Abstract
The existence of reservoirs of cells latently infected with human immunodeficiency virus (HIV) is a major obstacle to the elimination of HIV infection. We studied the changes in cellular gene expression that accompany the reactivation and completion of the lytic viral cycle in cell lines chronically infected with HIV-1. We found that several genes exhibited altered expression in the chronically infected cells compared to the uninfected parental cells prior to induction into lytic replication. A number of gene classes showed increased expression in the chronically infected cells, notably including genes encoding proteasomes, histone deacetylases, and many transcription factors. Following induction of the lytic replication cycle, we observed ordered, time-dependent changes in the cellular gene expression pattern. Approximately 1,740 genes, many of which fall into 385 known pathways, were differentially expressed (P < 0.001), indicating that completion of the HIV replication cycle is associated with distinct, temporally ordered changes in host cell gene expression. Maximum changes were observed in the early and intermediate phases of the lytic replication cycle. Since the changes in gene expression in chronically infected cells suggested that cells latently infected with HIV have a different gene expression profile than corresponding uninfected cells, we studied the expression profiles of three different chronically infected cell lines to determine whether they showed similar changes in common cellular genes and pathways. Thirty-two genes showed significant differential expression in all cell lines studied compared to their uninfected parental cell lines. Notable among them were cdc42 and lyn, which were downregulated and are required for HIV Nef binding and viral replication. Other genes previously unrelated to HIV latency or pathogenesis were also differentially expressed. To determine the effects of targeting products of the genes that were differentially expressed in latently infected cells, we treated the latently infected cells with a proteasome inhibitor, clastolactacystin-beta-lactone (CLBL), and an Egr1 activator, resveratrol. We found that treatment with CLBL and resveratrol stimulated lytic viral replication, suggesting that treatment of cells with agents that target cellular genes differentially expressed in latently infected cells can stimulate lytic replication. These findings may offer new insights into the interaction of the latently infected host cell and HIV and suggest therapeutic approaches for inhibiting HIV infection and for manipulating cells latently infected with HIV so as to trigger lytic replication.
Human immunodeficiency virus (HIV) infection has dramatic effects on host cell physiology. Infection causes the host cell to produce large quantities of viral RNAs and proteins, alters the progression of the host cell cycle (39, 50), leads to interactions between viral and host cell proteins (22), and can result in profound alterations in host cell morphology, such as syncytium formation (41). Perhaps the most consistent, synchronous changes are observed during in vitro experiments when chronically infected cells are exposed to agents that stimulate the completion of the lytic replication cycle (26). It might be expected that such profound functional and morphological changes in the host cell would be associated with significant changes in the patterns of host cell gene expression.
Several studies have described changes in certain selected cellular genes due to the expression of HIV proteins (20, 60, 61). Other studies have described changes in cellular gene expression due to acute infection following the addition of infectious virus to cells (16, 29, 67), and additional studies have described changes in host cell gene expression that accompany infections by other viruses, offering interesting insights into viral pathogenesis (12, 33, 37, 51, 63, 72). While these studies provide insights into the extensive effects of specific viral proteins or acute HIV infection (reviewed in reference 4), the global gene expression changes that may accompany reactivation of cells latently infected with HIV and completion of the lytic phase of viral replication have not been described.
Current antiretroviral agents can control HIV replication but cannot eliminate latently infected cells. Models incorporating higher-order decay kinetics (30, 31, 53) suggest that therapy must last some 60 years to eradicate the virus (23, 49). The predictions of these models may reflect the slow loss of virus from “reservoirs.” Latent HIV infection can exist in many such reservoirs, such as macrophages and resting memory CD4+ T cells (14, 46), which appear to “archive” HIV species (55).
Several lines of evidence show that treating host cells carrying HIV provirus with different agents can end latency (24, 26, 40, 47, 66, 70). For example, HIV-infected cells can be activated in vitro by inducing agents such as phorbol myristyl acetate (PMA), which can induce lytic replication in chronically infected cell lines. However, many agents that affect latent HIV infection are toxic or may not eliminate latent reservoirs in patients (13). Since viral reactivation is necessary for targeting by antiviral drugs (9), identification of new approaches that eject HIV from latency when used together with effective antiviral therapy might lead to the reduction of latent HIV reservoirs in an infected patient.
We studied the pattern of host cell gene expression when cells latently infected with HIV were induced to undergo the rest of the viral replication cycle. We found that cellular gene expression patterns are altered in latently infected cells even before they are induced to undergo lytic infection. Following the emergence of HIV from the latent state and during the subsequent completion of the lytic infection cycle, the host cell exhibited carefully ordered changes in the expression of a subset of its genes, which shadowed the well-known, ordered changes in the pattern of viral gene expression characteristic of the HIV replication cycle. Reasoning that the altered expression of some of the host cell genes prior to induction of lytic replication might be related to the establishment and maintenance of latency in these cells, we treated the chronically infected cells with agents targeting the gene products of certain differentially expressed genes and found that some of these agents could activate lytic HIV replication. The differentially expressed cellular genes may thus offer novel cellular targets for agents that activate latent virus into a lytic replication cycle and may provide potential new therapeutic approaches to eliminate latent viral reservoirs.
MATERIALS AND METHODS
Cells.
ACH-2, A3.01, J1.1, and U1 cells (11, 15, 24, 25, 48) were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS. U-937 cells were obtained from the American Type Culture Collection (Manassas, Va.). ACH-2, J1.1, and U1 are chronically infected cell lines harboring the HIV-1 LAV strain, while A3.01, Jurkat, and U-937 are the corresponding parental uninfected cell lines. Cells were grown in RPMI 1640 (Invitrogen, San Diego, Calif.) with 10% fetal bovine serum (Invitrogen), 5% penicillin-streptomycin (Invitrogen), and 2 mM glutamine (Invitrogen). Cells were maintained at a concentration of 106 cells/ml in T-175 flasks. Cell concentrations and cell viability were monitored throughout the experiment at all time points studied.
Cells were induced by addition of 20 ng of phorbol myristyl acetate (PMA; Sigma, St. Louis, Mo.) per ml for 1 h, after which the cells were washed with phosphate-buffered saline. Cells were harvested by centrifugation at 1,000 rpm for 10 min at 0.5, 3, 6, 8, 12, 18, 24, 48, 72, and 96 h after induction. HIV-infected and uninfected cells maintained and harvested in parallel with the PMA-treated cells but not induced with PMA were also harvested. For the time course experiment, 3′-azido-3′-deoxythymidine (AZT; Sigma) was not added to the ACH-2 or A3.01 cells in order to keep conditions as close to those of an acute infection as allowed by the experimental model. Harvested cells were washed thrice with ice-cold phosphate-buffered saline to remove the medium; cell pellets were snap frozen with an ethanol-dry ice mixture and stored at −80°C for subsequent RNA extraction. Three independent time course experiments (biological replicates) were performed.
In order to compare the expression profiles of chronically infected cell lines, ACH-2, U1, and J1.1 and their uninfected parental cell lines, A3.01, U-937, and Jurkat cells, respectively, were grown under identical conditions but in the presence of AZT (250 nM) in growth medium, and the cells were harvested as described above. In these studies, no inducing agent was used in either chronically infected or uninfected parental cell lines so as to study changes in cellular gene expression cells latently infected with HIV and uninfected cells.
Flow cytometry.
To confirm viral replication following PMA induction, we measured the accumulation of intracellular p24 over a period of 48 h by measuring cell populations labeled with anti-p24 fluorescein isothiocyanate-labeled antibody by flow cytometry. Cells (ACH-2 and A3.01) were washed twice with ice-cold PBS and suspended in 50 μl of ice-cold permeabilization buffer (BD Biosciences, San Jose, Calif.), and incubated at 4°C in the dark for 30 min. The cells were fixed with the CytoFix/CytoPerm kit (BD Biosciences), and 5 μl of fluorescein isothiocyanate-labeled p24 antibody KC57 (Beckman Coulter) was added to detect intracellular p24. A3.01 samples labeled with fluorescein isothiocyanate-labeled p24 antibody served as controls to ensure that the parental cell line did not show any p24 accumulation over that in samples labeled with fluorescein isothiocyanate-labeled mouse immunoglobulin G1 (IgG1; Immunotech, Hialeah, Fla.), which was used as the isotype control. Following incubation on ice for 30 min in the dark, the cells were washed thrice with permeabilization buffer, resuspended in 300 μl of permeabilization buffer, and analyzed with a Becton Dickinson FACSCAN instrument (BD Biosciences) in conjunction with CellQuest software (BD Biosciences) for flow cytometric analysis.
Total RNA extraction.
Total RNA was extracted with RNEasy Midiprep kits per the manufacturer's protocol (Qiagen, Valencia, Calif.). RNA concentrations and purity were measured by spectrophotometry, and RNA quality (absence of RNA degradation) was assessed by gel electrophoresis. The RNA concentration was adjusted to the levels required for subsequent microarray experiment protocols by concentration in a SpeedVac (Savant Instruments, Holbrook, Calif.). RNA samples (6 to 7 μg/μl) were stored in 100 μl of Tris-EDTA buffer at −80°C.
Real-time RT-PCR quantitation of viral RNA and cellular RNA.
Quantitation of HIV viral mRNA was carried out by real-time PCR with an ABI Prism 7000 instrument (Applied Biosystems, Foster City, Calif.). A housekeeping gene, glyceraldehyde phosphate dehydrogenase (GAPDH), whose expression was unchanged in ACH-2 cells before and after PMA treatment was used as a normalization control. RNA from the samples was subjected to DNase treatment to remove contaminating DNA, and the DNase was inactivated with the DNase Free kit (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's protocols; 2 μg of RNA was reverse transcribed with the Taqman reverse transcription (RT) kit from Applied Biosystems per the manufacturer's specifications. Briefly, the reaction mixture (50 μl) was incubated at 65°C for 5 min, followed by 37°C for 45 min and 94°C for 5 min, and then cooled on ice, and 1/40th aliquots of the corresponding samples were used in a real-time PCR with Taqman probes labeled with reporter dye label FAM and quencher dye label TAMRA at the 5′ and 3′ ends, respectively.
