Abstract
The type III sodium-dependent phosphate (NaPi) cotransporter, Pit2, is a receptor for amphotropic murine leukemia virus (A-MuLV) and 10A1 MuLV. In order to determine what is sufficient for Pit2 receptor function, a deletion mutant lacking about the middle half of the protein was made. The mutant supported entry for both viruses, unequivocally narrowing down the identification of the sequence that is sufficient to specify the receptor functions of Pit2 to its N-terminal 182 amino acids and C-terminal 170 amino acids.
The type III sodium-dependent phosphate (NaPi) symporter, Pit2, from humans is a receptor for amphotropic murine leukemia virus (A-MuLV) and the related strain 10A1 MuLV (13, 22, 23, 25, 40, 44). The related Pit1 protein from humans (62% amino acid identity) is a receptor for gibbon ape leukemia virus, feline leukemia virus subgroup B, and 10A1 MuLV (22, 24, 39). The two human type III NaPi symporters show about 25% amino acid identity with the Neurospora crassa NaPi symporter Pho-4 (11, 40). Several studies have aimed at identifying sequences that are critical for the viral receptor functions of Pit1 and Pit2. These studies used chimeras between Pit1 and Pit2 orthologs, Pit1 orthologs, human Pit1 and Pho-4, human Pit2 and Pho-4, and human Pit1 and Pit2 and Pho-4 as well as Pit1 and Pit2 mutants (3, 5-8, 12, 14-16, 22, 26-28, 31-33, 38, 42, 43). The results suggest that amino acids and/or sequences present in human Pit2 positions 67 to 91 (8), 107 to 141 (14), 517 to 530 (15), and 522 to 530 (28) or corresponding positions in related proteins (15, 16, 22, 28, 33) specify or are involved in A-MuLV receptor function. Moreover, human and mouse Pit1 chimera studies suggest that amino acids in the mouse and human Pit1 regions corresponding to human Pit2 sequence 522 to 530 also are involved in 10A1 receptor function (16); these results have been confirmed in studies of human Pit2 mutants (unpublished results).
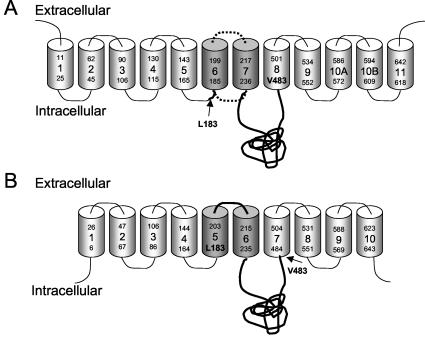
The design of the majority of the chimerical and mutant proteins (14-16, 22, 28, 33) was based on a membrane topology model for Pit1, Pit2, and Pho-4, which was suggested by Johann and coworkers on the basis of Kyte-Doolittle hydropathy plots (4, 11, 40) (Fig. 1B). Recently, new topology models that differ from the previous models have been proposed for Pit1 (7) and Pit2 (30) (Fig. 1A). If Pit1 and Pit2, however, exhibit different topologies in the membrane, the interpretation of which sequences are directly involved in receptor function for A-MuLV and 10A1 based on results obtained with Pit1/Pit2 chimeras may be incorrect. Moreover, one cannot in general exclude the possibility that amino acid exchanges in, for example, one part of Pit2 would lead to overall structural changes in another part of the protein, as shown for Pit1 by Farrell et al. (7). Results obtained with exchange mutants, therefore, may also potentially lead to incorrect interpretations concerning the identities of amino acids in Pit2 that are directly involved in A-MuLV and 10A1 receptor functions. It is obvious that these problems cannot be addressed simply by extending the use of chimeric Pit1/Pit2 proteins or exchange mutants of Pit2; however, functional deletion mutants can better identify which Pit2 sequences are directly involved in specifying receptor function.
FIG. 1.
