Abstract
The identification and characterization of new human monoclonal antibodies (hMAbs) able to neutralize primary human immunodeficiency virus type 1 (HIV-1) isolates from different subtypes may help in our understanding of the mechanisms of virus entry and neutralization and in the development of entry inhibitors and vaccines. For enhanced selection of broadly cross-reactive antibodies, soluble HIV-1 envelope glycoproteins (Envs proteins) from two isolates complexed with two-domain soluble CD4 (sCD4) were alternated during panning of a phage-displayed human antibody library; these two Env proteins (89.6 and IIIB gp140s), and one additional Env (JR-FL gp120) alone and complexed with sCD4 were used for screening. An antibody with relatively long HCDR3 (17 residues), designated m14, was identified that bound to all antigens and neutralized heterologous HIV-1 isolates in multiple assay formats. Fab m14 potently neutralized selected well-characterized subtype B isolates, including JRCSF, 89.6, IIIB, and Yu2. Immunoglobulin G1 (IgG1) m14 was more potent than Fab m14 and neutralized 7 of 10 other clade B isolates; notably, although the potency was on average significantly lower than that of IgG1 b12, IgG1 m14 neutralized two of the isolates with significantly lower 50% inhibitory concentrations than did IgG1 b12. IgG1 m14 neutralized four of four selected clade C isolates with potency higher than that of IgG1 b12. It also neutralized 7 of 17 clade C isolates from southern Africa that were difficult to neutralize with other hMAbs and sCD4. IgG1 m14 neutralized four of seven primary HIV-1 isolates from other clades (A, D, E, and F) much more efficiently than did IgG1 b12; for the other three isolates, IgG b12 was much more potent. Fab m14 bound with high (nanomolar range) affinity to gp120 and gp140 from various isolates; its binding was reduced by soluble CD4 and antibodies recognizing the CD4 binding site (CD4bs) on gp120, and its footprint as defined by alanine-scanning mutagenesis overlaps that of b12. These results suggest that m14 is a novel CD4bs cross-reactive HIV-1-neutralizing antibody that exhibits a different inhibitory profile compared to the only known potent broadly neutralizing CD4bs human antibody, b12, and may have implications for our understanding of the mechanisms of immune evasion and for the development of inhibitors and vaccines.
Major problems in prevention and treatment of human immunodeficiency virus type 1 HIV-1 infections are the ability of the virus to rapidly generate mutants resistant to immune responses and drugs and the side effects of antiretroviral drugs currently in use. Several human monoclonal antibodies (hMAbs) exhibit potent and broad HIV-1-neutralizing activity in vitro and can prevent HIV-1 infection in animal models (11, 25, 67). A recent clinical trial suggested that two of these broadly HIV-1-neutralizing hMAbs, 2F5 and 2G12, are without side effects in humans (1, 59). However, the potency of 2F5 and 2G12 used in combination in this clinical trial was significantly lower than that of currently used highly active antiretroviral therapy regimens, and relapses did occur (59). Further increases in the potency of the currently available broadly HIV-1-neutralizing hMAbs and development of new neutralizing hMAbs might help in the development of better approaches to the prevention and treatment of HIV-1 infection.
The molecular mechanisms that determine the antibody potency and breadth of HIV-1 neutralization have been studied most extensively with b12, which competes with CD4 and antibodies with epitopes that overlap the CD4 binding site (CD4bs) on gp120 (4, 12, 14, 44, 49, 53, 76). Many human CD4bs antibodies have been characterized, including Fab b6 (49, 53), 15e (27), F105 (64) F91 (40), 1125H (65), 21h (27, 62), and 654-30D (32). These CD4bs antibodies frequently exhibit broad reactivity with monomeric gp120 but do not neutralize many HIV-1 isolates from different clades as b12 does (19, 20, 53). The difference in neutralizing activity was ascribed to the ability of b12 but not other CD4bs antibodies to bind well to the trimeric Env on the virion surface. It appears that b12 binds with high affinity to both monomeric and trimeric forms of gp120 whereas other CD4bs antibodies bind well predominantly to the monomeric form. B12 is the only CD4bs human MAb known to exhibit broad and potent HIV-1-neutralizing activity. Identification and characterization of additional broadly neutralizing CD4bs antibodies could help to determine key molecular properties required for such an activity.
Here we described the identification of a new CD4bs hMAb, m14 that was selected by a method termed sequential antigen panning (SAP), which is based on alternating the antigen during the panning of phage display libraries. SAP leads to enhanced selection of antibodies that bind epitopes conserved among the isolates used for panning and screening (70). M14 was selected from a human Fab phage display library by using SAP against gp14089.6-soluble CD4 (sCD4) and gp140IIIB-sCD4 followed by screening with gp14089.6, gp120JR-FL, and gp140IIIB and their complexes with sCD4. The antibody inhibited HIV-1 entry mediated by Env proteins of more than 20 primary HIV-1 isolates from several clades with varying potencies and exhibited a different inhibitory profile from that of immunoglobulin G1 (IgG1) b12.
MATERIALS AND METHODS
Cells, viruses, plasmids, sCD4, gp120, gp140, and antibodies.
293T cells were purchased from American Type Culture Collection. HIV-1 isolates were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP). Alanine-scanning mutants were obtained by introducing single alanine substitutions into the pSVIIIexE7pAJR-CSF background (58). Two-domain sCD4 was obtained from the NIH ARRRP. Recombinant gp140s from primary isolates were produced as described previously (70). Purified gp12089.6, gp14089.6, and gp140IIIB were produced by using recombinant vaccinia virus (a gift of R. Doms, University of Pennsylvania, Philadelphia, Pa.) and a combination of lentil lectin affinity chromatography and size exclusion chromatography. Recombinant gp120JR-FL was provided by A. Schultz and N. Miller (National Institute of Allergy and Infections Diseases, Bethesda, Md.). The hMAbs b12 (14), X5 (41), m16 (72), and m18 (70) were produced in our laboratories; 17b (63), 48d (63), A32, and 48e were gifts from J. Robinson (Tulane University Medical Center, New Orleans, La.); 2G12 was a gift from H. Katinger (Institute of Applied Microbiology, University for Agricultural Sciences, Vienna, Austria); and F105 (50) was obtained through the NIH ARRRP from M. Posner and L. Cavacini. The following antibodies were purchased: polyclonal sheep anti-gp120 antibody D7324 (Sigma), horseradish peroxidase (HRP)-conjugated mouse anti-M13 MAb (Pharmacia, Uppsala, Sweden), and HRP-conjugated polyclonal anti-human IgG F(ab′)2 antibodies (Jackson ImmunoResearch, Westgrove, Pa.).
SAP and screening.
The phage library was constructed using pComb3H phagemid vector and 30 ml of bone marrow obtained from three long-term nonprogressors whose sera exhibited the broadest and most potent HIV-1 neutralization among 37 HIV-infected individuals (provided by T. Evans, University of California, Davis, Calif. [70]). Phage (5 × 1012 CFU/ml) were preadsorbed on streptavidin-M-280 Dynabeads in phosphate-buffered saline (PBS) for 1 h at room temperature and then subjected to depletion for 1 h at 37°C in an immunotube (Nunc, Roskilde, Denmark) coated with 10 μg of sCD4 per ml. The depleted phage library was incubated with 50 nM biotinylated HIV-1 envelope glycoprotein gp14089.6 complexed with sCD4 in solution (molar ratio of gp14089.6 to sCD4, 1:1) for 2 h at room temperature with gentle agitation. Phages that bound to biotinylated Env were separated from the phage library by using streptavidin-M280-Dynabeads and a magnetic separator (Dynal). After being washed 20 times with 1 ml of PBS containing 0.1% Tween 20 and another 20 times with 1 ml of PBS, bound phage were eluted from the beads using by 100 mM triethanolamine and neutralized with 1 M Tris- HCl (pH 7.5). For the second round of panning, 10 nM (2 nM for the third round) biotinylated gp14089.6 complexed with sCD4 (1:1 at the molar level) was used as the antigen. For the fourth round of panning, 2 nM biotinylated HIV-1 envelope glycoprotein gp140IIIB complexed with sCD4 (1:1 at the molar level) was used as the antigen. After the fourth round, 20 individual clones were screened for binding to gp14089.6, gp120JR-FL, and gp140IIIB and their complexes with sCD4 by using phage enzyme-linked immunosorbent assay (ELISA).
