Abstract
GB virus B (GBV-B), the virus most closely related to hepatitis C virus (HCV), infects tamarins and causes acute hepatitis. The 3′ untranslated region (UTR) of an infectious GBV-B clone (pGBB) has a proximal short sequence followed by a poly(U) tract and a 3′ terminal sequence. Our investigators previously demonstrated that the 3′ terminal sequence was critical for in vivo infectivity. Here, we tested the effect of deleting the short sequence and/or the poly(U) tract from pGBB; infectivity of each mutant was tested by intrahepatic transfection of two tamarins with transcribed RNA. A mutant lacking both regions was not viable. However, mutants lacking either the short sequence or the poly(U) tract were viable. All four tamarins had a wild-type-like acute infection and developed acute hepatitis. Whereas we found that five tamarins transfected with the wild-type clone pGBB had acute resolving infection, one tamarin transfected with the poly(U) deletion mutant became persistently infected. This animal had viremia and hepatitis until its death at week 90. The genomes recovered at weeks 2, 7, 15, 20, 60, and 90 lacked the poly(U) stretch. Eight amino acid changes were identified at week 90. One change, in the putative p7 protein, was dominant at week 15. Thus, persistence of GBV-B, like persistence of HCV, was associated with the emergence of virus variants. Four tamarins inoculated with serum collected at weeks 2 and 90 from the tamarin with persistent infection had an acute resolving infection. Nonetheless, the demonstration that GBV-B can persist in tamarins strengthens its relevance as a surrogate model for the study of HCV.
The study of GB virus B (GBV-B) has generated considerable interest because it is the virus most closely related to hepatitis C virus (HCV) (16, 18). In humans, infection with HCV is one of the leading causes of chronic liver disease, which in many cases will progress to liver cirrhosis and liver cancer (9). The natural host of GBV-B remains unknown, but it was demonstrated that this virus does not infect chimpanzees, a surrogate of humans, suggesting that it is not a human virus (3). However, experimental infection of tamarins with GBV-B causes acute hepatitis. Thus, experimental GBV-B infection of tamarins could potentially serve as a surrogate model for the study of HCV, which can only be studied in chimpanzees.
The GBV-B virus contains a positive-sense, single-stranded RNA genome of about 9.5 kb (4, 24). Like the other members of the Flaviviridae virus family, it has a single long open reading frame (ORF), bracketed by 5′ and 3′ untranslated regions (UTR). Based on hydropathy plots and known motifs, it was predicted to encode the same kinds of proteins as HCV (16). However, it should be emphasized that the overall homology of the predicted polyproteins between GBV-B and HCV is only about 25 to 30% (16). Significant homology was observed between GBV-B and HCV in the NS3 serine protease, the NS3 RNA helicase, and the NS5 RNA-dependent RNA polymerase regions (16, 34, 35). Furthermore, functional similarity between the GBV-B and HCV NS3 serine proteases was demonstrated (6, 19, 22). Finally, both viruses contain an internal ribosome entry site (IRES) with similar structure and function in the 5′ UTR (8, 17).
Our investigators previously identified the 3′ terminal sequence of GBV-B (4). Subsequently, this sequence was reported in two other studies (15, 20). Its discovery enabled us to generate an infectious clone (pGBB) of GBV-B (4). Interestingly, the 3′ UTR of GBV-B had a similar organization as the 3′ UTR of HCV, with a short proximal sequence after the ORF stop codon followed by a polypyrimidine tract and a 3′ terminal sequence. In pGBB the proximal short sequence consists of 27 nucleotides (nt), the poly(U) tract consists of 23 nt, and the 3′ terminal sequence consists of 309 nt. Our group previously demonstrated that the 3′ terminal sequence was critical for viability of GBV-B (4). The 3′ terminal sequence of members of the Flaviviridae is believed to be critical for initiation of negative-strand RNA synthesis during RNA replication (7, 33), and this region was found to be critical for viability of HCV in vivo (12, 31). Although the function of the two proximal regions of the HCV 3′ UTR is not known, it was demonstrated that the poly(U/UC) tract, but not the short proximal sequence, was critical for viability in vivo (31). The goal of the present study was to determine the importance of the short proximal sequence and the poly(U) tract of GBV-B.
In contrast to HCV infection, which frequently progresses to chronicity in humans and in experimentally infected chimpanzees (9), all reported GBV-B infections in tamarins initiated with wild-type virus have resolved during the acute phase (1, 3, 4, 21, 23). Recently, however, it was reported that an animal transfected with RNA transcripts from a full-length GBV-B clone had viremia for more than 2 years before it cleared the infection (15). In the present study, we studied the natural history and evolution of GBV-B in an animal that became persistently infected following transfection with RNA transcripts from a 3′ UTR deletion mutant of our infectious molecular clone.
MATERIALS AND METHODS
Construction of mutants of an infectious GBV-B clone.
The mutants tested in the present study (Fig. 1) were all constructed from pGBB (4). This clone contains the core sequence of the T7 promoter, a 5′ guanosine residue, and the consensus sequence of GBV-B with an XhoI site at the 3′ terminus. To facilitate the cloning process, we first constructed the vector pSP73/GBB by inserting the BglII and XhoI fragment of pGBB into the pSP73 vector (Promega). In order to construct pGBB-ΔV, we hybridized two oligonucleotides (Va/S-ODN and Va/R-ODN) corresponding to the following plus-strand sequence: GTT TTT GCC CTA GGG CTC ATT GCT GTT GGA TTA GCC ATC AGC TGA TTT TTT TTT TTT TTT TTT TTT TTA GGG CAG CGG CAA CAG GGG AGA CCC CGG GCT TAA CGA. The AvrII- and XmaI-digested hybridization product was cloned into the digested pSP73/GBB clone and, finally, the BglII and XhoI fragment was cloned into the digested pGBB plasmid. To construct pGBB-ΔU, we hybridized the oligonucleotides U/S-ODN and U/R-ODN corresponding to the following plus-strand sequence: GTT TTT GCC CTA GGG CTC ATT GCT GTT GGA TTA GCC ATC AGC TGA ACC CCC AAA TTC AAA ATT AAC TAA CAG TTT AGG GCA GCG GCA ACA GGG GAG ACC CCG GGC TTA ACG. The product was cloned into pGBB as described above. For the pGBB-ΔVU construct, hybridization was performed with oligonucleotides VU/S and VU/R, which correspond to the following plus-strand sequence: GTT TTT GCC CTA GGG CTC ATT GCT GTT GGA TTA GCC ATC AGC TGA AGG GCA GCG GCA ACA GGG GAG ACC CCG GGC TTA ACG. The resulting product was cloned into pGBB as described above. However, sequence analysis revealed that the obtained clones all had deletions or insertions in the region corresponding to the hybridization product. Thus, to obtain a pGBB-ΔVU clone with only the desired deletion, we performed PCR-directed mutagenesis of one of the clones by using primers that spanned the BglII and XmaI sites. Finally, to construct pGBB-GFP, the encephalomyocarditis virus (EMCV) IRES-GFP (green fluorescent protein) fragment was first PCR amplified from pIRES2-EGFP (Clontech) by using the primers 2F/ApoI/IRES (5′-GCG GCC AAT AAA TTC CCT CTC CCT CCC CCC-3′) and GFP-ApoI/R1 (5′-GAG CTC TGA ATT TCT ATT ACT TGT ACA GC-3′). The ApoI-digested amplicon was next subcloned into pSP73/GBB, and the BglII and XhoI fragment was cloned into pGBB. For each construct, DH5α competent cells (GIBCO BRL) were transformed and selected on Luria-Bertani agar plates containing 100 μg of ampicillin (SIGMA or Invitrogen)/ml and amplified in Luria-Bertani liquid cultures at 30°C for 18 to 20 h (30). Large-scale preparation of plasmid DNA was performed with a QIAGEN plasmid Endofree Maxi kit as described previously (30). The authenticity of each clone was confirmed by sequence analysis of the entire GBV-B sequence.