Putative membrane topology models of Pit2 based on Salaün et al. (30) (A) and Johann et al. (11, 40) (B); see references for details on the models. The membrane topology model of Pit2 after Salaün et al. (30) was based on (i) epitope tagging of N- and C-terminal ends, (ii) orientation of microsomal membrane-inserted in vitro transcribed and translated C-terminal-truncation mutants, and (iii) glycosylation of wild-type human Pit2 and lack of glycosylation of a human Pit2N81V mutant in whole-cell extracts from CHO K1 cells overexpressing Pit2 and Pit2N81V. Salaün and coworkers did not present functional studies (e.g., viral receptor function) of the C-terminal truncation mutants (30). For comparison, the first proposed membrane topology model of Pit2 based on Kyte-Doolittle hydropathy plots is shown (11, 40) in panel B. The middle part of human Pit2 including amino acid 183 to amino acid 483 (the sequences highlighted in both A and B) is deleted in the mutant Pit2ΔL183-V483.
We hypothesized that if the A-MuLV-and possibly 10A1-receptor determinants are indeed positioned in the N- and/or C-terminal ends of Pit2, it may be possible to delete the middle part of Pit2 and still retain its receptor functions. Accordingly, we made a mutant human Pit2 protein, Pit2ΔL183-V483, in which the part between arginine 182 and histidine 484 was deleted in the 652-amino-acid protein.
The Pit2ΔL183-V483 mutant was made from the pcDNA1ARtkpA-derived expression plasmid pOJ74 (Wyeth-Ayerst Research, Pearl River, N.Y.) encoding human Pit2 (40) by using the forward primer ATGGCTGGGGAAGTTAGTGC and the reverse primer GGGTTACCGGAGGCCCGTGTGGAGGACAAGGTA; the latter was used to create the link between the 5′ sequence encoding PNGLRA182 and the 3′ sequence encoding H484LLFH (Fig. 1). The PCR amplification product was digested with Bsu36I and Sse8387I and used to replace the corresponding fragment in plasmid pOJ74, resulting in a plasmid encoding the mutant Pit2ΔL183-V483 protein. The authenticity of the nucleotide sequence was confirmed. The ability of Pit2ΔL183-V483 to support A-MuLV and 10A1 entry was compared to that of human Pit2 by using a transient transfection-infection assay and A-MuLV and 10A1 vector pseudotypes derived from the packaging cell lines PA317 (19, 21) and PT67 (20), respectively, both carrying the G1BgSvN transfer vector (18) as previously described (2, 28). The G1BgSvN transfer vector is a Moloney MuLV-based vector that expresses LacZ and neomycin phosphotransferase (18). The titers of A-MuLV and 10A1 vector stocks were determined on dog D17 cells (ATCC CCL 183) as previously described (28) and were 1.4 × 105 to 4.7 × 105 and 0.7 × 105 to 6.0 × 105 CFU per ml, respectively; however, the stocks were diluted to 40,000 infectious vectors per 1.5 ml before use. Briefly, hamster CHO K1 cells (ATCC CCL 61), before the fifth passage and kept subconfluent during cultivation, were seeded in dishes with a diameter of 60 mm. These cells were transfected with 1 μg of plasmid DNA encoding human Pit2 or equimolar amounts of plasmid encoding Pit2ΔL183-V483. Mock-treated cells were transfected with empty vector DNA. The total amounts of DNA in the transfection solutions were kept constant by using plasmid pUC19 as carrier DNA. At 48 h posttransfection, 1.5 ml of diluted vector stocks was added per dish in the presence of Polybrene. After 48 h, the dishes were fixed and stained, and the number of β-galactosidase-positive (infected) cells per dish was either counted (Table 1) or visualized (Fig. 2). Note that in each experiment shown in Table 1, three independent transfection solutions were made per construct, and each solution was evaluated for its ability to support both A-MuLV and 10A1 infection (experiments 1 and 2) or 10A1 infection (experiment 3); thus, the numbers shown in Table 1 represent the average levels of infection from three 60-mm dishes that received independent transfection solutions.
TABLE 1.