Primer probe pairs were designed with PrimerExpress (Applied Biosystems). The reactions were carried out in triplicate for each time point, and the changes observed were normalized to expression of GAPDH for each time point. Melting-curve analysis and an assessment of the efficiency of the PCRs over a given concentration range were performed to determine if any errors occurred during the reactions, for example, primer dimer formation or poor priming effects. Real-time PCRs were set up as described above with primers and probes specific for early (multiply spliced mRNA) and late (unspliced mRNA). The sequences of the 5′ and 3′ primers used to quantitate early mRNA were 5′-CGAAGAGCTCATCAGAACAGTCA-3′ and 5′-TTGGGAGGTGGGTCTGCTT-3′. The sequence of the labeled probe was 5′-CTTCTCTATCAAAGCAGACCCACCTCC-3′, and it overlapped the splice site of the HIV-1 Rev sequence. The sequence detection primers for unspliced, late RNA were SK38 and SK39 from the HIV-1 Gene Amplimer kit (Applied Biosystems). A TAMRA-labeled probe identical in sequence to SK19 (Applied Biosystems) was used for real-time PCR quantitation of the late viral RNA species. Standards from the kit were diluted to calculate the copy number of virus based on gag mRNA concentrations.
Real-time RT-PCR analysis was also carried out for selected cellular genes with gene-specific primer-probe pairs and Taqman detection primers. Differences in mRNA expression in uninduced ACH-2 samples and the corresponding A3.01 samples were determined with the protocol described for quantitation of viral mRNA. Real-time RT-PCR quantitation was performed for genes PSMC5, p44s10 (proteasome subunits) Egr1 (early growth response 1), HDAC1 (histone deacetylase 1), NK4, EIF4, and SFRS3 to confirm that these genes were differentially expressed in the latently infected ACH-2 cells compared to the uninfected A3.01 parental cells. Primer-probe pairs specific for each gene were designed with PrimerExpress (Applied Biosystems). The sequences of the detection primers and probes for each gene are available as supplemental material (Table S1).
Microarray studies.
Total RNAs obtained from induced chronically infected and the corresponding uninfected parental cells were used for microarray experiments. For each time point, RNA from the induced chronically infected ACH-2 cells and RNA from the corresponding induced, uninfected A3.01 cells were compared to minimize effects due to PMA induction. Microarrays were obtained from the National Cancer Institute Microarray Facility, Advanced Technology Center (Gaithersburg, Md.). The microarrays (Hs. UniGem2) contained 10,395 cDNA spots on each glass slide. The cDNAs were selected for spotting on the slides based on their known or probable involvement in oncogenesis, signal transduction, apoptosis, immune function, inflammatory pathways, cellular transport, transcription, protein translation, and other important cellular functions. A number of expressed sequence tags (ESTs) from unknown genes homologous to known genes and cDNAs encoding housekeeping genes were also included in these gene sets.
For each time point, 50 μg of total RNA from PMA-induced ACH-2 cells and 70 μg of total RNA from PMA-induced A3.01 cells was labeled with indocarbocyanine (Cy3)-dUTP and indodicarbocyanine (Cy5)-dUTP, respectively, as previously described (34, 60). Larger amounts of RNA were used for Cy5 labeling to minimize the disparities in dye incorporation. Each sample of RNA from PMA-induced, infected cells from a particular time point was compared with RNA from the corresponding PMA-induced, uninfected cells from the same time point for subsequent hybridization to the same array to ensure accurate comparisons and to eliminate interarray variability. The labeled cDNAs were then combined and purified with MicroCon YM-30 (Millipore, Bedford, Mass.) spin column filters to remove any unincorporated nucleotides; 8 to 10 μg each of Cot-1 DNA (Boehringer Mannheim, Indianapolis, Ind.), Saccharomyces cerevisiae tRNA (Sigma), and poly(A) (Amersham Biosciences) were added to the reaction mixture and heated at 100°C for 1 min. Hybridization of the labeled cDNA to the microarray was carried out at 65°C overnight, followed by washes with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2× SSC, and 0.05× SSC. The slides were dried by centrifugation at 1,000 rpm for 3 min and then scanned as described below. RNA samples from three identical but independently conducted time course experiments were tested. Microarray experiments were performed at least twice for each time point (technical replicates) of each experiment. We also compared AZT-treated ACH-2 cells to untreated ACH-2 cells to determine whether any differences in gene expression might be due solely to AZT.
Microarray experiments for each chronically infected cell line (J1.1, U1, and ACH-2) were also conducted as per the protocol described above. Samples from eight independent experiments per cell line were used for studying the gene expression patterns in chronically infected cell lines. To compensate for dye labeling bias that might be due to differences in Cy5 and Cy3 labeling efficiency and preferential dye incorporation by some mRNA species, RNA from the same samples labeled with Cy5 (70 μg of RNA) and Cy3 (50 μg of RNA) were cohybridized to the same array and scanned, data were analyzed for all the cell lines studied with identical filtering and statistical tests, and genes showing dye incorporation bias were eliminated from further analysis as described below.
Microarray scanning and data analysis.
The slides were scanned with an Axon GenePix 4000 scanner (Axon Instruments, Union City, Calif.). The photomultiplier tube values were adjusted to obtain equivalent intensities at both wavelengths used, 635 and 532 nm for the indodicarbocyanine and indocarbocyanine channels, respectively. Image analysis was performed with GenePix analysis software (Axon Instruments), and data analysis was performed with the MicroArray Database (mAdb) system hosted by the Center for Information Technology and Center for Cancer Research at NIH (http://nciarray.nci.nih.gov). Each array was normalized with Lowess normalization (71). Normalization across arrays over the time course was not feasible because no particular time point could be established as a median for cellular gene expression data, since viral gene expression and associated host cell gene expression were different for different time points.
For each time point, there were at least six data sets (two technical replicates times three biological replicates). For each time period (0.5 to 8 h, 12 to 24 h, and 48 to 96 h postinduction), there were at least 18 datasets (at least three time points per time period). Only arrays that met initial spot size and intensity criteria and whose normalization ratio (between the two signal intensities) was close to 1 (0.85 to 1.15) were analyzed. Filtering criteria were as follows. For each spot, signal intensity must be at least twice that of the background intensity; each gene must have values in at least 70% of the arrays; and each array must have values for at least 70% of the gene spots. Genes that showed dye labeling bias in a particular cell line after normalization were excluded from that gene set prior to further analysis. This was determined with a one-sample t test on mean log ratios for replicate arrays with the same sample labeled with both Cy3 and Cy5.
The raw data from these experiments have been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE1441 and GSE1443.
Statistical analysis.
Comparison of the expression profiles of infected versus uninfected (both induced by PMA) cell lines at a given time point was performed with univariate parametric and multivariate permutation tests based on the one-sample random variance t statistic in BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools) (62). Since RNAs from infected and uninfected cell lines corresponding to the same time point were paired and cohybridized on the same array, interarray sources of variation were minimized and differential expression could be detected by a statistically significant nonzero mean log ratio in biologically independent replicates. All biological replicates that passed the filtering criteria described above were used in the analyses. Technical replicates were averaged. The random-variance model enabled variance information to be shared across genes without assuming that all genes had the same variance (69). For comparison of expression of latently infected versus uninfected cell lines, significance was based on P < 0.001 for a parametric one-sample random variance t test. For evaluation of differential expression between infected and uninfected cell lines at fixed times after induction, a multivariate permutation test based on the one-sample random variance t statistic was used in which the proportion of false discoveries was limited to 0.10 with 90% confidence (36, 62).
Hierarchical clustering analyses of the resulting data sets were done with the mAdb system as well as the Cluster and TreeView software programs (Stanford University, Stanford, Calif.). Since a gene exhibiting statistically significant differences in expression need not alter the physiology of a cell in a biologically meaningful way, an additional pathway analysis of the genes that showed significant differential expression was performed with analysis tools provided by the NIH mAdb database (http://nciarray.nci.nih.gov) and querying the database of the Cancer Genome Anatomy Project (CGAP) (http://cgap.nci.nih.gov/) with pathway information provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.genome.ad.jp/kegg/) and Biocarta (www.biocarta.com) pathway databases. Gene ontology summary analyses, which allow grouping of genes based on their molecular function, were also performed to ensure that the changes observed in our studies were not due to general dysregulation of all genes studied. Where possible, the observed/expected ratio (O/E) for each functional class of genes was determined to ensure that observed changes in the number of genes differentially expressed within a gene class were greater than that expected by chance (O/E > 1). This provided stringent thresholds for pathway and functional classification of the differentially expressed genes.
Latency reactivation studies.
Cells (ACH-2, J1.1, and U1) were seeded at a concentration of 2 × 105 cells/ml in 24-well plates in a volume of 1 ml. Clastolactacystin-beta-lactone, a proteasome inhibitor; resveratrol, an Egr1 activator; or trichostatin, a histone deacetylase inhibitor (Biomol Research Laboratories, Plymouth Meeting, Pa.) was dissolved in sterile dimethyl sulfoxide and further diluted with medium to obtain the desired final concentrations. The final concentration of dimethyl sulfoxide in contact with the cells was never greater than 0.001% at any dose tested. AZT (250 nM) was added to the chronically infected cells in order to inhibit p24 production that may be caused by low levels of actively replicating virus present along with the chronically infected cells and to ensure that any increases in p24 expression would be attributable to activation of latent provirus and not due to subsequent amplification via additional rounds of viral replication. Cells were incubated with different concentrations of either clastolactacystin-beta-lactone, resveratrol, or trichostatin at 37°C, and 200-μl samples of cell supernatant were collected at 24 h after treatment. Cells incubated with tumor necrosis factor alpha (0.5 μg/ml) in the presence of AZT served as a positive control. Cells treated with AZT alone served as a negative control. Cells not treated with AZT were also examined.