Preparation of Soluble Fab and Binding Assay.
Soluble Fab was produced as described previously (3). Protein G columns were used for purification. Phage ELISA has been described previously (25). Binding ELISA with soluble Fab m14 and recombinant HIV-1 gp120s or gp140s from different isolates were performed by using 96-well Nunc- Immuno Maxisorp surface plates (Nalge Nunc International, Roskilde, Denmark) as described previously (41). Competition ELISAs of Fab m14, Fab b12, and IgG 17b with other anti-gp120 antibodies and sCD4 were carried out using the D7324 capture assay as follows. gp120JRFL (1 μg/ml) was captured by polyclonal sheep anti-gp120 antibody D7324 (5 μg/ml) coated on 96-well plates. Following the addition of threefold serially diluted Fab m14, m16, m18, IgG b12, IgG 17b, Fab X5, F105, 48d, 48e, 2G12, A32, and sCD4, an equal volume of biotinylated Fab m14 at a concentration which led to 70% maximum binding was simultaneously added to each well. Bound biotinylated Fab m14 was detected by streptavidin-HRP and measured via its optical density at 405 nm.
The kinetics of m14 and b12 binding was measured by using a Biacore 1000 optical instrument (Pharmacia, Piscataway, NJ). An anti-gp41 hMAb Fab (m32) developed recently in our laboratory (M.-Y. Zhang and D. S. Dimitrov, unpublished data) was immobilized on a (CM5) sensor chip using carbodiimide coupling chemistry. gp140s (200 nM) was injected at a flow rate of 10 μl/min, and different concentrations of antibodies (Fabs) were then injected at a flow rate of 30 μl/min using PBS buffer (pH 7.4) with 0.05% Tween 20. The control surface was prepared similarly, and the experiment was performed by injecting solutions containing different concentrations of antibodies, using running buffer in lieu of gp140s. All the sensograms were corrected by subtracting the low signal from the control reference surface. The association and dissociation phase data were fitted simultaneously to a 1:1 Langumir global model by using the nonlinear data analysis program BIAevaluation 3.2.
HIV-1 Env clones and pseudovirus preparation.
Viruses pseudotyped with Envproteins from HIV-1 primary isolates representing HIV-1 group M, clades A to F, and laboratory-adapted HIV-1 isolates HxB2 and JRCSF were used in this study. Plasmids used for expression of various Env proteins were obtained through the NIH ARRRP from B. Hahn (University of Alabama at Birmingham), and the remaining clones are described below. NYU1545, 93BR029, and CA1-136 (CRF 11_cpx) were obtained by nested PCR using genomic DNA from infected human peripheral blood mononuclear cells (PBMCs) provided by S. Zolla-Pazner and cloned into pSV7d using the methodo described previously (47, 74). Clone XJ-C19 was obtained similarly from human PBMCs provided by T. Shao (Centers for Disease Control and Prevention, Beijing, China). Clone MJ4 was previously described (43) and obtained by reverse transcription-PCR using RNA extracted from supernatant from culture of SHIV-MJ4 propagated in human PBMCs, provided by M. Lewis (Southern Research Institute) followed by cloning into pSV7d. The following clones were described previously: GX-C44 and GX-E14 (22); Z2Z6 (74); VI14004, VI1399, VI1273, and VI423 (70); MACS#6, MACS#8, MACS#4, and MACS# (74), R2 (52, 73); MN-TCLA (45, 46); and MN-P (34). The clade C viruses from Southern Africa are described in reference 9.
Cloning of HIV-1 envelope genes and preparation of pseudoviruses have been described previously (72). Briefly, pseudotyped viruses were prepared by cotransfection of 70 to 80% confluent 293T cells with pNL4-3.luc.E-R- and pSV7d-env plasmid using the calcium phosphate/HEPES buffer technique, as specified by the manufacturer (Promega). At 16 h after the transfection, the medium was removed and replaced with medium supplemented with 0.1 mM sodium butyrate (Sigma). Cells were allowed to grow for an additional 24 h. The supernatant was harvested, centrifuged at 16,000 rpm for 5 min at 4°C in an SS-34 rotor, filtered through a 0.45-μm-pore-size filter, and either used fresh or kept frozen at −80°C.
HIV-1 neutralization assays.
Four HIV-1 neutralization assays were used in this study. In the first assay format, single-round infectious molecular clones, produced by envelope complementation as described above, were used. The degree of virus neutralization by antibody was achieved by measuring luciferase activity as described previously (assay 2 in reference 71). Briefly, 2 × 104 U87.CD4.CCR5.CXCR4 cells in 100 ml of Dulbecco minimal essential medium were added to microplate wells and incubated for 24 h at 37°C in 5% (vol/vol) CO2. A 100-μl volume of medium containing virus was mixed with various amounts of antibody, incubated for 1 h at 37°C, added to the cells, and incubated for a further 3 days. The wells were aspirated and washed once with PBS, and 60 μl of luciferase cell culture lysis reagent (Promega, Madison, Wis.) was added. The wells were scraped, the lysate was mixed by pipetting, 50 μl was transferred to a round-bottom plate, and the plate was centrifuged at 1,800 × g for 10 min at 4°C. A 20-μl volume was transferred to an opaque assay plate (Corning), and the luciferase activity was measured with a luminometer by using luciferase assay reagent (Promega). The second neutralization assay is based on infection of PBMCs with infectious viruses and measurement of the amount of reverse transcriptase 7 days after infection. The procedure is as follows. Antibodies (100 μl) diluted in complete RPMI 1640 with interleukin-2 were incubated for 30 min at 37°C with 50 μl of virus containing 100 50% tissue culture infective doses and added to 50 μl of phytohemagglutinin-activated PBMC (1 × 106) in complete RPMI 1640 with interleukin-2. The calculated neutralization titers refer to the antibody concentration present during this incubation step. Triplicate samples were taken on day 7 for the reverse transcriptase assay. The third assay, the pseudotype virus neutralization assay, was performed in triplicate by using a luciferase reporter HIV-1 Env pseudotyping system and HOS CD4+ CCR5+ or HOS CD4+ CXCR4+ cells as previously described (70).
In the fourth assay, neutralization was measured as a function of the reductions in luciferase reporter gene expression after multiple rounds of virus replication in 5.25.EGFP.Luc.M7 cells (39). This cell line is a genetically engineered clone of CEMx174 that expresses multiple entry receptors (CD4, CXCR4, and GPR15/Bob) and was transduced to express CCR5 (7). The cells also possess Tat-responsive reporter genes for luciferase (Luc) and green fluorescence protein. The cells were maintained in growth medium (RPMI 1640, 12% heat-inactivated fetal bovine serum, 50 μg of gentamicin/ml) containing puromycin (0.5 μg/ml), G418 (300 μg/ml), and hygromycin (200 μg/ml) to preserve the CCR5 and reporter gene plasmids. For neutralization assays, 5,000 50% tissue culture infective doses of virus was incubated with serial dilutions of test samples in triplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottom culture plates. The cells were suspended at a density of 5 × 105/ml in growth medium containing DEAE-dextran (10 μg/ml) but lacking puromycin, G418, and hygromycin. Then cells (100 μl) were added to each well. One set of control wells received cells plus virus (virus control), and another set received cells only (background control). The plates were incubated until approximately 10% of the cells in the virus control wells were positive for green fluorescence protein expression by fluorescence microscopy (approximately 3 days). At this time, a 100-μl suspension of cells was transferred to a 96-well white solid plate (Costar) for measurement of luminescence using Bright Glo substrate solution (Promega) and a model Victor 2 luminometer (Perkin-Elmer, Shelton, Conn.). Neutralization titers are defined as the dilution at which relative luminescence units (RLU) were reduced by 50% compared to virus control wells after subtraction of background RLU.
