, have been described previously (5, 16). Plasmids pD15-UL44ΔC260, pD15-UL44ΔC315, and pD15-UL44ΔC340, which were used for the expression of GST fusion proteins with the N-terminal 260, 315, and 340 residues of UL44, respectively, were constructed in a manner analogous to that reported previously for pD15-UL44ΔC290 (16). A list of the primers used to create these constructs is available at http://coen.med.harvard.edu.
All UL44 connector loop mutants were obtained with the Quick-Change mutagenesis kit (Stratagene), amplifying the pD15-UL44ΔC290 plasmid with primer pairs containing appropriate nucleotide changes. A list of the mutagenic primers is available at http://coen.med.harvard.edu. All constructs were sequenced by the Biopolymers Laboratory in the Department of Biological Chemistry and Molecular Pharmacology at Harvard Medical School to confirm the presence of the engineered mutation and the absence of undesired mutations.
Proteins and peptide.
Purified baculovirus-expressed HCMV UL54, prepared as described previously (15), was generously provided by H. S. Marsden (Institute of Virology, Glasgow, United Kingdom). GST and wild-type and mutant GST-UL44ΔC290 fusion proteins were purified from Escherichia coli BL21(DE3)pLysS harboring the appropriate plasmid, as described previously (16). Concentrations of all proteins were determined by amino acid analysis at the Molecular Biology Core Facility, Dana-Farber Cancer Institute.
The peptide corresponding to the last 22 residues of UL54 (here termed UL54 peptide 1, as in references 15 and 16) was synthesized by the Biopolymers Laboratory in the Department of Biological Chemistry and Molecular Pharmacology at Harvard Medical School. The peptide was dissolved in water, and the concentration was determined by quantitative amino acid analysis performed by the Molecular Biology Core Facility, Dana-Farber Cancer Institute. The peptide was then lyophilized prior to use.
dsDNA-cellulose chromatography.
Double-stranded DNA (dsDNA)-cellulose chromatography was performed as described previously (6). Briefly, in vitro expression of GST or wild-type or mutant GST-UL44ΔC290 was performed from the appropriate plasmid with the TNT T7-coupled reticulocyte lysate system from Promega. The translation products were labeled with [35S]methionine (Amersham Pharmacia Biotech), and 10-μl aliquots of in vitro expression reactions were diluted into 100 μl of TM buffer (20 mM Tris-HCl [pH 7.6], 5 mM MgCl2) containing 50 mM NaCl and 2 μl of RNace-it (Stratagene). After 20 min of incubation at room temperature, the mixture was applied to a dsDNA-cellulose (Sigma) column with a 0.5-ml bed volume. The column was then washed stepwise with 2 bed volumes each of TM buffer containing various concentrations of NaCl. The eluates were ethanol precipitated in the presence of 40 μg of bovine serum albumin and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were dried and used to expose film and phosphorescence screens.
GST pulldown assays.
GST pulldown assays with in vitro-expressed HCMV UL54 and E. coli-expressed purified wild-type or mutant GST-UL44ΔC290 fusion proteins were performed as described previously (16). Briefly, GST or wild-type or mutant GST-UL44ΔC290 fusion proteins (2.7 nmol) were incubated in a final volume of 550 μl with 50 μl of in vitro expression reactions on ice for 2 h and then loaded onto 0.2-ml glutathione columns. Bound proteins were eluted with wash buffer containing 15 mM glutathione and visualized by SDS-PAGE and autoradiography.
DNA polymerase assays.
Stimulation of UL54 activity by GST-UL44 or wild-type or mutant GST-UL44ΔC290 was assayed by measuring the rate of incorporation of labeled dTTP into either a poly(dA)-oligo(dT) or a calf thymus DNA template as previously described (14). Long-chain DNA synthesis by UL54 in the presence of GST-UL44 or of wild-type or mutant GST-UL44ΔC290 fusion proteins was assayed as reported previously (15), with 200 fmol of baculovirus-expressed purified UL54 and 400 or 800 fmol of GST-UL44 or GST-UL44ΔC290 in a final volume of 25 μl.
Isothermal calorimetry.
Isothermal calorimetry (ITC) experiments were performed as described previously (16), with protein concentrations of 5 to 10 μM and UL54 peptide 1 concentrations of 100 to 250 μM. The peptide was titrated into the protein-containing sample cell in 10-μl injections at 25°C with a mixing speed of 270 rpm. The data were analyzed as previously described (2).
RESULTS
UL44ΔC290 retains all known biochemical activities of full-length UL44.
During attempts to express and purify from E. coli GST fused to full-length UL44, we found that this protein was subject to a small but noticeable level of proteolytic cleavage. Furthermore, we noted that full-length GST-UL44 is prone to aggregation under certain conditions of low ionic strength. As UL44 contains five glycine-rich strings, beginning at residue 291 (7), this segment is predicted to be unstructured, which could make it susceptible to proteolysis and aggregation. It has been reported that the region corresponding to approximately the C-terminal 28% of UL44 (residues 310 to 433) is not necessary for UL44 to bind DNA or to stimulate the catalytic activity of UL54 (28).
