Abstract
The aim of this study was to evaluate metal binding and antioxidant activities of hydrolyzed oat bran proteins followed by the determination of peptide sequences. Protamex oat bran protein hydrolysates (OBPH) were separated by reverse-phase HPLC into eight peptide fractions (F1–F8) and their abilities to either chelate metals (Fe2+, Ca2+) or prevent the oxidation of lipids were investigated. In the Fe2+ chelation assay, OBPH had significantly (p < 0.05) higher activity (39.7 %) than the best performed fraction F7 (22.8 %). The second most active was F5 with 12.1 % chelating activity and this was higher than the activity of the tripeptide glutathione (5.8 %) used as control. The two most Fe2+ chelating fractions (F5, F7) however had weak calcium binding (0.6–1.0 %) properties at peptide concentration ranging from 0.2 to 1.0 mg/mL. In the lipid peroxidation assay, OBPH and all HPLC fractions prevented the oxidation of linoleic acid. More than 60 peptides mainly derived from globulin and avenin proteins were identified using tandem mass spectrometry.
Keywords: Oat bran, Protein hydrolysate, Metal chelation, Calcium binding, Mass spectrometry
Introduction
The reduction of molecular oxygen under various conditions (e.g. respiration, light, presence of metals, irradiation) can lead to the formation of reactive oxygen species like superoxide anion, peroxyl and hydroxyl radicals in addition to non-radicals such as hydrogen peroxide and singlet oxygen (Schneider and Reischak de Oliveira 2009). Further reactions of partially reduced oxygen (i.e. superoxide anion radical) can also lead to the formation of nitrogen-containing reactive species including nitric oxide, peroxynitrite and nitric dioxide (Reuter et al. 2010). Under normal conditions, reactive species are essential for several physiological functions such as control of cells proliferation, growth, immune system modulation, and infection (Brieger et al. 2012). Excessive production of reactive species in vivo can however result in oxidative stress and subsequent damage to DNA, proteins, enzymes and lipids (Pinchuk et al. 2012). In foods, damages to molecules lead to rancidity and reduce shelf life. Antioxidant molecules have been proven to be useful in preventing or minimizing the harmful effect of reactive species by converting them to stable entities (Kumar 2011). Many works have been done to demonstrate the role that polyphenols from fruits, legumes and grains can have in preventing oxidation of foods and body molecules thereby reducing the risk of chronic diseases or increasing shelf life (Hu and Willett 2002; Lehtinen et al. 2003).
Research has also been done to determine the biological function of peptides naturally occurring in foods and from the breakdown of proteins. For example, antioxidant activities have been found for peptides or hydrolysed proteins from milk (Meisel 2004), fish (Girgih et al. 2013), chickpeas (Chang et al. 2011), walnuts (Chen et al. 2012), wheat germ (Zhu et al. 2006), or soybeans (Rho et al. 2007). Food peptides, like those from milk and soy (Bao et al. 2007) can also enhance the absorption of calcium, a metal important for proper bone density and muscle function (Chen et al. 2014). Recently, it was demonstrated that the protease Protamex at various concentrations yielded digested oat bran proteins with radical scavenging properties (Gao et al. 2014). The activity of fractions from chromatographic separations has not yet been investigated. The objective of this study was then to investigate the mineral binding capacity and the antioxidant activity of fractions from Protamex digested oat bran proteins in addition to the determination of the sequence of peptides.
Methodology
Materials and chemicals
Oat bran (sample i.d. 112-001) was supplied by Richardson Milling (Portage La Prairie, Manitoba MB). Viscozyme L® (V2010), Protamex® (P0029), Ferrozine®, L-glutathione (GSH), linoleic acid (98 %), calcium chloride, ferrous chloride, ammonium thiocyanate, sodium hydroxyde, were obtained from Sigma Aldrich (Oakville, ON). Potassium phosphate, ethanol, ethylenediaminetetraacetic acid and sodium dodecyl sulfate were purchased from Fisher Scientific (Nepean, ON). Spectrophotometric measurements were performed on the BioTek® Epoch™ UV–Vis microplate reader, controlled by Gen5™ data analysis software. Incubations were done on a MaxQ™ 5000 shaker model (Fisher Scientific, Nepean, ON).
Extraction and hydrolysis of proteins
Proteins were extracted from defatted brans at pH 4.5 in the presence of vscozyme 3 FBG/g as previously described (Jodayree et al. 2012). To perform the hydrolysis, protein isolates (19 g) were mixed with water (1:11 w/v) ratio. The pH was adjusted to 6.5 with 0.1 M NaOH. The protease, Protamex was added at 1:20 (E/S) ratio. The mixture was incubated at 50 °C, 130 rpm for 2 h on the shaker incubator and the enzyme was inactivated by heating at 90 °C for 10 min. The hydrolyzed proteins were obtained after centrifugation to remove insoluble proteins. It was then freeze dried and stored at −20 °C until separation and testing.
