Abstract
Human immunodeficiency virus type 1 (HIV-1), feline immunodeficiency virus (FIV), and Moloney murine leukemia virus (MoMLV) integrases were stably expressed to determine their intracellular trafficking. Each lentiviral integrase localized to cell nuclei in close association with chromatin while the murine oncoretroviral integrase was cytoplasmic. Fusions of pyruvate kinase to the lentiviral integrases did not reveal transferable nuclear localization signals. The intracellular trafficking of each was determined instead by the transcriptional coactivator LEDGF/p75, which was required for nuclear localization. Stable small interfering RNA expression eliminated detectable LEDGF/p75 expression and caused dramatic, stable redistribution of each lentiviral integrase from nucleus to cytoplasm while the distribution of MoMLV integrase was unaffected. In addition, endogenous LEDGF/p75 coimmunoprecipitated specifically with each lentiviral integrase. In vitro integration assays with preintegration complexes (PICs) showed that endogenous LEDGF/p75 is a component of functional HIV-1 and FIV PICs. However, HIV-1 and FIV infection and replication in LEDGF/p75-deficient cells was equivalent to that in control cells, whether cells were dividing or growth arrested. Two-long terminal repeat circle accumulation in nondividing cell nuclei was also equivalent to that of LEDGF/p75 wild-type cells. Virions produced in LEDGF/p75-deficient cells had normal infectivity. We conclude that LEDGF/p75 fully accounts for cellular trafficking of diverse lentiviral, but not oncoretroviral, integrases and is the main lentiviral integrase-to-chromatin tethering factor. While lentiviral PIC nuclear import is unaffected by LEDGF/p75 knockdown, this protein is a component of functional lentiviral PICs. A role in HIV-1 integration site distribution merits investigation.
The least understood interval in the retroviral life cycle is the series of trafficking and maturation steps that follows the entry of the viral core into the target cell cytoplasm and culminates with integration. These steps must involve interactions with cellular proteins and macromolecular assemblies (51), but the timing and spatial details of particle uncoating, the evolving molecular structure of the preintegration complex (PIC), its passage into the nuclear environment, and in particular, intranuclear preintegration trafficking are all poorly defined. Lentiviruses, a group of species-specific complex retroviruses that cause progressive degenerative diseases, are especially interesting and complex in this regard. Their ability to achieve integration in nondividing cells, e.g., macrophages, stands in intriguing and pathogenetically important contrast to the requirement that genetically simpler gammaretroviruses (e.g., murine oncoretroviruses) have for host cell mitosis (40, 63). A consensus about the underlying mechanisms—in particular, how lentiviral PICs transit the nucleopores of mitotically inactive cells—has been elusive (27). Karyophilic properties of PIC components have deservedly attracted considerable interest and also debate. Candidate effectors of PIC nuclear translocation have included signal-mediated transport directed by peptide determinants within the matrix (MA) (3, 4, 22-24, 28, 31, 73), Vpr (19, 31, 61), and integrase (22). Divergent views exist on the particular roles of nuclear localization signals (NLSs) in MA (21, 24), and MA-deficient viruses can infect nondividing cells with approximately wild-type efficiency under some circumstances (62). Vpr is dispensable in most nondividing cell targets but is needed for efficient replication in macrophages (9, 31, 72). The central DNA flap (18, 74, 78) has also been implicated functionally (18, 78), again with countervailing views (14, 43).The dependence of PIC import on importin-7 has been suggested in semipermeabilized cell assays (17). For reviews, see references 27 and 71.
The role of integrase in PIC trafficking is similarly unresolved. This 32-kDa virion-incorporated enzyme has conceptual appeal as a PIC-targeting determinant because it is an obligate constituent until integration. A suggestive experimental observation is that the human immunodeficiency virus type 1 (HIV-1) integrase protein localizes to cell nuclei (7, 13, 22, 57, 58), a property that it confers to some but not all (37) recombinant fusion proteins. A number of studies have sought to identify discrete NLSs in HIV-1 integrase. A canonical bipartite NLS comprised of two basic amino acid stretches has been reported (22), but contrasting data exist (12, 57, 70). A different peptide initially appeared to be a transferable NLS (1), but follow-up studies clarified that it is not (14, 42). In addition, nonprimate lentivirus integrase proteins lack sequences homologous to these putative peptide NLSs (see reference 64 for an alignment of feline immunodeficiency virus [FIV] and HIV-1 integrases). An NLS has been reported in the integrase of avian sarcoma virus (55), an alpharetrovirus that can infect but cannot productively replicate in nondividing cells (34), and in the integrase of the yeast retrotransposon Ty1 (36, 53).