Permissivity for infection of CHO K1 cells transiently transfected with vector expressing Pit2 or Pit2ΔL183-V483 or empty vectora
| Constructb | No. (%) of cells infectedc
|
||||
|---|---|---|---|---|---|
| A-MuLV
|
10A1 MuLV
|
||||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | Expt 3 | |
| Pit2 (pOJ74) | 100 ± 22 | 100 ± 8 | 100 ± 5 | 100 ± 8 | 100 ± 9 |
| Pit2ΔL183-V483 | 25 ± 1 | 13 ± 2 | 10 ± 1 | 12 ± 4 | 24 ± 3 |
| Empty vectord | <0.0008 | <0.002 | <0.002 | <0.001 | <0.001 |
The experimental setup is described in the text. A-MuLV and 10A1 vector pseudotypes were tested on the same DNA precipitates of a given construct in experiment 1 (one independent set of DNA preparations) and in experiment 2 (another independent set of DNA preparations). In experiment 3, only the 10A1 vector pseudotype was tested (same DNA preparations as in experiment 1).
Receptor and mutant receptor sequences were cloned into pcDNA1ARtkpA.
The data are averages of three independent transfections ± standard errors of the means. The average number of blue cells per three 60-mm dishes transfected with a plasmid expressing Pit2 was assigned a value of 100% (42,000, 19,000, 18,000, 32,000, and 32,000 blue cells per dish for A-MuLV in experiments 1 and 2 and for 10A1 in experiments 1, 2, and 3, respectively).
Values are based on the detection limit of 1 blue cell per three 60-mm-diameter dishes.
FIG. 2.
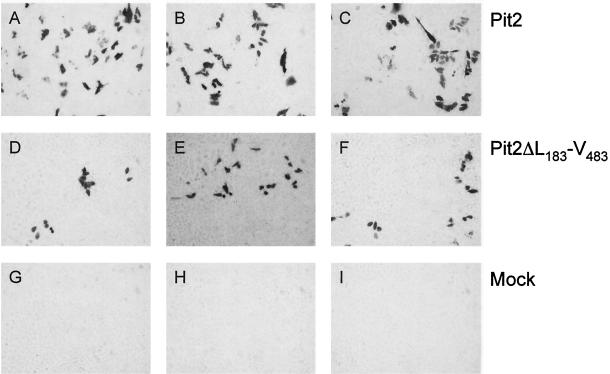
Susceptibilities of transfected CHO K1 cells. Transfections were performed as described in the text. Panels A to C show three dishes receiving independent DNA precipitates of the plasmid encoding Pit2, panels D to F show three dishes receiving independent DNA precipitates of the plasmid encoding Pit2ΔL183-V483, and panels G to I show three dishes receiving independent DNA precipitates of empty expression vector (Mock). The experiment was performed according to the method shown in Table 1; however, a third independent set of DNA preparations was used. At 48 h posttransfection, cells were challenged with PT67-derived vector pseudotypes (10A1 MuLV pseudotypes) carrying the LacZ-expressing G1BgSvN transfer vector. Forty-eight hours after vector exposure, cells were fixed and stained for the presence of β-galactosidase-positive (infected) cells; infected cells are blue (dark). No blue cells were present in the mock-transfected cultures. Random fields from each plate in the triplicate setups are shown at a magnification of ×200.
Even though approximately the middle half of the Pit2 protein is deleted in Pit2ΔL183-V483, the mutant sustained receptor function for both A-MuLV and 10A1 in the ranges of 13 to 25% and 10 to 24%, respectively, of the infection levels supported by wild-type human Pit2 (Table 1). No infection by 10A1 or A-MuLV vector pseudotypes was observed in mock-transfected CHO K1 cells (Table 1 and Fig. 2). We occasionally observe a low background level of infection with these vector pseudotypes on our CHO K1 cells (<2 to 6 blue cells per 60-mm dish or <2 CFU/ml) (reference 15 and our unpublished observations) although such an occurrence is the exception (Table 1 and Fig. 2) (2, 15, 28). It should be noted, however, that there exist CHO K1 subpopulations or subclones that are susceptible to 10A1 infection (9, 22).