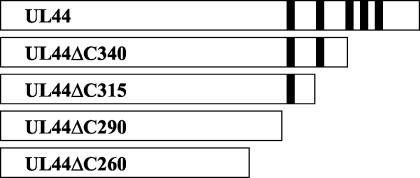
To determine whether the C-terminal glycine-rich region is required for any known biochemical activity of UL44, we constructed a series of plasmids encoding C-terminal truncation mutants of the GST-UL44 fusion protein (GST-UL44ΔC290, GST-UL44ΔC315, and GST-UL44ΔC340), lacking all, four, or three of the glycine strings (Fig. 1). As a control, we also created a GST-UL44 fusion in which UL44 is truncated at residue 260 (GST-UL44ΔC260), which was previously shown not to bind DNA (28). We then assayed the ability of each mutant to bind dsDNA. Plasmids carrying the appropriate GST-UL44 construct were transcribed in vitro, and the transcripts were translated in a rabbit reticulocyte system in the presence of [35S]methionine. Labeled proteins were applied to a dsDNA-cellulose column and eluted stepwise with 2 bed volumes of buffer containing increasing salt concentrations.
FIG. 1.
UL44 C-terminal truncations. UL44 contains five glycine-rich strings in the C terminus, which begin at residue 291. The truncation mutants studied here are depicted as white bars, with the location of each glycine string shown as a black bar.
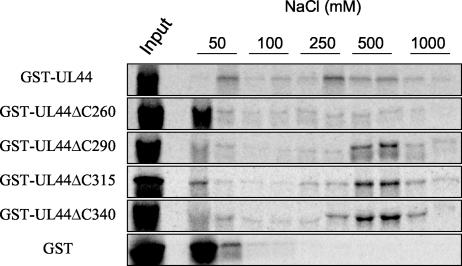
As shown in Fig. 2, most of the wild-type protein was retained on the dsDNA column and eluted at approximately 250 to 500 mM NaCl. Only a small fraction was present in the first 50 mM fraction. UL44ΔC290, UL44ΔC315, and UL44ΔC340 also bound to the dsDNA column efficiently, eluting at approximately 250 to 1,000 mM NaCl (Fig. 2). In contrast, the majority of GST and of GST-UL44ΔC260, which do not bind to DNA, eluted at 50 mM NaCl. These findings confirm previous observations (28) and extend them by demonstrating that the region of UL44 downstream of residue 290 is not required for DNA binding.
FIG. 2.
DNA-binding activity of C-terminal truncation mutants of UL44. Radiolabeled full-length and truncated GST-UL44 fusion proteins (as indicated to the left of the panels) were expressed in vitro and applied to a dsDNA-cellulose column equilibrated in buffer containing 50 mM NaCl. Elution was carried out by washing with buffer containing the concentrations of NaCl indicated above the panels. Fractions were analyzed by SDS-PAGE and autoradiography. Input, unfractionated input protein.
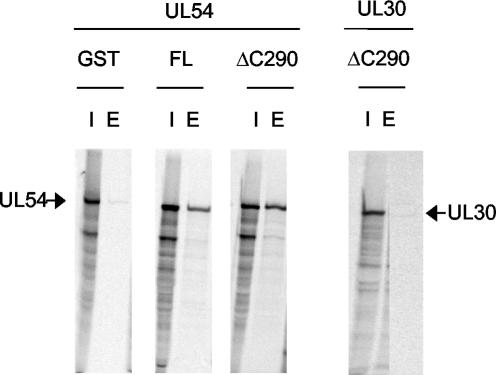
Subsequent studies were performed with the smallest active construct, GST-UL44ΔC290. In order to test whether 290 residues of UL44 are sufficient to bind UL54, we used a previously described GST pulldown assay (16). Purified GST-UL44 or GST-UL44ΔC290 fusion proteins were incubated with in vitro-translated and radiolabeled UL54 or, as a negative control, in vitro-expressed HSV-1 UL30 and then loaded onto glutathione columns. The columns were washed, and bound proteins were eluted with glutathione. The results are shown in Fig. 3. UL44ΔC290 bound UL54 as well as did full-length UL44, confirming that the truncated protein maintained the ability to interact with UL54. Control experiments indicated that the binding was specific, as GST did not bind UL54 and GST-UL44ΔC290 did not bind HSV-1 UL30 (Fig. 3).
FIG. 3.
UL44ΔC290 binds to UL54. The physical binding of GST-full-length UL44 and of GST-UL44ΔC290 proteins to UL54 was tested by GST pulldown experiments. Purified GST, GST-full-length UL44 (FL), and GST-UL44ΔC290 (ΔC290) proteins, as indicated at the tops of the figure panels, were incubated with in vitro-expressed and radiolabeled HCMV UL54 (leftmost six lanes) or HSV-1 UL30 (rightmost two lanes) and allowed to bind to glutathione columns. The columns were washed, and the proteins were eluted with 15 mM glutathione. The radiolabeled proteins were visualized by autoradiography following electrophoresis on an SDS-7.5% polyacrylamide gel. The positions of UL54 and UL30 are marked by arrows. I, input; E, eluted by glutathione.