RESULTS
Selection of a phage Fab (m14) by SAP.
We hypothesized that by alternating antigens during panning of phage display libraries and screening the panned libraries using different antigens, the selected phage will display Fabs against conserved epitopes shared among all antigens used during the entire selection process. Complexes of two different recombinant soluble Env proteins (gp14089.6 and gp140IIIB) with two-domain sCD4 were used as antigens for phage library panning as described in Materials and Methods. After four rounds of panning, screening of individual phage clones was performed by phage ELISA with gp14089.6, gp120JR-FL, and gp140IIIB and their complexes with sCD4. Two clones were selected based on their significant binding to all six antigens used for screening, including sCD4-gp120 and gp120 from an HIV-1 isolate (JR-FL) which was not used for panning and has significant sequence differences compared to 89.6 and IIIB. One phage clone, designated m14, was selected for further characterization because of technical problems with the production of the other clone (m12). In a control experiment to assess the efficiency of the SAP methodology, the panning was performed with only one antigen (sCD4-gp14089.6). In this case, none of the clones tested bound to every one of gp140IIIB, sCD4-gp140IIIB, gp120JR-FL, and sCD4-gp120JR-FL.
Potent neutralizing activity of m14 against selected isolates.
To compare the potency (concentrations required to achieve 50% [IC50] and 90% [IC90] inhibition) of Fab m14 and Fab b12 for some commonly studied clade B isolates, we first used an assay based on a single-round infectious virus and U87 cells (Table 1). In this assay, Fab m14 exhibited high potency, with IC50s and IC90s ranging from 1 to 8 and from 7 to 50 μg/ml, respectively. Fab b12 was as potent as Fab m14 for the primary isolate Yu2, severalfold more potent for two other clade B primary isolates, 89.6 and JRCSF, and much more potent for the laboratory-adapted strain HxB2. Note that m14 neutralizes virus entry mediated by the Env proteins from two of the isolates (89.6 and HxB2) that were used for the SAP procedure. These findings suggest that m14 can potently neutralize several clade B isolates, although on average Fab b12 and Fab X5 were more potent for the tested clade B isolates (Tables 1 and 2 and data not shown).
TABLE 1.
Neutralization of selected HIV-1 isolates by Fabs m14 and b12a
| Virus | Subtype | Fab b12
|
Fab m14
|
||
|---|---|---|---|---|---|
| IC50 (μg/ml) | IC90 (μg/ml) | IC50 (μg/ml) | IC90 (μg/ml) | ||
| HxB2 | B | 0.01 | 0.2 | 1 | 7 |
| 89.6 | B | 0.3 | 5 | 8 | 25 |
| JRCSF | B | 0.5 | 15 | 5 | 50 |
| Yu2 | B | 8 | 33 | 7 | 33 |
A neutralization assay using U87 and single-round infectious virus in a luciferase assay, as described in Materials and Methods (assay 1), was used.
TABLE 2.
Neutralization of subtype B HIV-1 primary isolates by b12 and m14a
| HIV-1 isolate | IC50 (μg/ml) of:
|
||||
|---|---|---|---|---|---|
| Fab m14 | IgG1 m14 | IgG1 b12 | IgG1 m18 | Fab X5 | |
| R2 | 6 | 3 | 25 | 3 | 3.6 |
| MN-P | 200 | 120 | 2.1 | >200 | NDb |
| MACS#6 | 100 | 200 | 6 | >200 | ≥100 |
| MACS#8 | >200 | 200 | 7 | >200 | 1.9 |
| MACS#4 | 200 | 100 | <3 | 100 | 2.8 |
| MACS#9 | >200 | 200 | 50 | >200 | 100 |
| VI1399 | >200 | >200 | <3 | >200 | 3.9 |
| VI1273 | >200 | >200 | <3 | >200 | 6.2 |
| VI423 | >200 | >200 | 6 | >100 | 12 |
| MN-TCLA | >100 | 30 | 3 | 15 | ND |
A neutralization assay using HOS cells and single-round infectious virus in a luciferase assay, as described in Materials and Methods, was used.
ND, not done.
Differential neutralization activity of Fab m14, IgG1 b12, and Fab X5 against primary HIV-1 isolates from different clades.
To evaluate the possibility for differential neutralization activity of m14 and b12, we measured their ability to inhibit virus entry mediated by Env proteins of primary isolates from different clades. The higher potency of b12 than m14 for the selected clade B viruses described above prompted us to evaluate the inhibitory activity of m14 for some isolates from other clades for which b12 was previously shown to be of low inhibitory activity. We specifically evaluated the Fab m14 inhibitory activity against one isolate from clade A (RW009) and one from clade C (BR025), which were only weakly (IC50 > 200 μg/ml for RW009 and IC90 > 200 μg/ml for BR025) neutralized by IgG1 b12 (41). For this evaluation, the experiments were performed by an assay based on spreading infection of PBMCs, as used in our previous study (41). We found that at 90 μg/ml, m14 inhibited 83 and 92% of RW009 and BR025 infectivity respectively (data not shown). For another clade B isolate (Bal), m14 was weakly inhibitory (21% at 90 μg/ml) (data not shown), while in our previous study (41) we found that IgG1 b12 potently (IC50 = 4 μg/ml) inhibits infection by Bal. Fab X5 was used as a “calibrating” control antibody that potently (>95% at 100 μg/ml) inhibited all three isolates, in agreement with the results from our previous study (IC90 = 20 to 56 μg/ml) (41). These findings suggested that m14 can exhibit a different inhibitory profile from b12.
Higher neutralizing activity of IgG1 m14 than that of Fab m14 and a differential inhibitory profile with IgG1 b12.
To further increase the potency of Fab m14 and compare its inhibitory activity to other antibodies in the IgG1 format, we constructed IgG1 m14 and compared its activity with those of Fab m14 and IgG1 b12 for several primary isolates from different clades. The IgG1 m14 was on average more efficient (lower IC50) than Fab m14, probably due to the increase in avidity (Table 3).
TABLE 3.
Differential neutralization of HIV-1 group M isolates by IgG1 m14 and IgG1 b12a
| HIV-1 isolate | Subtype | Concn resulting in 90% neutralizationb (μg/ml)
|
||
|---|---|---|---|---|
| Fab m14 | IgG1 m14 | IgG1 b12 | ||
| MJ4 | C | 6 | 8 | 50 |
| XJ-C19 | C | >100 | 100 | >200 |
| GX-C44 | C | 100 | 60 | >100 |
| Z2Z6 | D | >200 | 100 | 6 |
| NYU1545 | D | >200 | >200 | <3 |
| GX-E14 | E | >200 | >200 | 20 |
| VI14004c | F | 25 | 6 | >100 |
| 93Br | B/F | 4 | <3 | >200 |
| CA1-136 | Complex | 4 | 5 | >200 |
Neutralization assays were carried out in triplicate wells by preincubation of serial dilutions of Fab m14 and X5 with pseudotyped viruses for 1 h at 4°C followed by infection of 1 × 104 to 2 × 104 HOS CD4+ CCR5+ (CXCR4+) cells. Luminescence was measured after 3 days. The mean luminescence readings for triplicate wells were determined, and the end point was considered to be the last dilution of Fabs at which the mean results from the test samples were less than 50% of the nonneutralized control mean.