Interactions of integrase with cellular factors could also influence intranuclear PIC trafficking as well, although such roles have not been established (5). The determinants of PIC trafficking after nuclear import are unknown but are of considerable clinical importance, since recent data have established that HIV-1 integration is not entirely random. HIV-1 integrates preferentially in regions downstream of promoters of active genes (66, 76). In contrast, Moloney murine leukemia virus (MoMLV) integrations show a predilection for promoter regions of genes (76). Integration site preference has important implications for HIV-1 latency and recrudescence after antiretroviral therapy (32) as well as for insertional mutagenesis risks in human gene therapy.
Despite the sufficiency of the purified enzyme to catalyze the early integration reaction steps in vitro (10), interactions of HIV-1 integrase with host proteins in cells have been demonstrated (5, 33, 52). Examples include BAF (39, 44), HMG I(Y) (15), INI-1 (33, 77), DNA-PK (11), hRad18 (54), and most recently, lens epithelium-derived growth factor/p75 (LEDGF/p75) (6, 50). LEDGF/p75 is a widely expressed transcriptional coactivator known to interact with components of the general transcription machinery and with the transcription activation domain of VP16. It was originally identified as a 75-kDa protein that copurified with the coactivator PC4 (26). The acronym derives from an independent isolation from a lens epithelial cell library (68), which, along with other studies, has suggested roles in cellular stress responses and as an auto-antigen in inflammatory diseases (67). An alternatively spliced 52-kDa product of the same gene, which lacks the 205 C-terminal amino acids of p75, acts more potently and with broader specificity as a transcriptional coactivator. Both p75 and p52 display predominantly nuclear localization (56). LEDGF/p75 interacts with HIV-1 integrase in cells (6). Transient small interfering RNA (siRNA) knockdown was reported to produce a diffuse, pancellular distribution of an otherwise nuclear green fluorescent protein (GFP)-integrase fusion protein (50); the cellular distribution of HIV-1 integrase protein itself in the absence of LEDGF/p75 has not been determined. It is also not known if LEDGF/p75 is a component of PICs, if it interacts with integrases of other retroviruses, or what the functional implications of this interaction are for the viral life cycle.
We observed that FIV and other nonprimate lentiviral integrases lack putative peptide NLS sequences proposed for HIV-1 integrase and decided to analyze the comparative karyophilic properties of the integrase proteins of three biologically diverse retroviruses: HIV-1, FIV, and MoMLV. Since the two lentiviruses infect nondividing human cells (40, 59) and MoMLV, a gammaretrovirus, does not (40), we initially sought to correlate karyophilic properties of integrase proteins with viral phenotypes. Stable, Rev-independent expression of the integrases was enabled to facilitate analyses of interactions with endogenous cellular proteins and to avoid the overexpression artifacts to which integrase proteins are prone. Potential transferable NLSs were investigated with pyruvate kinase (PK) fusions (60). Stable siRNA knockdowns of LEDGF/p75 were performed, and the relevance of this protein to the karyophilic properties of the various integrases was studied. Finally, whether LEDGF/p75 is a component of lentiviral PICs and its potential role in the viral life cycle were analyzed.
MATERIALS AND METHODS
Integrase plasmids.