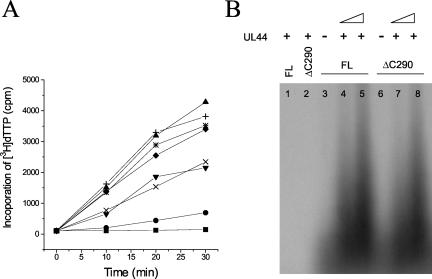
We then tested the ability of UL44ΔC290 to stimulate the DNA polymerase activity of UL54 by assaying the rate of incorporation of labeled dTTP into different templates with a filter-based assay. In this assay, UL44ΔC290 stimulated the incorporation activity of UL54 with either poly(dA)-oligo(dT) (Fig. 4A) or calf thymus DNA (data not shown) as a template. Comparing different amounts of either full-length UL44 or UL44ΔC290, the stimulatory activities of the two proteins were indistinguishable. We also tested the ability of UL44ΔC290 to stimulate long-chain DNA synthesis by UL54, by measuring the incorporation of radionucleotides into an oligo(dT)/poly(dA) primer/template and analyzing the resulting products on an alkaline agarose gel (Fig. 4B). As previously observed (16), only limited synthesis of short DNA products was detectable after incubation of this template with 200 fmol of UL54 in the absence of UL44 (Fig. 4B, lanes 3 and 6), and no extension of the oligo(dT) primer was observed when only GST-UL44 or GST-UL44ΔC290 was included in the reaction (Fig. 4B, lanes 1 and 2). Stimulation of long-chain DNA synthesis was only detected when both UL54 and GST-UL44 or GST-UL44ΔC290 were included in the reaction. This stimulation was dependent on the amount of GST-UL44ΔC290 added (Fig. 4B, lanes 7 and 8), in a manner similar to that observed when equal amounts of full-length UL44 were added (lanes 4 and 5).
FIG. 4.
UL44ΔC290 stimulates DNA synthesis by UL54. (A) The DNA polymerase activity of purified baculovirus-expressed UL54 alone (•) and in the presence of 200 (▾), 500 (*), or 1,000 (▴) fmol of GST-UL44 (full length) or of 200 (×), 500 (⧫), or 1,000 (+) fmol of GST-UL44ΔC290 was measured by incorporation of [3H]dTTP into a poly(dA)-oligo(dT) DNA template. As a control, the activity of 1,000 fmol of GST-UL44ΔC290 alone (▪) was also assayed. Samples were taken after 0, 10, 20, and 30 min of incubation at 37°C and spotted onto DE81 filters. The filters were washed, and radioactivity was counted. (B) Stimulation of UL54-mediated long-chain DNA synthesis by GST-full-length UL44 (FL) or GST-UL44ΔC290 (ΔC290) was assayed by measuring the incorporation of labeled TTP on a poly(dA)-oligo(dT) template. The reaction products were visualized by autoradiography following electrophoresis on a 4% alkaline agarose gel. Lane 1 contains 800 fmol of GST-UL44 (full-length) alone; lane 2 contains 800 fmol of GST-UL44ΔC290 alone; lanes 3 and 6 contain 200 fmol of UL54 alone; lanes 4 and 5 contain UL54 plus 400 and 800 fmol of GST-UL44 (full-length), respectively; lanes 7 and 8 contain UL54 plus 400 and 800 fmol of GST-UL44ΔC290, respectively.
Taken together, these results indicate that the UL44ΔC290 truncated protein, which lacks all five C-terminal glycine strings, retains the ability to bind DNA, to interact with UL54, and to stimulate DNA synthesis by UL54. Furthermore, we observed that UL44ΔC290 exhibits less aggregation and proteolysis than the full-length protein when expressed from E. coli (data not shown).
Effects of mutations in the connector loop of UL44 on physical binding to UL54.
The recent crystal structure of the UL44ΔC290 truncated protein revealed that UL44 possesses a structural element, from residues 129 to 140, analogous to the so-called connector loop of HSV-1 UL42 protein (1). This region of UL42, which stretches from residues 160 to 175, establishes specific interactions with the C terminus of UL30 (29), and substitution of a single residue in this segment, Q171, crucially affects the interaction of UL42 with UL30 (2). Therefore, the presence of a connector loop in UL44 led us to hypothesize that this structural element might play a role in the interaction with UL54. To analyze the importance of single residues of the UL44 connector loop for UL54 binding, we replaced each of the amino acids of this segment with alanine (Fig. 5). Since full-length UL44 and UL44ΔC290 display no obvious differences in biochemical activities, we chose to clone all mutations in the UL44ΔC290 backbone and avoid potential problems with aggregation or proteolysis.
FIG. 5.
UL44 mutants. The sequence of the UL44 connector loop (amino acids 129 to 140), in single-letter code, is reported on the top. A series of substitutions were engineered in this region of UL44 as described in Materials and Methods. For each mutant, the sequence of the region containing the mutation is shown, with the mutated residue indicated by an underlined boldface letter.
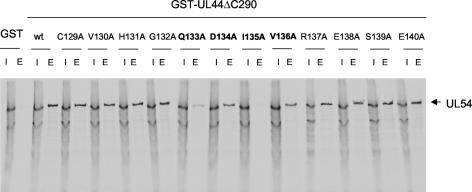
In order to test the ability of the UL44 mutants to bind UL54, the GST pulldown assay described above was employed. We found that substitution of residue I135 with an alanine resulted in undetectable binding of UL44 to UL54 (Fig. 6). We also observed that substitutions at positions Q133, D134, and V136 of UL44 reduced binding to UL54. This was obvious for the Q133A mutant, although less obvious for the D134A and V136A mutants. However, when the ratio of bound protein to input UL44 in the two latter mutants was compared to that of wild-type protein, it was consistently lower. All of the other UL44 mutants bound UL54 in a manner similar to that of wild-type UL44.