The Fab concentration that resulted in 90% neutralization was two- to eightfold (usually fourfold) higher than that which produces 50% neutralization. Neutralization assays for each envelope clone against the antibody were carried out two or three times, and the averages are shown.
Patient whose sera demonstrated broad cross-neutralizing activity.
IgG1 m14 exhibited a differential inhibitory activity when evaluated in parallel with IgG1 b12 (Table 3). IgG1 b12 neutralized two clade D and one clade E isolates much more efficiently than did IgG1 m14. However, for two isolates from clade F and one with a complex genotype, IgG1 m14 was much more potent than IgG1 b12. It is of interest that the clade F VII4004 isolate was obtained from a patient whose serum had been shown to demonstrate broad cross-neutralizing-antibody responses. The VI14004 Env was neutralized by m14 much more efficiently than by IgG1 b12 and Fab X5 (Table 3 and data not shown). Note also the IgG1 m14 also neutralized the three clade C isolates, especially MJ4 (43), more efficiently than IgGI b12.
To further evaluate the inhibitory activity of m14 for clade C isolates, we used a panel of 17 isolates from southern Africa. It has been previously shown for 16 of these isolates (all except TV1) that they are largely resistant to neutralization by sCD4, 2G12, and 2F5; only IgG1 b12 was effective and neutralized 9 of them, but at relatively high concentrations (9). IgG1 m14 neutralized 4 of the isolates with IC50 less than 10 μg/ml and 4 with IC50 of 60 μg/ml or less; for the remaining 9 isolates the IC50 was higher than 68 μg/ml which was the highest concentration tested (Table 4). Interestingly, the m14 inhibitory profile was different from that of b12. For example, IgG1 m14 neutralized Du172, S180, and S017 at much lower concentrations than IgG1 b12 did; IgG1 b12 was much more potent than IgG1 m14 for S018 and S103. Indeed, there were only three isolates (Du179, S021, and S007) that were not efficiently neutralized by either antibody. A note of caution is that IgG1 b12 was tested in an assay based on infection of PBMCs (9) although the M7-Luc cell-based assay was shown to resemble the PBMCs assays quite closely. These results suggest that IgG1 m14 is a potent broadly HIV-1-neutralizing antibody and that access restriction size effects (sterical hindrance) that could decrease the potency of some IgGs for some isolates (33) do not exist or are small compared to the effect of avidity. In addition, it exhibits a differential inhibitory profile compared to that of IgG1 b12 for a panel of primary isolates from different clades, mostly clades C and F.
TABLE 4.
Neutralization of HIV-1 subtype C isolates from South Africa by IgG1 m14a
| HIV-1 isolatec | Neutralization concn (μg/ml)b of:
|
|
|---|---|---|
| IgG1 m14 | IgG1 b12d | |
| Du123 | 43 | 16 |
| Du151 | >68 | 50 |
| Du156 | 60 | >50 |
| Du172 | 2.7 | 29 |
| Du174 | 49 | >50 |
| Du179 | >68 | >50 |
| Du368 | >68 | 30 |
| Du422 | 32 | 23 |
| S080 | >68 | 33 |
| S021 | >68 | >50 |
| S009 | >68 | 50 |
| S018 | >68 | 2 |
| S007 | >68 | >50 |
| S180 | 6.3 | >50 |
| S017 | 9.1 | >50 |
| S103 | >68 | <2 |
| TV1 | 7.1 | NDe |
Samples were assayed at multiple concentration in triplicate in M7-Luc cells.
Values are the concentrations needed to reduce RLU by 50% compared to the RLU in virus control wells (no test sample).
All Du isolates and TV1 are from southern Africa, and the rest from Malawi. All isolates are R5 except Du179, which is R5X4.
Note that the IgG1 b12 inhibitory activity was determined in a PBMC-based assay previously and the numbers represent ID80 (9).
ND, not determined.
High-affinity binding of Fab m14 to gp120 and gp140 that is reduced by sCD4.
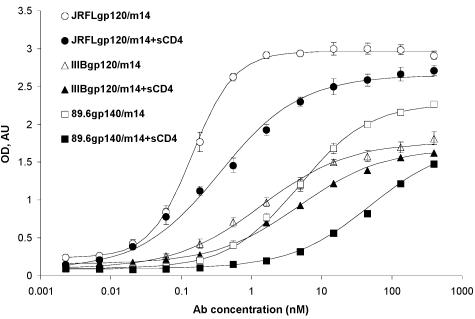
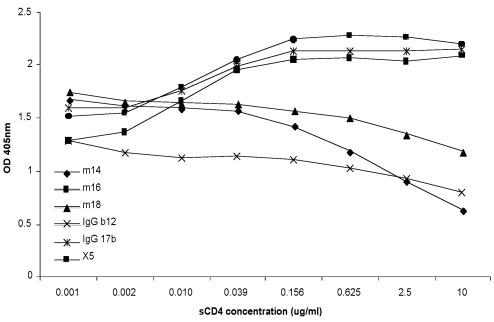
To further characterize m14 and begin to elucidate the mechanisms of its cross-reactivity and HIV-1 neutralization, we measured its binding to gp120 and gp140 under various conditions. M14 bound to gp140 from 89.6 and to gp120 from JR-FL and IIIB with high (nanomolar and subnanomolar) affinity as measured by an ELISA (Fig. 1). Binding of m14 to gp120 was inhibited at all sCD4 concentrations tested (Fig. 2); binding of IgG1 b12 and Fab m18 was also inhibited, while binding of Fab m16, X5, and IgG1 17b was increased, as expected (Fig. 2). The observation that m14 competes with CD4 but was selected by using gp120-CD4 complexes could be explained by a very high affinity to gp120 of the phage-associated m14 that, under the panning conditions used, was able to displace sCD4. These results suggest that m14 strongly binds to gp120 and gp140 and behaves as a CD4bs antibody, although the possibility for sCD4-induced, conformational changes leading to decreased binding of m14 to an epitope that does not overlap the CD4 binding site cannot be excluded based on these data.
FIG. 1.
Binding of m14 to gp140/120 and sCD4-gp140/120. Env proteins were captured by the polyclonal sheep anti-gp120 antibody D7324, which was coated (at 1 μg/ml) on microplates. After addition of gp14089.6, gp120IIIB, or gp120JR-FL at 1 μg/ml, threefold serially diluted m14 was added to the wells in the presence or absence of sCD4 (2 μg/ml). Bound m14 was detected by anti-human IgG F(ab')2-HRP and measured via its optical density at 405 nm (OD). The background was estimated from the amount of Fabs bound to bovine serum albumin and was subtracted. The data were fitted to the Langmuir adsorption isotherm: (B − Bbg)/(Bmax − Bbg) = F/(EC50 + F), where B is the amount of bound Fab, Bbg and Bmax are the background level and maximal amount of bound Fab, respectively, F is its bulk concentration, and EC50 is the (ELISA) concentration of half-maximal binding approximately equal to the equilibrium dissociation constant. Mean EC50 ± standard deviation are 4.7 ± 0.3, 0.14 ± 0.01, and 1.4 ± 0.3 nM for gp14089.6, gp120JR-FL, and gp120IIIB in the absence of sCD4 and 54 ± 7.6, 0.82 ± 0.13, and 4.5 ± 0.3 nm for gp14089.6, gp120JR-FL, and gp120IIIB in the presence of sCD4.
FIG. 2.
sCD4 effect on m14 binding to gp120 in comparison with other CD4bs and CD4i anti-gp120 antibodies. gp120JR-FL was captured by the polyclonal sheep anti-gp120 antibody D7324 (5 μg/ml) coated on 96-well plates. Following the addition of fourfold serially diluted sCD4, Fab m14, m16, m18, IgG b12, IgG 17b, or Fab X5 at a constant concentration that led to 70% of maximum binding was simultaneously added to the wells. Bound antibodies were detected by anti-human IgG F(ab')2-HRP and measured via their optical densities at 405 nm (OD).