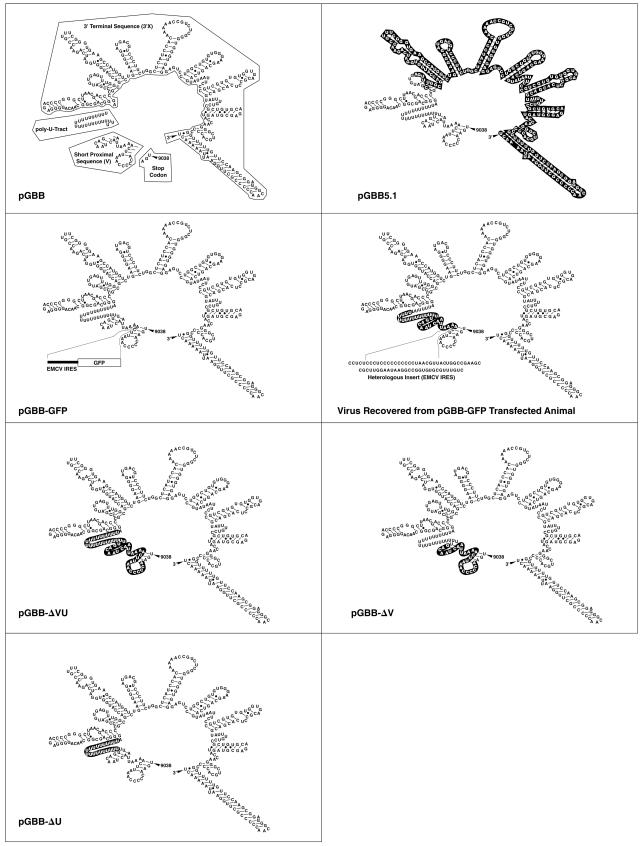
FIG. 1.
3′ UTR deletion mutants of an infectious cDNA clone of GBV-B (pGBB) (4). Deleted sequences are shown in black boxes. The 3′ UTR of GBV-B has 362 nt consisting of the ORF stop codon, a short proximal sequence of 27 nt (V region), a poly(U) tract of 23 nt, and a 3′-terminal sequence of 309 nt (3′X region). In the pGBB-GFP construct, EMCV IRES and GFP sequences were inserted into the short proximal sequence of the 3′ UTR. The genome sequence recovered from an animal transfected with pGBB-GFP is indicated. Note that in this sequence a portion of the V region and the poly(U) region were deleted, but 68 nt of the original IRES-GFP insert were retained.
Intrahepatic transfection of tamarins with transcribed GBV-B RNA.
Animal studies were carried out under a protocol which was approved by the NIAID Animal Care and Use Committee and by the BIOQUAL, Inc., Animal Care and Use Committee. Animals were housed and procedures were performed at BIOQUAL, Inc., an AAALAC International-accredited facility, and in accordance with the 1996 Guide for the Care and Use of Laboratory Animals (10). In 100-μl reaction mixtures, RNA was transcribed in vitro with T7 RNA polymerase (Promega) from 10 μg of XhoI-digested pGBB or mutants thereof (4). The integrity of the RNA was checked by electrophoresis through an agarose gel stained with ethidium bromide. Each transcription mixture was diluted with 400 μl of ice-cold phosphate-buffered saline without calcium or magnesium and then immediately frozen on dry ice and stored at −80°C. Within 24 h, two transcription mixtures were injected into each tamarin (Saguinus mystax [SM]) by percutaneous intrahepatic injection guided by ultrasound (31, 32). The viability of each construct was tested in two animals. For transmission of GBV-B present at weeks 2 and 90 in animal SM890, 0.5 ml of a 10−2 serum dilution was inoculated intravenously into additional tamarins.
All animals used in the present study were negative for GBV-A and GBV-A variants (2). Serum samples were collected weekly from the tamarins and monitored for liver enzyme levels (alanine aminotransferase, gamma-glutamyltranspeptidase, and isocitrate dehydrogenase [ICD]) by standard methods. Serum samples were also tested for the presence of GBV-B RNA (see below). There is no standardized test for detecting antibodies to GBV-B. Additionally, in most animals weekly liver biopsy samples were collected in a percutaneous procedure. For histological analysis, liver biopsy specimens were fixed in a 10% formalin solution. Paraffin-embedded liver biopsies were sectioned and stained with hematoxylin and eosin and examined for necroinflammatory changes within the lobules by using the following scoring criteria: 0, normal (no necrosis); 1+, mild (at least 1 focus of necrosis per 10× field); 2+, mild to moderate (2 to 5 foci per 10× field); 3+, moderate to severe (6 to 10 foci per 10× field); 4+, severe (>10 foci per 10× field). In addition, liver biopsies collected during the chronic phase in SM890 were scored using the Ishak system, where the total necroinflammatory score includes periportal, lobular (confluent and spotty), and portal inflammation, a system used for scoring chronic HCV in humans. The maximum score is 18. Finally, portal fibrosis was scored on a scale from 0 (normal) to 6 (cirrhosis).
Testing for GBV-B RNA and determination of the GBV-B genome titer by real-time PCR.
Selected serum samples were tested for GBV-B RNA in a reverse transcriptase (RT) nested PCR assay (3, 4). In short, viral RNA was extracted from 100 μl of serum with the TRIzol system (GIBCO/BRL). The RNA pellet was resuspended in 10 mM dithiothreitol containing 5% (vol/vol) RNasin (20 to 40 U/μl; Promega). The RT-nested PCR assay was performed with primers deduced from the 5′ UTR of GBV-B. The reverse transcription was performed with avian myeloblastosis virus RT (Promega) and the external antisense primer. Nested PCR was performed with AmpliTaq Gold DNA polymerase (Perkin-Elmer). Specificity was confirmed by sequence analysis of selected DNA products. Each set of experiments included a positive control sample (a 10−6 and/or a 10−7 dilution of the GB 8/93 serum pool [4], with an estimated titer of approximately 10 to 100 genome equivalents [GE]).
