Summary
The archaeological documentation of the development of sedentary farming societies in Anatolia is not yet mirrored by a genetic understanding of the human populations involved, in contrast to the spread of farming in Europe [1, 2, 3]. Sedentary farming communities emerged in parts of the Fertile Crescent during the tenth millennium and early ninth millennium calibrated (cal) BC and had appeared in central Anatolia by 8300 cal BC [4]. Farming spread into west Anatolia by the early seventh millennium cal BC and quasi-synchronously into Europe, although the timing and process of this movement remain unclear. Using genome sequence data that we generated from nine central Anatolian Neolithic individuals, we studied the transition period from early Aceramic (Pre-Pottery) to the later Pottery Neolithic, when farming expanded west of the Fertile Crescent. We find that genetic diversity in the earliest farmers was conspicuously low, on a par with European foraging groups. With the advent of the Pottery Neolithic, genetic variation within societies reached levels later found in early European farmers. Our results confirm that the earliest Neolithic central Anatolians belonged to the same gene pool as the first Neolithic migrants spreading into Europe. Further, genetic affinities between later Anatolian farmers and fourth to third millennium BC Chalcolithic south Europeans suggest an additional wave of Anatolian migrants, after the initial Neolithic spread but before the Yamnaya-related migrations. We propose that the earliest farming societies demographically resembled foragers and that only after regional gene flow and rising heterogeneity did the farming population expansions into Europe occur.
Keywords: ancient DNA, archaeogenomics, Neolithic, Anatolia, population genetics, genetic diversity
Highlights
-
•
Pre-pottery farmers had low genetic diversity, akin to Mesolithic hunter-gatherers
-
•
Genetic diversity levels are higher in the subsequent Pottery Neolithic
-
•
Central Anatolian farmers belonged to the same gene pool as early European farmers
-
•
Copper Age genetic affinities suggest a second wave of Anatolian gene flow
Kılınç et al. study ancient genomes from the earliest farmers of central Anatolia, one of the first areas where farming appears outside the Fertile Crescent. Genetic diversity increases as the Neolithic develops, indicating rising mobility. Similarities between Anatolian and European farmers suggest two gene flow events from Anatolia into Europe.
Results and Discussion
The causes, effects, and mechanisms of the transition from foraging to farming in western Eurasia are key issues in understanding the development of our species, especially in understanding the development of larger, more dense, and more socially complex populations. Over the past decade, archaeogenetic studies have largely focused on processes that drove the spread of farming practices, particularly the introduction of farming and sedentism into Europe [2, 3, 5, 6, 7, 8, 9]. However, the demographic aspects of the transformation of forager communities in Southwest Asia into communities practicing substantial-scale mixed farming and the full extent of the role of Anatolian populations in the spread of farming into Europe have remained unclear. Here, we investigate human remains excavated from two different Neolithic settlements in central Anatolia, Boncuklu and Tepecik-Çiftlik, between circa (ca.) 8300 and 5800 calibrated (cal) BC to explore the demographic processes during the earliest (Aceramic) phase of the Neolithic transition, as well as the later Pottery Neolithic period in Anatolia.
Archaeological records show that the Neolithic era in Anatolia spanned more than 3,000 years—from around 9500 cal BC to around 6000 cal BC [4]. Farming practices were first established in the Fertile Crescent in the tenth and early ninth millennium cal BC [10] and in central Anatolia by 8300 cal BC [11, 12], or possibly earlier [12]. Between ca. 8000 cal BC and 6600 cal BC, farming spread west of central Anatolia, reaching the Aegean coast before 6600 cal BC and northwest Anatolia by 6600 at the latest [13, 14]. Debate exists as to whether this may have been a slow, steady process over those 1,400 years or relatively rapid between ca. 7000 and 6600 cal BC. Boncuklu, the earliest Anatolian site in our sample, and with evidence of very early crop cultivation in central Anatolia, is a small settlement mound dating between ca. 8300 and 7500 cal BC in the Aceramic Neolithic [11]. The excavators suggest that the Boncuklu community consisted of indigenous foragers who adopted small-scale cultivation and possibly experimented with animal herding alongside substantial traditional foraging practices [4, 11]. Tepecik-Çiftlik is a village with mixed and complex plant and animal exploitation practices, including notable elements of farming, located in the volcanic Cappadocian region of central Anatolia, dating between ca. 7500 and 5800 cal BC, from the latter Pre-Pottery Neolithic into the Pottery Neolithic [15, 16]. The evidence from Tepecik-Çiftlik indicates more substantial scale mixed farming relative to Boncuklu, although both hunting and gathering played a part in plant and animal exploitation. Both Boncuklu and Tepecik-Çiftlik show evidence of significant scale regional and inter-regional interactions, in the Tepecik-Çiftlik case especially with communities in the Fertile Crescent possibly related to the widespread distribution of obsidian [11, 15, 16]. The differences in subsistence patterns between these two settlements reflect a larger regional pattern seen in several other Aceramic and Pottery Neolithic sites in Anatolia [4, 13].