FIG. 6.
Binding of mutant UL44 proteins to UL54. The physical binding of wild-type (wt) and mutant GST-UL44ΔC290 proteins, as indicated at the tops of the figure panels, to UL54 was tested by GST pulldown assays as described in the legend to Fig. 3. I, input; E, eluted by glutathione. The position of UL54 is indicated by an arrow to the right of the panel. The mutants that exhibited reduced binding are indicated in boldface.
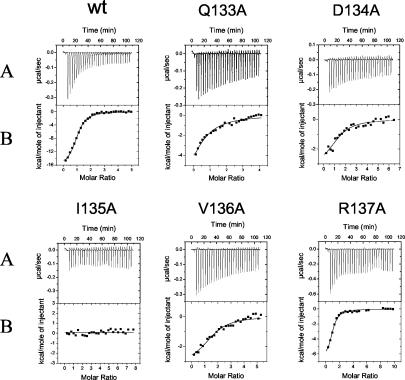
To examine this interaction quantitatively, we used ITC, which measures heat generated or absorbed upon binding and allows one to obtain values for the stoichiometry, the dissociation constant (Kd), the change in enthalpy (ΔH), the free energy (ΔG), and the entropic term TΔS of the interaction. We performed ITC on the interaction of wild-type and mutant GST-UL44ΔC290 with a peptide corresponding to the C-terminal 22 residues of UL54, which are necessary and sufficient for interaction with UL44 (16). This peptide (termed UL54 peptide 1) has previously been shown to specifically bind to full-length GST-UL44, GST-UL44ΔC290, and cleaved UL44ΔC290 but not to GST alone or to a maltose-binding protein (MBP)-UL42ΔC340 fusion which contains the N-terminal 340 residues of HSV-1 UL42 (16).
A typical titration experiment for binding of purified wild-type GST-UL44ΔC290 fusion protein to the UL54 C-terminal peptide is shown in Fig. 7. Analysis of the binding data indicated a stoichiometry of 1 molecule of peptide per molecule of GST-UL44ΔC290 fusion protein, and a Kd value of 0.7 ± 0.1 μM (Table 1), in agreement with the values obtained previously (16). No release of heat was detected when GST-UL44 or GST-UL44ΔC290 was titrated with a peptide corresponding to the last 10 residues of UL54 (16) (data not shown), a region previously shown not to be sufficient for binding UL44 (16). Consistent with the GST pulldown data, we observed that mutants Q133A, D134A, and V136A still bound the UL54 peptide but with affinities 4- to 10-fold lower than that of wild-type UL44 (see Fig. 7 and Kd values in Table 1). Mutant I135A, which did not detectably associate with full-length UL54 in GST pulldown assays, exhibited no measurable binding to UL54 peptide 1 (Fig. 7). As a control, we also measured the binding to UL54 peptide 1 of R137A, a UL44 mutant that does interact with full-length UL54 in GST pulldown assays. We found that this mutant possessed a Kd for the peptide (1.0 μM) similar to that of wild-type protein, indicating that the R137A substitution does not meaningfully affect the binding of UL44 to the C terminus of UL54.
FIG. 7.
Results of ITC experiments binding wild-type and mutant UL44 to a synthetic peptide (UL54 peptide 1) corresponding to the C-terminal 22 residues of UL54. (A) Raw data for the titration of UL54 peptide 1 into a sample cell containing wild-type (wt) or mutant GST-UL44ΔC290 protein, as indicated on the top of each panel. (B) The heat of dilution of both protein and peptide in panel A was subtracted, and the area under each injection curve was integrated to generate the points, which represent heat exchange in kilocalories per mole, which are plotted against the cumulative peptide-to-protein ratio for each injection. The solid line is the best-fit curve for the data.
TABLE 1.
Parameters for binding of wild-type UL44 and mutant GST-UL44ΔC290 to the C-terminal UL54 peptide 1a
| Protein | Stoichiometry | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Wild type | 1.1 ± 0.1 | 0.7 ± 0.1 | −8.3 ± 1.1 | −16.2 ± 1.9 | −8.1 ± 1.0 |
| Q133A | 0.7 ± 0.2 | 7.4 ± 0.3 | −6.9 ± 0.8 | −10.3 ± 0.4 | −3.4 ± 0.6 |
| D134A | 1.2 ± 0.1 | 4.1 ± 0.2 | −7.3 ± 1.2 | −10.2 ± 2.1 | −2.9 ± 0.4 |
| I135A | —b | — | — | — | — |
| V136A | 1.3 ± 0.2 | 3.1 ± 0.4 | −7.5 ± 1.0 | −10.9 ± 1.8 | −3.4 ± 0.7 |
| R137A | 0.9 ± 0.1 | 1.0 ± 0.2 | −8.2 ± 1.4 | −16.5 ± 1.9 | −8.3 ± 0.8 |
Titrations were performed with 10-μl peptide injections. The heat of dilution of both peptide and protein was subtracted from the raw data prior to analysis. Nonlinear least-squares analysis of the raw data was performed with the fitting algorithm provided with the calorimeter. Values are means of three determinations ± standard deviation.
—, no binding was detected.
Mutations in the UL44 connector loop do not affect binding to DNA.