The genome titer of samples collected during the acute infection was determined in a real-time PCR assay. We used TaqMan to quantitate GBV-B RNA with a primer-probe set previously described by Beames et al. (1). The primer-probe set is specific for the capsid gene of GBV-B. We used final concentrations of 900 nM for the forward (558F) and reverse (626R) primers and a final concentration of 90 nM for the probe (579P). Viral RNA was isolated from 10 μl of serum with the QIAmp viral RNA mini kit (QIAGEN). RT-PCR was performed by using the TaqMan one-step RT-PCR kit (Applied Biosystems) in an ABI Prism 7900HT sequence detection system. The profile included a 30-min RT step at 48°C, followed by 10 min of AmpliTaq Gold activation at 95°C and 45 cycles of PCR that consisted of 15 s of denaturing at 95°C and 1 min of annealing and extending at 60°C.
A well-characterized S. mystax tamarin serum pool, designated GB 2/94, was selected for our standard preparation (4). Viral RNA was extracted as described above from 10 μl of the serum pool, and 10-fold dilutions of the extract were made to cover a 6-log dynamic range (107 to 102 GE/ml), aliquoted, and stored at −80°C. Optimization of the test prompted us to exclude from the standard curve the highest dilution, since it did not consistently register a value. Every TaqMan run included at least duplicates of the standards that were used to develop the assay standard curve. The standard line R2 fit values ranged from 0.974 to 0.998 (percent coefficient of variation = 0.9). The interassay precision (percent coefficient of variation) of standards ranged from 3 to 4%. In addition, we included four sets of controls with each extraction series to allow examination of sensitivity and specificity. These included high- and low-positive, negative, and no-template controls in duplicate. The high-positive control was a serum pool previously shown to contain 108 GE of GBV-B/ml by RT-nested PCR on 10-fold-serially diluted RNA, and we used a 10−2 dilution. In the test runs, the mean quantity of RNA at this dilution was 106.0 GE/ml. The low-positive control was another serum pool previously shown to contain 108 GE of GBV-B/ml by the in-house RT-nested PCR, and we used it at a 10−4 dilution. In the test runs, the mean quantity of RNA at this dilution was 104.3 GE/ml. Since data points below the 103 GE/ml mark were outside of the dynamic linear range and inconsistent, we set our lower detection cutoff at 103 GE/ml.
Sequence analysis of GBV-B.
The consensus genome sequence of GBV-B virus recovered from serum of infected animals was determined by direct sequencing of RT-PCR amplicons. The near-full-length genome was first amplified in a long RT-PCR procedure (26, 27), using GBV-B-specific primers from the 5′ and 3′ UTRs (Table 1). The primers in Table 1 were deduced from the sequence originally published by Simons et al. (24). The reverse transcription reaction was performed at 42°C for 1 h by using Superscript II RT (Invitrogen). The transcription mixture was next incubated with RNase T1 (Gibco BRL) and RNase H (Invitrogen) for 20 min. The PCRs were performed with the Advantage cDNA polymerase mix (Clontech) and a Robocycler thermal cycler (Stratagene). The following cycling parameters were used: denaturation at 99°C for 35 s, annealing at 67°C for 30 s, and elongation at 68°C for 10 min during the first 5 cycles, for 11 min during the next 10 cycles, for 12 min during 10 additional cycles, and for 13 min during the last 10 cycles. Nine overlapping subgenomic regions of GBV-B covering the entire ORF were next amplified by nested PCR with GBV-B-specific primers (Table 1) using the same protocol as for the first round of PCR except that the elongation time was 6 min. The nine amplicons were purified and sequenced directly by standard procedures. The genome region analyzed corresponded to nt 36 to 9107 of pGBB. In addition, a genome region (nt 8765 to 9359), including all but the 3′ 40 nt of the 3′ UTR, was amplified in the RT-nested PCR (see above) with GBV-B-specific primers as follows: I PCR, 8510S_GBV-B (5′-TGT TTG TGG GTT AGC CGT CTG TTG-3′) and 9399R_GBB (5′-AGT TTT TAA TTC CAA GCG GGG G-3′); II PCR, 8745S_GBV-B (5′-CCC TGC GAG CCT GGC GAA AGA AAG-3′) and 9380R_GBB (5′-GGG TTG CCC TCC GCT TGG AAC-3′).
TABLE 1.
Primers used for long RT-nested PCR for GBV-Ba
| Use and primer | Sequence |
|---|---|
| cDNA synthesis | |
| SacI_BamHI_GB9143R (22) | 5′-TGCATGAGCTCGGATCCACATCGCGGGGTCGTTAAGCC-3′ |
| First round of long PCR | |
| Sense: NotI_T7_G_GB1S (30) | 5′-TTTTTTTTGCGGCCGCTAATACGACTCACTATAGACCACAAACACTCCAGTTTGTTACACTCCG-3′ |
| Reverse: SacI_BamHI_GB9143R (22) | Same as cDNA primer |
| Second round of long PCR | |
| Product 1: 5′ S-35_GBV-B | 5′-ACCACAAACACTCCAGTTTGTTACACTCCGCTAGG-3′ |
| 1150R_GBV-B | 5′-GTCACAGGTCACAAGAGCGCCC-3′ |
| Product 2: 999S_GBV-B | 5′-CTTGCCTACACGAGCCTGGTTGTG-3′ |
| 2160R_GBV-B | 5′-GCACCCGTGGCGGAATATAAGACC-3′ |
| Product 3: 1997S_GBV-B | 5′-CCGGAGAGGTGGGCTAGGTTGC-3′ |
| 3152R_GBV-B | 5′-CATTCCTATAGACCCTTGCCTTGC-3′ |
| Product 4: 3017S_GBV-B | 5′-TTAGTGTTTGGTGAGAATGGTGTG-3′ |
| 4149R_GBV-B | 5′-GCATCGGTAGCATGGCATTCGTC-3′ |
| Product 5: 3999S_GBV-B | 5′-CGTACGGCGTGAATCCAAATTGC-3′ |
| 5145R_GBV-B | 5′-GCGAGTGCGGCTGTCCCAGAAG-3′ |
| Product 6: 5008S_GBV-B | 5′-CCGGCTTGGGAAAAAACCTTGTGG-3′ |
| 6152R_GBV-B | 5′-AGAAAGGACAACCAGGAATGTTAACC-3′ |
| Product 7: 5998S_GBV-B | 5′-CATCCGTTGGCTCCACACCCCGAC-3′ |
| 7148R_GBV-B | 5′-TTGTAGTCGTTGACCAACTTCCG-3′ |
| Product 8: 7004S_GBV-B | 5′-CTGTTGGGAGCAGGTGAGTGTAAC-3′ |
| 8151R_GBV-B | 5′-TTCAGCCAGCAGGTCAAACTGTTG-3′ |
| Product 9: 8000S_GBV-B | 5′-CAACACCGAGCTGGCATTCACACC-3′ |
| 9133R_GBV-B | 5′-GGTCGTTAAGCCCGGGGTCTCC-3′ |
See the text for details of the protocol used.
RESULTS
Course of GBV-B infection in tamarins infected with the wild-type molecular clone pGBB.