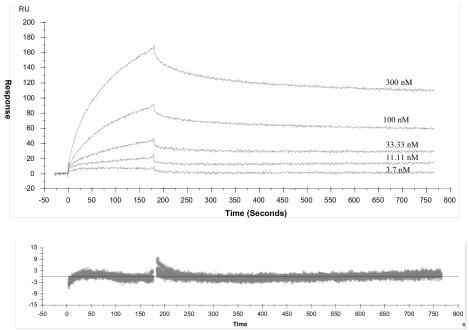
The kinetics of interaction of Fab m14 and Fab b12 with different Env proteins was measured by using an optical biosensor system (Biacore) based on surface plasmon resonance. We found that for three different Env ectodomains (tethered gp14089.6 [16] [clade B], gp140R2 [clade B], and gp140GX-C44 [clade C]), there were no major differences between m14 and b12 in the kinetics of their interactions with Env proteins from three different isolates (Fig. 3, Table 5).
FIG. 3.
Interaction of m14 with gp14089.6 (tethered) envelope, as determined by surface plasmon resonance. Sensorgram overlays of m14 injected at a rate of 30 μl/min at different concentrations (reported above the sensorgrams) over a CM5 sensor chip, containing captured gp14089.6 (tethered), are shown. The surface plasmon resonance response was recorded as a function of time, rate constants were calculated by using a 1:1 Langmuir global model, and the quality of the fitting was demonstrated by the relative residual plots (bottom panels), where the difference between the experimental and fitted data is shown on the x axis.
TABLE 5.
Binding kinetics of m14 to gp120/gp140 measured by Biacorea
| HIV-1 gp140 |
ka (105 M−1 s−1)
|
kd (10−4 s−1)
|
Kd (nM)
|
|||
|---|---|---|---|---|---|---|
| m14 | b12 | m14 | b12 | m14 | b12 | |
| gp14089.6 (tethered) (clade B) | 3.4 | 2.0 | 5.0 | 4.8 | 1.4 | 2.4 |
| gp140GXC-44 (clade C) | 1.0 | 2.2 | 2.0 | 3.4 | 2.1 | 1.6 |
| gp140R2 (clade B) | 1.2 | 2.4 | 2.3 | 3.2 | 1.9 | 1.3 |
The experiments were performed as described in Materials and Methods and in the legend to Fig. 3 but with different Env proteins on the chip. The association (ka) and dissociation (kd), rate constants were calculated by fitting both association and dissociation phases with a 1:1 Langmuir global model, and the equilibrium dissociation constant (Kd) was determined from their ratio.
Binding of Fab m14 to gp140s from diverse primary isolates.
Fab m14 also bound to several gp140s from primary isolates that were not used in the panning and screening procedures, suggesting that its epitope is exposed in gp120s of isolates from different clades, although the affinity of binding varied widely (Table 6).
TABLE 6.
Binding of m14 to gp120/gp140 from primary isolatesa
| HIV-1 gp140 | Clade | EC50 (μg/ml)b |
|---|---|---|
| UG037.8 | A | 8.95 ± 0.9 |
| 89.6 (tethered) | B | 2.37 ± 0.21 |
| R2 | B | 0.04 ± 0.002 |
| R2gp120 | B | 0.12 ± 0.02 |
| HT593.1 | B | 0.02 ± 0 |
| Bal-L | B | 40.75 ± 3.71 |
| GXC/44 | C | 0.19 ± 0.01 |
| MW965.26 | C | 0.11 ± 0.01 |
| CR1192UG024.2 | D | 0.11 ± 0.01 |
| GXE/14 | E | 110 ± 101.05 |
| 93BR019.10 | F/B | 110 ± 73.67 |
| 92UG975.10 | G | 57.17 ± 6.61 |
gp140s were captured by the polyclonal sheep anti-gp120 antibody D7324 (5 μg/ml) coated on 96-well plates and washed, and Fab m14 was added at the indicated concentrations. HIV immunoglobulin was used for calibration to ensure that equal amount of gp140s from different primary isolates were captured. Bound antibodies were detected by anti-human IgG F(ab′)2-HRP and measured via their optical densities at 405 nm. The data were fitted as described in Fig. 1.
The Kd values (approximately equal to the 50% effective concentration [EC50]) of m14 for gp140 from primary isolates are summarized. Results are given as mean ± standard deviation.
Characterization of the m14 epitope.
To approximately localize the m14 epitope, we evaluated the competitive binding activity of Fab m14 and Fab b12 in the presence of sCD4 and other anti-gp120 antibodies. M14 competed significantly with sCD4 and b12 and two other CD4bs antibodies (F105 and m18) but only weakly with 17b and other CD4i antibodies (X5, 48d, and 48e); it did not compete with 2G12 and A32 (data not shown). Therefore, its epitope does not overlap significantly with the epitopes of 2G12, A32. The competition profile was similar to that of b12, further suggesting that m14 is a CD4bs antibody.
To further characterize the m14 epitope, we measured its binding to alanine-scanning mutants of gp120JR-CSF. The mutated residues are in major regions of gp120 and are presumably solvent exposed. Most of the mutations that significantly (more than fivefold) decreased the affinity of m14 binding to gp120 were in the conserved C3 and C5 regions, including P363, D368, P369, E370, Y384, N36b, G472, D474, M475, R476, and W479 (Table 7). Mutations in C2 (T257), C4 (K421), and V5 (P470), as well as deletion of both the V1 and V2 loops but not deletion of V1 alone, also significantly decreased m14 binding to gp120. Some of those mutations also significantly decreased the binding b12 to gp120, (e.g., D368, E370, Y384, N386, K421, P470, G472, R476, and W479) and decreased the binding of CD4 to gp120, (e.g., T257, D368, E370, P470, G472, D474, and W479) (44). Interestingly, several mutations, mostly in C4 (R419, I420, and E429) but also in C2 (V200 and A204) and C3 (W338), as well as deletion of the V3 loop, resulted in a more than twofold increase in the affinity of m14 binding to gp120. It is interesting that V3 loop deletion enhanced m14 binding to gp120 fourfold, in contrast to the finding that V3 loop deletion decreased b12 binding to gp120 by threefold (44). These results demonstrated that the m14 epitope significantly overlaps with the CD4 binding site and the b12 epitope, although a note of caution is that mutations could indirectly affect binding. In addition, they suggest that engineering of the m14 epitope could lead to higher binding affinity, with possible implications for the design of vaccine immunogens.
TABLE 7.