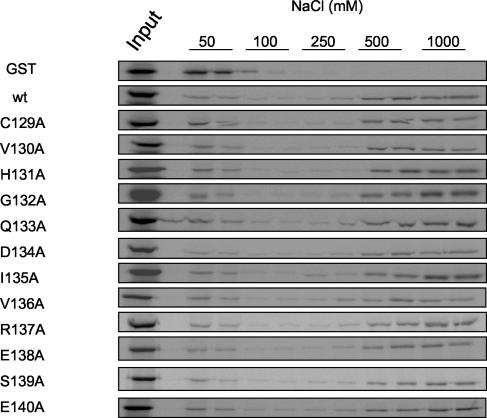
To determine whether the effects of the mutations on UL54 binding were due to global changes in protein structure, we tested the ability of UL44 mutants to bind DNA. Wild-type or mutated GST-UL44ΔC290 was expressed and labeled in vitro and subjected to DNA-cellulose chromatography as described above. As observed for wild-type UL44ΔC290, for all of the mutants, the majority of the protein was found in the fractions eluted with 500 to 1,000 mM NaCl (Fig. 8). This suggests that the mutations in the UL44 connector loop do not impair the DNA-binding activity of UL44. Moreover, the I135A mutant possessed an affinity for dsDNA similar to that of wild-type UL44ΔC290 in both electrophoretic mobility shift assays and filter-binding assays (A. Loregian and D. M. Coen, unpublished results), indicating that this mutation does not quantitatively affect the binding of UL44 to DNA.
FIG. 8.
UL44 mutants are able to bind DNA. The DNA-binding activity of UL44 mutants was tested by comparing the elution profiles of wild-type (wt) and mutant GST-UL44ΔC290 proteins and GST, as indicated to the left of the panels, from a dsDNA-cellulose column as described in the legend to Fig. 2. The concentrations of NaCl used to elute the proteins are indicated above the panels. Input, unfractionated input protein.
Since these mutants were not impaired in their DNA-binding activity, we conclude that their defect in UL54 binding is specific and not due to global misfolding of the protein.
Effect of UL44 substitutions on long-chain DNA synthesis.
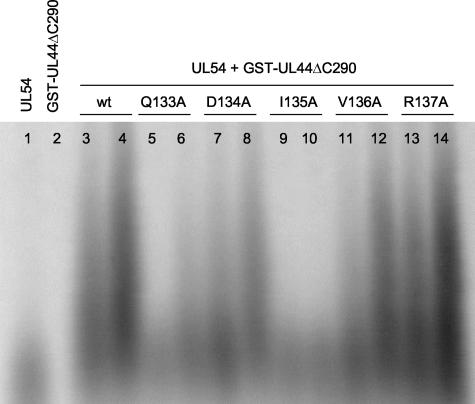
We next tested the ability of mutant GST-UL44ΔC290 proteins to stimulate long-chain DNA synthesis by UL54. Of all UL44 substitutions, the I135A substitution had the greatest effect on long-chain DNA synthesis, completely abolishing the ability of UL44 to stimulate synthesis of long DNA products by UL54 (<1% of wild-type activity; Fig. 9, lanes 9 and 10). The three other mutations, Q133A, D134A, and V136A, which reduced interaction with UL54 (Fig. 6 and 7), also partially impaired long-chain DNA synthesis (Fig. 9, lanes 5 and 6, 7 and 8, and 11 and 12, respectively). When the stimulation of UL54-mediated long-chain DNA synthesis by these mutants was quantified, the Q133A, D134A, and V136A mutants were found to have about 12, 35, and 57% of wild-type UL44 activity, respectively. As a control, we assayed the ability of the R137A mutant, which interacted with UL54 in the GST pulldown and with the UL54 peptide in the ITC assays similarly to wild-type UL44, to stimulate UL54 activity. This mutant stimulated long-chain DNA synthesis as strongly as did wild-type UL44 (Fig. 9, lanes 13 and 14). Similar results were obtained by assaying the rate of incorporation of labeled dTTP into a poly(dA)-oligo(dT) template with a filter-based assay (data not shown).
FIG. 9.
Stimulation of long-chain DNA synthesis by UL54. Experiments were performed with a poly(dA) template and an oligo(dT) primer and labeled TTP as described in the legend to Fig. 4B. Lane 1, 200 fmol of UL54 alone; lane 2, 800 fmol of wild-type GST-UL44ΔC290 alone. The remaining lanes contained 200 fmol of UL54 plus 400 (lane 3) and 800 (lane 4) fmol of wild-type (wt) GST-UL44ΔC290, 400 (lane 5) and 800 (lane 6) fmol of GST-UL44ΔC290 Q133A, 400 (lane 7) and 800 (lane 8) fmol of GST-UL44ΔC290 D134A, 400 (lane 9) and 800 (lane 10) fmol of GST-UL44ΔC290 I135A, 400 (lane 11) and 800 (lane 12) fmol of GST-UL44ΔC290 V136A, and 400 (lane 134) and 800 (lane 14) fmol of GST-UL44ΔC290 R137A.
The properties of selected UL44 mutants are summarized in Table 2. While the Q133A, D134A, I135A, and V136A mutants were able to bind to DNA, they were impaired for binding to UL54 and for stimulating long-chain DNA synthesis by UL54. The inhibitory effect of the Q133A, D134A, I135A, and V136A substitutions on UL44 stimulatory activity quantitatively correlates with the effect of each substitution on the binding affinity of the mutants as measured by ITC.. This correlation strongly suggests the concept that the role of this segment of UL44 in stimulation of UL54 activity is via its physical interaction with UL54.