Peptide fractionation by RP-HPLC
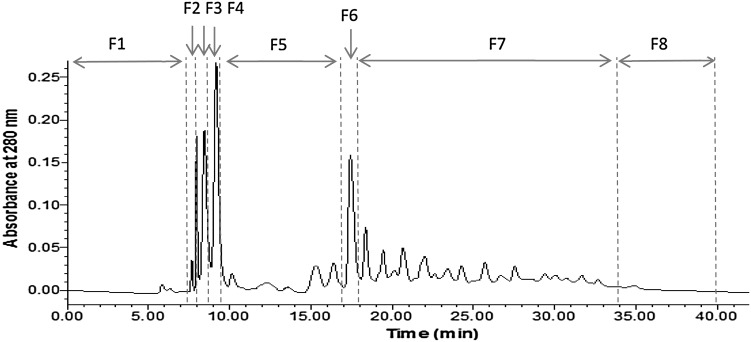
The fractionation of hydrolyzed oat bran proteins was carried out on a semi-preparative HPLC system that included 1525 binary pump, 2998 photodiode array detector (set at 280 nm), 2707 auto-sampler maintained at 8 °C, and fraction collector III from Waters (Montreal, QC, Canada). The lyophilized oat bran protein hydrolysate was dissolved in 0.1 % acetic acid in water (Solvent A) at a concentration of 100 mg/mL and filtered through 0.45 μm nylon membrane. For each run, 2 mL were injected onto a Waters Prep XBridge BEH C18, 130 Å, 10 μm, 19 × 150 mm column. Peptides were eluted with a linear gradient of 95–5 % of solvent A over 54 min. Solvent B was 100 % methanol. Eluates were collected and pooled into eight fractions as shown in Fig. 1. The solvent in pooled fractions was evaporated using a BUCHI Rotavapor® R-215 at 45 °C, re-suspended in water and freeze dried. Protein contents of the oat bran hydrolysate and fractions were measured using a modified Lowry method (Markwell et al. 1978).
Fig. 1.
HPLC chromatogram of hydrolyzed oat bran proteins on C18 column using a linear gradient of 95–5 % water containing 0.1 % acetic acid and methanol. Eight fractions (F1–F8) were collected as indicated
Iron chelating assay
The metal chelating assay was measured according to a previously reported procedure (Xie et al. 2008) with some modifications. Oat protein hydrolysate, peptide fraction or GSH (1.425 mL of, 0.5 mg/mL) was mixed with 25 μL of FeCl2 (2 mmol/L) and then 50 µL of 5 mM Ferrozine (3-(2-Pyridyl)-5,6-diphenyl-1,2,4-triazine-4′,4′′-disulfonic acid sodium salt) followed by vortex mixing. The mixture was incubated for 10 min at room temperature, after which 200 µL were transferred into a 98-well clear microplate and the absorbance was recorded at 562 nm. The chelating effect was calculated using the following equation:
Chelating effect (%) = [(Ac − As)/Ac)] × 100, where Ac and As are the absorbance of the control and sample respectively.
Calcium binding capacity
The calcium binding capacity was measured based on a previously reported procedure (Jung et al. 2006). Oat protein hydrolysate and peptide fractions were dissolved in 20 mM sodium phosphate buffer (pH 7.8). In 1:1 ratio, seven different concentrations 0.2–1.0 mg/mL were mixed with 10 mM CaCl2 in doubled distilled water. The reaction mixture was shaken and incubated at 25 °C for 30 min. After the incubation, pH was adjusted to 7.8 using NaOH 0.1–0.5 M. The sample mixture was filtered using 0.45 µm nylon membrane (DIMA Glass Inc., Richmond Hill, ON) to remove non-soluble phosphate-calcium complexes. Calcium binding capacity was measured using a calcium ion-selective electrode and the Go-link interface (Merlan Scientific Ltd, Mississauga, ON).
Inhibition of linoleic acid oxidation
The in vitro lipid peroxidation inhibition activity was determined according to literature (Girgih et al. 2013). Oat bran protein hydrolysates, peptide fractions and GSH, 0.5 mg/ml each were prepared in phosphate buffer (50 mM, pH 7.0). Each sample (0.4 mL) was added to 0.4 mL of linoleic acid (75 mM, 95 % ethanol) and then allowed to incubate at 60 °C for 5 days. The degree of oxidation was determined at 24 h intervals, by measuring the ferric thiocyanate values as follows. An amount of each incubated sample (50 μL) was added to 2.35 ml of 75 % ethanol, 50 μL of 30 % ammonium thiocyanate and 50 μL of 20 mM ferrous chloride solution prepared in 1 M HCl. Mixtures were immediately vortexed and 200 µL were then transferred into a 96-well clear microplate and the absorbance was measured at 500 nm. The increase in absorbance is associated with the increase formation of linoleic acid hydroperoxides.