Binding of m14 to alanine-scanning mutants of JR-CSF gp120
| Mutanta | gp120 domainb | Conservation (%)c | Relative affinityd |
|---|---|---|---|
| Wild type | 100 | ||
| D113A | C1 | 94 | 62 |
| V120A | C1 | 98 | 54 |
| K121A | C1 (V1/V2-stem) | 91 | 140 |
| L122A | C1 (V1/V2-stem) | 94 | 111 |
| T123Af | C1 (V1/V2-stem) | 99 | 90 |
| L125Af | C1 (V1/V2-stem) | 98 | 96 |
| V127A | C1 (V1/V2-stem) | 99 | 165 |
| T198A | C2 (V1/V2-stem) | 86 | 167 |
| S199Af | C2 (V1/V2-stem) | 94 | 69 |
| V200A | C2 (V1/V2 stem) | 42 | 202 |
| I201A | C2 (V1/V2 stem) | 89 | 83 |
| T202A | C2 (V1/V2 stem) | 76 | 81 |
| Q203A | C2 | 99 | 88 |
| A204G | C2 | 97 | 280 |
| K207Ae | C2 | 98 | 31 |
| S256Af | C2 | 97 | 48 |
| T257Af | C2 | 99 | 1 |
| R298A | C2 | 99 | 141 |
| W338A | C3 | 98 | 1,038 |
| N339A | C3 | 72 | 50 |
| P363A | C3 | 31 | 10 |
| S365Af | C3 | 85 | 94 |
| G366Af | C3 | 98 | 109 |
| G367Af | C3 | 99 | 110 |
| D368Af | C3 | 99 | 10 |
| P369Af | C3 | 42 | 5 |
| E370Af | C3 | 99 | 1 |
| Y384A | C3 | 98 | 3 |
| N386A | C3 | 90 | 6 |
| N392A | V4 | 93 | 45 |
| P417A | C4 | 79 | 113 |
| R419A | C4 | 81 | 209 |
| I420A | C4 | 97 | 234 |
| K421A | C4 | 91 | 4 |
| Q422A | C4 | 98 | 175 |
| I423A | C4 | 92 | 167 |
| I424A | C4 | 65 | 191 |
| N425Af | C4 | 85 | 116 |
| M426Af | C4 | 82 | |
| W427Af | C4 | 98 | 121 |
| Q428Af | C4 | 95 | 124 |
| E429Af | C4 | 40 | 422 |
| V430Af | C4 | 86 | 150 |
| G431A | C4 | 86 | 131 |
| M432A | C4 | 42 | 119 |
| M434A | C4 | 84 | 87 |
| Y435Af | C4 | 99 | 146 |
| P437A | C4 | 94 | 92 |
| R469A | V5 | 97 | 47 |
| P470A | V5 | 98 | 16 |
| G471A | V5 | 84 | 97 |
| G472A | C5 | 98 | 14 |
| G473Af | C5 | 98 | 124 |
| D474A | C5 | 71 | 14 |
| M475A | C5 | 90 | 6 |
| R476A | C5 | 74 | 1 |
| D477A | C5 | 94 | 39 |
| W479A | C5 | 99 | 0 |
| ΔV1 | Δ134–154 | 43 | |
| ΔV1/2 | Δ134–154/Δ160–193 | 4 | |
| ΔV3 | Δ303–324 | 427 |
Residue numbering scheme is based on the sequence of prototypic HxBc2 gp120 glycoprotein. Mutants with more than a twofold decrease in m14 binding are highlighted in bold type.
C, constant domain; V, variable loop.
Conservation was calculated as a percentage of the HIV-1 isolates which had the same residue at the same position with respect to a total of 380 isolates sequenced.
Calculated using the formula [apparent affinity (wild type)/apparent affinity (mutant)] × 100%, where apparent affinities were calculated as the antibody concentration at 50% of maximal binding.
Residues involved in maintaining the overall structure of gp120.
Residues that exhibit decreased solvent accessibility in the presence of sCD4 (D1D2) in the ternary complex.
Sequence analysis of m14 and its footprint defined by alanine-scanning mutagenesis.
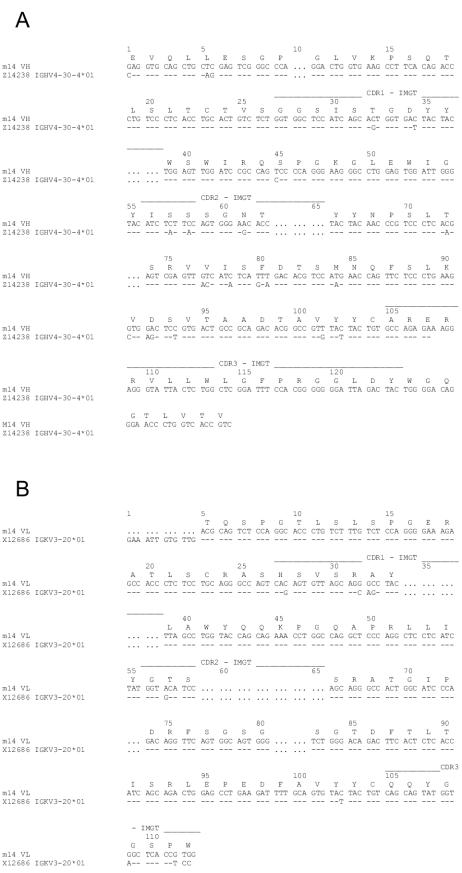
In an initial attempt to correlate the neutralization and binding activity of m14 with the primary structure of its paratope and epitope, we analyzed its sequence and the sequences of a variety of gp120s. Phagemid DNA of m14 was prepared and sequenced. M14 uses a V segment of the VH4 family and a V segment of the VKIII subgroup. Genomic analysis identified the m14 heavy-chain precursor as VH4-30-4; 13 of its 115 amino acids mutated during affinity maturation (Fig. 4A). The kappa light-chain precursor is VL3-20; 6 of its 91 residues mutated during maturation (Fig. 4B). The CDR H3 of m14 has 17 amino acids and contains three Arg residues, two hydrophobic residues (Trp and Phe), and one Tyr that could be part of its paratope. Experiments are in progress to determine the role of these and other residues in m14 binding.
FIG. 4.
Genomic analysis of m14. Alignment of the m14 DNA with the germ line sequence, as determined by using the IMGT sequence database, is shown. The m14 deduced amino acid sequences are shown above the aligned DNA sequences. (A) Heavy chain. (B) Light chain.
We also analyzed the extent of conservation of amino acid residues in 380 isolates that could contribute to the energy of binding to gp120 as identified by alanine-scanning mutagenesis. Most of the residues that lead to a more than fivefold decrease in binding affinity are highly (>90%) conserved, indicating a possible explanation for the broad cross-reactivity of m14 (Table 7). There are few residues for which the extent of conservation is lower. Note, however, that the extent of conservation was calculated as a percentage of the residues that are not mutated, and that mutations not only to alanine but also to any other residue were counted; obviously, many of the mutated residues could still directly or indirectly mediate binding. Because the gp120 conformation depends in a complex way on sequence, the conservation of most of the residues is only indicative of, but not conclusive for, the basis of the m14 cross-reactivity.
DISCUSSION
Identification of novel broadly cross-reactive HIV-1-neutralizing hMAbs has major implications for the development of vaccines, inhibitors, and tools to study mechanisms. The existence of broadly cross-reactive hMAbs (IgG1 b12 [14], 2G12 [8, 66], 2F5 [51], 447-52D [17, 26], 4E10 [61], Z13 [75], Fab X5 [41], and m9 [71]) that potently neutralize HIV-1 in vitro and can prevent infection in vivo suggests their potential as antiretroviral drugs, prophylactics, and microbicides (11, 24, 25, 54, 55, 60, 67). The first clinical study performed with an hMAb (F105) did not show beneficial effects for HIV-1-infected patients following infusion of a single dose of 500 mg/m2 (15). However, this antibody has only modest neutralizing activity in vitro. The use of a single 10-mg/kg dose of a more potent inhibitor, a fusion protein of IgG2 and sCD4 (PRO 542), reduced the viral concentration in HIV-1-infected adults (29). Treatment with a higher single dose (25 mg/kg) of PRO 542 resulted in a statistically significant mean reduction of about 0.5 log unit in HIV-1 RNA concentrations in plasma; a significant correlation was observed between antiviral effects observed in vivo and potency in vitro (28). Recent clinical trials evaluated the potential of a combination of two hMAbs, 2G12 and 2F5, for the treatment of chronic HIV-1 infections in humans (1, 59). However, although these antibodies can clearly decrease the plasma HIV-1 concentration in infected humans, it appears that their potency is not sufficient to significantly reduce the HIV-1 load, and the long-term virological response is poor (59). Thus, the identification of new neutralizing hMAbs and new combinations including these antibodies could be critical for their efficacy. Anti-HIV-1 MAbs have also proven to be very powerful tools not only for the establishment of the conditions for protection in vivo (2, 18, 23, 37, 38, 48, 54, 67) but also as probes of the structure and function of Env proteins (5, 21, 40, 69) and for investigation of their antigenicity (10, 13, 68).