Samples were mixed with lysing buffer (10% Triton X-100; Sigma) to inactivate virus and diluted fivefold with sample diluent (1% bovine serum albumin, 0.2% Tween 20 in RPMI 1640). p24 expression was assayed by enzyme-linked immunosorbent assay with HIV-1 p24 antigen capture kits (AIDS Vaccine Program, Frederick, Md.) per the manufacturer's specifications. Briefly, plates were washed with plate wash buffer, and samples were added in duplicate wells. The samples (100 μl) were incubated for 2 h at 37°C. The plates were washed, and rabbit anti-HIV p24 antibody was added at a 1:400 dilution. Following incubation for 1 h, the plates were washed, and goat anti-rabbit IgG peroxidase-labeled antibody at a 1:300 dilution was added. The plates were incubated for 1 h at 37°C, followed by washing and addition of a two-component substrate. The substrate solution consisted of equal volumes of tetramethylbenzedine peroxidase substrate and peroxidase solution B (Kirkegaard and Perry Laboratories, Gaithersburg, Md.). Samples were incubated for 30 min at room temperature, and reactions were stopped by addition of 1 N hydrochloric acid solution. The absorbance was measured at 450 nm with a SpectraMax 250 spectrophotometer (Molecular Devices Corporation, Sunnyvale, Calif.). The samples were assayed in duplicate, and experiments were performed at least thrice with independent cell samples.
RESULTS
We studied cellular gene expression patterns in ACH-2 cells, a cell line chronically infected with HIV, before and during activation into a lytic viral replication cycle. We also studied the global expression patterns in two other chronically infected cell lines J1.1, a T-lymphocytic cell line, and U1, a promonocytic cell line, prior to induction.
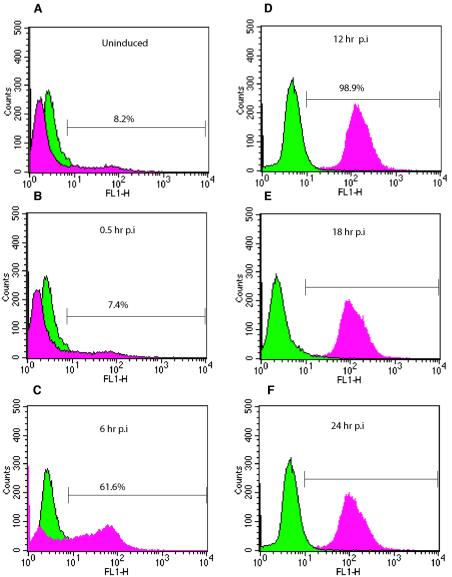
We treated the ACH-2 chronically infected cell line with PMA to trigger the initiation and completion of the lytic replication cycle. Virus production was confirmed by flow cytometry for the late HIV protein p24 (Fig. 1). p24 production was low in the absence of PMA (8.2%) in ACH-2 cells not treated with AZT. A3.01, the parental uninfected cell line, did not show any p24-specific staining over that of the isotype control (data not shown). At 6 h postinduction, 62% of the ACH-2 cells showed p24 production, and from 12 h up to 96 h postinduction, nearly all cells were positive for p24 production, indicating that a lytic HIV infection was under way in essentially all the cells in the culture (Fig. 1). By 48 h postinduction, flow cytometry analysis showed high levels of p24 production but with increased cell death due to cytopathic effects (data not shown).
FIG. 1.
Flow cytometric analysis of chronically infected ACH-2 cells before and after induction. Uninduced cells and cells from serial time points were fixed and permeabilized for intracellular p24 labeling. As an isotype control, cell samples were labeled with mouse IgG1. For each sample, 100,000 events were collected. Each sample histogram labeled for p24 (red) is overlaid with the control histogram labeled for the isotype control (green). (A) Uninduced ACH-2 cells, showing minimal p24 accumulation with 8.2% of cells infected. (B) ACH-2 cells at 0.5 h postinduction (p.i.) with 7.4% of cells positive for p24. (C) ACH-2 cells at 6 h postinduction, with 61.6% of cells infected. (D, E, and F) ACH-2 cells at 12, 18, and 24 h postinduction, respectively, showing complete infection. Flow cytometric analysis was performed on all batches of cells to ensure active replication of HIV following induction with PMA. Data from one induction experiment are shown. The data indicate that viral replication occurs in an ordered manner postinduction and that complete infection of cells is achieved within 12 h postinduction of chronically infected ACH-2 cells.
Cell viability before and after induction of lytic replication was carefully monitored to ensure that changes in gene expression could be associated with the process of lytic replication and not with excessive cell death due to HIV replication. From the start of PMA induction to up to 24 h postinduction, the cell viability of induced ACH-2 cells remained at levels similar to that of uninfected, induced parental A3.01cells and also to that of uninduced ACH-2 cells, indicating that cell death due to HIV lytic replication was not a significant factor in changes in gene expression for that period. Beyond 24 h postinduction (48 to 96 h postinduction), cytopathic effects and increased cell death were observed in the induced ACH-2 cells (also confirmed by flow cytometry), indicating that changes in cellular gene expression for 48 to 96 h postinduction may be attributed to a combination of viral replication and cellular mechanisms involved in cell death. RNA yields decreased for this time period. However, RNA quality and purity were similar to the values at the previous time points, suggesting that changes in cellular gene expression were not due to RNA degradation in these cells. Since nearly all cells undergoing lytic replication were positive for p24 antigen by 24 h postinduction, when cellular viability was unaffected, the changes in cellular gene expression from 0.5 h to 24 h postinduction are most likely associated with the process of lytic replication in ACH-2 cells.
Viral mRNA expression by real-time RT-PCR.
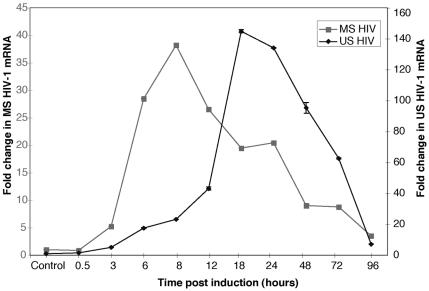
In order to confirm that production of early and late viral mRNAs was under way in lytically induced cells and to determine the relative proportions of early (multiply spliced; tat, rev, and nef) and late (unspliced; gag, pol, and env) HIV-1 mRNAs over the time course, we used real-time RT-PCR to quantitate the message classes. In our system, multiply spliced mRNA expression increased after lytic induction and showed a maximum 40-fold increase over uninduced ACH-2 cells at 8 h postinduction. Unspliced mRNA concentrations showed a gradual increase, with a 148-fold increase at 18 h postinduction (Fig. 2). The results indicate that viral RNA expression in our lytically induced cells followed known kinetic expression patterns (6, 35). There was a clear distinction in the peak expression of the early and late viral mRNAs, indicating that viral RNA expression followed a discrete temporal pattern in these cells.
FIG. 2.
Levels of expression of multiply spliced (MS) and unspliced (US) HIV-1 mRNA pre- and postinduction of chronically infected ACH-2 cells. Real-time RT-PCRs were carried out with Taqman probes specific for early (multiply spliced) and late (unspliced) transcripts of HIV-1 and tagged with FAM and TAMRA fluorescent dyes at the 5′ and 3′ ends, respectively. Reactions were performed in triplicate for each time point as described in Materials and Methods, and average values are shown. Maximal changes in mRNA levels for early transcripts (multiply spliced) were observed at 8 h postinduction. The changes for late transcripts (unspliced) showed a maximal increase at 18 h postinduction.
Effects on ACH-2 cellular gene expression before and after induction.
For the cellular gene expression studies, we compared the cellular gene expression pattern of ACH-2 with that of its uninfected parental line, A3.01. Both the infected and uninfected parental lines were subjected to identical PMA treatments and sampling procedures over a 96-h time course, and RNA samples from three separate time course experiments were each tested in duplicate. In all, 66 arrays were examined. The filtering criteria yielded 9,122 analyzable gene spots out of the 10,395 gene spots printed on each microarray. The expression of most cellular genes was similar in the uninfected and chronically infected cells and did not change during activation into a lytic infection cycle. We also performed experiments to detect any gene spots that were false-positives due to bias in dye labeling under our experimental conditions. A small set of genes that showed dye bias (43 genes) in our dye-labeling bias experiments were excluded from the data set.
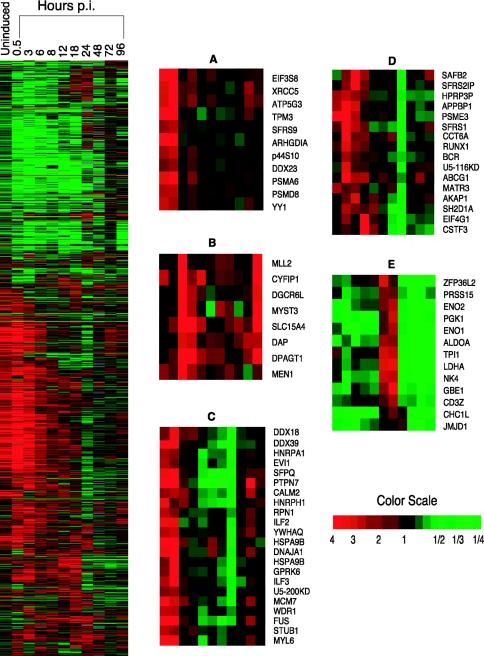
Data from the technical and biological replicates for each time point were normalized for each array as described in Materials and Methods. Statistical analyses to identify genes that were significantly differentially expressed were performed with univariate and multivariate random-variance one-sample t tests with all the replicates that passed the filtering criteria. Based on the statistical analyses of the genes showing altered expression, 131 genes showed altered expression even prior to induction; 1,740 of all the analyzable spots showed statistically significant (P < 0.001) altered gene expression at some point, either before induction or over the entire period of the lytic replication cycle (Fig. 3).
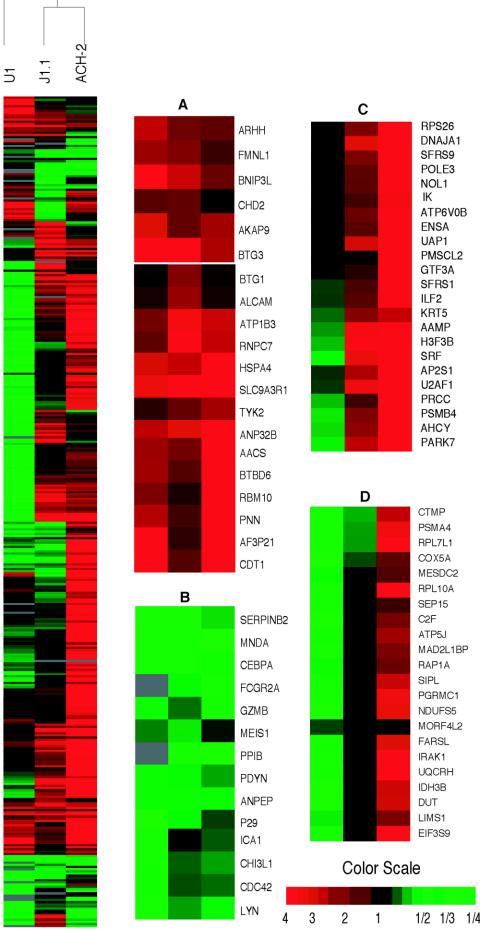
FIG. 3.