We investigated a total of nine ancient individuals excavated from Boncuklu (n = 4) and Tepecik-Çiftlik (n = 5) (Data S1). We generated genome sequence data from these individuals with a mean coverage between 0.03-fold and 6-fold per individual, using a combination of whole-genome capture and direct shotgun sequencing strategies (Supplemental Experimental Procedures; Table 1; Data S2; Figures S1A and S1B). We authenticated the sequence data using multiple well-established approaches (Supplemental Experimental Procedures; Data S1; Figure S1C). Mitochondrial genome coverages were between 66- and 2,379-fold (Table 1), and all five Tepecik-Çiftlik and three Boncuklu individuals carried the haplogroups previously found in Neolithic farmers in Europe (haplogroups K and N) (Table 1; Data S2; Figure S1D) [17]. One of the Boncuklu individuals carried the haplogroup U3, which has also been observed in a later northwest Anatolian (Pottery) Neolithic site, Barcın (Figure 1), and in early Neolithic European farmers [8, 17, 18], but not among Eurasian hunter-gatherers [19]. We identified four individuals as females and the other five as males (Table 1; Data S1).
Table 1.
Summary Statistics of the Sequencing Data for Nine Ancient Individuals
| Sample | Genome Coverage | mtDNA Coverage | Read Length (Mean) | mtDNA Haplogroup | Genetic Sex |
|---|---|---|---|---|---|
| Bon001 | 0.166 | 654.604 | 63.208 | U3 | XY |
| Bon002 | 6.688 | 2,379.090 | 69.841 | K1a | XX |
| Bon004 | 0.243 | 351.234 | 70.703 | N1a1a1 | XY |
| Bon005 | 0.039 | 68.615 | 71.021 | N1a1a1 | XX |
| Tep001 | 0.023 | 66.812 | 80.863 | K1a | XY |
| Tep002 | 0.721 | 730.833 | 60.814 | K1a12a | XX |
| Tep003 | 0.694 | 281.963 | 60.849 | N1b1a | XY |
| Tep004 | 0.473 | 391.608 | 61.473 | N1a1a1 | XX |
| Tep006 | 0.267 | 259.879 | 83.585 | N1a1a1 | XY |
See Data S2 for summary statistics for each library and for SNPs used for haplogroup classification.
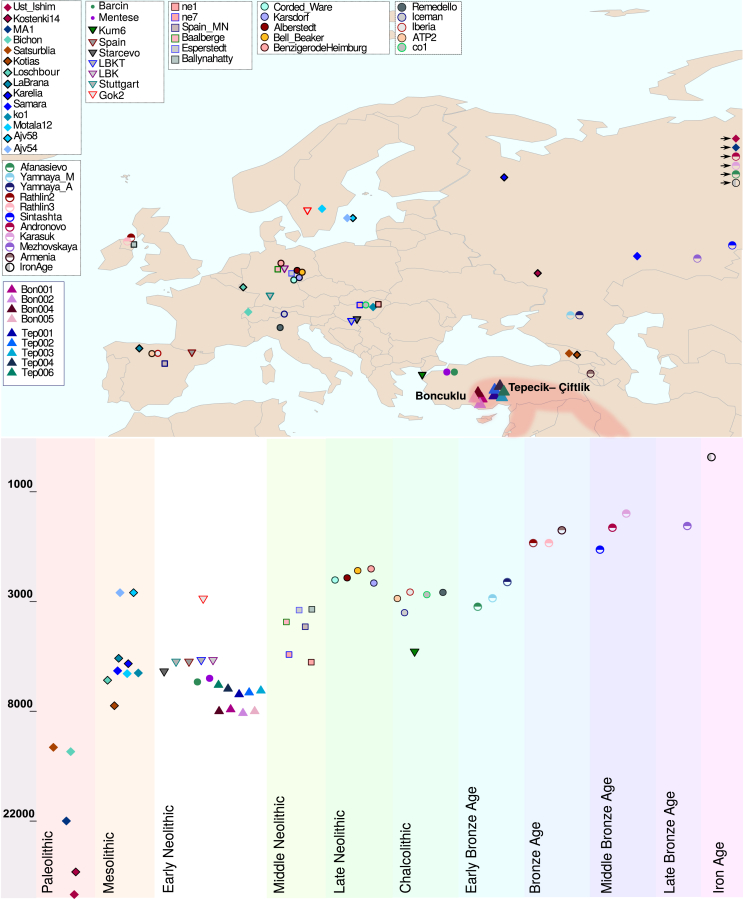
Figure 1.
Geographical Location and Timeline of Ancient Individuals Included in This Study
Map showing the geographical distribution and timeline showing the approximate log-scaled time period (BC) of the ancient individuals used in this study. The colors and symbols for each individual are same with the principal component analysis (PCA). The regions where the Neolithic first emerged and was established are shaded. See Figure S1 for deamination patterns, sequencing efficiency using different methods for the individuals sequenced in this study, and an mtDNA haplogroup network. See also Data S1.