The major finding of our study is the identification of a new hMAb Fab, m14, which neutralizes a range of primary HIV-1 isolates from different clades. Importantly, the m14 neutralization profile is different from that of IgG1 b12 and possibly from those of other HIV-1-neutralizing antibodies. The ability of m14 to potently neutralize representative isolates from clades A, B, C, and F suggests that it can be used in combination with other hMAbs that have poor neutralizating activity or lack activity against such isolates.
To further improve the potency of Fab m14, we constructed a whole-antibody molecule, IgG1 m14, which exhibited significantly higher neutralizing activity. Because m14 competes with CD4 for binding to gp120, it is likely that its mechanism of neutralization involves interactions with Env proteins before binding to CD4. Thus, restriction access effects due to size, as observed for IgG1 X5 (33), are unlikely to play a role. These results suggest that m14 can neutralize a broad range of HIV-1 primary isolates and that the IgG1 format would ensure a long half-life in vivo and biological effector functions.
Recently, a broadly HIV-neutralizing hMAb (IgG1 b12) was shown to protect macaques from vaginal challenge with SHIV-SF162P4 (67), supporting the concept that antibodies can be used for protection against sexually transmitted virus. Further studies are needed to determine whether m14 has protective activity in vivo. Its potency could be improved by constructing fusion proteins with other molecules, e.g., hMAbs and toxins, and using them in combination with other drugs and hMAbs. Only experiments with animal models and clinical trials of the effects of the drugs in humans will show whether this new antibody has potential as HIV-1 therapeutic and prophylactics.
The molecular mechanism of the high-affinity binding of Fab m14 to gp120 is currently under investigation. It is possible that by analogy to b12, the long CDR H3 plays a critical role (57). This loop also contains aromatic residues that could mimic Phe 43 of CD4.
Fab m14 binds to Env proteins from a number of primary isolates. Alanine-scanning mutagenesis suggested that most of the residues involved in binding to gp120 are conserved. Therefore, it is reasonable to assume that its epitope is conserved. Conserved epitopes could serve as targets for small-molecule inhibitors and immunogens for elicitation of protective immunity against viruses that show a high degree of genetic polymorphism. Until now, experiments to develop immunogens that are able to elicit any of the known broadly HIV-1-neutralizing hMAbs have not been successful (11). The identification of m14 may provide new opportunities to develop antigens that could elicit such broadly neutralizing antibodies in vivo. In this respect, it is the existence of gp120 mutations and deletions that result in a significant increase of m14 binding to gp120 is especially remarkable (Table 7). The W338A mutation increased m14 binding 10-fold; deletion of the V3 loop increased m14 binding 4-fold (Table 7). One can speculate that deletion of the V3 loop uncovers portions of the m14 epitope, resulting in an enhancement of binding. In this regard, it is interesting that Env proteins with a deleted V3 loop (and other variable loops) can elicit relatively broad neutralizing-antibody responses (30, 35), although in other cases such responses were not observed despite the exposure of new epitopes (31, 36). The identification of a new cross-reactive hMAb and the characterization of its epitope might help in the design of vaccine immunogens, perhaps in combination with other immunogens to elicit potent broadly reactive neutralizing hMAbs.
Of the large number of MAbs and Fabs that have been generated against HIV-1 Env proteins, until recently only three MAbs were identified that exhibited broad and potent HIV-1-neutralizing activity (19): two against gp120 (IgG1 b12 [14, 53] and 2G12 [56, 58, 66]) and one against gp41 (2F5 [42]). Four other antibodies, two against gp120 (Fab X5 [41] and IgG 447-52D [26]) and two against gp41 (4E10 [61, 75] and Fab Z13 [75]) are also known to potently neutralize a variety of HIV-1 primary isolates from different clades. The identification of a new broadly cross-reactive HIV-1-neutralizing MAb Fab suggests that such antibodies could play an even more important role in vivo than anticipated. A note of caution is that we have no evidence yet that IgG m14 does exist in patients or plays any role in virus neutralization in vivo.
Acknowledgments
We thank T. Evans, A. Schultz, and N. Miller for reagents; M. Yarchoan for helping with the data fitting; and S. Phogat, Y. Shu, A. F. Labrijn, J. M. Binley, R. Pantophlet, and P. Kwong for interesting discussions.
This project was supported by the NIH Intramural AIDS Targeted Antiviral Program (IATAP) and CPA from CCR, NCI, to D.S.D., DHHS NO1-CO-12400 to M.-Y.Z., and NIH grants AI47438 and AI48280 to G.V.Q. and AI48380 to C.C.B.
REFERENCES
- 1.Armbruster, C., G. M. Stiegler, B. A. Vcelar, W. Jager, N. L. Michael, N. Vetter, and H. W. Katinger. 2002. A phase I trial with two human monoclonal antibodies (hMAb 2F5, 2G12) against HIV-1. AIDS 16:227-233. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Barbas, C. F., D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., H. J. Ditzel, C. F. 3. Barbas, N. Sullivan, J. Sodroski, P. W. Parren, and D. R. Burton. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retroviruses 12:911-924. [DOI] [PubMed] [Google Scholar]
- 6.Bouma, P., M. Leavitt, P. F. Zhang, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 77:8061-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt, S. M., R. Mariani, A. U. Holland, T. J. Hope, and N. R. Landau. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291-17299. [DOI] [PubMed] [Google Scholar]
- 8.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, and A. Jungbauer. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359-369. [DOI] [PubMed] [Google Scholar]
- 9.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 12.Burton, D. R., C. F. Barbas, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 14.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 15.Cavacini, L. A., M. H. Samore, J. Gambertoglio, B. Jackson, M. Duval, A. Wisnewski, S. Hammer, C. Koziel, C. Trapnell, and M. R. Posner. 1998. Phase I study of a human monoclonal antibody directed against the CD4-binding site of HIV type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 14:545-550. [DOI] [PubMed] [Google Scholar]
- 16.Chow, Y. H., O. L. Wei, S. Phogat, I. A. Sidorov, T. R. Fouts, C. C. Broder, and D. S. Dimitrov. 2002. Conserved structures exposed in HIV-1 envelope glycoproteins stabilized by flexible linkers as potent entry inhibitors and potential immunogens. Biochemistry 41:7176-7182. [DOI] [PubMed] [Google Scholar]
- 17.Conley, A. J., M. K. Gorny, J. A. Kessler, Jr., L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conley, A. J., J. A. Kessler, Jr., L. J. Boots, P. M. McKenna, W. A. Schleif, E. A. Emini, G. E. Mark III, H. Katinger, E. K. Cobb, S. M. Lunceford, S. R. Rouse, and K. K. Murthy. 1996. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 70:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, and the AIDS Clinical Trials Group Antibody Selection Working Group. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 20.D'Souza, M. P., G. Milman, J. A. Bradac, D. McPhee, C. V. Hanson, and R. M. Hendry. 1995. Neutralization of primary HIV-1 isolates by anti-envelope monoclonal antibodies. AIDS 9:867-874. [DOI] [PubMed] [Google Scholar]
- 21.Ditzel, H. J., P. W. Parren, J. M. Binley, J. Sodroski, J. P. Moore, C. F. Barbas III, and D. R. Burton. 1997. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J. Mol. Biol. 267:684-695. [DOI] [PubMed] [Google Scholar]