Mass spectrometry and identification of peptides
Analysis was performed at the Quebec Genomics Center (Sainte-Foy, QC, Canada). Peptides were identified in one of the HPLC fraction (F5) with lipid peroxidation and metal chelating activities. Experiments were performed with an Agilent nanoscale capillary liquid chromatography (nanoLC) coupled to a triple TOF 5600 plus mass spectrometer (AB Sciex, USA) equipped with a nano-electrospray ion source in positive mode. Peptides were separated for 60 min on PicoFrit 15-μm tip, BioBasic C18, 10 cm × 75 μm column (New Objective, USA) using a linear gradient of 5–80 % of formic acid (0.1 %) in acetonitrile (Solvent B). Spectra were acquired using a data-dependent acquisition mode (Analyst software version 1.6). Full scan mass spectrum (400–1250 m/z) was followed by collision-induced dissociation (MS/MS) of multiple charged peaks (2+–5+). The Protein Pilot version 4.5 software (AB Sciex, USA) was used to generate MS/MS peak lists which were then analyzed using Mascot (Matrix Science, London, UK; version 2.4.1) and X! Tandem [The Global Proteome Machine, thegpm.org; version CYCLONE (2010.12.01.1)]. Search was limited to the TAX_Poeae_147387_20141216 database assuming non-specific enzyme digestion. Fragment and parent ion mass tolerances were set to 0.100 Da. Scaffold version 4.4.1.1 (Proteome Software Inc., Portland, OR) was used to validate MS/MS data. Peptide identifications were accepted if they could be established at greater than 99 % probability to achieve a False Discovery Rate (FDR) less than 1.0 % by the Scaffold Local FDR algorithm.
Statistical analysis
Data were expressed as mean ± standard deviation from triplicates. One way ANOVA and Fisher’s least significant difference test were used to determine significant differences between groups (p < 0.05). IBM SPSS Statistics 21 (SPSS Inc., Chicago, IL) were used for all analyses.
Results and discussion
Fractionation of hydrolyzed proteins by HPLC and protein content of fractions
HPLC was used to separate oat bran protein hydrolysates into different fractions. The stationary phase was C18 column, while the mobile phase was a gradient of water and methanol. Less hydrophobic molecules have little interaction with the C18 stationary phase, resulting in rapid elution with high water percentage solvent mixtures. In contrast, more hydrophobic samples stick on the stationary phase longer and are eluted later with increasing content of methanol and decreasing water. Eight peptide fractions were collected (Fig. 1). Protein contents of fractions F3–F8 ranged from 70.0 to 104.31 %. Oat bran protein hydrolysate had 68.04 % protein which was lower compared to the fractions (F3–F8). Fractionation of oat protein hydrolysate then increased the protein content of most fractions. Most of non-proteins molecules were recovered in fractions 1 and 2 (less than 25 % proteins). These two early eluting fractions certainly contained sugars that have weak binding to the C18 column or salts used in various pH adjustments. Similar finding have been shown for fractions obtained after separation of other hydrolyzed food proteins (Girgih et al. 2013). F1 and F2 were not further investigated because of their low protein contents.
Iron chelating activity
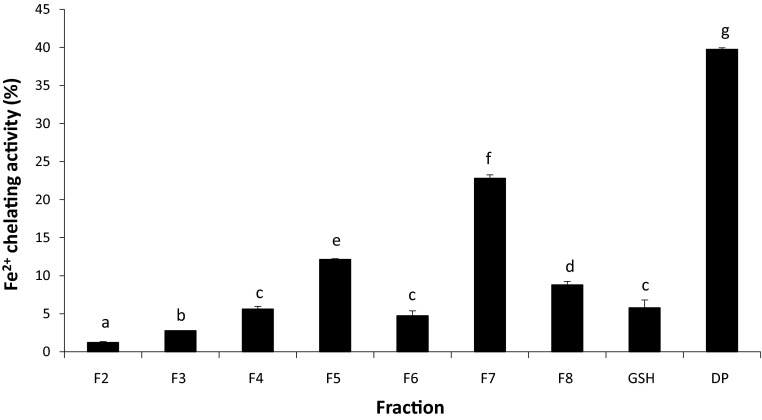
The hydrolyzed proteins, OBPH had the highest (p < 0.05) ferrous iron chelating activity (39.7 %) in comparison to peptide fractions F2–F8 (1.2–22.8 %; Fig. 2). The chelating activity of OBPH was similar to that of walnut protein hydrolysate (40 %; Chen et al. 2012). Cysteine has been shown to play a role in the chelating activity of peptides. However, its effects alone may be limited, as GSH a cysteine containing peptide used as control had only 5.8 % ferrous ion chelating activity. Other studies have also reported a low ferrous ion chelating activity for GSH (Xie et al. 2008). Amongst the fractions F7 had the highest chelating (22.8 %) followed by F5 with 12.1 % chelating activity. Negatively charged groups like carboxylic moieties in acidic amino acids (AAs) have excess electrons that enhance electrostatic and ionic interactions with iron and can result in an increase of chelating activity for peptides that contain these AAs (Zhu et al. 2008). Histidine located at the N-terminal of peptides can also enhance metal ion chelation because of higher electron density of the imidazole ring (Prior et al. 2005; Girgih et al. 2013). A number of peptides identified in fraction F5 (Table 1) contain AAs with carboxylate groups on their side chain or histidine that may have increased its chelating activities compared to F3, F4, F6 or F8. Data are however only qualitative and the true contribution of these peptides cannot be determined. In addition, if was not possible to obtain information of the sequences of peptides present in other fractions.
Fig. 2.
Metal chelating of oat protein hydrolysate (DP) and its HPLC fractions (F2–F8) obtained using a linear gradient of water and methanol containing 1 % acetic acid. GSH: glutathione. Values are means of triplicates ± SD. Bars with same letters are not significantly different and bars with different letters are significantly different (p < 0.05)
Table 1.