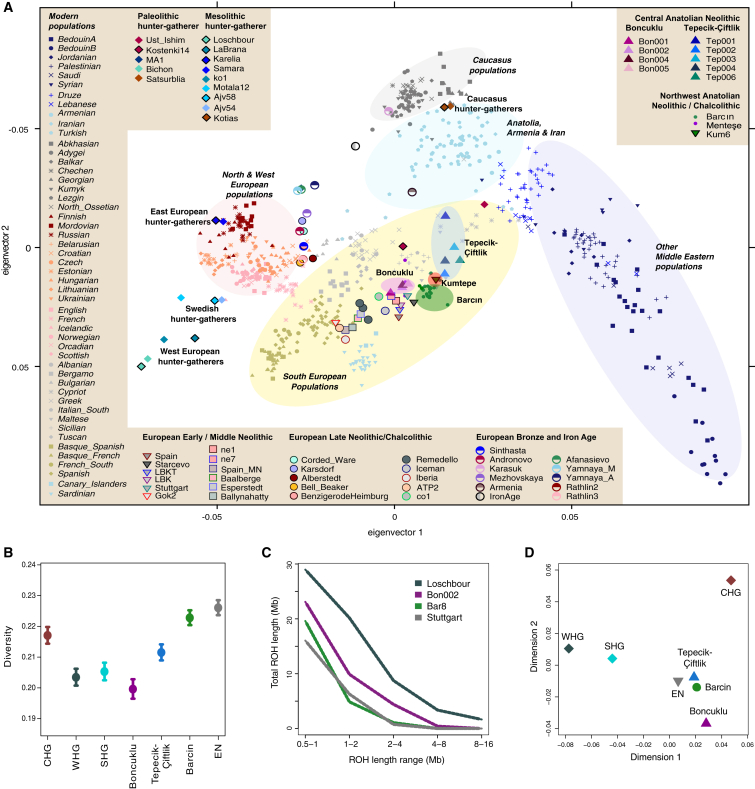
We analyzed the new sequence data in the context of published ancient genetic variation (Figure 1). To discover the genetic affinities among ancient and modern-day individuals, we carried out principal component analysis (PCA). We calculated the principal components from 55 modern-day west Eurasian populations and projected the Boncuklu and Tepecik-Çiftlik individuals, as well as 85 published ancient individuals (Supplemental Experimental Procedures; Table S1), onto the first two principal components (Figure 2A). All individuals from the central Anatolian Neolithic, both the Aceramic Boncuklu group and the Pottery Neolithic Tepecik-Çiftlik group, were positioned within the genetic variation of present day southern European populations, consistent with outgroup f3 statistics (Figure S2; Data S3). Our central Anatolian Neolithic individuals (Boncuklu and Tepecik-Çiftlik), together with later (Pottery) Neolithic and Chalcolithic (Copper Age) individuals from northwest Anatolia (Barcın, Menteşe, and Kumtepe) and with early and middle Neolithic individuals from Europe, formed a distinct cluster to the exclusion of hunter-gatherers from western and eastern Europe (WHG and EHG, respectively), Sweden (SHG), and the Caucasus (CHG) (Figure 2A). Consistent with the PCA, D tests confirmed a clustering of Neolithic and Chalcolithic Anatolians to the exclusion of hunter-gatherers from Europe and the Caucasus. Hunter-gatherers from Europe and the Caucasus also share more alleles with their own groups than with Neolithic Anatolians (Figure S3A; Data S3). Interestingly, although geographically close, the Anatolian Neolithic populations from different time phases each formed discrete but proximate clusters in the PCA. Boncuklu individuals, representing the earliest phase of the Neolithic transition on the central Anatolian plateau, clustered tightly together, implying low genetic diversity within the population. In contrast, Tepecik-Çiftlik individuals, representing the later phase of the Neolithic in central Anatolia, were positioned at a peripheral position within the whole cluster and displayed high within-group diversity (Figure 2A). Pairwise f3 statistics between populations also showed significant differentiation between Boncuklu and Tepecik-Çiftlik populations (permutation test p < 0.05) (Data S3).
Figure 2.
Genetic Structure and Diversity of Central Anatolian Neolithic Populations
(A) PCA on contemporary west Eurasian populations onto which a total of 85 ancient individuals are projected from this study and previous studies. See Table S1 for number of SNPs per individual. Neighboring modern populations and ancient Anatolian populations are shown encircled. Modern population names are in italics.
(B) Conditional nucleotide diversity calculated as the average pairwise mismatches between individuals. Diversities for each group were calculated using the SNPs identified in Yoruba individuals. We used two individuals per group, which yields the highest number of SNPs. Western European, eastern European, Swedish, and Caucasus hunter-gatherers are represented as WHG, EHG, SHG, and CHG, respectively. The European early Neolithic population is denoted with EN. Note that the diversities calculated for CHG and WHG are overestimates, as the individuals representing CHG are separated by three millennia and those representing WHG are separated by >1,000 km (Supplemental Experimental Procedures; Table S1). The error bars represent ±2 SEMs.