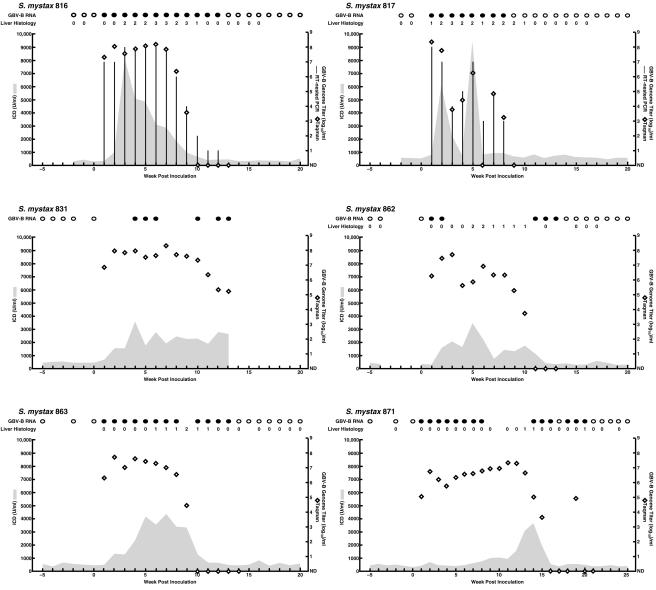
Our investigators previously developed an infectious clone of GBV-B (pGBB) (4). RNA transcripts from this clone, which represented a consensus sequence of GBV-B, were infectious after intrahepatic transfection of two tamarins (SM816 and SM817) (Fig. 2). Furthermore, the viral genome sequence (nt 36 to 9359), including the entire ORF, recovered from one of the animals was identical to the sequence of pGBB. In the present study four additional animals served as positive controls and were transfected intrahepatically with RNA transcripts from pGBB. All became infected (Fig. 2), and the course of GBV-B infection following transfection with RNA transcripts was similar to that observed after intravenous inoculation of wild-type virus. Taken together, the six animals transfected with the pGBB plasmid all had viremia at week 1 postinoculation (p.i.), with titers of 105.0 to 108.4 GE/ml. The peak titers ranged from 107.3 to 108.4 GE/ml. Viral clearance was observed during weeks 9 to 21 (one animal died prior to clearance). Finally, all six animals had evidence of acute viral hepatitis with elevated liver enzymes (peak ICD, 3,454 to 9,500 U/ml). Necroinflamatory changes in liver biopsy samples indicative of acute hepatitis were observed in all five animals examined (ranging from mild [1+] to moderate-severe [3+]).
FIG. 2.
Course of GBV-B infection in six S. mystax tamarins following intrahepatic transfection with RNA transcripts from a molecular clone (pGBB). Results of qualitative RT-nested PCR for GBV-B RNA in serum (filled circles, positive; empty circles, negative) and examination of liver biopsy samples (necroinflammatory changes within the lobules graded as 0 [no change], 1 [mild], 2 [mild-moderate], 3 [moderate-severe], or 4 [severe]) are shown at the top. Only selected samples were tested in the qualitative RT-PCR assay. Serum levels of ICD (in units per milliliter; shaded areas) and the estimated log10 GBV-B GE titer per ml, determined in a TaqMan assay (diamonds), were plotted against time. Samples found to be below the assay cutoff of 103 GE/ml in the TaqMan assay are shown as not detected (ND). Only selected samples were tested in the TaqMan assay. Vertical columns indicate the genome titer previously determined for SM816 and SM817 in an RT-nested PCR on 10-fold-serially diluted RNA (4). The GBV-B titers for the other animals were not determined with this test. SM816 and SM817 had not previously been tested in the TaqMan assay.
Failed attempt to insert the GFP gene into the proximal short sequence of the GBV-B 3′ UTR.
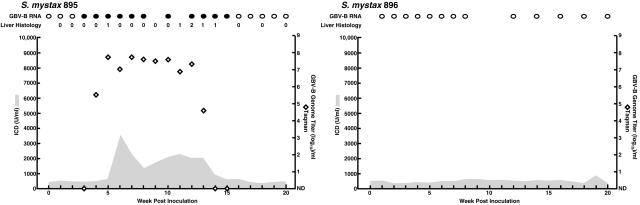
The 3′ UTR of pGBB begins with a short proximal sequence of 27 nt immediately after the ORF stop codon, followed by a 23-nt-long poly(U) tract and a 3′ terminal sequence of 309 nt. Our group previously demonstrated that the 3′ terminal sequence is essential for GBV-B, since RNA transcripts from the consensus pGBB clone lacking most of this sequence (Fig. 1) were nonviable in vivo (4). In the present study, we examined the importance of the two other regions of the GBV-B 3′ UTR. In an attempt to express GFP in a replicating GBV-B virus, we inserted the EMCV IRES and GFP into the short proximal sequence of the pGBB 3′ UTR at nucleotide position 9052 (Fig. 1). Two tamarins were transfected intrahepatically with RNA transcripts from the resulting clone, pGBB-GFP (Fig. 3). One animal (SM896) remained GBV-B RNA negative. The other animal (SM895) had late appearance of viremia, at week 3 p.i., but subsequently had a typical acute GBV-B infection with peak titers of ∼108 GE/ml and evidence of mild to moderate acute hepatitis. The sequence of a region corresponding to nt 36 to 9359 of pGBB was determined for virus recovered from the animal at week 5. We found that all but 68 nt of the IRES of the EMCV IRES-GFP insert had been deleted (Fig. 1). In addition, the carboxy-terminal 15 nt of the short proximal sequence and all but 6 U bases of the poly(U) tract had been deleted (Fig. 1). It should be noted, however, that 6 of the 12 bases of the EMCV IRES preceding this short U tract are U (Fig. 1). No other mutations were observed in the remainder of the genome. These data suggested that the proximal short sequence and the poly(U) tract of the 3′ UTR might not be critical for viability of GBV-B.
FIG. 3.
Intrahepatic transfection of tamarins with RNA transcripts of pGBB-GFP. In this construct, the EMCV IRES and GFP sequences were inserted into the short proximal sequence of the 3′ UTR of pGBB. SM896 did not become infected with this mutant. See also the legend to Fig. 2.
A mutant with deletion of both the proximal short sequence and the poly(U) tract of the GBV-B 3′ UTR is not viable.
We found that two tamarins transfected intrahepatically with RNA transcripts of pGBB-ΔVU (Fig. 1) remained negative for GBV-B RNA in weekly serum samples collected during 18 weeks of follow-up and had normal liver enzyme values. One of these animals (SM862) was subsequently transfected with the wild-type clone pGBB and became infected with a typical GBV-B infection (Fig. 2). At the same time, the other animal (SM880) was retransfected with RNA transcripts of pGBB-ΔVU but remained uninfected. However, this animal became infected following subsequent intravenous inoculation with GBV-B virus (see below). Thus, each of the animals was susceptible to infection with GBV-B, and deletion of both the proximal short sequence and the poly(U) tract rendered GBV-B noninfectious.
Mutants with deletion of the proximal short sequence or of the poly(U) tract of the GBV-B 3′ UTR are viable in vivo.