While the results presented here do not provide new information on the membrane topology of Pit2 per se, the observation that Pit2ΔL183-V483 supports A-MuLV and 10A1 infection shows that the N-terminal 182 amino acids and C-terminal 170 amino acids of human Pit2 are sufficient for specifying A-MuLV and 10A1 receptor functions. Thus, the results of the present study narrow the viral binding domains and other possible sequences directly involved in A-MuLV and/or 10A1 entry to sequences positioned in these N- and C-terminal regions of human Pit2. The regions include all Pit2-specific sequences previously identified as influencing Pit2 receptor function for A-MuLV and 10A1 (8, 14-16, 22, 28, 33), and the Pit2ΔL183-V483 mutant may prove to be a powerful tool in identifying whether all of these sequences, indeed, are directly involved in A-MuLV binding and entry. The mutant lacks a sequence originally identified as a topogenic determinant in Pit1 (region B) and shown to affect the results of Pit1/Pit2 chimerical studies (7); however, a number of related putative phosphate symporters from archaea and bacteria (e.g., U15187, AL939110, AP001512, AE000978, AE013582 [National Center for Biotechnology Information accession numbers]) resemble Pit2ΔL183-V483, which suggests that the mutant constitutes a structural unit here shown to specify viral receptor function. It is therefore also possible that the mutant will sustain NaPi transport. It should be noted, however, that there is no correlation between the ability of Pit2 to transport phosphate and support viral entry (2), and the uncoupling of transport and receptor functions has also been shown for two other transporters with retroviral receptor function (37, 41) as thoroughly discussed in a recent review (36). Indeed, like Pit2, a number of the identified receptors for retroviruses are polytopic solute transporters (1, 10, 17, 24, 29, 34, 35). The present study shows that it is possible to use a deletion mutant of Pit2 for studying its viral receptor function and raises the possibility that deletion mutants of other polytopic proteins may also provide insight into which receptor sequences are directly involved in receptor functions for their cognate viruses.
Acknowledgments
We thank Bryan O’Hara for plasmid pOJ74 and Maribeth V. Eiden for the PA317GBN cell line.
This study was supported by the Lundbeck Foundation, the Novo Nordisk Foundation, the Danish Medical Research Council (grant 9802349), the Karen Elise Jensen Foundation, and the Danish Cancer Society (grant DP00092).
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Bøttger, P., and L. Pedersen. 2002. Two highly conserved glutamate residues critical for type III sodium-dependent phosphate transport revealed by uncoupling transport function from retroviral receptor function. J. Biol. Chem. 277:42741-42747. [DOI] [PubMed] [Google Scholar]
- 3.Chaudry, G. J., and M. V. Eiden. 1997. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J. Virol. 71:8078-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien, M.-L., J. L. Foster, J. L. Douglas, and J. V. Garcia. 1997. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J. Virol. 71:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyer, K., F. S. Pedersen, and L. Pedersen. 2000. A 13-amino-acid Pit1-specific loop 4 sequence confers feline leukemia virus subgroup B receptor function upon Pit2. J. Virol. 74:2926-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiden, M. V., K. B. Farrell, and C. A. Wilson. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J. Virol. 70:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell, K. B., J. L. Russ, R. K. Murthy, and M. V. Eiden. 2002. Reassessing the role of region A in Pit1-mediated viral entry. J. Virol. 76:7683-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman, S. A., K. B. Farrell, R. K. Murthy, J. L. Russ, and M. V. Eiden. 2004. Identification of an extracellular domain within the human PiT2 receptor that is required for amphotropic murine leukemia virus binding. J. Virol. 78:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, J. Y., P. M. Cannon, K. M. Lai, Y. Zhao, M. V. Eiden, and W. F. Anderson. 1997. Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J. Virol. 71:8103-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hein, S., V. Prassolov, Y. Zhang, D. Ivanov, J. Lohler, S. R. Ross, and C. Stocking. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J. Virol. 77:5926-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johann, S. V., J. J. Gibbons, and B. O'Hara. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66:1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johann, S. V., M. van Zeijl, J. Cekleniak, and B. O'Hara. 1993. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 67:6733-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavanaugh, M. P., D. G. Miller, W. Zhang, W. Law, S. L. Kozak, D. Kabat, and A. D. Miller. 1994. Cell surface receptors for gibbon ape leukemia virus and amphotropic murine leukemia virus are inducible sodium-dependent phosphate transporters. Proc. Natl. Acad. Sci. USA 91:7071-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leverett, B. D., K. B. Farrell, M. V. Eiden, and C. A. Wilson. 1998. Entry of amphotropic murine leukemia virus is influenced by residues in the putative second extracellular domain of its receptor, Pit2. J. Virol. 72:4956-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundorf, M. D., F. S. Pedersen, and L. Pedersen. 1999. Amphotropic murine leukemia virus entry is determined by specific combinations of residues in receptor loops 2 and 4. J. Virol. 73:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundorf, M. D., F. S. Pedersen, B. O’Hara, and L. Pedersen. 1998. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J. Virol. 72:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manel, N., F. J. Kim, S. Kinet, N. Taylor, M. Sitbon, and J.-L. Battini. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115:449-459. [DOI] [PubMed] [Google Scholar]
- 18.McLachlin, J. R., N. Mittereder, M. B. Daucher, M. Kadan, and M. A. Eglitis. 1993. Factors affecting retroviral vector function and structural integrity. Virology 195:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70:5564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-990. [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 68:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunn, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 25.Olah, Z., C. Lehel, W. B. Anderson, M. V. Eiden, and C. A. Wilson. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 269:25426-25431. [PubMed] [Google Scholar]
- 26.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen, L., M. van Zeijl, S. V. Johann, and B. O'Hara. 1997. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J. Virol. 71:7619-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, L., S. V. Johann, M. van Zeijl, F. S. Pedersen, and B. O'Hara. 1995. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J. Virol. 69:2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasko, J. E., J. L. Battini, R. J. Gottschalk, I. Mazo, and A. D. Miller. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA 96:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salaün, C., P. Rodrigues, and J. M. Heard. 2001. Transmembrane topology of PiT-2, a phosphate transporter-retrovirus receptor. J. Virol. 75:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommerfelt, M. A. 1999. Retrovirus receptors. J. Gen. Virol. 80:3049-3064. [DOI] [PubMed] [Google Scholar]
- 33.Tailor, C. S., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tailor, C. S., A. Nouri, Y. Zhao, Y. Takeuchi, and D. Kabat. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tailor, C. S., B. J. Willett, and D. Kabat. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 73:6500-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tailor, C. S., D. Lavillette, M. Marin, and D. Kabat. 2003. Cell surface receptors for gammaretroviruses. Curr. Top. Microbiol. Immunol. 281:29-106. [DOI] [PubMed] [Google Scholar]
- 37.Tailor, C. S., M. Marin, A. Nouri, M. P. Kavanaugh, and D. Kabat. 2001. Truncated forms of the dual function human ASCT2 neutral amino acid transporter/retroviral receptor are translationally initiated at multiple alternative CUG and GUG codons. J. Biol. Chem. 276:27221-27230. [DOI] [PubMed] [Google Scholar]
- 38.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Zeijl, M., S. V. Johann, E. Closs, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., M. P. Kavanaugh, and D. Kabat. 1994. A critical site in the cell surface receptor for ecotropic murine retroviruses required for amino acid transport but not for viral reception. Virology 202:1058-1060. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and non-susceptible murine cells. J. Gen. Virol. 75:1901-1908. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J. Virol. 68:7697-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, C. A., M. V. Eiden, W. B. Anderson, C. Lehel, and Z. Olah. 1995. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J. Virol. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]