(C) Distribution of runs of homozygosity (ROH) for Loschbour (European Mesolithic), Bon002 (Anatolian Aceramic), Bar8 (Anatolian Pottery Neolithic), and Stuttgart (early European Neolithic).
(D) Multidimensional scaling analysis based on the Weir and Cockerham’s Fst calculated between populations using transversions overlapping with African Yoruba individuals. See Data S3 for f3 statistics, D statistics, pairwise mismatch estimates, and Fst estimates; Figure S2 for outgroup f3 statistics with present-day populations; and Figure S3 for D statistics, mean pairwise f3 statistics, and MDS analysis based on pairwise f3 statistics.
To directly gauge levels of genetic diversity in Anatolian Neolithic populations, we calculated conditional nucleotide diversity in Boncuklu, Tepecik-Çiftlik, and Barcın, as well as in European Neolithic and hunter-gatherer populations (Data S3). Herein, we restricted the analysis to transversions identified in Yoruba as in [5] to avoid ascertainment bias, sequencing errors, and post-mortem degradation effects (Supplemental Experimental Procedures; Table S1). The Boncuklu population had remarkably low diversity relative to later ancient Anatolian populations, Tepecik-Çiftlik and Barcın, and European early Neolithic individuals from Hungary (Figure 2B). Comparison of the mean pairwise f3 statistics within populations also supported this result, with conspicuously higher genetic similarity within the Boncuklu group compared to Barcın and Tepecik-Çiftlik (Figure S3B; Data S3; 100% jackknife support). We further investigated short and intermediate runs of homozygosity (0.5–1.6 Mb); this is an indicator of historical effective population size and is expected to be influenced by geographic isolation and bottlenecks, but not recent inbreeding [20]. Our highest quality genome, Bon002 of Boncuklu, had 30% fewer such runs than the central European forager Loschbour, but 25%–40% more such runs relative to high-quality genomes from the Pottery Neolithic, Bar8 of Barcın and Stuttgart of Germany (Supplemental Experimental Procedures; Figure 2C). This supports the notion of a small ancestral population size in the Boncuklu population.
We further evaluated genetic differentiation among Boncuklu, Tepecik-Çiftlik, Barcın, European Mesolithic, and Neolithic populations by calculating Fst (Supplemental Experimental Procedures; Data S3). The results were consistent with the pattern of differentiation in the PCA; particularly, Boncuklu appeared to be distinct from both Tepecik-Çiftlik and Barcın (Fst = 0.020 and 0.030, respectively; Z > 4). A multidimensional scaling (MDS) plot summarizing pairwise Fst values revealed clustering of Tepecik-Çiftlik and Barcın with European Neolithic populations, whereas Boncuklu attained a peripheral location (Figure 2C). This peripheral location is most likely due to high genetic homogeneity and drift in Boncuklu, as such a pattern was not observed in an MDS analysis of mean f3 statistics (Figure S3C).
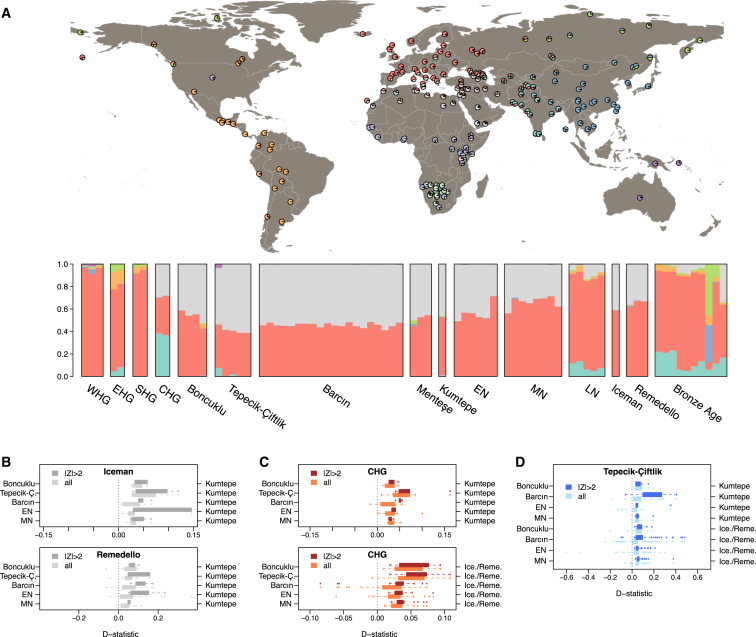
We next conducted ADMIXTURE analysis [21], inferring ancestral clusters from modern-day worldwide populations and estimating the ancestry proportions of each ancient individual based on the inferred ancestral cluster allele frequencies (Figures 3A and S4). With ten clusters (K = 10), ancestry proportions of all Anatolian (Boncuklu, Tepecik-Çiftlik, Barcın, Menteşe, and Kumtepe) and European Neolithic individuals consisted of two components, a “northern component” associated with European hunter-gatherers (WHG, SHG, and EHG) and found in modern-day northern Europe at highest frequency (orange), and a “southern component” found in the modern-day Middle East and North Africa (gray). Notably, Boncuklu displayed lower amounts of this “southern component” compared to individuals from Tepecik-Çiftlik and Barcın (Mann-Whitney U test, p < 0.001; Data S3), implying an influx of “southern component” alleles into late Aceramic and/or Pottery Neolithic settlements in Anatolia. This finding was also in line with higher genetic diversity in the later Neolithic Anatolian populations compared to Boncuklu (Figures 2B and 2C). D statistics results revealed genetic affinity between Caucasus hunter-gatherers (CHGs) and one of the individuals from Tepecik-Çiftlik, Tep003, which was greater than the rest of the individuals from Tepecik-Çiftlik and other Neolithic individuals from central Anatolia, northwest Anatolia, and Europe (Data S3). An admixture graph fitted by modeling gene flow from CHG to Tep003 using TreeMix [22] further confirmed the genetic relationship between Tep003 and CHG individuals (admixture proportion = 0.012, p = 0.002) (Figure S3D). These results show the buildup of genetic diversity during the development of the Neolithic in Anatolia.