Hierarchical clustering of differentially expressed cellular genes before and after induction of chronically infected ACH-2 cells. The figure shows the hierarchical clustering of the cellular genes that showed significant differential expression (P < 0.001) across the time course (before induction up to 96 h postinduction) following reactivation of chronically infected ACH-2 cells as described in the text. Genes shown in red showed upregulation, those in green were downregulated, while those that did not show any change with respect to a normalized matched control are shown in black. The gray areas indicate missing data for the given gene and time point. The magnified panels indicate selected kinetic profiles that were seen before and following induction into active viral replication. (A) Upregulation of selected genes observed before induction; (B) upregulation of genes immediately following induction; (C) genes that are upregulated prior to induction and downregulated 12 to 24 h postinduction; (D) genes that are upregulated in the early stage following reactivation but downregulated in the intermediate stage; (E) genes that are downregulated before induction but upregulated in the intermediate stage followed by downregulation in the late stage (48 to 96 h postinduction).
Cellular gene expression profiles before induction and during lytic replication.
The changes observed during lytic replication following induction occurred in an orderly, time-dependent manner. To better appreciate the time-dependent changes in cellular gene expression that accompanied the portion of the lytic replication cycle occurring after activation of the chronically infected cells with PMA, we grouped our observations into three time periods after induction: early (0.5 to 8 h postinduction), intermediate (12 to 24 h postinduction), and late (48 to 96 h postinduction), roughly corresponding to the times during lytic replication when the early and late viral mRNAs peak and the end of the lytic cycle, respectively. Statistical analyses and hierarchical clustering of the data also grouped the various cellular genes into these time periods (data not shown).
The changes in gene expression observed during the late period (48 to 96 h postinduction) cannot be solely associated with the process of lytic replication because during this time period the cells showed cytopathic effects. However, the data are presented in order to provide the most complete possible view of the cellular environment following lytic replication. Some genes showed differential expression only during the early stages of lytic replication and returned to the levels observed in the uninfected cells, while certain other genes were initially unaltered and showed differential expression at later time points.
Certain groups of genes showed similar patterns of regulation following induction (Fig. 3). In the early time period, 1,334 genes were differentially expressed. In the intermediate time period, 756 genes were differentially expressed. The late time period (48 to 96 h postinduction) showed the least change, with 566 genes exhibiting significantly altered expression (P < 0.001). Many of the genes that were differentially expressed in the early time period also showed either a similar or the opposite trend in their expression patterns during the other time periods; hence, some genes were included in the analysis of both time periods.
A number of discrete patterns of gene regulation were observed. Several cellular genes showed distinct temporal expression patterns during the lytic replication cycle, an expected finding, but more interestingly, a smaller number of genes appeared to be differentially expressed in the latently infected ACH-2 cells compared to their parental uninfected cells even before induction of the lytic cycle. A total of 131 genes showed a significant change (P < 0.001) in their expression prior to induction. They included genes encoding transcription factors, components of proteasomes, factors that control immune function, apoptosis, and other functional classes. For gene classes that were annotated in the gene ontology database (www.geneontology.org) (5), the O/E ratio for the number of genes within a functional class that were differentially expressed was set at greater than 1 (O/E > 1) to eliminate functional classes where the number of differentially expressed genes was not greater than that randomly expected. However, not all genes that were significantly differentially expressed are annotated in the database. Extensive literature studies for functional significance and classification were conducted in such cases to ensure that important classes of genes were still included in the analyses. An abbreviated listing of the genes grouped according to known functions that were differentially expressed before induction is given in Table 1.
TABLE 1.
Functionally related genes that were differentially expressed prior to induction in chronically infected ACH-2 cellsa
| Functional group | Unigene identification | Gene | Gene description | Change (fold) |
|---|---|---|---|---|
| Proteasome | Hs.350939 | p44S10 | Proteasome regulatory particle subunit p44S10 | 1.98 |
| Hs.251531 | PSMA4 | Proteasome (prosome, macropain) subunit, alpha type, 4 | 1.89 | |
| Hs.76913 | PSMA5 | Proteasome (prosome, macropain) subunit, alpha type, 5 | 1.84 | |
| Hs.374499 | PSMA6 | Proteasome (prosome, macropain) subunit, alpha type, 6 | 2.32 | |
| Hs.9661 | PSMB10 | Proteasome (prosome, macropain) subunit, beta type, 10 | 1.66 | |
| Hs.89545 | PSMB4 | Proteasome (prosome, macropain) subunit, beta type, 4 | 2.05 | |
| Hs.250758 | PSMC3 | Proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 1.74 | |
| Hs.79387 | PSMC5 | Proteasome (prosome, macropain) 26S subunit, ATPase, 5 | 2.75 | |
| Hs.279554 | PSMD13 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | 2.11 | |
| Hs.9736 | PSMD3 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 2.01 | |
| Hs.78466 | PSMD8 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 2.05 | |
| Hs.75348 | PSME1 | Proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | 1.43 | |
| Hs.434081 | PSME2 | Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 1.46 | |
| Hs.152978 | PSME3 | Proteasome (prosome, macropain) activator subunit 3 (PA28 gamma) | 2.60 | |
| Hs.119563 | PSME4 | Proteasome (prosome, macropain) activator subunit 4 | 1.80 | |
| Nuclear transport | Hs.414565 | CLIC1 | Chloride intracellular channel 1 | 2.20 |
| Hs.434408 | IPO7 | Importin 7 | 2.01 | |
| Hs.439683 | KPNB1 | Karyopherin (importin) beta 1 | 3.22 | |
| Hs.172108 | NUP88 | Nucleoporin 88 kDa | 2.02 | |
| Hs.356630 | NUTF2 | Nuclear transport factor 2 | 1.81 | |
| Hs.10842 | RAN | RAN, member RAS oncogene family | 3.09 | |
| Hs.446673 | THOC4 | THO complex 4 | 2.19 | |
| Splicing factors | Hs.181368 | PRPF8 | PRP8 pre-mRNA processing factor 8 homolog (yeast) | 0.36 |
| Hs.77897 | SF3A3 | Splicing factor 3a, subunit 3, 60 kDa | 3.20 | |
| Hs.68714 | SFRS1 | Splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) | 2.32 | |
| Hs.77608 | SFRS9 | Splicing factor, arginine/serine-rich 9 | 1.90 | |
| Hs.180610 | SFPQ | Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | 2.37 | |
| Hs.356549 | SNRPD3 | Small nuclear ribonucleoprotein D3 polypeptide 18 kDa | 2.14 | |
| Transcription and translation | Hs.135643 | C2F | C2f protein | 2.71 |
| Hs.15591 | COPS6 | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | 1.75 | |
| Hs.371001 | EIF3S9 | Eukaryotic translation initiation factor 3, subunit 9 eta; 116 kDa | 3.00 | |
| Hs.129673 | EIF4A1 | Eukaryotic translation initiation factor 4A, isoform 1 | 3.41 | |
| Hs.406408 | EIF4EBP1 | Eukaryotic translation initiation factor 4E binding protein 1 | 1.94 | |
| Hs.445977 | GTF3A | General transcription factor IIIA | 1.88 | |
| Hs.89525 | HDGF | Hepatoma-derived growth factor (high-mobility group protein 1-like) | 2.20 | |
| Hs.75117 | ILF2 | Interleukin enhancer binding factor 2, 45 kDa | 2.40 | |
| Hs.5215 | ITGB4BP | Integrin beta 4 binding protein | 2.47 | |
| Hs.448398 | MAZ | Myc-associated zinc finger protein (purine-binding transcription factor) | 2.64 | |
| Hs.444086 | SRF | Serum response factor (c-fos serum response element-binding transcription factor) | 2.32 | |
| Hs.60679 | TAF9 | TAF9 RNA polymerase II, TATA box binding protein associated factor, 32 kDa | 1.97 | |
| Hs.365116 | U2AF1 | U2(RNU2) small nuclear RNA auxiliary factor 1 | 2.33 | |
| Hs.388927 | YY1 | YY1 transcription factor | 2.18 | |
| Chaperone function | Hs.1708 | CCT3 | Chaperonin containing TCP1, subunit 3 (gamma) | 2.23 |
| Hs.1600 | CCT5 | Chaperonin containing TCP1, subunit 5 (epsilon) | 2.79 | |
| Hs.368149 | CCT7 | Chaperonin containing TCP1, subunit 7 (eta) | 3.01 | |
| Hs.446481 | DNAJC7 | DnaJ (Hsp40) homolog, subfamily C, member 7 | 1.94 | |
| Hs.151903 | GRPEL1 | GrpE-like 1, mitochondrial (Escherichia coli) | 1.66 | |
| Hs.184233 | HSPA9B | Heat shock 70kDa protein 9B (mortalin-2) | 2.97 | |
| Hs.74335 | HSPCB | Heat shock 90kDa protein 1, beta | 3.01 | |
| Hs.79037 | HSPD1 | Heat shock 60kDa protein 1 (chaperonin) | 2.31 | |
| Hs.36927 | HSPH1 | Heat shock 105-kDa/110-kDa protein 1 | 3.07 | |
| Hs.434937 | PPIB | Peptidylprolyl isomerase B (cyclophilin B) | 0.61 | |
| Signal transduction | Hs.25277 | ARAP3 | ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin homology domain- containing protein 3 | 2.01 |
| Hs.3109 | ARHGAP4 | Rho GTPase activating protein 4 | 0.45 | |
| Hs.159161 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 2.73 | |
| Hs.292738 | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 1.72 | |
| Hs.433888 | RAB11B | RAB11B, member RAS oncogene family | 0.57 | |
| Hs.32217 | RAB32 | RAB32, member RAS oncogene family | 0.64 | |
| Hs.24763 | RANBP1 | RAN binding protein 1 | 2.15 |
List of selected classes of genes, based on known function, that are differentially expressed in latently infected ACH-2 cells relative to the uninfected parental cell line A3.01. A number of genes involved in similar cellular functions previously not associated with the presence of proviral HIV were altered coordinately even during the latent nonreplicative stage.