List of peptides identified in F5, a fraction with higher metal chelating activities from HPLC separation of Protamex digested oat brans proteins on a C18 column
| Sequence | Observed | Actual mass | Protein ID from Uniprot |
|---|---|---|---|
| FEPLRQVRSQAGITE | 577.64 | 1729.90 | P12615, Q38779, Q38780 |
| ILSPYWNINAH | 664.34 | 1326.66 | P12615, P14812 |
| LGISQQAAQRIQSQKEQRGEI | 592.82 | 2367.26 | O49257, O49258 |
| LGISQQVAQKIQSQNDQRGEI | 780.74 | 2339.19 | P14812 |
| RVQVVNNHGQTVF | 499.93 | 1496.77 | P12615, P14812 |
| RVQVVNNNGQTVF | 737.89 | 1473.77 | O49257, O49258, Q38779 |
| YRISRQEARNLKNNRGEE | 559.04 | 2232.14 | Q38779, Q38780 |
| YRISRQESQNLKNNRGEE | 556.03 | 2220.09 | P12615, Q38781 |
| FNREGLPLFILANFR | 603.04 | 1806.09 | G9FL41 |
| FGAFTPKF | 457.74 | 913.4676 | P12615 |
| LVEHQAYQPIQSQEGQSTQ | 724.35 | 2170.03 | O49258 |
| DVNNNANQLEPRQKEF | 639.31 | 1914.92 | O49257, O49258, P12615, P14812 |
| DVNNNANQLEPRQKEFL | 677.00 | 2027.98 | O49257, P12615, P14812, Q38780 |
| KTNPNSMVSHIAGKSSI | 590.98 | 1769.91 | O49257, O49258 |
| KTNPNSMVSHIAGKSSIL | 628.67 | 1882.99 | O49257, O49258 |
| KTNPNSMVSHIAGKSSILRA | 532.54 | 2126.13 | O49257, Q38780 |
| NDILRRGQL | 542.81 | 1083.61 | P12615, Q38780 |
| TQTGSQSYQDEGESSSTEKASE | 779.33 | 2334.96 | P12615, P14812 |
| TQTSFQPYPEGEDES | 857.86 | 1713.70 | O49257 |
| TQTSFQPYPEGEDESSLTN | 710.64 | 2128.91 | O49258 |
| VIRRVIEPQGLL | 464.96 | 1391.85 | O49258, P12615, P14812 |
| EIIRVSQGLQ | 571.83 | 1141.64 | P12615, P14812 |
| LVLPQYHNAPAL | 668.37 | 1334.73 | Q38779 |
| YVFDVNNNANQLEPRQKE | 726.69 | 2177.05 | O49257, P12615, P14812, Q38780 |
| LAGNNKREQQFGQNIF | 932.47 | 1862.93 | P14812 |
| LAGNNKREQQSGNNIFSGL | 683.01 | 2046.02 | Q38779 |
| LAGNNKREQQSGNNIFSGLS | 712.02 | 2133.05 | Q38779 |
| LPQYHNAPGLVY | 686.35 | 1370.69 | O49257, P12615, P14812 |
| NSKNFPTL | 460.75 | 919.4756 | P14812 |
| PAGIVHWGYNDGDA | 736.33 | 1470.65 | O49257 |
| PQYHNAPGL | 498.74 | 995.4752 | O49257, P12615, P14812 |
| PQYHNAPGLV | 548.28 | 1094.54 | O49257, P12615, P14812 |
| PQYHNAPGLVY | 629.81 | 1257.60 | O49257, P12615, P14812 |
| QAFEPLRQ | 494.76 | 987.5114 | O49257, O49258, P12615, P14812, Q38780 |
| QFLKPTMSQQE | 668.83 | 1335.64 | O49258 |
| QFLKPTMSQQEL | 725.37 | 1448.72 | O49258 |
| QQQQQQQQQQF | 717.27 | 1432.52 | G8ZCV2, G8ZCV3 |
| QQVFNQPQQQAQF | 795.88 | 1589.76 | G8ZCT7, Q09114, G8ZCT6, G8ZCT5, L0L6K5 |
| QQVFNQPQQQAQFEGMRA | 717.67 | 2150.00 | G8ZCT7, Q09114, G8ZCT6, G8ZCT5, L0L6K5 |
| RALPIDVL | 448.78 | 895.5464 | Q38780 |
| RALPVDVLA | 477.29 | 952.5646 | P12615, P14812 |
| SPFWNINAHS | 586.77 | 1171.53 | O49258 |
| SPYWNINAHSVM | 717.82 | 1433.63 | P14812 |
| VEHQAYQPIQSQEGQSTQ | 686.65 | 2056.92 | O49258 |
| VQMSATRVNL | 559.80 | 1117.58 | O49257, P12615, P14812, Q38779, Q38780 |
| VYILQGRGF | 526.80 | 1051.58 | O49257, P12615, P14812, Q38780 |
| VYILQGRGFTG | 605.83 | 1209.65 | O49257, P12615, P14812, Q38779 |
| IQGHARVQVVNNNGQTVF | 661.01 | 1980.01 | O49257, Q38779 |
| VSHIAGKSSIL | 556.32 | 1110.63 | O49257, Q38780 |
| FLKPIVSQQVPVE | 742.43 | 1482.84 | Q38779 |
| FLKPIVSQQVPVEQ | 806.45 | 1610.89 | Q38779 |
| GPVEHQAYQPIQSQE | 570.94 | 1709.80 | P14812 |
| GPVEHQAYQPIQSQQ | 570.94 | 1709.80 | P12615, Q38781 |