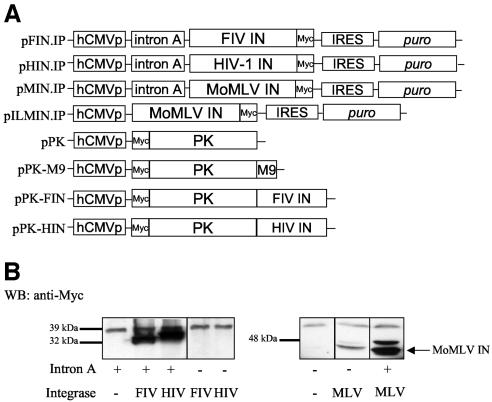
pFIN, pHIN, and pMIN express C-terminally Myc epitope-tagged versions of integrase proteins from FIV 34TF10, HIV-1 YU-2, and MoMLV, respectively. These were constructed by PCR amplification. Primers for FIV were 5′-ATATTCTAGAATGTCTTGGGTTGACAGAATTGAG-3′ and 5′-ATATGGGCCCCTCATCCCCTTCAGGAAGAG-3′. Primers for HIV-1 were 5′-ATATTCTAGAATGTTTTTAGATGGAATAGATAAGGCCC-3′ and 5′-ATATGGGCCCATCCTCATCCTGTCTACCTGCC-3′. The amplicons were inserted as Xba/ApaI fragments into pCMV-Myc, which contains the promoter and intron A of the human cytomegalovirus (CMV) immediate early gene (45). MoMLV integrase was amplified with 5′-ATATGAATTCATGATAGAAAATTCATCACC-3′ and 5′-TATAGAATTCTTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGGGGCCTCGCGG-3′ and inserted as EcoRI fragments into pCMV and pCMV2, which lacks intron A. For use in generating stable cell lines, plasmids pFIN.ip, pHIN.ip, and pMIN.ip were created by inserting into the homonymous plasmids an internal ribosome entry site (IRES)-puromycin resistance gene (pac) cassette distal to the integrase C termini.
pPK-HIN and pPK-FIN express fusion proteins in which PK (Myc tagged) is fused to the amino termini of HIV-1 and FIV integrases, respectively. These plasmids were constructed by inserting integrase cDNAs into pMyc-PK (69), which was a gift of Gideon Dreyfuss (University of Pennsylvania) and is a pcDNA3-based version. The FIV integrase cDNA insert was synthesized with PCR primers 5′-ATATCTCGAGTCACTCATCCCCTTCAGGAAGAG-3′ and 5′-ATATGGTACCGCTGGGTTGACAGAATTGAGGAAGC-3′. The HIV-1 (YU-2) integrase insert was synthesized with 5′-ATATGGTACCGCTTTTTAGATGGAATAGATAAGGCC-3′ and 5′-ATATCTCGAGCTAATCCTCATCCTGTCTACCTG-3′. Both were inserted as KpnI-XhoI fragments into pMyc-PK. pMyc-PK-M9 (69) was used as a positive control.
siRNA expression plasmids.
The LEDGF/p75 siRNA target sequence 1340 (50) is 5′-AAAGACAGCATGAGGAAGCGA-3′, with the numbering indicating the position from the start codon of the mRNA. We also selected an siRNA target sequence at nucleotide (nt) 1428 (5′-AAGACTCTAAATGGAGGATCT-3′). Both are chosen to be distal to the p52/p75-differentiating splice junction to target only LEDGF/p75 while leaving the LEDGF/p52 mRNA unaffected. To construct cells lines stably expressing these anti-LEDGF/p75 siRNAs, 64-nt sense and antisense oligonucleotides with the internal loop of Brummelkamp et al. (2) were synthesized (1340, 5′-GATCCCAGACAGCATGAGGAAGCGATTCAAGAGATCGCTTCCTCATGCTGTCTTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAAGACAGCATGAGGAAGCGATCTCTTGAATCGCTTCCTCATGCTGTCTGG-3′; 1428, 5′-GATCCCGACTCTAAATGGAGGTTTCAAGAGAAGATCCTCCATTTAGAGTCTTTTTTGGAA-3′ and 5′-AGCTTTTCCAAAAAAGACTCTAAATGGAGGATCTTCTCTTGAAAGATCCTCCATTTAGAGTCG-3′). These were annealed to form adaptors and placed downstream of an H1 or a U6 Pol III promoter by ligation between the BamHI and HindIII sites of pSilencer3.1-H1puro and pSilencer2.1-U6hygro (Ambion), resulting in pSi1340 and pSi1428. psiScram, a control derived from pSilencer2.1-U6hygro, expresses a scrambled siRNA (Ambion) validated to avoid silencing of human cellular mRNAs, was used to make control stable cell lines.