Figure 3.
Admixture Analysis and Genetic Affinities among Neolithic/Chalcolithic Populations
(A) ADMIXTURE ancestry components (K = 10) for present-day world populations and for ancient individuals. Admixture fractions are shown on map for modern-day individuals and as bar charts for ancient individuals. See Figure S4 for K = 2 to K = 20 plots with all individuals. Western European, eastern European, Swedish, and Caucasus hunter-gatherers are represented as WHG, EHG, SHG, and CHG, respectively. European early, middle, and late Neolithic populations are denoted with EN, MN, and LN, respectively.
(B–D) Distributions of D statistics calculated as (B) D(Denisova,Iceman;X,Kumtepe) and D(Denisova,Remedello;X,Kumtepe), (C) D(Denisova,CHG;X,Kumtepe) and D(Denisova,CHG;X,Iceman/Remedello), and (D) D(Denisova,Tepecik;X,Kumtepe) and D(Denisova,Tepecik;X,Iceman/Remedello), where X stands for an ancient Anatolian or European early Neolithic (EN) or middle Neolithic (MN) individual, indicated on the left-hand y axis. (See Figure S3 for a plot of D statistics of comparisons of CO1, EHG, and WHG.) In brief, D < 0 indicates higher genetic affinity between the test population (name indicated on the top) and X, and D > 0 indicates higher genetic affinity between the test population and the second population (name indicated on the right-hand y axis). In each comparison, lighter-color boxplots show all D statistics calculated using all available individuals in the populations compared, and darker-color boxplots show only nominally significant D statistics with |Z| ≥ 2. The numbers in the middle indicate the percentage of comparisons where the test population resembles the population indicated on the right-hand y axis (i.e., D > 0). See Data S3 for D statistics.
We next used our data to investigate a more recent case of possible regional migration. Previous work [6] had noted genetic affinity between Kumtepe from northwest Anatolia and the Tyrolean Iceman [23] from northern Italy. We found that the three Remedello individuals from Chalcolithic northern Italy [24], largely contemporary and possibly genetically and culturally affiliated with the Iceman, also had high affinity to Kumtepe in D statistics (Figure 3B; Data S3). A similar tendency for Kumtepe allele sharing was seen for a Chalcolithic individual from Hungary, CO1 [7], but was non-significant (Figure S3E; Data S3). Intriguingly, the Iceman/Remedello group was more similar to Kumtepe than to Boncuklu, Barcın, Tepecik-Çiftlik, or European Neolithic individuals. We further found that both Kumtepe and the Iceman/Remedello group carried more CHG alleles than other Neolithic populations (Figure 3C). This pattern of additional CHG allele sharing simultaneously observed in Iceman/Remedello and in Kumtepe is not mirrored in convergent allele sharing with other European hunter-gatherers (Figures S3F and S3G). We also found that Tepecik-Çiftlik individuals were consistently closer to Iceman/Remedello and to Kumtepe than to any other Anatolian or European early Neolithic population, including their contemporary Barcın and the neighboring Boncuklu (Figure 3D). These results point to gene flow from an eastern source into Chalcolithic Kumtepe and later into Europe, which could have crossed central Anatolia already before the Chalcolithic.
Archaeogenetic studies have shown the existence of two distinct Mesolithic hunter-gatherer gene pools in west Eurasia: hunter-gatherers from Europe, ranging from Iberia to Scandinavia and to the Urals, and hunter-gatherers from the Caucasus [3, 5, 25]. The whereabouts of the so-called “early/first European farmer” gene pool [3], however, had remained unclear. Here we show that the genomes of Aceramic and Pottery Neolithic populations in central Anatolia belonged to the same group as northwestern Neolithic Anatolians and the first European farmers but were distinct from European and Caucasus foragers. The adoption of farming in central Anatolia by indigenous foragers, as suggested for Boncuklu [4, 11], would safely link the “early/first European farmer” gene pool to Anatolian foragers. However, the full geographic range of this forager population still remains to be described.