Genes and pathways affected prior to induction.
While a large number of pathways are altered during lytic replication, a smaller number of pathways are also altered prior to induction of a lytic replication cycle. This observation was interesting and is important for understanding the mechanisms involved in latency maintenance; hence we conducted further analyses on this data set. Pathways involved in cell-cell signaling, signal transduction, inhibition of T-cell receptor signaling, protein translation, and cell cycle transition-regulation showed altered expression prior to induction. Most notably, a number of genes encoding different subunits of proteasomes, including those that constitute the 20S/26S core complex (PSMB4, PSMA6, and PSMA5) as well as regulatory subunits like PSMD13 (11S regulatory subunit) were upregulated prior to induction. PSMB4 has peptidase activity, which is inhibited by Tat during viral replication. Tat competes with the 11S regulatory subunit for binding to the 20S core complex due to the presence of a common binding site in Tat and the 11S regulator alpha subunit (32, 59). Proteasomes are also involved in processing certain regions of HIV-1 Nef preferentially, which leads to production of Nef-specific cytotoxic T-lymphocytes (44). Many other classes of genes encoding immune response modulators, integrins, cell cycle modulators (such as Egr1), nuclear import factors, and G-protein-signaling molecules were also differentially expressed. A listing of genes that were differentially expressed prior to induction, based on their functional classification, is given in Table 1. A list of pathways that were affected in the uninduced, chronically infected cells is given in the supplemental material (Table S2).
Real-time RT-PCR analysis was performed on a selected set of cellular genes with gene-specific sequence detection primers and probes to determine if the microarray results correlated with the actual normalized differences in the corresponding ACH-2 and A3.01 samples prior to induction. For selected genes that were found to be statistically and/or biologically significant, differential expression was confirmed by RT-PCR quantitation (supplemental material, Table S3).
Trends seen in pathway profiles during lytic replication.
While a number of genes that had altered expression during lytic replication could be classified based on their functionality, an alternative method was also used to determine if the genes that showed differential expression grouped into known cellular pathways. Genes encoding components of several distinct pathways were regulated in a coordinated fashion during lytic viral replication. Of the 1,740 genes that were differentially expressed over the lytic infection cycle in ACH-2 cells, 697 were assigned to various known pathways with the CGAP pathway databases, used in conjunction with the MicroArray database mAdb data analysis tools (http://nciarray.nci.nih.gov). Based on the information in the database, these genes were found to lie in a total of 385 known pathways. The remainder of the genes could not be classified into any known pathways listed in the CGAP database.
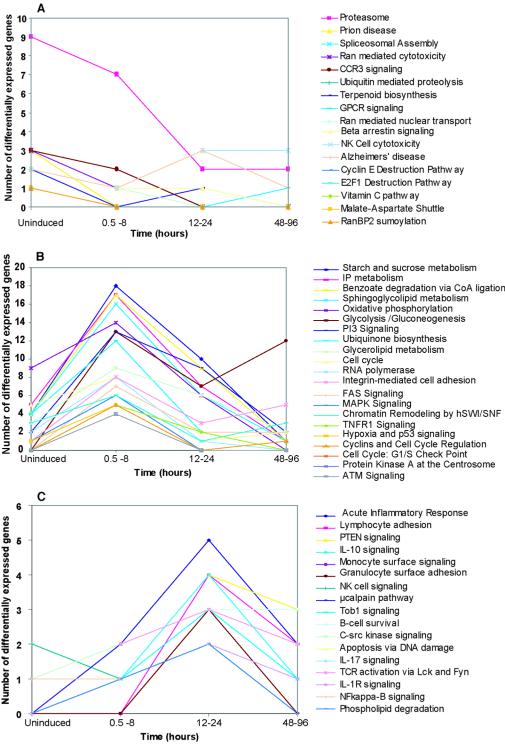
The principal pathways affected over the time course and classified based on the number of genes involved in a particular pathway that are differentially expressed are shown in Fig. 4 (and in the supplemental material, Table S2). We observed that a number of pathways involved in signaling, cell cycle, and transcription showed maximum changes even prior to induction. During the early stage (0.5 to 8 h postinduction), a number of metabolic pathways and signaling pathways showed similar patterns. The intermediate stage (12 to 24 h postinduction) showed maximal changes in pathways involved in immune response modulation and cell survival. These patterns correlated well with the levels of early (multiply spliced) and late (unspliced) HIV protein gene expression and known viral replication effects on the host cell (32, 44, 59).
FIG. 4.
Trends seen in pathways that show differential expression before and after induction of chronically infected ACH-2 cells. Pathway profiles observed prior to induction and following reactivation of ACH-2 cells with PMA over a period of 96 h. The figure shows the number of genes in each pathway that were differentially expressed in a particular pathway. (A) Pathways that were maximally altered prior to induction. (B) Pathways that showed maximum change during the early phase of the lytic cycle (0.5 to 8 h postinduction). (C) Pathways that showed maximal change during from 12 to 24 h postinduction. Most pathways did not show any change during the period from 48 to 96 h postinduction. The groups above are a selected representation of the various pathways that changed differentially prior to induction and/or over the time course studied. Classification of the altered genes into various pathways was performed with the CGAP pathway databases.
Microarray analysis of chronically infected cell lines.
Our initial experiments with the ACH-2 cell line were aimed at studying the changes in cellular host gene expression profiles during a lytic infection following activation. However, our results showed that even prior to induction, many cellular genes were differentially expressed. This led us to study cellular gene expression in other chronically infected cell lines to assess whether they also showed alterations in cellular gene expression in the absence of active viral replication and, if so, whether different cell lines showed similar patterns of altered gene expression. Different chronically infected HIV cell lines, including J1.1, a chronically infected T-lymphocytic cell line derived from Jurkat cells, and U1, a promonocytic chronically infected cell line derived from U937 cells, were studied with microarrays to determine the similarities and differences in their expression profiles. p24 expression in all the latently infected cell lines was below 1 ng/ml (0.2 to 0.8 ng/ml), indicating that the cells were not lytically active at the time of harvest.
Experiments were performed on eight independent cell samples for each cell line, and similar parameters were applied for filter criteria, gene selection, and statistical analysis as with the ACH-2 cell line, as described in Materials and Methods. In these sets of experiments, 24 arrays were analyzed, and 8,902 of the 10,395 gene spots passed the selection criteria. Statistical analysis of expression ratios of AZT-treated ACH-2 cells and untreated ACH-2 cells over their respective A3.01 controls did not show any significant differences in gene expression profiles, indicating that changes in gene expression were not due to low levels of actively replicating viral population (data not shown). Genes that had shown differential dye incorporation in our dye labeling bias experiments were excluded from each cell line data (43 genes in ACH-2, 18 genes in Jurkat, and 22 genes in U937 cells). Upon analyzing the resulting data sets, we found that 131 genes were differentially expressed in ACH-2, 65 genes were differentially expressed in J1.1, and 155 genes were differentially expressed in U1 cells compared to their uninfected AZT-treated parental cell lines. While stringent statistical thresholds were used for genes that were differentially expressed in each cell line, we reasoned that if a gene showed up as significantly differentially expressed (P < 0.001) in at least one cell line, then the expression data for that gene in the other cell lines may be important even if not found statistically significant for that cell line. We found that ACH-2 and J1.1 were more similar in their profiles, which may be due to their common T-cell lineage, compared to U1, which is a promonocytic cell line (Fig. 5).
FIG. 5.
Hierarchical clustering of genes that showed differential expression across three chronically infected cell lines prior to induction. Hierarchical clustering of differentially expressed genes that showed a significant change in expression (P < 0.001) in the chronically infected cell lines ACH-2, U1, and J1.1. Genes shown in red were upregulated, those in green exhibited downregulation, and black indicates normal expression. Gray areas indicate missing values. Many genes were altered similarly across the cell lines. Each cell line also showed some unique patterns of cellular expression. Data are the averages of values from eight independent samples per cell line. The magnified portions of the cluster highlight some of the patterns of gene expression across the cell lines. (A) Genes that were upregulated in all three cell lines; (B) genes that were downregulated in all three cell lines; (C) genes that were upregulated in ACH-2 and J1.1 and downregulated in U1 cells; (D) genes which showed no significant similarity in their expression in the three cell lines.
Gene and pathway profile analysis of chronically infected cell lines.
An analysis of the gene expression profiles showed a limited number of genes that changed similarly across the three cell lines tested (Fig. 5). These genes included cdc42, Lyn, MNDA, CEBPalpha, and Meis1, which were downregulated in all cell lines. Genes that showed upregulation included those encoding BTG1, BTG3, CDT1, and pinin. Cdc42 is critical for activation of Nef-associated kinase (PAK2), while Lyn is required for binding to the PXXP motif of HIV Nef (43, 57). MNDA, CEBPalpha, and Meis1 are all tightly clustered. The proteins encoded by these genes are known to be critical in the pathogenesis of certain leukemias (18, 54, 65) but have not been hitherto related to HIV latency. Certain genes showed similar differential expression in ACH-2 and J1.1 but not in U1 cells. Also, some genes showed opposite trends in the cell lines tested. For example, proteasome subunits were upregulated in ACH-2 but downregulated in U1 cells. A list of common pathways affected and some pathways that change selectively is given in the supplemental material (Table S4). The list of differentially expressed genes common to all three cell lines is given along with their expression ratios (supplemental material, Table S5).
Reactivation of chronically infected cells by targeting specific gene classes.