| GPVEHQAYQPIQSQQEQSTQ | 762.02 | 2283.04 | P12615, Q38781 |
| QQLQQQLIQPQL | 724.41 | 1446.81 | I4EP64, F4MJY2, F2Q9W3 |
| QQQPFMQQQQPFMQQQQ | 728.28 | 2181.82 | I4EP85, I4EP86, I4EP78, F4MJY6, F4MJY5 |
| VRSQAGITEY | 562.29 | 1122.56 | P12615, Q38779 |
| FTPWQSSRQ | 568.78 | 1135.54 | O48257, P12615, P14812 |
| FTPWQSSRQGGL | 682.34 | 1362.66 | O48257, P12615, P14813 |
| VIRRVIEPQGLVL | 497.98 | 1490.93 | Q38779 |
| FDVNNNANQLEPRQKEFL | 726.02 | 2175.05 | O49257, P12615, P14812, Q38780 |
| FDVNNNANQLEPRQKK | 639.31 | 1914.92 | O49257, P12615, P14812, Q38779 |
| IRRVIEPQGLL | 431.94 | 1292.79 | O49257, P12615, P14812 |
| IRRVIEPQGLLL | 469.63 | 1405.88 | O49257, P12615, P14812 |
| SVIRRVIEPQG | 418.58 | 1252.72 | O49257, P12615, P14812, Q38779 |
| YILQGRGF | 477.26 | 952.5058 | O49257, P12615, P14812, Q38779 |
| YILQGRGY | 485.26 | 968.5014 | O49258 |
| ISFKTNPNSM | 569.78 | 1137.54 | O49257, P14812, Q38779, Q38780 |
| VFDVNNNANQLEPRQKE | 672.34 | 2013.99 | O49257, P12615, P14812, Q38779 |
Scaffold software was used to validate MS/MS data. Peptide identifications were accepted if they could be established at greater than 98 % probability
Q38779 and Q3780: 11S globulin; Q38781: oat storage 12S globulin; O49257 O49258: 12S globulin; P12615: 12S seed storage globulin 1; P14812: 12S seed storage globulin 2; I4EP64, I4EP78, I4EP85 and I4EP86: avenin; F2Q9W3: avenin protein; F4MJY2, F4MJY5, F4MJY6: avenin protein; G8ZCV2 and G8ZCV3: avenin protein; G8ZCT5 and G8ZCT6: avenin protein; Q09114: avenin-E; L0L6K5: Gliadin-like avenin; G9FL41: plastid acetyl-CoA carboxylase
Calcium binding capacity
The iron chelating assay revealed that Fractions F5 and F7 were the two most efficient. It was then of interest to determine their calcium binding properties. It is known that peptides that bind calcium can increase their absorption and therefore their biological function (Chen et al. 2014). The binding activity of fractions F5 and F7 ranged from 0.6 to 1.0 mg/L for peptide concentrations between 0.2 and 1.0 mg/L. These values were very weak as they were close to the control (i.e. no peptide) value. Calcium binding properties of hydrolyzed proteins from some foods have been investigated and in general those from animal derived proteins appeared to perform better. At peptides concentration with the ranged of those tested here, calcium binding activities of 36.3 and 32.0 mg/L for milk and fish peptides, respectively were reported (Jiang and Mine 2000; Jung et al. 2006). In both cases, the activity was attributed to the presence of phosphoryl groups, serine or aspartic acid that are electron rich. Soluble calcium concentration in the presence of fish peptide fractions from membrane separation was reported to vary from 6 to 25 mg/L (Jung and Kim 2007). Hydrolyzed proteins from wheat germ were demonstrated to bind 5–16 mg of calcium/g depending on the protease used after 2 h digestion (Liu et al. 2013). These results indicate that the type of enzyme used for proteolysis has an effect on the calcium binding capacity and different proteases may produce different results for oat bran proteins. Fractionation based on charge may also enhance the binding activity of food peptides (Jung et al. 2006).