The low genetic diversity of the Boncuklu population, resembling the low diversity in European hunter-gatherers [5, 25] is interesting (Figures 2B and 2C). It suggests that the population sizes at the very early stages of the Neolithic were not different from those of hunter-gatherers. This accords well with the view of indigenous forager adoption of cultivation and possible local initiation of herding in central Anatolia [4, 11]. Nearly 1,500 years later, Tepecik-Çiftlik and Barcın, fully established Neolithic populations practicing mixed farming (and within 200 km east and 400 km northwest of Boncuklu, respectively), were significantly more diverse (Figure 2B). Part of this increased genetic diversity could be linked to (1) putative southern gene flow (Figure 3A) that could be related to the Aceramic Neolithic to Pottery Neolithic transition in the Neolithic Levant or could be related to widespread interactions in the late Aceramic Neolithic between central Anatolia and the Fertile Crescent in the late Pre-Pottery Neolithic B [26]; (2) migration from the east related to similar factors of inter-regional exchanges (Figure S3D); and (3) admixture among local populations. Southern and eastern gene flow into Tepecik-Çiftlik is consistent with the site’s presumed role as an obsidian hub and its cultural links with the Levant and might have started already before the Pottery Neolithic [15, 16]. For Barcın, these results are also in line with archaeological evidence indicating cultural influx from central Anatolia [27]. This diverse Neolithic population most likely served as one of the sources for the well-documented wave of Neolithic migration to Europe [8, 9].
Post-Neolithic contacts between parts of Anatolia and central Europe are a matter of discussion. Genetic affinity between a Chalcolithic group in northwest Anatolia represented by Kum6 of Kumtepe and by a group represented by the Tyrolean Iceman was earlier explained by gene flow post-dating the earlier stages of the Neolithic in Europe [6]. But it has alternatively been interpreted as the Iceman representing a relic of the first migratory event from Anatolia [9]. As we have shown in this paper, individuals of the Chalcolithic Remedello group [24] from northern Italy also share strong affinity with Kumtepe. This pattern may be explained with one out of four scenarios: (1) Iceman/Remedello representing a relict population stemming from an early farmer migratory event, (2) late-Neolithic/Chalcolithic back-migration from central Europe into Anatolia, (3) a third source-population admixing with both the population represented by Iceman/Remedello and the population represented by Kumtepe, and (4) secondary late-Neolithic/Chalcolithic migration from Anatolia. Because the Iceman/Remedello group is genetically closer to Chalcolithic Kumtepe than to earlier Anatolian Neolithic populations, including Boncuklu and Barcın, the first scenario seems unlikely. The fact that both Iceman/Remedello and Kumtepe display shared drift with Caucasus hunter-gatherers, independent of the Bronze Age Yamnaya expansions [24, 28], also argues against Iceman/Remedello being a relict population. Second, as Kumtepe predates the Iceman/Remedello group by some 1,300 years, back migration is an unlikely explanation. Finally, the Tepecik-Çiftlik population shows significant affinity to the Iceman/Remedello group and Kumtepe relative to other Anatolian and European Neolithic populations (Figure 3D); but Tepecik-Çiftlik also predates Iceman/Remedello by approximately 3,000 years. This implies gene flow events from Tepecik-Çiftlik-related populations into the Kumtepe-related west Anatolian populations, as predicted by archaeological evidence [29], and further gene flow that reached northern Italy by the fourth millennium BC. We propose an additional, yet undescribed, gene flow process from Anatolia into Europe as a better explanation than a contribution from a hypothetical third source into Neolithic central Anatolia, Chalcolithic northwest Anatolia, and Chalcolithic central Europe. Thus, Neolithic population dynamics that initiated in the Anatolian region resulted in multiple waves of expansion and admixture in west Eurasia.
Experimental Procedures
DNA was isolated from petrous bone and teeth samples of nine ancient individuals. Double-stranded libraries were prepared and sequenced on Illumina HiSeq2500 and X platforms. Paired-end reads were merged, and adapters were removed. Reads were mapped to the human reference genome version hg18 and hs37d5 using BWA 0.7.12 [30]. Published ancient genomes were also mapped with the same parameters. Data was authenticated using four different methods [31, 32, 33, 34]. Mitochondrial haplogroups were discovered using PhyloTree and Haplofind [35, 36]. Biological sex was determined using the Ry method [2, 37]. Principal component analysis was conducted using Eigensoft [38], and model-based clustering was performed using ADMIXTURE [21]. For ADMIXTURE analysis, ancestral components were determined using modern populations, and cluster memberships of each ancient individual were then inferred on the basis of these ancestral allele frequencies as in [39]. Outgroup f3 statistics were computed using popstats.py (https://github.com/pontussk/popstats). D statistics were calculated using qpDstat program of ADMIXTOOLS [40]. For computation of conditional nucleotide diversity, two approximately contemporaneous individuals with the highest quality genomes were selected to represent each group, and the average number of mismatches per each site overlapping with African Yoruba population between two individuals was calculated as in [41]. Weir and Cockerham’s Fst was calculated using popstats.py (https://github.com/pontussk/popstats). Runs of homozygosity for four high-quality genomes were calculated using PLINK [42]. See the Supplemental Experimental Procedures for details.