TABLE 2.
Summary of the biochemical properties of selected UL44 mutantsa
| Mutation | UL54 binding | UL54 peptide 1 binding | DNA binding | Stimulation of UL54 activity |
|---|---|---|---|---|
| Q133A | +/− | +/− | + | +− |
| D134A | +/− | +/− | + | +/− |
| I135A | − | − | + | − |
| V136A | +/− | +/− | + | +/− |
| R137A | + | + | + | + |
+, wild type levels of activity; +/−, partially impaired activity; −, no detectable activity. Binding to UL54 was determined in GST pulldown experiments (Fig. 6); binding to the peptide corresponding to the C-terminal 22 residues of UL54 (UL54 peptide 1) was measured by Isothermal calorimetry (Fig. 7). DNA binding was assayed by column chromatography (Fig. 8). Stimulation of DNA synthesis by UL54 was determined both by analyzing the products on alkaline agarose gels (Fig. 9) and by filter assays (not shown).
DISCUSSION
All known biochemical activities of UL44 as a polymerase subunit reside in the N-terminal 290 residues.
The C terminus of UL44 is peculiar, as it contains several glycine-rich strings which are not present in any other human herpesvirus counterpart studied (7). A previous study suggested that this region of UL44 may be dispensable for DNA- and UL54-binding activities and stimulation of long-chain DNA synthesis, as deletion of residues 341 to 433 or 310 to 398 of UL44 did not significantly reduce its ability to bind DNA or its capacity to stimulate UL54 (28). However, the effect of deletion of the entire C-terminal glycine-rich region of UL44 had not yet been examined. Here we show that a truncation mutant, UL44ΔC290, which lacks all five glycine-rich strings of the UL44 C terminus retains the ability to bind DNA, bind UL54, and stimulate UL54-mediated long-chain DNA synthesis in a manner similar to that of full-length UL44. This suggests that the C terminus of UL44 is not important for the role of this protein as a polymerase subunit, although it could have other, as yet unknown function(s).
Similarly, it has been shown that the C-terminal one-third of other herpesvirus processivity factors, including HSV-1 UL42, Epstein-Barr virus BMRF1, and human herpesvirus 8 PF-8, is dispensable for these proteins to bind DNA, bind their respective catalytic subunits, and stimulate long-chain DNA synthesis (3, 6, 11, 26). Consistent with these observations also is the prediction that the processivity fold, a structure shared by HSV-1 UL42 and sliding clamp processivity factors such as PCNA, is contained within the N-terminal two-thirds of these proteins (29). The recent crystal structure of UL44 has indeed shown that the N-terminal two-thirds of this protein have an overall fold strikingly similar to that of HSV-1 UL42 and of human PCNA (1).
UL54-binding site of UL44.
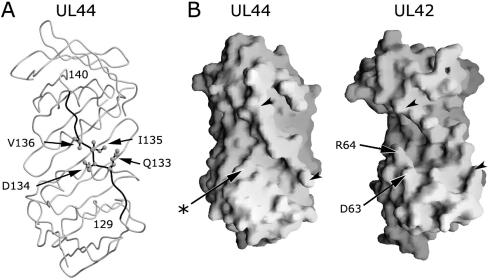
The observation that UL44 possesses a structural element analogous to the UL42 connector loop (1) suggested that a residue(s) of this region could play a role in UL54 binding. By carrying out an extensive mutational analysis, we identified residue I135 of the UL44 connector loop as crucial for UL54 binding, as substitution of this amino acid with an alanine completely disrupted the UL54-UL44 interaction. Three other residues of the UL44 connector loop, Q133, D134, and V136, were found to participate in the interaction but with a less important role. These residues lie roughly in the middle of the connector loop, and all appear to be accessible for binding to UL54 (Fig. 10A). However, which, if any, of these residues actually interact with UL54 remains to be determined.
FIG. 10.
Structural features of UL44. (A) The peptide backbone of UL44 is shown in grey. The connector loop of UL44 (residues 129 to 140) is highlighted in black, with the four residues identified in this study as important for binding to UL54 (Q133, D134, I135, and V136) shown as ball-and-stick models and indicated by arrows. It should be noted that the connector loop appears to be flexible, as the individual atoms display higher average temperature factors than the rest of the molecule. Thus, the side chains may adopt multiple orientations in solution, which could differ from the positions displayed in this figure. This figure was created with Molscript and Raster3D (12, 19). (B) The solvent-accessible surfaces of the face containing the connector loop of UL44 (left) and of UL42 (right) are displayed with Grasp (20). The connector loop of each protein is indicated by arrowheads. In UL42, a deep groove, which accommodates the C-terminal helix of UL30, is present between the connector loop and a projection formed by residues D63 and R64 (indicated by arrows). In contrast, the analogous region of UL44 displays a much shallower groove, as it lacks a noticeable projection below the connector loop. The corresponding position in UL44 of the UL42 projection is indicated by an arrow and an asterisk.