Inhibition of lipid peroxidation
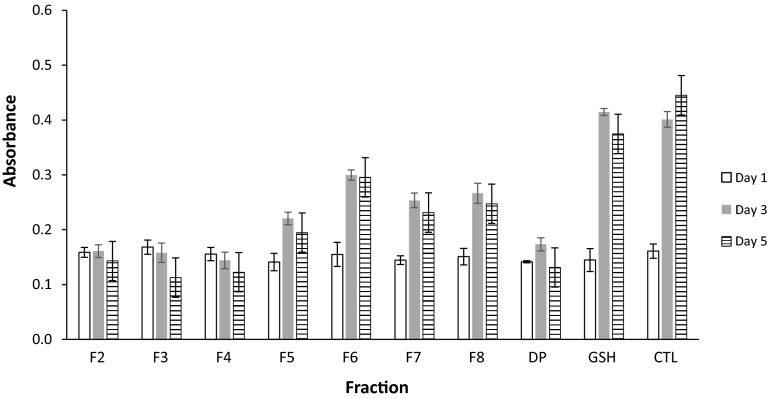
Fatty acids are the main building blocks of phospholipids and triglycerides and depending on their degree of unsaturation they can be readily oxidized under various conditions (Shahidi and Zhong 2010). Oxidized lipid in the form of hydroperoxides convert ferrous ions (Fe2+) to ferric ions (Fe3+) and the later react with ammonium thiocyanate to generate a colored complex whose absorbance is proportional to the degree of oxidation (Girgih et al. 2013). The presence of antioxidant molecules decrease the color intensity. Linoleic acid (LA) that can be autoxidized to generate hydroperoxides at position 9, 11 and 13 is often used to test the ability of molecules to prevent or retard the oxidation of lipids. After 24 h (i.e. day 1), there was no difference in peroxide values of sample control linoleic acid (LA) and peptides fractions (Fig. 3). As the incubation time increase, all fractions tested significantly inhibited LA hydroperoxides compared to control with no peptides. F3 and F4 were the most active and no increase of the formation of lipid peroxides was observed in fractions that contain them. The second most active was F5 and the non-fractionated hydrolysate. The tripeptide GSH had no effect. Hydrophobicity and amino acids like histidine or methionine are considered to be very important in the ability of peptides to inhibit lipid oxidation (Chen et al. 1998, 2012). In the present study hydrophobicity alone did not seem as important because fractions F6–F8 eluted with more proportion of organic solvent have the least activity. When lipid are oxidized especially in the model system like LA emulsion, the peroxide group moves to the surface where it can be reduced by molecules with more affinity for aqueous environment. This can explain the better activity of F3–F5 in the present work. Using synthetic peptides it was demonstrated that those with histidine at the N-terminal better inhibited the formation LA hydroperoxides compared to the ones with histidine at other positions (Chen et al. 1998).
Fig. 3.
Inhibition of linoleic acid oxidation by oat protein hydrolysate and peptide fractions. F2–F8 were obtained from after HPLC separation on C18 using a linear gradient of water and methanol containing 1 % acetic acid. DP digested proteins (no separation), GSH glutathione, CTL control (linoleic acid without addition of peptides). Values are means of triplicates ± SD
Identification of peptides
A hybrid quadrupole time-of-flight instrument was used for identification of peptides in F5 which appeared to possess good ferrous ions chelating activity and good inhibition of the formation of linoleic acids hydroperoxides. Multiple-charge ion scanning was conducted thereby, facilitating the identification of peptides over other molecules that may have been present. MS/MS data were automatically obtained for peaks with charges of 2+–5+. Peak lists were analyzed using Mascot™ and X! Tandem as described in the methodology section and were statistically interpreted using Scaffold, a software for validating MS/MS data (Searle 2010).
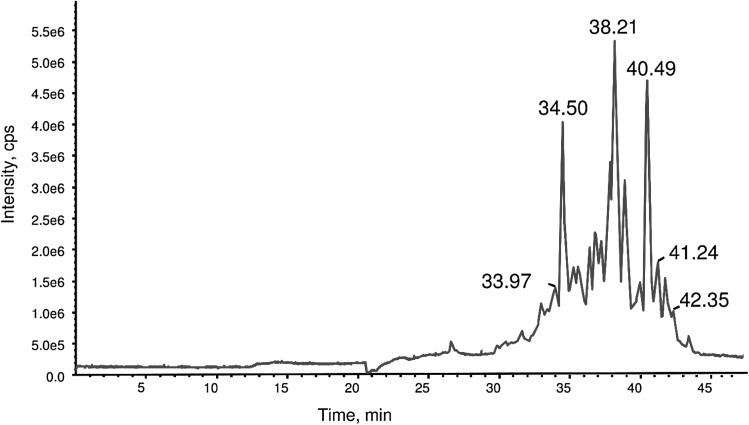
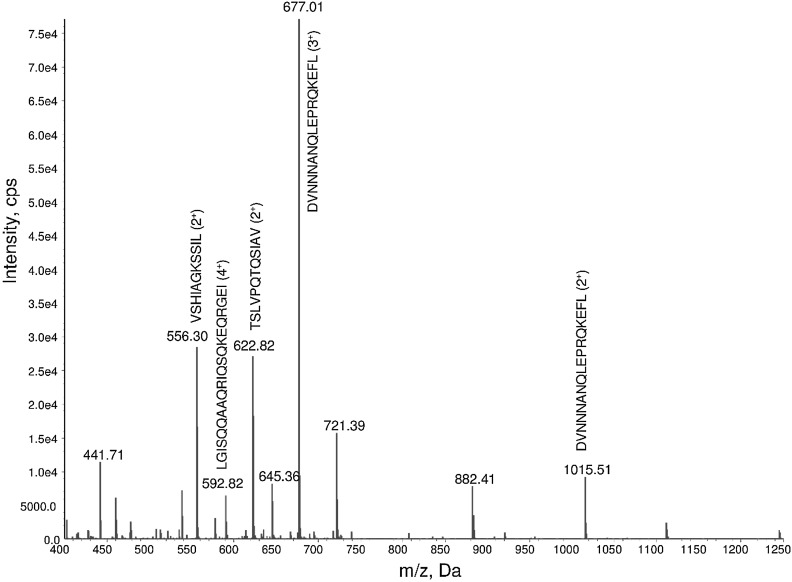
The total ion count chromatogram of F5 (Fig. 4) indicated that it was a complex mixture with ion peaks appearing between 25 and 45 min. An example of a TOF MS scan at 38.15 min is displayed in Fig. 5. Products spectra (MS/MS) and database searches allow the identification of the peptides with more than 99 % probably. Peaks at m/z 6.77.01 and 1015.51 correspond to DVNNNANQLEPRQKEFL that was triple and double charged, respectively; m/z 622.82 to TSLVPQTQSIAV; m/z 592.82 to LGISQQAAQRIQSQKEQRGEI; and m/z 556.30 to VSHIAGKSSIL. The major peak in Fig. 5 is attributed DVNNNANQLEPRQKEFL and this is because it is derived from five different oat proteins, four other peptide identified (Fig. 1) are also derived from its direct breakdown. Other peaks present in Fig. 5 were not assigned to peptides. The list of all identified peptides in this study is shown in Table 1. They are mainly derived from 11S and 12S globulins, 12S seed storage globulin, avenin and gliadin like avenin proteins. Twenty