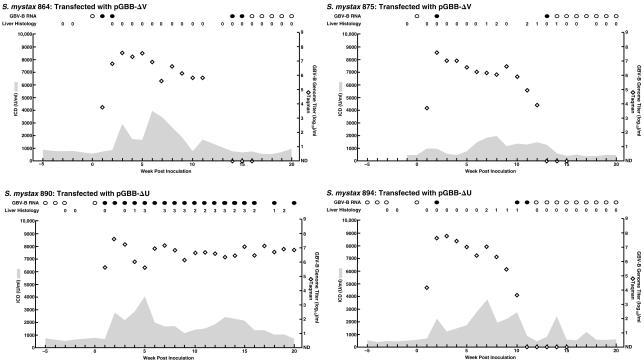
A mutant pGBB-ΔV lacking only the proximal short sequence was viable (Fig. 1 and 4). Both tamarins (SM864 and SM875) transfected with this mutant became viremic at week 1 (103.8 and 103.7 GE/ml, respectively), and the peak titers were 107 to 108 GE/ml. The genomes of virus recovered from both animals, at week 2 p.i., lacked the proximal short sequence of the 3′ UTR. In a more complete analysis of the virus recovered from SM864, we found that its sequence (corresponding to nt 36 to 9359 of pGBB) was identical with that of pGBB-ΔV. Both tamarins developed acute resolving hepatitis with elevated liver enzyme values, and the virus was cleared during weeks 16 and 14, respectively. However, necroinflammatory changes in liver biopsies were observed only in SM875.
FIG. 4.
Course of GBV-B infection in tamarins following intrahepatic transfection with RNA transcripts from deletion mutants of pGBB. SM864 and SM875 were transfected with pGBB-ΔV, a mutant lacking the proximal short sequence of the 3′ UTR. SM890 and SM894 were transfected with pGBB-ΔU, a mutant lacking the poly(U) tract of the 3′ UTR. See also the legend to Fig. 2.
The RNA transcripts of a deletion mutant of pGBB lacking the poly(U) tract (pGBB-ΔU) also were infectious (Fig. 1 and 4). Both transfected tamarins (SM890 and SM894) became viremic at week 1 (105.6 and 104.2 GE/ml, respectively) and had peak titers of 107 to 108 GE/ml. The genomes of virus recovered from each animal, at week 2 p.i., lacked the poly(U) tract. Analysis of a region corresponding to nt 36 to 9359 of pGBB in one animal (SM890) revealed no other mutations. Both tamarins developed acute hepatitis, with elevated liver enzyme values and necroinflammatory changes in liver biopsies. One tamarin (SM894) had an acute resolving infection with clearance during week 12. However, surprisingly, the other animal (SM890) remained infected (see below).
The mean week 1 genome titer of the four animals infected following transfection with the 3′ UTR deletion mutants (pGBB-ΔV and pGBB-ΔU) was ∼3 logs lower than the mean titer for the six animals transfected with the wild-type clone (105.0 and 107.6 GE/ml, respectively). However, this difference did not reach statistical significance (data not shown). There was also no significant difference between the mutant group and the wild-type group with respect to the mean peak titers (107.6 and 108.1 GE/ml, respectively) or the mean peak ICD levels (3,442 and 5,454 U/ml, respectively) (data not shown).
Persistent GBV-B infection in a tamarin transfected with a deletion mutant.
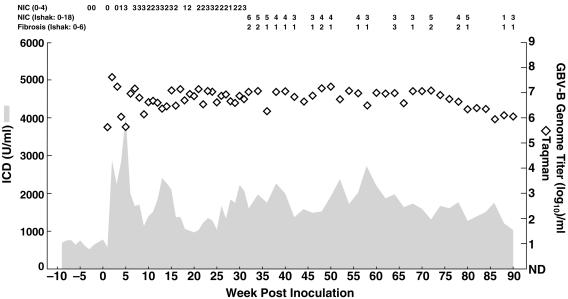
All GBV-B infections that our group had previously studied in S. mystax tamarins by using sensitive detection methods, including at least 10 infections initiated from wild-type virus (4; J. Bukh, unpublished data) and 5 infections initiated from RNA transcripts of our infectious clone pGBB (Fig. 2), were resolved. However, one of the S. mystax tamarins (SM890) transfected with the poly(U) tract deletion mutant virus remained infected until its death at week 90 (Fig. 5). A necropsy performed on the deceased animal did not determine the cause of death, and macroscopic and microscopic examinations of the liver suggested that it was not caused by liver disease. The animal was caught in the wild and we therefore did not know its age, but it had been in our facility for ∼5 years (under the best of circumstances a tamarin lives ∼10 years in captivity). Thus, most likely the death of this animal was related to old age and not complications caused by the GBV-B infection. Throughout the chronic phase of the infection in SM890, the GBV-B titers remained at high levels (106 to 107 GE/ml). Furthermore, during the entire follow-up period the animal had evidence of mild hepatitis with elevated liver enzyme levels and/or necroinflammatory changes in liver biopsies. In the Ishak scoring system, most biopsies taken during weeks 32 to 90 had necroinflammatory scores of 3 to 7 and fibrosis scores of 1 to 2. It is noteworthy that this animal had a marked degree of hemosiderosis, which was not observed in any of the other animals examined in the present study.
FIG. 5.
Persistent infection of GBV-B in SM890. This animal was transfected with pGBB-ΔU, a mutant lacking the poly(U) tract of the GBV-B 3′ UTR. The animal was followed until its death at week 90. The cause of death did not appear to be related to the ongoing GBV-B infection. Weekly samples were tested during the first 32 weeks of follow-up; biweekly samples were obtained thereafter. The results of histological examinations of liver biopsy samples are indicated on top. During the acute phase, necroinflammatory changes (NIC) within the lobules were graded as 0 (no change) to 4 (severe). During the chronic phase of the infection (weeks 32 to 90), liver biopsy samples were examined for necroinflammatory changes (on a scale from 0 [normal] to 18 [severe]) and fibrosis (on a scale from 0 [normal] to 6 [cirrhosis]) by using the Ishak scoring system (see Materials and Methods for details).