Author Contributions
A. Götherström, M.J., J.S., İ.T., and M.S. designed and supervised the study; A.O., F. Ö., R.Y., M.K., E.D., E.Y., and N.D.D. performed the experiments; G.M.K. analyzed population-genetic data with contributions from M.S., A.O., T.G., H.M.D., A. Ghalichi, D.K., S.C.A., P.P., R.Y., E.Y., and N.D.D.; D.B., A.M.B., A.F., J.P., G.M., Y.S.E., Y.G.Ç., and E.B. excavated the samples, performed osteological assessments, and provided archaeological interpretations; and G.M.K., A.O., F.Ö., T.G., D.B., A.M.B., R.Y., A. Ghalichi, E.B., A. Götherström, M.J., J.S., İ.T., and M.S. wrote the manuscript with input from all authors.
Acknowledgments
We thank C. Bilgin, A. Birand, R. Özbal, C. Knüsel, M. Özdoğan, F.S. Quinto, and two anonymous referees for helpful suggestions; İ.D. Toker and the Konya and Niğde Museums for support; A. Munters for assistance; and EMBO (Short-Term Fellowship to G.M.K.), Hacettepe University Scientific Research Projects Coordination Unit (project no. 13G602003 to A.M.B), İstanbul University Scientific Research Projects Unit (project no. 52349 to E.B), Australian Research Council (grant no. DP120100969 to A.F.), British Academy Research Development Award (grant no. BR100077 to D.B.), British Institute at Ankara grants 2012–2015 (to D.B.), National Geographic GEFNE 1–11 (to D.B.), Wainwright Fund University of Oxford (to D.B.), TÜBİTAK (grant no. 114Z927 to M.S.), TÜBA (GEBİP Prize to M.S), Sci. Acad. Turkey (BAGEP Prize to M.S.), METU (internal grants to İ.T. and M.S.), and ERC (starting grant no. 311413 to M.J.). Sequencing was conducted at the Uppsala University SNP&SEQ Technology Platform. Computational analyses were carried out at the Swedish National Infrastructure for Computing (SNIC-UPPMAX, projects b2013236, b2013240, b2015307, and b2015364).
Published: August 4, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and three data sets and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2016.07.057.
Contributor Information
İnci Togan, Email: togan@metu.edu.tr.
Mehmet Somel, Email: msomel@metu.edu.tr.
Jan Storå, Email: jan.stora@ofl.su.se.
Mattias Jakobsson, Email: mattias.jakobsson@ebc.uu.se.
Anders Götherström, Email: anders.gotherstrom@arklab.su.se.
Accession Numbers
The accession number for the genome data produced in this study is European Nucleotide Archive: PRJEB14675.
Supplemental Information
References
- 1.Bramanti B., Thomas M.G., Haak W., Unterlaender M., Jores P., Tambets K., Antanaitis-Jacobs I., Haidle M.N., Jankauskas R., Kind C.-J. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 2.Skoglund P., Malmström H., Raghavan M., Storå J., Hall P., Willerslev E., Gilbert M.T.P., Götherström A., Jakobsson M. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 3.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baird D. The Late Epipaleolithic, Neolithic, and Chalcolithic of the Anatolian Plateau, 13,000–4000 BC. In: Potts D., editor. A Companion to the Archaeology of the Ancient Near East. Wiley-Blackwell; 2012. pp. 431–466. [Google Scholar]
- 5.Skoglund P., Malmström H., Omrak A., Raghavan M., Valdiosera C., Günther T., Hall P., Tambets K., Parik J., Sjögren K.-G. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 6.Omrak A., Günther T., Valdiosera C., Svensson E.M., Malmström H., Kiesewetter H., Aylward W., Storå J., Jakobsson M., Götherström A. Genomic evidence establishes Anatolia as the source of the European neolithic gene pool. Curr. Biol. 2016;26:270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmanová Z., Kreutzer S., Hellenthal G., Sell C., Diekmann Y., Díez-Del-Molino D., van Dorp L., López S., Kousathanas A., Link V. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. USA. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asouti E., Fuller D.Q. A contextual approach to the emergence of agriculture in Southwest Asia: reconstructing early neolithic plant-food production. Curr. Anthropol. 2013;54:299–345. [Google Scholar]
- 11.Baird D., Fairbairn A., Martin L., Middleton C. The Boncuklu project: the origins of sedentism, cultivation and herding in central Anatolia. In: Özdoğan M., Başgelen N., Kuniholm P., editors. The Neolithic in Turkey: New Excavations and New Research, Volume 3, Central Turkey. Archaeology and Art Publications; 2012. pp. 219–244. [Google Scholar]