seven of the identified peptides contain tyrosine or tryptophan and amongst these fourteen also contain histidine. The presence of these are amino acids is addition to their position on the sequence are known to contribute to both chelating and lipid peroxidation activities (Chen et al. 1998; He et al. 2013; McDermott 2009). Although, some identified peptides also contain glutamic and aspartic acids which can interact with calcium (Cariolou and Morse 1988; Vavrusova and Skibsted 2013), F5 did not displayed calcium binding activities. This may be because their position or the length the sequence did not allow proper interaction. It is also possible that the amount of peptides containing these two AAs in F5 were small as MS/MS data were only qualitative.
Fig. 4.
Total ion count chromatogram of fraction 5 from LC-qTOF mass spectrometry. Data were collected for 65 min. Signals are concentrated between 25 and 45 min
Fig. 5.
TOF MS at 38.15 min. Full scan was performed from 400 to 1250 Da. Subsequence MS/MS analysis was used to determine the sequence of peaks at m/z 556.30, 592.82, 677.01 and 1015.51 as indicated on the figure. Other peaks were not assigned to peptides
Conclusion
Chromatographic separation of Protamex digested oat bran proteins and metal chelating testing showed that the two fractions (F5, F7) with higher ferrous ions chelating activities did not possess calcium binding properties suggesting different chelating mechanism. In the linoleic acid peroxidation assay, both fractions inhibited the formation of lipid peroxide with F5 being more active than F7. These fractions can be further tested for their ability to retard lipid oxidation in food systems. Many peptides were identified in F5 and it will be interesting to test the ones that contain amino acids known to contribute to chelating and radical scavenging properties.
Acknowledgments
This work was supported by a Grant from National Science and Engineering Research Council of Canada (Grant No. 371908) and a Saudi Arabia government’s King Abdulah Foreign Scholarship to M. M. Baakdah. The authors thank Mr. Daniel Defoy from the Quebec Proteomics Platform available at the Quebec Genomics Center for technical assistance.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Bao XL, Song M, Zhang J, et al. Calcium-binding ability of soy protein hydrolysates. Chin Chem Lett. 2007;18:1115–1118. doi: 10.1016/j.cclet.2007.07.032. [DOI] [Google Scholar]
- Brieger K, Schiavone S, Miller FJ, Krause K-H. Reactive oxygen species: from health to disease. Swiss Med Wkly. 2012;142:w13659. doi: 10.4414/smw.2012.13659. [DOI] [PubMed] [Google Scholar]
- Cariolou MA, Morse DE. Purification and characterization of calcium-binding conchiolin shell peptides from the mollusc, Haliotis rufescens, as a function of development. J Comp Physiol B. 1988;157:717–729. doi: 10.1007/BF00691002. [DOI] [Google Scholar]
- Chang Y-W, Alli I, Konishi Y, Ziomek E. Characterization of protein fractions from chickpea (Cicer arietinum L.) and oat (Avena sativa L.) seeds using proteomic techniques. Food Res Int. 2011;44:3094–3104. [Google Scholar]
- Chen HM, Muramoto K, Yamauchi F, et al. Antioxidative properties of histidine-containing peptides designed from peptide fragments found in the digests of a soybean protein. J Agric Food Chem. 1998;46:49–53. doi: 10.1021/jf970649w. [DOI] [PubMed] [Google Scholar]
- Chen N, Yang H, Sun Y, et al. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides. 2012;38:344–349. doi: 10.1016/j.peptides.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Chen D, Mu X, Huang H, et al. Isolation of a calcium-binding peptide from tilapia scale protein hydrolysate and its calcium bioavailability in rats. J Funct Foods. 2014;6:575–584. doi: 10.1016/j.jff.2013.12.001. [DOI] [Google Scholar]
- Gao Q, Smith JC, Tsopmo A. Optimized Protamex digested oat bran proteins: antioxidant properties and identification of new peptides. Austin J Nutr Food Sci. 2014;2:1053. [Google Scholar]
- Girgih AT, Udenigwe CC, Hasan FM, et al. Antioxidant properties of Salmon (Salmo salar) protein hydrolysate and peptide fractions isolated by reverse-phase HPLC. Food Res Int. 2013;52:315–322. doi: 10.1016/j.foodres.2013.03.034. [DOI] [Google Scholar]