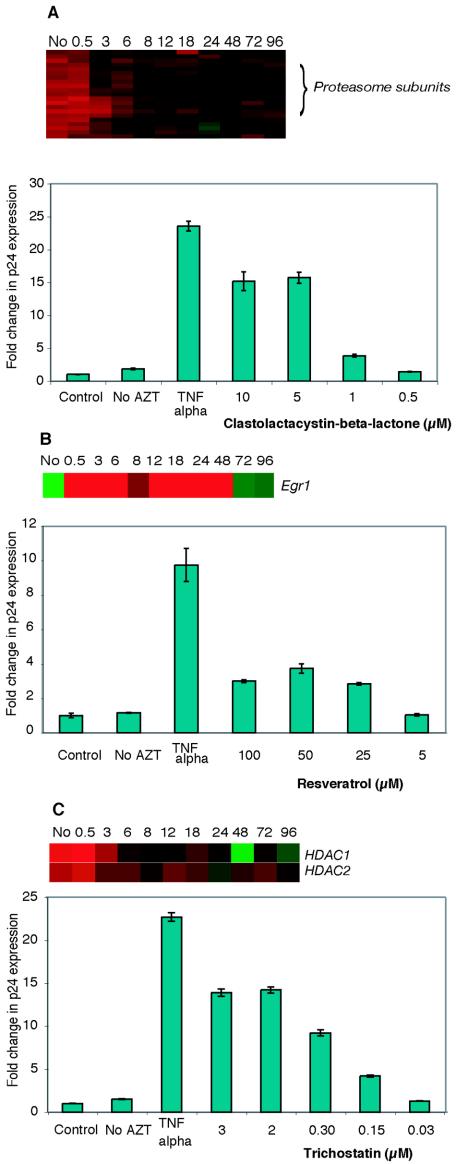
We reasoned that the differential expression of certain cellular genes in latently infected cells might be involved in maintaining the virus in latency and so hypothesized that treating latently infected cells with agents targeting the products of differentially expressed genes may force the virus out of latency. This may be important because reactivation of proviral HIV into lytic infection by targeting cellular factors, notably including cellular factors differentially expressed in latent, chronically infected cell lines, may provide new approaches to reduce or eliminate latent viral reservoirs. To test our hypothesis, we focused on the proteasome class of genes and Egr1 due to the availability of specific agents. We also found other differentially expressed genes such as the histone deacetylases, HDAC1 and HDAC2, which may represent good targets for reactivation of viral replication. Treatment of latently infected cells with agents that alter histone acetylation has already been shown to trigger lytic replication in latently infected cells (21, 40, 66).
We observed that the proteasome class of genes showed increased expression in ACH-2 cells even prior to induction but not in J1.1 and U1 cells. Hence, we sought to determine whether inhibition of proteasomes would induce latent provirus into lytic replication. We studied the effects of different concentrations of a proteasome inhibitor, clastolactacystin-beta-lactone (19), on p24 protein levels in chronically infected ACH-2 cells. A previous study has shown that proteasome inhibitors are capable of increasing the efficiency of an acute HIV infection (58). We therefore studied the effect of the proteasome inhibitor in the presence of 250 nM AZT (50% inhibitory concentration, 50 nM) to ensure that any increases in p24 levels were not due to increased efficiency of infection by actively replicating virus present in chronically infected cells or to subsequent rounds of viral replication. ACH-2 cells treated with AZT alone showed slightly lower p24 amounts than cells that were not treated with AZT, consistent with the observation that the chronically infected cells supported a low but detectable amount of ongoing viral replication.
Tumor necrosis factor alpha, a known activator of latently infected cells, caused a 20- to 24-fold increase in p24 expression in ACH-2 cells treated with AZT, and 5 μM clastolactacystin-beta-lactone caused a 10- to 15-fold increase in p24 expression compared to AZT-treated ACH-2 cells, indicating that clastolactacystin-beta-lactone activated latent provirus into lytic replication. The increases in p24 expression observed in cells treated with clastolactacystin-beta-lactone were dose dependent, indicating a drug-related response (Fig. 6A). This effect was not observed in similarly treated J1.1 and U1 cells (data not shown), which is consistent with the finding that the expression of a large number of proteasomal subunits was not upregulated in these chronically infected cells. We also tested PS-341, a highly specific proteasome inhibitor that binds to the beta-5 subunit of the proteasome and is approved for clinical treatment of multiple myeloma (1, 3). However, due to the extreme cytotoxicity of PS-341 even at a low concentration (100 nM), we were unable to observe any changes in p24 expression under our experimental conditions (1 to 1,000 nM) in latently infected cells (data not shown).
FIG. 6.
Effects of specific agents on HIV p24 production in chronically infected ACH-2 cells. Different concentrations of (A) the proteasome inhibitor clastolactacystin-beta-lactone, (B) resveratrol, an Egr1 activator, or (C) trichostatin, a histone deacetylase inhibitor were tested in chronically infected ACH-2 cells treated with 250 nM AZT. Samples were collected 24 h after addition of agent, and p24 concentrations were determined by enzyme-linked immunosorbent assay. p24 production in cells treated with tumor necrosis factor alpha (TNFα) was used as a positive control in ACH-2 cells compared to control (AZT-treated) cells. p24 production in untreated cells (no AZT) was also determined. Experiments were performed in triplicate, and values are representative of three independent experiments. Microarray data for the specific genes targeted are shown for each agent tested.
In our microarray studies, the gene Egr1 (early growth response 1) was downregulated in ACH-2 cells prior to induction and, interestingly, upregulated during lytic replication. Egr1 is involved in cell cycle regulation and cell differentiation, in response to a number of different growth factors (56, 64). Egr1 activity is suppressed in many cancers, including breast cancer and brain tumors, indicating that its activity is essential in cell cycle regulation (42, 38). It has recently been shown that resveratrol, an antioxidant stilbene (3,5,4′-trihydroxystilbene), can activate Egr1, thereby modulating p21cip expression and exerting an effect on cell cycle regulation. Egr1 expression also causes growth arrest (G1-S and G2-M phases) (52). We therefore tested the ability of resveratrol to activate lytic replication. Treatment of ACH-2 cells with resveratrol caused a dose-dependent increase in p24 expression indicative of viral reactivation (Fig. 6B). At concentrations greater than 50 μM, an increase in cell death was observed, which led to decreased p24 levels at higher concentrations.
Considerable prior work has been done on the influence of chromatin structure and histone acetylation status on HIV gene expression (21, 40, 66). These studies postulated that agents that can alter the histone acetylation state of the host cell would activate lytic HIV replication, and this was indeed found to be the case. However, there has been no indication that the cellular machinery that maintains histones in the deacetylated state might be upregulated in cells latently infected with HIV. In our expression data, we observed that HDAC1 and HDAC2 were upregulated more than twofold prior to induction. Also, the gene encoding YY1, known to repress the HIV-1 long terminal repeat through histone deacetylase recruitment (17) was also significantly upregulated prior to induction in ACH-2 cells. We confirmed that treating cells latently infected with HIV with trichostatin, an agent that targets this class of differentially expressed gene products, activates lytic HIV replication (Fig. 6C). Studies with agents that target specific cellular factors not only provide new approaches to cause viral reactivation, but also validate the biological relevance of these differentially expressed gene classes.
DISCUSSION
In chronically infected cell lines, each cell harbors at least one copy of the HIV provirus. Upon induction, most of the cells undergo viral replication, and the process is quite synchronous, in contrast to infection caused by addition of virus to cells. When infection is initiated by addition of virus to cells, there is greater variability in the time at which the viral replication cycle begins in the cells. The use of chronically infected cell models also circumvents the problems of using virus at a high multiplicity of infection, such as nonspecific effects due to interaction of host cells with cell debris, other nonspecific material in virus stocks, and defective viral particles. While inducing a lytic infection cycle in chronically infected cell lines provides a good model of viral replication and offers advantages over adding virus to host cells, it suffers from some disadvantages, such as the inability to study the cellular effects caused by events prior to proviral integration. In our studies, the effects caused by addition of PMA and by cells transitioning through the cell cycle were compensated for by using PMA-treated, matched, uninfected controls. Studies employing latently infected cells also offered an opportunity to study host cell expression in chronically infected cells prior to induction of a lytic viral cycle.
We studied the responses of 9,127 cellular genes following induction of a lytic replication cycle in cells chronically infected with HIV. Of these, 1,740 genes showed statistically significant differential expression at least once over the period of the lytic cycle. We classified the differentially expressed genes based on function and into known pathways by querying pathway databases hosted by CGAP. Classification of differentially expressed genes into known pathways in addition to functional classification provides valuable information regarding pathways that show differential expression in functionally unrelated genes but change similarly across the lytic replication cycle.
Our studies indicated that, following induction of cells latently infected with HIV, clear, temporally ordered changes in the cellular gene expression pattern accompanied the completion of the viral lytic cycle. Some of these changes may result from incidental changes extraneous to the processes of viral replication, for example, nonspecific cytopathic effects; some may reflect specific modifications to host cell physiology produced by the targeted action of viral gene products aimed at enhancing the ability of the host cell to support viral replication; and some may represent attempts by the host cell to blunt the effects of viral replication. Several genes previously implicated as having important roles in viral replication were differentially expressed, suggesting that at least some of the observed changes may be important for HIV pathophysiology. Other studies of acute infection have also shown some of these effects (for example, differential expression of genes encoding transcription factors and factors involved in apoptosis and signaling), indicating that the changes that accompany lytic replication are not too different from those associated with acute infection (29, 67).
While a number of gene classes did show similar trends during our lytic replication study, which correlated well with findings from other groups studying acute HIV infection, a direct and quantitative comparison of all gene classes may not be accurate due to the different controls employed in studies conducted by other groups, which may cause a skewed interpretation of the results. Also, since most other published studies looked at acute infection in a small number of cells against a background of a large population of uninfected cells, many gene classes that showed up as differentially expressed in our study may have failed to pass the filtering criteria due to high background noise in those studies.
In our microarray experiments, genes encoding histone deacetylases were upregulated prior to induction, correlating with studies that histone deacetylase inhibitors end viral latency. Genes involved in the mitogen-activated protein kinase signaling pathway are also upregulated during lytic replication, which corroborates studies that show that mitogen-activated protein kinase inhibitors suppress lytic replication (70). However, due to the innate complexity of many signaling pathways and the presence of feedback mechanisms and because various posttranslational regulatory mechanisms may be involved, a direct correlation may not always be present. The identification of differentially expressed gene classes may thus provide only a helpful starting point for further detailed research into the mechanisms that influence viral latency and the initiation of lytic replication. Assessing the precise significance of the many changes in host cell gene expression pattern that accompany the completion of the lytic infection cycle will necessarily require much additional work. However, the identification of many cellular genes that are differentially expressed during HIV lytic replication may offer additional targets for agents aimed at blocking HIV replication.