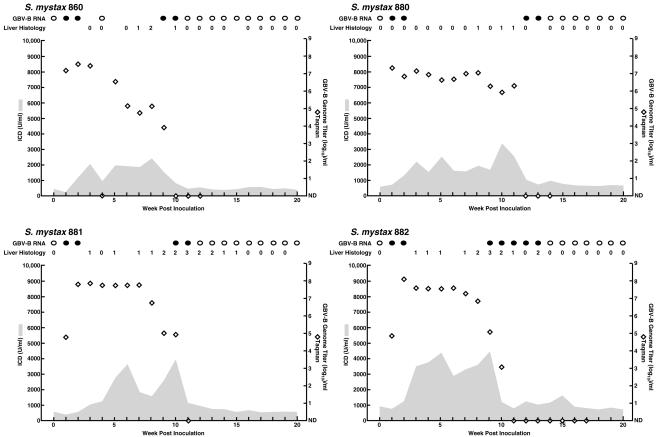
To determine whether the chronicity observed in this animal reflected a unique phenotype of the mutant pGBB-ΔU, we inoculated tamarins with serum collected from SM890 at weeks 2 (SM860 and SM880) and 90 (SM881 and SM882) (Fig. 6). All four animals became infected, with peak titers of 107 to 108 GE/ml. Viruses recovered from all four animals lacked the poly(U) tract. Furthermore, analysis of a region corresponding to nt 36 to 9359 of pGBB recovered at week 2 from SM860, which was inoculated with the SM890 week 2 sample, demonstrated that its sequence was identical to the sequence of virus recovered at week 2 from SM890. Similarly, analysis of nt 36 to 9359 from virus recovered at week 2 from SM881, which was inoculated with the SM890 week 90 sample, demonstrated it had the same sequence as the sequence of virus recovered at week 90 from SM890, although in this case there were a few quasispecies positions with a different dominant sequence (Tables 2 and 3). All four animals had acute resolving infection with viral clearance during weeks 11 to 14.
FIG. 6.
Course of GBV-B infection in tamarins inoculated with serum collected from the animal that became persistently infected with GBV-B. SM860 and SM880 were inoculated intravenously with serum collected from SM890 at week 2, whereas SM881 and SM882 were inoculated with serum collected at week 90. See also the legend to Fig. 2.
TABLE 2.
Evolution of GBV-B in SM890, which became persistently infected with GBV-Ba
| Animal and week p.i. | Nucleotide found at the indicated position, by region
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ UTR | E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
3′ UTR
|
|||||||||||
| 211T | 1585*T | 2198C | 2552A | 2752*C | 4295A | 4861*C | 5082G | 5321G | 6649*T | 6690C | 6993T | 7031T | 7050C | 7151G | 7248G | 8942A | 9035A | 9067G | |
| SM890 | |||||||||||||||||||
| 2 | T | T | C | A | C | A | C | G | G | T | C | T | T | C | G | G | A | A | G |
| 7 | T | T | C | A/g | C | A | C | G | G | T | C | T | T | C | G | G | A | A | G |
| 15 | T | T | C | G | C | A | C | G | G | T | C | T | C | C | G | A | A | A | G |
| 20 | T | T | C | G | C | A | C | G | G | T | C | T | C | C | G | A | A | A | G |
| 60 | T | T | C/a | G | C | T/a | C | A | G/a | T | A | C/t | T | C/g | G | G | A/g | A | G |
| 90 | T/C | T/c | A | G | C/t | T/A | C/T | A | A/g | T/c | A | C | T | G | G/a | G | G/a | A/g | A/g |
| SM881 | |||||||||||||||||||
| 2 | T/C | C/t | A | G | C/T | A/t | T/c | A | A | T/C | A | C | T | G | G/A | G | G | A/G | A/g |
| SM882 | |||||||||||||||||||
| 2 | G | A/G | A/g | ||||||||||||||||
This animal was transfected with pGBB-ΔU. The nucleotides and their positions in the molecular clone are shown on top. The differences from the pGBB-ΔU sequence in sequences recovered from SM890 are indicated below: dominant sequences are shown in capital letters; minor sequences are shown in lowercase letters. Also indicated are the sequences of viruses recovered at week 2 from SM881 and SM882 (Fig. 6), which were inoculated intravenously with serum collected from SM890 at week 90. For SM882, the sequence of only a subdomain (corresponding to nt 8765 to 9359 of pGBB) was determined. Included in this table are also positions at which the week 90 sequence from SM890 and/or the week 2 sequence from SM881 had a significant quasispecies, but at which the consensus sequence did not change in SM890 (positions 211, 1585, 2752, 4861, 6649, 7151, and 9035). *, silent mutation.
TABLE 3.
Changes observed in the polyprotein of GBV-B in SM890a
| Animal and week p.i. | Amino acid found at the indicated position, by region
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E2 | p7 | NS3
|
NS4B
|
NS5A
|
NS5B
|
||||||
| 585L | 703N | 1284T | 1546S | 1626A | 2082T | 2183M | 2196F | 2202A | 2268G | 2833I | |
| SM890 | |||||||||||
| 2 | L | N | T | S | A | T | M | F | A | G | I |
| 7 | L | N/d | T | S | A | T | M | F | A | G | I |
| 15 | L | D | T | S | A | T | M | L | A | E | I |
| 20 | L | D | T | S | A | T | M | L | A | E | I |
| 60 | L/i | D | S/t | N | A/t | K | T/m | F | A/g | G | I/v |
| 90 | I | D | T/S | N | T/a | K | T | F | G | G | V/i |
| SM881 | |||||||||||
| 2 | I | D | T/s | N | T | K | T | F | G | G | V |
The differences from the pGBB-ΔU polyprotein sequence in sequences recovered from SM890 are indicated: dominant sequences are shown in capital letters, and minor sequences are shown in lowercase letters. Also indicated are the sequences of viruses recovered at week 2 from SM881, which was inoculated intravenously with serum collected from SM890 at week 90. See also Table 2.
The persistence of GBV-B in a tamarin permitted analysis of virus evolution of GBV-B. We determined the sequence of the near-complete genome (corresponding to nt 36 to 9107 of pGBB), including the entire ORF of virus genomes recovered at weeks 2, 7, 15, 20, 60, and 90 (Tables 2 and 3). All lacked the poly(U) tract. The weeks 2 and 7 consensus sequences were identical with that of pGBB-ΔU. At weeks 15 and 20 the sequence had three nucleotide changes that all resulted in changes in the deduced amino acid sequence. The change in the putative p7 protein (N703D) was found as a minor quasispecies already at week 7 and was the only change maintained at weeks 60 and 90, since the two changes in NS5A (F2196L and G2268E) had the sequence of pGBB-ΔU at weeks 60 and 90. Seven additional nucleotide changes that all resulted in changes in the deduced amino acid sequence were identified at week 90. It is impressive that we did not observe silent mutations in the ORF. However, a single nucleotide change was detected at week 90 in the short proximal sequence of the 3′ UTR. Thus, a total of nine nucleotide changes, eight of which resulted in amino acid changes, had accumulated during 90 weeks of active infection. Based on these data we calculated a mutation rate of 0.6 × 10−3 nucleotide substitutions/site/year and 1.6 × 10−3 amino acid substitutions/site/year.
DISCUSSION
The GBV-B agent is the virus most closely related to HCV, and these viruses will probably be classified together within the same genus of the Flaviviridae family. The only animal model for HCV is the chimpanzee. However, since GBV-B infects tamarins and causes acute hepatitis, experimental infections of these small New World monkeys could potentially serve as a surrogate model for studies of HCV. One of the common characteristics of GBV-B and HCV genomes is the organization of the 3′ UTR. The importance of the genetic elements of the GBV-B 3′ UTR was determined by constructing deletion mutants of an infectious clone of GBV-B and testing their viability by injecting RNA transcripts directly into the livers of tamarins. The reproducibility of this in vivo transfection system was confirmed by the initiation of GBV-B infection in all six animals transfected with RNA transcripts from the wild-type clone pGBB. Our group previously demonstrated that the sequence 3′ of the poly(U) tract of the GBV-B 3′ UTR was essential for infectivity in vivo (4). In the present study, we demonstrated that the proximal short sequence or the poly(U) tract of the GBV-B 3′ UTR is not critical for the viability of GBV-B.