- 12.Stiner M.C., Buitenhuis H., Duru G., Kuhn S.L., Mentzer S.M., Munro N.D., Pöllath N., Quade J., Tsartsidou G., Özbaşaran M. A forager-herder trade-off, from broad-spectrum hunting to sheep management at Aşıklı Höyük, Turkey. Proc. Natl. Acad. Sci. USA. 2014;111:8404–8409. doi: 10.1073/pnas.1322723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Özdoğan M. Anatolia: from the pre-pottery neolithic to the end of the early bronze age (10,500–2000 BCE) In: Renfrew C., Bahn P., editors. The Cambridge World Prehistory. Cambridge University Press; 2014. pp. 1508–1544. [Google Scholar]
- 14.Weninger B., Clare L., Gerritsen F., Horejs B., Krauß R., Linstädter J., Özbal R., Rohling E.J. Neolithisation of the Aegean and Southeast Europe during the 6600–6000 calBC period of rapid climate change. Doc. Praehist. 2014;XLI:1–32. [Google Scholar]
- 15.Bıçakcı E. Tepecik-Çiftlik kazısı 2010 yılı çalışmaları. In: Donmez H., Otgun O., editors. 33. Kazı Sonucları Toplantısı 1.Cilt. Kültür Varlıkları ve Müzeler Genel Müdürlüğü; 2011. pp. 69–89. [Google Scholar]
- 16.Bıçakçı E., Godon M., Çakan Y.G. Tepecik-Çiftlik. In: Ozdoğan M., Başgelen N., Kuniholm P., editors. Neolithic in Turkey: New Excavations and New Research, Volume 3, Central Turkey. Archaeology and Art Publications; 2012. pp. 89–134. [Google Scholar]
- 17.Brandt G., Haak W., Adler C.J., Roth C., Szécsényi-Nagy A., Karimnia S., Möller-Rieker S., Meller H., Ganslmeier R., Friederich S., Genographic Consortium Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013;342:257–261. doi: 10.1126/science.1241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szécsényi-Nagy A., Brandt G., Haak W., Keerl V., Jakucs J., Möller-Rieker S., Köhler K., Mende B.G., Oross K., Marton T. Tracing the genetic origin of Europe’s first farmers reveals insights into their social organization. Proc. Biol. Sci. 2015;282:20150339. doi: 10.1098/rspb.2015.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A., Li J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A., Graefen A., Ball M., Matzas M., Boisguerin V., Maixner F., Leidinger P., Backes C., Khairat R., Forster M. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 24.Allentoft M.E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P.B., Schroeder H., Ahlström T., Vinner L. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 25.Jones E.R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R.L., Gallego Llorente M., Cassidy L.M., Gamba C. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Özdoğan M. An alternative approach in tracing changes in demographic composition. In: Bar-Yosef O., Bocquet-Appel J.P., editors. The Neolithic Demographic Transition and Its Consequences. Springer; 2008. pp. 139–178. [Google Scholar]
- 27.Thissen L., Özbal H., Türkekul A., Gerritsen F., Özbal R. The land of milk? Approaching dietary preferences of Late Neolithic communities in NW Anatolia. Leiden J. Pottery Stud. 2010;26:157–172. [Google Scholar]
- 28.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Özdoğan M. Archaeological evidence on the westward expansion of farming communities from Eastern Anatolia to the Aegean and the Balkans. Curr. Anthropol. 2011;52:S415–S430. [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skoglund P., Northoff B.H., Shunkov M.V., Derevianko A.P., Pääbo S., Krause J., Jakobsson M. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Q., Meyer M., Gao X., Stenzel U., Burbano H.A., Kelso J., Pääbo S. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. USA. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green R.E., Malaspinas A.S., Krause J., Briggs A.W., Johnson P.L.F., Uhler C., Meyer M., Good J.M., Maricic T., Stenzel U. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vianello D., Sevini F., Castellani G., Lomartire L., Capri M., Franceschi C. HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment. Hum. Mutat. 2013;34:1189–1194. doi: 10.1002/humu.22356. [DOI] [PubMed] [Google Scholar]
- 36.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 37.Skoglund P., Storå J., Götherström A., Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. [Google Scholar]
- 38.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikora M., Carpenter M.L., Moreno-Estrada A., Henn B.M., Underhill P.A., Sánchez-Quinto F., Zara I., Pitzalis M., Sidore C., Busonero F. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 2014;10:e1004353. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Günther T., Valdiosera C., Malmström H., Ureña I., Rodriguez-Varela R., Sverrisdóttir Ó.O., Daskalaki E.A., Skoglund P., Naidoo T., Svensson E.M. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. USA. 2015;112:11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.