While changes in cellular gene expression during the viral lytic cycle were expected, a more important finding was that a significant number of genes showed altered expression even prior to induction. These changes could result either from the direct action of low levels of certain HIV gene products expressed in latency or, more likely, because changes in the cellular pattern of gene expression were selected for as the chronically infected cell lines were isolated and cloned. However, regardless of the mechanisms through which the alterations in cellular gene expression developed, the differential expression of these cellular genes may play a role in HIV latency and the maintenance of latency. While the differential expression of cellular genes in the latently infected cells may not be directly caused by the latent provirus, it is reasonable to postulate that the products of at least some of these differentially expressed genes may collaborate in the maintenance of the latent state, even though those genes were differentially expressed in the chronically infected cell lines as a consequence of the development and cloning of the chronically infected cell lines. Treating latently infected cells with agents targeting the differentially expressed cellular genes may help eject HIV from the latent state, thus providing novel approaches for the reduction of latent viral reservoirs with agents that target the differentially expressed cellular genes.
The effects seen prior to induction are probably not due merely to low levels of HIV replication in the latently infected cell population. Many genes that were downregulated during the lytic phase showed increased expression prior to induction and vice versa, for example, some transcription factors and DEAD-box proteins. Our flow cytometry studies showed only a low level (8%) of active replication prior to induction. In microarray experiments, signals originating from such a small fraction of the cells are likely to go undetected. Consistent with this, our microarray data for AZT-treated, latently infected ACH-2 cells yielded the same gene sets as obtained for untreated ACH-2 cells, indicating that changes in expression ratios were due mainly to the latently infected population and not to the low level of cells harboring actively replicating virus. Also, most of the cellular genes that were differentially expressed prior to induction also showed a gradual decrease in the level of up- or downregulation as the lytic phase progressed. It is therefore unlikely that most of the changes in gene expression observed prior to induction in chronically infected cell lines are due to actively replicating virus present in only a small fraction of cells. Hence, the effects on cellular gene expression seen prior to induction are most likely associated with the state of latent infection in these cell lines.
Based on our hypothesis that targeting the products of genes that were differentially expressed during viral latency would lead to viral reactivation, we showed that specific targeting of certain genes and gene classes can indeed force the virus out of latency. The mechanisms contributing to the observed effects may not be clear in all cases. For example, the mechanisms by which proteasome inhibitors end viral latency are not immediately obvious, though some available data may suggest an explanation. Studies have shown that HIV TAR sequence (present in the 5′ untranslated region of all HIV mRNAs) serves as the substrate for proteasomal degradation (28). Also, Nef, an early protein which is present in the host cell even before proviral integration, is processed by proteasomes, which provides many of the cytotoxic T-lymphocyte epitopes of Nef (45). Studies have also shown that Tat has an inhibitory effect on proteasome function by physically binding to 20S proteasome, thereby reducing viral protein degradation (59).
We propose that in chronically infected ACH-2 cells, the higher expression of proteasomes may lead to increased degradation of HIV mRNA. This effect, along with insufficient levels of full-length Tat and Nef proteins, would tend to inhibit HIV lytic replication. In such a case, addition of a proteasome inhibitor may release the suppressive effect of proteasomal degradation, potentiate the function of Tat by binding to the active sites on the proteasome, and thus allow accumulation of sufficient early viral proteins to trigger viral reactivation. Proteasome inhibition also inhibits NF-κB activity, which should decrease viral replication. However, it must be noted that even during active viral replication, use of a proteasome inhibitor increases viral replication efficiency (58). Binding of Tat to proteasomes has been shown to affect the peptidase activity (32). This may explain why we did not observe viral reactivation with PS-341 (2), which specifically inhibits the chymotryptic activity of proteasomes (3). A broader range of proteasomal inhibition may be required for viral reactivation. Clastolactacystin-beta-lactone, which caused viral reactivation, affects all three catalytic activities of the proteasome. The broad range of effects that the different active sites of proteasomes have on different pathways will necessarily make understanding the crucial mechanisms underlying these phenomena much more difficult.
Egr1 was downregulated during viral latency in ACH-2 cells. Resveratrol, an Egr1 activator, caused viral reactivation. Increased levels of Egr1 lead to growth arrest (G1/S and G2/M) (52), similar to that observed during viral replication. Downregulation of Egr1 may lead to decreased Egr1 levels, preventing growth arrest and creating conditions unfavorable for viral replication. The effect observed with resveratrol may be a combination of manipulating the latent cellular environment and mimicking the effects of active HIV replication on the cell cycle, providing favorable conditions for viral replication. Resveratrol has been shown to not affect the levels of NF-κB in other cell lines studied (52), suggesting that mechanisms other than NF-κB modulation may contribute to viral latency and reactivation. Our studies with specific agents that target cellular factors encoded by genes that are differentially expressed during viral latency not only provide new approaches to target viral latency but also provide support for the biological relevance of the genes that are differentially expressed.
Our studies in latently infected cell lines identified a common set of genes that were differentially expressed in all three cell lines studied. Notable among them was the downregulation of Cdc42 and Lyn, which are necessary for viral replication and maturation. Cdc42 and Lyn have been shown to be critical for activation of Nef-associated kinase (PAK2) and for binding of the PXXP motif, respectively (43, 57). Their downregulation in cell lines of different origins indicates that they may be critical for the cellular maintenance of viral latency. We were unable to test the effects of targeting the gene products of genes that were commonly differentially expressed in all cell lines because no specific agents for Cdc42 or Lyn are currently available.
Interestingly, three genes associated with acute myeloid leukemia, MNDA, CEBPalpha, and Meis1, were also downregulated in all the latently infected cell lines, similar to cdc42 and Lyn, indicating a new set of genes previously not related to HIV pathogenesis that may be involved in latency maintenance. MNDA also has been shown to bind with LANA (latency-associated nuclear antigen), a human herpesvirus 8 protein, upon viral reactivation by alpha interferon (27). There may be interactions between HIV proteins and MNDA too, which may be inhibited in latently infected cells due to downregulation of MNDA, thus preventing viral replication.
A small set of 20 genes were also upregulated in all three cell lines. While none of these has been previously linked to HIV infection or latency, some of them have been shown to interact with other viruses. For example, CDT1 inhibition is observed in active cytomegalovirus infection, which has been proposed as a mechanism by which cellular dysregulation due to cytomegalovirus occurs (7, 68). In our studies, CDT1 expression was upregulated in all three cell lines, suggesting that the cell may be maintaining latency by modulating certain cellular replication check points. Further research into the mechanisms of latency maintenance and reactivation and future therapeutic advances may be facilitated by the development of agents that target the genes that are similarly differentially expressed in these cell lines.
The existence of long-lasting HIV reservoirs is the principal barrier preventing the eradication of HIV infection (8, 49). Since there are currently no known phenotypic differences between latently infected and uninfected cells (10), gene expression profiling may provide crucial evidence of differences in the cellular environment between the two populations. Our data also suggest that specific targeting of genes showing differential expression in latently infected cells may force viral reactivation.
In summary, our studies show that a number of cellular genes and pathways are altered in chronically infected cell lines during viral latency. Following induction of a lytic replication, a number of genes and pathways are altered in a temporally ordered manner in the presence of actively replicating HIV. Some of these genes have been implicated in viral pathogenesis previously. However, a number of the differentially expressed genes have not been previously associated with HIV replication and/or viral latency. Our studies show that specific targeting of cellular gene products based only on carefully chosen gene expression data can be used to force HIV out of latency. In a clinical setting, triggering lytic replication in latently infected cells while maintaining good control of viral replication with antiretroviral drugs may potentially eliminate viral reservoirs. While the compounds used in our studies to eject virus from latency may not have immediate clinical utility, other agents targeting host cell gene products differentially expressed in latently infected cells may prove clinically useful.
Our studies provide a set of genes that were similarly differentially expressed in all the latently infected cell lines, which may yield a greater understanding of the functions of certain known cofactors and expand our knowledge of the factors involved in latency maintenance. Our studies also indicate new classes of cellular genes that may constitute good targets for agents that inhibit lytic viral replication. Such agents targeting host cell functions may be particularly helpful in blocking HIV replication by virus that is resistant to existing agents that target viral gene products. Our work thus may suggest new approaches to inhibit viral replication and new approaches to activate lytic infection, which may help decrease or eliminate latent viral reservoirs.