Both tamarins transfected with a mutant lacking the proximal short sequence of the GBV-B 3′ UTR became infected, and analysis of virus genomes recovered from the infected animals confirmed that they lacked the proximal short sequence and that they did not contain other compensating mutations. The corresponding region of the HCV 3′ UTR also was not critical for infectivity in vivo (31) and, furthermore, was found not to be critical for replication in the HCV replicon system (7, 33). At least in the replicon system, however, replication appeared to have been significantly reduced by deletion of this sequence. The only evidence of impaired replication in tamarins of GBV-B without the short proximal sequence of the 3′ UTR was lower titers at week 1 posttransfection, compared with those seen following transfection with the wild-type molecular clone, but with no evidence of compensating mutations the titers rapidly increased to peak titers similar to those of wild-type infections.
Among positive-strand RNA viruses, the inclusion of a poly(U) tract within the 3′ UTR is unique to GBV-B and HCV (11, 24, 25). The poly(U) tract of GBV-B is shorter than the corresponding poly(U) tract of HCV. Here, we found that RNA transcripts of a deletion mutant of pGBB lacking the poly(U) tract were infectious and replicated to wild-type-like titers in the animals. Sequence analysis of viruses recovered from the transfected tamarins confirmed the absence of the poly(U) tract and, furthermore, that compensatory mutations were not required for the viability of this deletion mutant. This is in contrast to HCV, since our investigators previously demonstrated that the poly(U/UC) tract of HCV was critical for infectivity in vivo (31). Recent studies performed with HCV replicons have confirmed this finding, since a poly(U) tract of at least 26 nt, or a poly(U/UC) tract of at least 52 nt, was required for replication of HCV (7, 33). It was suggested that more efficient replication of HCV occurred in the presence of a pure poly(U) sequence (33). Although the function of the poly(U) tracts of GBV-B and HCV is not known, it is clear from our data that, even though the GBV-B 3′ UTR contains such a pure poly(U) tract, its function is dispensable.
Although we found that both the proximal short sequence and the poly(U) tract were not essential for GBV-B and that these regions did not appear to affect significantly the replication capacity of GBV-B, we also found that a deletion mutant lacking both of these regions was not infectious. This finding could be explained by a similar or duplicative function of the proximal short sequence and the poly(U) tract. However, it is more likely that the composition of this sequence is not critical. This was evidenced by the composition of the 3′ UTR of viruses recovered from an animal transfected with a mutant in which we had attempted to insert GFP into the proximal short sequence of the GBV-B 3′ UTR. In the viruses recovered, about half of the proximal short sequence and all but six residues of the poly(U) tract of the 3′ UTR had been deleted. Furthermore, 68 nt of a heterologous sequence (from the EMCV IRES) were retained. Thus, a total of 86 nt were present between the ORF stop codon and the 3′ terminal sequence, whereas in the wild-type clone only 50 nt exist. Only one strain of GBV-B has been identified, and so the natural variation of the proximal sequence of the GBV-B 3′ UTR has not been determined. However, in other members of the Flaviviridae family of viruses, including flaviviruses, pestiviruses, and HCV, this sequence is highly variable and not necessarily required for RNA replication (reviewed in references 7 and 31).
An important difference between the natural history of HCV in chimpanzees and humans and the natural history of GBV-B in tamarins relates to their chronicity rate. HCV persists in most instances, whereas reported GBV-B infections initiated from wild-type virus always resolved. A recent study, however, reported that a Saguinus oedipus tamarin infected from a molecular clone became persistently infected and developed chronic hepatitis (15). This animal was originally transfected with RNA transcripts from a GBV-B clone that differed from our infectious GBV-B clone (pGBB) at three amino acid positions. However, two of these changed to the sequence found in pGBB during the first 10 weeks of follow-up. The virus persisted for more than 2 years but was eventually cleared. We found that one S. mystax tamarin transfected with the poly(U) deletion mutant became persistently infected and also developed chronic hepatitis. In this animal, GBV-B persisted until the animal died at week 90. In the animals that became persistently infected, peak titers during the early acute infection were equivalent to those observed in other GBV-B-infected animals. In the animal reported by Martin et al. (15), there was a >5-log decrease in titers during the late acute-phase infection, and thereafter the viral titers increased over time. In our animal the titers decreased only by 2 to 3 logs and remained at relatively high levels throughout follow-up. Thus, the course of infection of GBV-B in persistent infection varies, and this is also the case for HCV (28).
We demonstrated that the persistence of GBV-B in an animal transfected with the poly(U) tract deletion mutant did not reflect a unique phenotype of this particular mutant. Similarly, it was found by Martin et al. (15) that virus transmitted from their persistently infected animal also did not result in persistent infection of the recipient. The numbers are too small to make any definitive conclusions about whether these findings reflect a low chronicity rate or are related to the fact that one virus might have been attenuated due to the presence of two nonconsensus amino acids and the other virus was attenuated because it was a mutant lacking the poly(U) tract of the 3′ UTR. Another possibility is that the transmission mode influences the outcome, since both animals were transfected from molecular clones. However, five animals that we transfected with the wild-type clone all had acute resolving infections, as did two animals transfected with another molecular GBV-B clone (21). Lanford et al. (13) observed that transient immunosuppression of four tamarins could prolong GBV-B viremia in some cases, but it did not lead to persistent infections. Overall, the more likely explanation for the observed findings is a low inherent chronicity rate of GBV-B, determined primarily by the individual host response combined with the ability of GBV-B to escape host immune defenses.