- 22.Dong, M., P. F. Zhang, F. Grieder, J. Lee, G. Krishnamurthy, T. VanCott, C. Broder, V. R. Polonis, X. F. Yu, Y. Shao, D. Faix, P. Valente, and G. V. Quinnan, Jr. 2003. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J. Virol. 77:3119-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrantelli, F., R. Hofmann-Lehmann, R. A. Rasmussen, T. Wang, W. Xu, P. L. Li, D. C. Montefiori, L. A. Cavacini, H. Katinger, G. Stiegler, D. C. Anderson, H. M. McClure, and R. M. Ruprecht. 2003. Post-exposure prophylaxis with human monoclonal antibodies prevented SHIV89.6P infection or disease in neonatal macaques. AIDS 17:301-309. [DOI] [PubMed] [Google Scholar]
- 24.Ferrantelli, F., R. A. Rasmussen, R. Hofmann-Lehmann, W. Xu, H. M. McClure, and R. M. Ruprecht. 2002. Do not underestimate the power of antibodies—lessons from adoptive transfer of antibodies against HIV. Vaccine 20(Suppl. 4):A61-A65. [DOI] [PubMed] [Google Scholar]
- 25.Ferrantelli, F., and R. M. Ruprecht. 2002. Neutralizing antibodies against HIV—back in the major leagues? Curr. Opin. Immunol. 14:495-502. [DOI] [PubMed] [Google Scholar]
- 26.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson, J. M., R. J. Israel, I. Lowy, N. A. Ostrow, L. S. Vassilatos, M. Barish, D. N. Tran, B. M. Sullivan, T. J. Ketas, T. J. O'Neill, K. A. Nagashima, W. Huang, C. J. Petropoulos, J. P. Moore, P. J. Maddon, and W. C. Olson. 2004. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 48:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson, J. M., I. Lowy, C. V. Fletcher, T. J. O'Neill, D. N. Tran, T. J. Ketas, A. Trkola, M. E. Klotman, P. J. Maddon, W. C. Olson, and R. J. Israel. 2000. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J. Infect. Dis. 182:326-329. [DOI] [PubMed] [Google Scholar]
- 30.Jeffs, S. A., C. Shotton, P. Balfe, and J. A. McKeating. 2002. Truncated gp120 envelope glycoprotein of human immunodeficiency virus 1 elicits a broadly reactive neutralizing immune response. J. Gen. Virol. 83:2723-2732. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y. B., D. P. Han, C. Cao, and M. W. Cho. 2003. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology 305:124-137. [DOI] [PubMed] [Google Scholar]
- 32.Laal, S., S. Burda, M. K. Gorny, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leavitt, M., E. J. Park, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 77:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorin, C., L. Mollet, F. Delebecque, C. Combredet, B. Hurtrel, P. Charneau, M. Brahic, and F. Tangy. 2004. A single injection of recombinant measles virus vaccines expressing human immunodeficiency virus (HIV) type 1 clade B envelope glycoproteins induces neutralizing antibodies and cellular immune responses to HIV. J. Virol. 78:146-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retroviruses 14:151-155. [DOI] [PubMed] [Google Scholar]
- 37.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 39.Montefiori, D. C. Evaluating neutralizing antibodies against HIV, SIV and SHIV in a luciferase reporter gene assay, in press. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 40.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantophlet, R., S. E. Ollmann, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, E. J., M. K. Gorny, S. Zolla-Pazner, and G. V. Quinnan, Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J. Virol. 74:4183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, E. J., and G. V. Quinnan. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 73:5707-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, E. J., L. K. Vujcic, R. Anand, T. S. Theodore, and G. V. Quinnan, Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parren, P. W., H. J. Ditzel, R. J. Gulizia, J. M. Binley, C. F. Barbas III, D. R. Burton, and D. E. Mosier. 1995. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 9:F1-F6. [DOI] [PubMed] [Google Scholar]
- 49.Parren, P. W., P. Fisicaro, A. F. Labrijn, J. M. Binley, W. P. Yang, H. J. Ditzel, C. F. Barbas, and D. R. Burton. 1996. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type 1 envelope. J. Virol. 70:9046-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 51.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, and A. Jungbauer. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 52.Quinnan, G. V., P. F. Zhang, D. W. Fu, M. Dong, and H. J. Alter. 1999. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retroviruses 15:561-570. [DOI] [PubMed] [Google Scholar]
- 53.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruprecht, R. M., F. Ferrantelli, M. Kitabwalla, W. Xu, and H. M. McClure. 2003. Antibody protection: passive immunization of neonates against oral AIDS virus challenge. Vaccine 21:3370-3373. [DOI] [PubMed] [Google Scholar]
- 55.Safrit, J. T., R. Ruprecht, F. Ferrantelli, W. Xu, M. Kitabwalla, K. Van Rompay, M. Marthas, N. Haigwood, J. R. Mascola, K. Luzuriaga, S. A. Jones, B. J. Mathieson, and M. L. Newell. 2004. Immunoprophylaxis to prevent mother-to-child transmission of HIV-1. J Acquir. Immune Defic. Syndr. 35:169-177. [DOI] [PubMed] [Google Scholar]
- 56.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 58.Scanlan, C. N., R. Pantophlet, M. R. Wormald, S. E. Ollmann, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stiegler, G., C. Armbruster, B. Vcelar, H. Stoiber, R. Kunert, N. L. Michael, L. L. Jagodzinski, C. Ammann, W. Jager, J. Jacobson, N. Vetter, and H. Katinger. 2002. Antiviral activity of the neutralizing antibodies 2F5 and 2G12 in asymptomatic HIV-1-infected humans: a phase I evaluation. AIDS 16:2019-2025. [DOI] [PubMed] [Google Scholar]
- 60.Stiegler, G., and H. Katinger. 2003. Therapeutic potential of neutralizing antibodies in the treatment of HIV-1 infection. J Antimicrob. Chemother. 51:757-759. [DOI] [PubMed] [Google Scholar]
- 61.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 62.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tilley, S. A., W. J. Honnen, M. E. Racho, M. Hilgartner, and A. Pinter. 1991. A human monoclonal antibody against the CD4-binding site of HIV1 gp120 exhibits potent, broadly neutralizing activity. Res. Virol. 142:247-259. [DOI] [PubMed] [Google Scholar]
- 66.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veazey, R. S., R. J. Shattock, M. Pope, J. C. Kirijan, J. Jones, Q. Hu, T. Ketas, P. A. Marx, P. J. Klasse, D. R. Burton, and J. P. Moore. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343-346. [DOI] [PubMed] [Google Scholar]
- 68.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 69.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, M. Y., Y. Shu, S. K. Phogat, X. Xiao, F. Cham, A. Choudhary, Y. R. Feng, I. Sanz, S. Rybak, C. C. Broder, G. V. Quinnan, Jr., T. Evans, and D. S. Dimitrov. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 283:17-25. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, M. Y., Y. Shu, D. Rudolph, P. Prabakaran, A. F. Labrijn, M. B. Zwick, R. B. Lal, and D. S. Dimitrov. 2004. Improved breadth and potency of an HIV-1-neutralizing single-chain antibody by random mutagenesis and sequential antigen panning. J Mol. Biol. 335:209-219. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, M. Y., Y. Shu, I. Sidorov, and D. S. Dimitrov. 2004. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res. 61:161-164. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, P. F., P. Bouma, E. J. Park, J. B. Margolick, J. E. Robinson, S. Zolla-Pazner, M. N. Flora, and G. V. Quinnan, Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 76:644-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, P. F., X. Chen, D. W. Fu, J. B. Margolick, and G. V. Quinnan, Jr. 1999. Primary virus envelope cross-reactivity of the broadening neutralizing antibody response during early chronic human immunodeficiency virus type 1 infection. J. Virol. 73:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zwick, M. B., P. W. Parren, E. O. Saphire, S. Church, M. Wang, J. K. Scott, P. E. Dawson, I. A. Wilson, and D. R. Burton. 2003. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]