In contrast, although the residues of UL42 that bind to the C terminus of UL30 have been identified (29), a detailed mutational analysis to determine which of these residues are important for binding to UL30 has not yet been undertaken. Initial attempts to identify regions in UL42 responsible for interaction with and stimulation of UL30 were unsuccessful (6, 21). Random peptide display studies coupled with mutational and calorimetric analyses finally identified residue Q171, which is within the UL42 connector loop, as important for binding to UL30 (2). Indeed, the Q171A substitution of UL42 drastically reduced both binding to and long-chain DNA synthesis by UL30 (2). A cocrystal structure of UL42 bound to a UL30 C-terminal peptide then showed the presence of a hydrogen-bonding network which connects Q171 to the side chain of UL30 residue R1229 (29).
Although residues of both UL44 and UL42 that are important for binding to the cognate catalytic subunit lie in the connector loop, whether the role played by these residues is similar or different in the two systems remains to be determined. Two observations hint at differences. First, weak binding of UL30 could be detected with the UL42 Q171A mutant in maltose-binding protein pulldown assays and a small release of heat was measured by ITC when a large excess of a UL30 C-terminal peptide was added to the Q171A mutant (2), whereas the I135A substitution of UL44 completely impaired binding to UL54 in GST pulldown assays and reduced the affinity for the UL54 C-terminal peptide to unquantifiable levels in ITC experiments. Second, Q171 is a polar residue, while I135 is nonpolar.
Interactions of processivity factors with cognate polymerases.
The similarity in overall folding between HCMV UL44 and HSV UL42, together with the fact that residues involved in binding to the respective catalytic subunit are in the same structural element, could lead to the hypothesis that the interaction of UL44 with UL54 is analogous to that of UL42 with UL30. Moreover, the interaction of UL54 with UL44 has been mapped to the extreme C terminus of UL54 (15, 16), and the interaction of HSV-1 UL30 with UL42 had been mapped to the extreme C terminus of UL30 (5, 13, 18, 24, 25). However, the molecular details of the HCMV UL54-UL44 interaction are likely to be different from those of the interaction between the HSV counterparts, as recent studies, including this report, have highlighted important differences between the two systems. In particular, the UL44-UL54 interaction is likely to be more dependent upon hydrophobic interactions, since the most important of the residues of UL44 connector loop for binding (I135) and another important residue (V136) are hydrophobic. Similarly, UL54 residues important for UL44 binding are hydrophobic (16), whereas those of UL30 important for interaction with UL42 are basic (2, 29).
Consistent with this idea are the larger ΔH values for the HCMV interaction than for the HSV-1 interaction (2, 16), which may relate to the more crucial role of hydrophobic versus hydrophilic residues in the two systems. This contrasts with the UL30-UL42 interaction, where a few specific hydrogen bonds between polar residues constitute the crucial sequence-specific determinants for binding (1a, 2).
Given the α-helical propensity displayed by the C terminus of UL54 (15), it is tempting to speculate that this region might adopt an α-β-α structure, as seen for the analogous region of UL30 (29), and that an UL54 C-terminal α-helix might bind in a groove of UL44, as the extreme C-terminal helix of UL30 is accommodated in a deep groove of UL42. This groove lies between the connector loop and a projection formed by residues D63 and R64 (Fig. 10B, right) (29). However, the region below the connector loop of UL44 is much flatter, as it lacks a projection analogous to that formed by D63 and R64 of UL42 (Fig. 10B) (1).
A comparison of the interaction between UL54 and UL44 with the interactions of prokaryotic and eukaryotic sliding clamps with peptides deriving from their binding partners reveals intriguing similarities. The majority of the interactions between the gp45 sliding clamp and a polymerase C-terminal peptide from bacteriophage RB69 are hydrophobic (23). Similarly, a few C-terminal residues of the cell-cycle checkpoint protein p21WAF1/CIP1 make hydrophobic interactions with human PCNA (10, 27). An alignment of the p21WAF1/CIP1 and the RB69 Pol C-terminal peptides shows that the two peptides have a very similar overall conformation, which is partially α-helical (23). It is therefore possible that the C terminus of UL54 folds in a manner more similar to the p21WAF1/CIP1 and RB69 Pol peptides, which bind to the same face of their partners as does the HSV UL30-derived peptide to UL42 but do not adopt an α-β-α structure (10, 23). A crystal structure of UL44 bound to the C terminus of UL54 is needed to test these ideas.
The differences between HCMV and HSV DNA polymerase subunit interactions likely account for the fact that even though the residues most important for binding lie in analogous regions, i.e., the C terminus of the catalytic subunit and the connector loop of the accessory protein, the interaction between the subunits of HSV and of HCMV DNA polymerases is specific, since noncognate partners do not bind (15).
Implications for drug discovery.
New antiherpesvirus drugs are needed, especially for HCMV infections, as currently approved antivirals are frequently toxic and have pharmacological drawbacks and/or problems with viral resistance. As the interaction of UL44 with UL54 is specific and essential for long-chain DNA synthesis, it is an attractive target for antiviral compounds (14).
The demonstration that substitution of a single residue of UL44 is sufficient to completely disrupt the physical and functional interaction between UL54 and UL44 heralds the prospect that small molecules could also interfere with such interactions. Encouragement for this approach comes from the recent identification of small inhibitory molecules able to block the HSV-1 UL30-UL42 interaction in vitro as well as virus replication (21a). Thus, it is our hope that either high-throughput screening of small molecules or structure-based design will identify compounds that can specifically inhibit the UL54-UL44 interaction and HCMV replication.