- He X, Cao W, Zhao Z, Zhang C. Analysis of protein composition and antioxidant activity of hydrolysates from Paphia undulate. J Food Nutr Res. 2013;1:30–36. [Google Scholar]
- Hu FB, Willett WC. Optimal diets for prevention of coronary heart disease. JAMA. 2002;288:2569–2578. doi: 10.1001/jama.288.20.2569. [DOI] [PubMed] [Google Scholar]
- Jiang B, Mine Y. Preparation of novel functional oligophosphopeptides from hen egg yolk phosvitin. J Agric Food Chem. 2000;48:990–994. doi: 10.1021/jf990600l. [DOI] [PubMed] [Google Scholar]
- Jodayree S, Smith JC, Tsopmo A. Use of carbohydrase to enhance protein extraction efficiency and antioxidative properties of oat bran protein hydrolysates. Food Res Int. 2012;46:69–75. doi: 10.1016/j.foodres.2011.12.004. [DOI] [Google Scholar]
- Jung WK, Kim S-K. Calcium-binding peptide derived from pepsinolytic hydrolysates of hoki (Johnius belengerii) frame. Eur Food Res Technol. 2007;224:763–767. doi: 10.1007/s00217-006-0371-4. [DOI] [Google Scholar]
- Jung WK, Karawita R, Heo SJ, et al. Recovery of a novel Ca-binding peptide from Alaska Pollack (Theragra chalcogramma) backbone by pepsinolytic hydrolysis. Process Biochem. 2006;41:2097–2100. doi: 10.1016/j.procbio.2006.05.008. [DOI] [Google Scholar]
- Kumar S. Free radicals and antioxidants: human and food system. Adv Appl Sci Res. 2011;2:129–135. [Google Scholar]
- Lehtinen P, Kiiliainen K, Lehtomaki K, Laakso S. Effect of heat treatment on lipid stability in processed oats. J Cereal Sci. 2003;37:215–221. doi: 10.1006/jcrs.2002.0496. [DOI] [Google Scholar]
- Liu F-R, Wang L, Wang R, Chen Z-X. Calcium-binding capacity of wheat germ protein hydrolysate and characterization of peptide-calcium complex. J Agric Food Chem. 2013;61:7537–7544. doi: 10.1021/jf401868z. [DOI] [PubMed] [Google Scholar]
- Markwell MAK, Haas SM, Bieber LL, Tolbert NE. Modification of Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- McDermott A. Bioactive peptides. New York: Springer Science & Business Media; 2009. [Google Scholar]
- Meisel H. Multifunctional peptides encrypted in milk proteins. BioFactors. 2004;21:55–61. doi: 10.1002/biof.552210111. [DOI] [PubMed] [Google Scholar]
- Pinchuk I, Shoval H, Dotan Y, Lichtenberg D. Evaluation of antioxidants: scope, limitations and relevance of assays. Chem Phys Lipids. 2012;165:638–647. doi: 10.1016/j.chemphyslip.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Prior R, Wu X, Kschaich K. Standardized methods for the determination of antioxidant capacity and phenolics in foods and dietary supplements. J Agric Food Chem. 2005;53:4302. doi: 10.1021/jf0502698. [DOI] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho SJ, Park S, Ahn C-W, et al. Dietetic and hypocholesterolaemic action of black soy peptide in dietary obese rats. J Sci Food Agric. 2007;87:908–913. doi: 10.1002/jsfa.2808. [DOI] [Google Scholar]
- Schneider CD, Reischak de Oliveira A. Oxygen free radicals and exercise: mechanisms of synthesis and adaptation to the physical training. Rev Bras Med Esporte. 2009;10:314–318. [Google Scholar]
- Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- Shahidi F, Zhong Y. Lipid oxidation and improving the oxidative stability. Chem Soc Rev. 2010;39:4067–4079. doi: 10.1039/b922183m. [DOI] [PubMed] [Google Scholar]
- Vavrusova M, Skibsted LH. Calcium binding to dipeptides of aspartate and glutamate in comparison with orthophosphoserine. J Agric Food Chem. 2013;61:5380–5384. doi: 10.1021/jf400741e. [DOI] [PubMed] [Google Scholar]
- Xie Z, Huang J, Xu X, et al. Antioxidant activity of peptides isolated from alfalfa leaf protein hydrolysate. Food Chem. 2008;111:370–376. doi: 10.1016/j.foodchem.2008.03.078. [DOI] [PubMed] [Google Scholar]
- Zhu K, Zhou H, Qian H. Antioxidant and free radical-scavenging activities of wheat germ protein hydrolysates (WGPH) prepared with alcalase. Process Biochem. 2006;41:1296–1302. doi: 10.1016/j.procbio.2005.12.029. [DOI] [Google Scholar]
- Zhu L, Chen J, Tang X, et al. Reducing, radical scavenging, and chelation properties of in vitro digests of alcalase-treated zein hydrolysate. J Agric Food Chem. 2008;56:2714–2721. doi: 10.1021/jf703697e. [DOI] [PubMed] [Google Scholar]