The persistence of GBV-B in a tamarin (SM890) permitted analysis of virus evolution of GBV-B. We observed a lower nucleotide mutation rate than that reported by Martin et al. (15), although the rate of amino acid changes was similar. We found that all dominant nucleotide changes that occurred in the ORF during 90 weeks of follow-up resulted in changes in the deduced amino acid sequence. This finding suggests that these mutations occurred as the result of strong selective pressure, most likely as a consequence of host immune responses. Eight amino acid changes had accumulated by week 90. All of these changes were also present in virus recovered from an animal inoculated with a serum sample collected from SM890 at week 90. The first amino acid change (N703D), located in the putative p7 polypeptide, was present as a minor species already at week 7 and had completely replaced the original sequence by week 15. This mutation was retained throughout follow-up. It is noteworthy that this is one of the positions at which our group previously found heterogeneity (residues N and D) in GBV-B from a GBV-B pool consisting of serum samples from three acutely infected animals (4) and that at this position the originally published GBV-B sequence had the D residue (24). Possibly this residue is within an immunodominant T-cell epitope. Mutations in the HCV p7 are often among the first to develop in chimpanzees that become persistently infected with HCV and have been demonstrated to represent T-cell escape mutants (5, 14, 29; J. Bukh, unpublished). Two changes located in NS5A (F2196L and G2268E) were present at weeks 15 and 20, but not at weeks 60 and 90. These two changes also evolved in the persistently infected animal reported previously (15), but in that case they were present late during the first year of follow-up. We observed additional mutations in NS3 and NS5A at week 40. Finally, we observed changes in E2, NS4B, NS5A, and NS5B at week 90. Two of the changes that we observed at week 40 (M2183T) and 90 (I2833V) were also observed by Martin et al. (15). Thus, 4 of the 11 amino acid changes we observed in a persistent GBV-B infection were also found in the only other reported animal with persistent infection. At two additional amino acid sites (positions 703 and 2082), our group had previously observed heterogeneity of GBV-B in an acute-phase plasma pool (4). Considering that the GBV-B polyprotein consists of 2,864 amino acids, such a pattern of recurrent mutations suggests that most, if not all, of the observed amino acid changes might be located in important immunodominant epitopes. The only mutation found in the envelope proteins occurred after week 60, suggesting that escape from neutralizing antibodies does not play an important role in the establishment of a persistent GBV-B infection. Thus, persistence of GBV-B, like persistence of HCV, was associated with the emergence of virus variants that most likely represented T-cell escape mutants.
Acknowledgments
We thank staff members at BIOQUAL, Inc., for providing excellent animal care and Suzanne U. Emerson and Robert H. Purcell for reviewing the manuscript.
REFERENCES
- 1.Beames, B., D. Chavez, B. Guerra, L. Notvall, K. M. Brasky, and R. E. Lanford. 2000. Development of a primary tamarin hepatocyte culture system for GB virus-B: a surrogate model for hepatitis C virus. J. Virol. 74:11764-11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukh, J., and C. L. Apgar. 1997. Five new or recently discovered (GBV-A) virus species are indigenous to New World monkeys and may constitute a separate genus of the Flaviviridae. Virology 229:429-436. [DOI] [PubMed] [Google Scholar]
- 3.Bukh, J., C. L. Apgar, S. Govindarajan, and R. H. Purcell. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 65:694-697. [DOI] [PubMed] [Google Scholar]
- 4.Bukh, J., C. L. Apgar, and M. Yanagi. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470-478. [DOI] [PubMed] [Google Scholar]
- 5.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butkiewicz, N., N. Yao, W. Zhong, J. Wright-Minogue, P. Ingravallo, R. Zhang, J. Durkin, D. N. Standring, B. M. Baroudy, D. V. Sangar, S. M. Lemon, J. Y. Lau, and Z. Hong. 2000. Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3. J. Virol. 74:4291-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grace, K., M. Gartland, P. Karayiannis, M. J. McGarvey, and B. Clarke. 1999. The 5′ untranslated region of GB virus B shows functional similarity to the internal ribosome entry site of hepatitis C virus. J. Gen. Virol. 80:2337-2341. [DOI] [PubMed] [Google Scholar]
- 9.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott Raven, Philadelphia, Pa.
- 10.Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council, National Academy of Sciences. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 11.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanford, R. E., D. Chavez, L. Notvall, and K. M. Brasky. 2003. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology 311:72-80. [DOI] [PubMed] [Google Scholar]
- 14.Major, M. E., K. Mihalik, J. Fernandez, J. Seidman, D. Kleiner, A. A. Kolykhalov, C. M. Rice, and S. M. Feinstone. 1999. Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J. Virol. 73:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, A., F. Bodola, D. V. Sangar, K. Goettge, V. Popov, R. Rijnbrand, R. E. Lanford, and S. M. Lemon. 2003. Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proc. Natl. Acad. Sci. USA 100:9962-9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muerhoff, A. S., T. P. Leary, J. N. Simons, T. J. Pilot-Matias, G. J. Dawson, J. C. Erker, M. L. Chalmers, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 69:5621-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijnbrand, R., G. Abell, and S. M. Lemon. 2000. Mutational analysis of the GB virus B internal ribosome entry site. J. Virol. 74:773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shini, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 19.Sbardellati, A., E. Scarselli, V. Amati, S. Falcinelli, A. S. Kekule, and C. Traboni. 2000. Processing of GB virus B non-structural proteins in cultured cells requires both NS3 protease and NS4A cofactor. J. Gen. Virol. 81:2183-2188. [DOI] [PubMed] [Google Scholar]
- 20.Sbardellati, A., E. Scarselli, L. Tomei, A. S. Kekule, and C. Traboni. 1999. Identification of a novel sequence at the 3′ end of the GB virus B genome. J. Virol. 73:10546-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sbardellati, A., E. Scarselli, E. Verschoor, A. De Tomassi, D. Lazzaro, and C. Traboni. 2001. Generation of infectious and transmissible virions from a GB virus B full-length consensus clone in tamarins. J. Gen. Virol. 82:2437-2448. [DOI] [PubMed] [Google Scholar]
- 22.Scarselli, E., A. Urbani, A. Sbardellati, L. Tomei, R. De Francesco, and C. Traboni. 1997. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J. Virol. 71:4985-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlauder, G. G., G. J. Dawson, J. N. Simons, T. J. Pilot-Matias, R. A. Gutierrez, C. A. Heynen, M. F. Knigge, G. S. Kurpiewski, S. L. Buijk, T. P. Leary, et al. 1995. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J. Med. Virol. 46:81-90. [DOI] [PubMed] [Google Scholar]
- 24.Simons, J. N., T. J. Pilot-Matias, T. P. Leary, G. J. Dawson, S. M. Desai, G. G. Schlauder, A. S. Muerhoff, J. C. Erker, S. L. Buijk, M. L. Chalmers, et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. USA 92:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 26.Tellier, R., J. Bukh, S. Emerson, and R. H. Purcell. 2003. Long PCR methodology. Methods Mol. Biol. 226:175-179. [DOI] [PubMed] [Google Scholar]
- 27.Tellier, R., J. Bukh, S. U. Emerson, R. H. Miller, and R. H. Purcell. 1996. Long PCR and its application to hepatitis viruses: amplification of hepatitis A, hepatitis B, and hepatitis C virus genomes. J. Clin. Microbiol. 34:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson, M., M. Nascimbeni, S. Gonzales, K. K. Murthy, B. Rehermann, and T. J. Liang. 2001. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology 121:1226-1233. [DOI] [PubMed] [Google Scholar]
- 30.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 33.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong, W., P. Ingravallo, J. Wright-Minogue, A. Skelton, A. S. Uss, R. Chase, N. Yao, J. Y. Lau, and Z. Hong. 1999. Nucleoside triphosphatase and RNA helicase activities associated with GB virus B nonstructural protein 3. Virology 261:216-226. [DOI] [PubMed] [Google Scholar]
- 35.Zhong, W., P. Ingravallo, J. Wright-Minogue, A. S. Uss, A. Skelton, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. RNA-dependent RNA polymerase activity encoded by GB virus-B nonstructural protein 5B. J. Viral Hepat. 7:335-342. [DOI] [PubMed] [Google Scholar]