Acknowledgments
We thank P. F. Ertl for kindly providing the pRSET-Pol plasmid, H. S. Marsden for purified baculovirus-expressed UL54 protein, C. E. Dahl and colleagues for synthesis of UL54 peptide 1 and sequencing, and personnel of the Molecular Biology Core Facility, Dana-Farber Cancer Institute, for peptide and protein quantification.
This work was supported by NIH grant AI19838.
REFERENCES
- 1.Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. [DOI] [PubMed] [Google Scholar]
- 1a.Bridges, K. G., Q. Hua, M. R. Brigham-Burke, J. D. Martin, P. Hensley, C. E. Dahl, P. Digard, M. A. Weiss, and D. M. Coen. 2000. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 275:472-478. [DOI] [PubMed] [Google Scholar]
- 2.Bridges, K. G., C. S. Chow, and D. M. Coen. 2001. Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches. J. Virol. 75:4990-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. R., and B. Chandran. 2000. Characterization of human herpesvirus 8 ORF59 protein (PF-8) and mapping of the processivity and viral DNA polymerase-interacting domains. J. Virol. 74:10920-10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. [DOI] [PubMed] [Google Scholar]
- 5.Digard, P., W. Bebrin, K. Weisshart, and D. M. Coen. 1993. The extreme C-terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J. Virol. 67:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Digard, P., C. S. Chow, L. Pirrit, and D. M. Coen. 1993. Functional analysis of the herpes simplex virus UL42 protein. J. Virol. 67:1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 66:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallo, M. L., D. H. Jackwood, M. Murphy, H. S. Marsden, and D. S. Parris. 1988. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J. Virol. 62:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of P21(WAF/CIP1) complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 11.Kiehl, A., and D. I. Dorsky. 1995. Bipartite DNA-binding region of Epstein-Barr virus BRMF1 product essential for DNA polymerase accessory function. J. Virol. 69:1669-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystrallogr. 24:946-950. [Google Scholar]
- 13.Loregian, A., E. Papini, B. Satin, H. S. Marsden, T. R. Hirst, and G. Palù. 1999. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc. Natl. Acad. Sci. USA 96:5221-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loregian, A., H. S. Marsden, and G. Palù. 2002. Protein-protein interactions as targets for antiviral chemotherapy. Rev. Med. Virol. 12:239-262. [DOI] [PubMed] [Google Scholar]
- 15.Loregian, A., R. Rigatti, M. Murphy, E. Schievano, G. Palù, and H. S. Marsden. 2003. Inhibition of human cytomegalovirus DNA polymerase by C-terminal peptides from the UL54 subunit. J. Virol. 77:8336-8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78:158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mar, E. C., J. F. Chiou, Y. C. Cheng, and E. S. Huang. 1985. Human cytomegalovirus-induced DNA polymerase and its interaction with the triphosphates of 1-(2′-deoxy-2′-fluoro-beta-d-arabinofuranosyl)-5-methyluracil, -5-iodocytosine, and -5-methylcytosine. J. Virol. 56:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsden, H. S., M. Murphy, G. L. McVey, K. A. MacEachran, A. M. Owsianka, and N. D. Stow. 1994. Role of the carboxy terminus of herpes simplex virus type 1 DNA polymerase in its interaction with UL42. J. Gen. Virol. 75:3127-3135. [DOI] [PubMed] [Google Scholar]
- 19.Merritt, E. A., and D. J. Bacon. 1997. Raster3D photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 20.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 21.Owsianka, A. M., G. Hart, M. Murphy, J. Gottlieb, R. Boehme, M. Challberg, and H. S. Marsden. 1993. Inhibition of herpes simplex virus type 1 DNA polymerase activity by peptides from the UL42 accessory protein is largely nonspecific. J. Virol. 67:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Pilger, B. D., C. Cui, and D. M. Coen. 2004. Identification of a small molecule that inhibits herpes simplex virus DNA polymerase subunit interactions and viral replication. Chem. Biol. 11:647-654. [DOI] [PubMed] [Google Scholar]
- 22.Randell, J. C. W., and D. M. Coen. 2004. The herpes simplex virus processivity factor UL42 binds DNA as a monomer. J. Mol. Biol. 335:409-413. [DOI] [PubMed] [Google Scholar]
- 23.Shamoo, Y., and T. A. Steitz. 1999. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99:155-166. [DOI] [PubMed] [Google Scholar]
- 24.Stow, N. D. 1993. Sequences at the C terminus of the herpes simplex virus type 1 UL30 protein are dispensable for DNA polymerase activity but not for viral origin-dependent DNA replication. Nucleic Acids Res. 21:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenney, D. J., P. A. Micheletti, J. T. Stevens, R. K. Hamatake, J. T. Matthews, A. R. Sanchez, W. W. Hurlburt, M. Bifano, and M. G. Cordingley. 1993. Mutations in the C terminus of herpes simplex virus type 1 DNA polymerase can affect binding and stimulation by its accessory protein UL42 without affecting basal polymerase activity. J. Virol. 67:543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenney, D. J., W. W. Hurlburt, M. Bifano, J. T. Stevens, P. A. Micheletti, R. Hamatake, and M. G. Cordingley. 1993. Deletions of the carboxy terminus of herpes simplex virus type 1 UL42 define a conserved amino-terminal functional domain. J. Virol. 67:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1995. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr. Biol. 5:275-282. [DOI] [PubMed] [Google Scholar]
- 28.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34:191-206. [DOI] [PubMed] [Google Scholar]
- 29.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]