Supplementary Material
Acknowledgments
This research was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
We are very grateful to Richard Simon for advice on statistical analyses of microarray data. We thank Mike Lu and Ann Gordon for assistance. We thank John Coffin, Kuan-Teh Jeang, Paul Meltzer, and Richard Koup for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Adams, J. 2003. Potential for proteasome inhibition in the treatment of cancer. Drug Discov. Today 8:307-315. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. 2001. Proteasome inhibition in cancer: development of PS-341. Semin. Oncol. 28:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Adams, J. 2003. The proteasome: structure, function, and role in the cell. Cancer Treat. Rev. 29(Suppl. 1):3-9. [DOI] [PubMed] [Google Scholar]
- 4.Arendt, C. W., and D. R. Littman. 2001. HIV: master of the host cell. Genome Biol. 2:1030.1-1030.4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagnarelli, P., A. Valenza, S. Menzo, R. Sampaolesi, P. E. Varaldo, L. Butini, M. Montroni, C. F. Perno, S. Aquaro, D. Mathez, J. Leibowitch, C. Balotta, and M. Clementi. 1996. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J. Virol. 70:7603-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, N., V. Sanchez, and D. H. Spector. 2003. Hum. cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. J. Virol. 77:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, D. G., D. H. Hamer, P. A. Arlen, L. Gao, G. Bristol, C. M. Kitchen, E. A. Berger, and J. A. Zack. 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19:413-423. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, D. G., and J. A. Zack. 2002. Effect of latent human immunodeficiency virus infection on cell surface phenotype. J. Virol. 76:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buttke, T. M., and T. M. Folks. 1992. Complete replacement of membrane cholesterol with 4,4′,14-trimethyl sterols in a human T cell line defective in lanosterol demethylation. J. Biol. Chem. 267:8819-8826. [PubMed] [Google Scholar]
- 12.Chang, Y. E., and L. A. Laimins. 2001. Interferon-inducible genes are major targets of human papillomavirus type 31: insights from microarray analysis. Dis. Markers 17:139-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun, T. W., and A. S. Fauci. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. USA 96:10958-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clouse, K. A., D. Powell, I. Washington, G. Poli, K. Strebel, W. Farrar, P. Barstad, J. Kovacs, A. S. Fauci, and T. M. Folks. 1989. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 142:431-438. [PubMed] [Google Scholar]
- 16.Corbeil, J., D. Sheeter, D. Genini, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, R. Sasik, D. Huang, J. Drenkow, D. D. Richman, and T. Gingeras. 2001. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 11:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coull, J. J., F. Romerio, J. M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cousar, J. B., and R. C. Briggs. 1990. Expression of human myeloid cell nuclear differentiation antigen (MNDA) in acute leukemias. Leuk. Res. 14:915-920. [DOI] [PubMed] [Google Scholar]
- 19.Craiu, A., M. Gaczynska, T. Akopian, C. F. Gramm, G. Fenteany, A. L. Goldberg, and K. L. Rock. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 272:13437-13445. [DOI] [PubMed] [Google Scholar]
- 20.de la Fuente, C., F. Santiago, L. Deng, C. Eadie, I. Zilberman, K. Kehn, A. Maddukuri, S. Baylor, K. Wu, C. G. Lee, A. Pumfery, and F. Kashanchi. 2002. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Kharroubi, A., G. Piras, R. Zensen, and M. A. Martin. 1998. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 18:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 23.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 24.Folks, T. M., K. A. Clouse, J. Justement, A. Rabson, E. Duh, J. H. Kehrl, and A. S. Fauci. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA 86:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folks, T. M., J. Justement, A. Kinter, C. A. Dinarello, and A. S. Fauci. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800-802. [DOI] [PubMed] [Google Scholar]
- 26.Folks, T. M., J. Justement, A. Kinter, S. Schnittman, J. Orenstein, G. Poli, and A. S. Fauci. 1988. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol. 140:1117-1122. [PubMed] [Google Scholar]
- 27.Fukushi, M., M. Higuchi, M. Oie, T. Tetsuka, F. Kasolo, K. Ichiyama, N. Yamamoto, H. Katano, T. Sata, and M. Fujii. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with human myeloid cell nuclear differentiation antigen induced by interferon alpha. Virus Genes 27:237-247. [DOI] [PubMed] [Google Scholar]
- 28.Gautier-Bert, K., B. Murol, A. S. Jarrousse, L. Ballut, S. Badaoui, F. Petit, and H. P. Schmid. 2003. Substrate affinity and substrate specificity of proteasomes with RNase activity. Mol. Biol. Rep. 30:1-7. [DOI] [PubMed] [Google Scholar]
- 29.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van 't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 30.Graziosi, C., G. Pantaleo, L. Butini, J. F. Demarest, M. S. Saag, G. M. Shaw, and A. S. Fauci. 1993. Kinetics of human immunodeficiency virus type 1 (HIV-1) DNA and RNA synthesis during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 90:6405-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 32.Huang, X., U. Seifert, U. Salzmann, P. Henklein, R. Preissner, W. Henke, A. J. Sijts, P. M. Kloetzel, and W. Dubiel. 2002. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 323:771-782. [DOI] [PubMed] [Google Scholar]
- 33.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan, J., R. Simon, M. Bittner, Y. Chen, S. B. Leighton, T. Pohida, P. D. Smith, Y. Jiang, G. C. Gooden, J. M. Trent, and P. S. Meltzer. 1998. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 58:5009-5013. [PubMed] [Google Scholar]
- 35.Kim, S., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korn, E. L., J. F. Troendle, L. M. McShane, and R. Simon. 2004. Controlling the number of false discoveries: Application to high-dimensional genomic data. J. Statist. Planning Inference 124:379-398. [Google Scholar]
- 37.Kramer, M. F., W. J. Cook, F. P. Roth, J. Zhu, H. Holman, D. M. Knipe, and D. M. Coen. 2003. Latent herpes simplex virus infection of sensory neurons alters neuronal gene expression. J. Virol. 77:9533-9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krones-Herzig, A., E. Adamson, and D. Mercola. 2003. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc. Natl. Acad. Sci. USA 100:3233-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kundu, M., S. Sharma, A. De Luca, A. Giordano, J. Rappaport, K. Khalili, and S. Amini. 1998. HIV-1 Tat elongates the G1 phase and indirectly promotes HIV-1 gene expression in cells of glial origin. J. Biol. Chem. 273:8130-8136. [DOI] [PubMed] [Google Scholar]
- 40.Laughlin, M. A., S. Zeichner, D. Kolson, J. C. Alwine, T. Seshamma, R. J. Pomerantz, and F. Gonzalez-Scarano. 1993. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology 196:496-505. [DOI] [PubMed] [Google Scholar]
- 41.Lifson, J. D., G. R. Reyes, M. S. McGrath, B. S. Stein, and E. G. Engleman. 1986. AIDS retrovirus induced cytopathology: giant cell formation and involvement of CD4 antigen. Science 232:1123-1127. [DOI] [PubMed] [Google Scholar]
- 42.Liu, C., V. M. Rangnekar, E. Adamson, and D. Mercola. 1998. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 5:3-28. [PubMed] [Google Scholar]
- 43.Lu, X., X. Wu, A. Plemenitas, H. Yu, E. T. Sawai, A. Abo, and B. M. Peterlin. 1996. CDC42 and Rac1 are implicated in the activation of the Nef-associated kinase and replication of HIV-1. Curr. Biol. 6:1677-1684. [DOI] [PubMed] [Google Scholar]
- 44.Lucchiari-Hartz, M., V. Lindo, N. Hitziger, S. Gaedicke, L. Saveanu, P. M. van Endert, F. Greer, K. Eichmann, and G. Niedermann. 2003. Differential proteasomal processing of hydrophobic and hydrophilic protein regions: contribution to cytotoxic T lymphocyte epitope clustering in HIV-1-Nef. Proc. Natl. Acad. Sci. USA 100:7755-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucchiari-Hartz, M., P. M. van Endert, G. Lauvau, R. Maier, A. Meyerhans, D. Mann, K. Eichmann, and G. Niedermann. 2000. Cytotoxic T lymphocyte epitopes of HIV-1 Nef: Generation of multiple definitive major histocompatibility complex class I ligands by proteasomes. J. Exp. Med. 191:239-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikovits, J. A., N. C. Lohrey, R. Schulof, J. Courtless, and F. W. Ruscetti. 1992. Activation of infectious virus from latent human immunodeficiency virus infection of monocytes in vivo. J. Clin. Investig. 90:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 48.Perez, V. L., T. Rowe, J. S. Justement, S. T. Butera, C. H. June, and T. M. Folks. 1991. An HIV-1-infected T cell clone defective in IL-2 production and Ca2+ mobilization after CD3 stimulation. J. Immunol. 147:3145-3148. [PubMed] [Google Scholar]
- 49.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poon, B., K. Grovit-Ferbas, S. A. Stewart, and I. S. Chen. 1998. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science 281:266-269. [DOI] [PubMed] [Google Scholar]
- 51.Radhakrishnan, S., J. Otte, S. Enam, L. Del Valle, K. Khalili, and J. Gordon. 2003. JC virus-induced changes in cellular gene expression in primary human astrocytes. J. Virol. 77:10638-10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ragione, F. D., V. Cucciolla, V. Criniti, S. Indaco, A. Borriello, and V. Zappia. 2003. p21Cip1 gene expression is modulated by Egr1: a novel regulatory mechanism involved in the resveratrol antiproliferative effect. J. Biol. Chem. 278:23360-23368. [DOI] [PubMed] [Google Scholar]
- 53.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82-85. [DOI] [PubMed] [Google Scholar]
- 54.Rozovskaia, T., E. Feinstein, O. Mor, R. Foa, J. Blechman, T. Nakamura, C. M. Croce, G. Cimino, and E. Canaani. 2001. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene 20:874-878. [DOI] [PubMed] [Google Scholar]
- 55.Ruff, C. T., S. C. Ray, P. Kwon, R. Zinn, A. Pendleton, N. Hutton, R. Ashworth, S. Gange, T. C. Quinn, R. F. Siliciano, and D. Persaud. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J. Virol. 76:9481-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto, K. M., C. Bardeleben, K. E. Yates, M. A. Raines, D. W. Golde, and J. C. Gasson. 1991. 5′ upstream sequence and genomic structure of the human primary response gene, EGR-1/TIS8. Oncogene 6:867-871. [PubMed] [Google Scholar]
- 57.Saksela, K., G. Cheng, and D. Baltimore. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seeger, M., K. Ferrell, R. Frank, and W. Dubiel. 1997. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 272:8145-8148. [DOI] [PubMed] [Google Scholar]
- 60.Shaheduzzaman, S., V. Krishnan, A. Petrovic, M. Bittner, P. Meltzer, J. Trent, S. Venkatesan, and S. Zeichner. 2002. Effects of HIV-1 Nef on cellular gene expression profiles. J. Biomed. Sci. 9:82-96. [DOI] [PubMed] [Google Scholar]
- 61.Simmons, A., V. Aluvihare, and A. McMichael. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763-777. [DOI] [PubMed] [Google Scholar]
- 62.Simon, R. M., E. L. Korn, L. M. McShane, M. D. Radmacher, G. M. Wright, and Y. Zhao. 2003. Design and analysis of DNA microarray investigations-An artificial intelligence milestone. Springer Verlag, New York, N.Y.
- 63.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sukhatme, V. P., X. M. Cao, L. C. Chang, C. H. Tsai-Morris, D. Stamenkovich, P. C. Ferreira, D. R. Cohen, S. A. Edwards, T. B. Shows, T. Curran, et al. 1988. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 53:37-43. [DOI] [PubMed] [Google Scholar]
- 65.Tenen, D. G. 2001. Abnormalities of the CEBP alpha transcription factor: a major target in acute myeloid leukemia. Leukemia 15:688-689. [DOI] [PubMed] [Google Scholar]
- 66.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 67.van't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiebusch, L., R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 4:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wright, G. W., and R. M. Simon. 2003. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19:2448-2455. [DOI] [PubMed] [Google Scholar]
- 70.Yang, X., Y. Chen, and D. Gabuzda. 1999. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J. Biol. Chem. 274:27981-27988. [DOI] [PubMed] [Google Scholar]
- 71.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.