Cell culture, transfections, and selection of stable cell lines.
Jurkat, HeLa, and 293T human embryonic kidney cells were obtained from the American Type Tissue Culture Collection and grown in 10% RPMI and Dulbecco's modified Eagle's medium (Gibco BRL), respectively, supplemented with 10% fetal calf serum, penicillin, and streptomycin. Other transfections were performed by the calcium phosphate coprecipitation method with a total of 2 μg of DNA per well of a six-well plate or 1 μg of DNA per chamber in a two-chamber LabTek II glass chamber slide (Nalge Nunc, Naperville, Ill.). Briefly, cells were transfected 24 h after being plated in 2 ml of medium at 0.45 × 106 cells/well or 1 ml of medium at 0.8 × 106 cells/chamber. After 14 to 16 h, the transfection mix was replaced with fresh culture medium. Cells were harvested or used for indirect immunofluorescence 40 to 48 h after the transfection mix was added. For stable cell lines, 3 × 106 293T cells were plated in 75-cm2 flasks and cotransfected the next day with 7 μg of DNA that had been linearized at a restriction site in the prokaryotic backbone. After 14 to 16 h, the transfection mix was replaced with fresh culture medium. To generate Jurkat stable cell lines, 107 cells were electroporated with 10 μg of linearized plasmid and 20 μg of carrier pCDNA3. At 48 h after transfection, puromycin (3 to 0.6 μg/ml) or hygromycin (200 to 800 μg/ml) was added.
Immunoblotting and confocal immunofluorescence microscopy.
For Western blotting, cells were lysed in 150 mM NaCl, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40, and 150 mM Tris-HCl (ph 8.0) plus a protease inhibitor cocktail (Complete-mini; Boehringer). Proteins (30 μg/lane) were resolved in SDS-10% polyacrylamide gels and transferred to Immobilon P membranes (Millipore). Blocked membranes were incubated overnight at 4°C with anti-Myc monoclonal antibody (MAb) (clone 9E10; Covance), anti-α-tubulin MAb (clone B-5-1-2; Sigma), and anti-LEDGF/p75 MAb (BD Transduction Laboratories) in Tris-buffered saline-5% nonfat milk plus 0.05% Tween 20. After washing, membranes were incubated with the appropriate horseradish peroxidase-tagged secondary antibody. Bound antibodies were detected by enhanced chemiluminescence (Amersham Pharmacia Biotech). Indirect immunofluorescence detection of Myc-tagged proteins was performed by confocal fluorescence microscopy with an anti-Myc epitope MAb (clone 9E10; Covance) or a polyclonal anti-Myc antibody (Santa Cruz) when costained. Endogenous LEDGF/p75 was detected by using anti-LEDGF/p75 MAb (BD Transduction Laboratories). MoMLV IN was detected with a rabbit polyclonal antiserum (a gift from S. Goff). Cells grown in LabTek II chamber slides (Nalge Nunc) were then fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min at 37°C, washed with PBS, and then permeabilized with ice-cold methanol for 2 min at room temperature. Fixed cells were blocked in 10% fetal calf serum, 20 mM ammonium chloride, and PBS for 30 min at room temperature and then incubated with the appropriate antibodies, followed by Alexa-488- or Texas Red-conjugated goat anti-mouse antibody or goat anti-rabbit antibody (Molecular Probes, Eugene, Oreg.). Nuclear DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma).
Immunoprecipitation.
Cytoplasmic and nuclear fractions were isolated by using the method of Cherepanov et al. (6). Briefly, 107 cells stably expressing HIV or FIV integrase were lysed for 10 min on ice in modified CSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 6.8), 10% (wt/vol) sucrose, 1 mM dithiothreitol, and 1 mM MgCl2 plus an EDTA-free protease inhibitor mixture (Roche Molecular Biochemicals)] supplemented with 0.5% NP-40 and 100 mM NaCl. Nuclear and cytoplasmic fractions were separated by centrifugation for 5 min at 500 × g. The supernatant (cytoplasmic fraction) was then centrifuged at 6,000 × g for 2 min and used in immunoprecipitation experiments. The nuclear fraction was washed twice in the lysis buffer and incubated for 5 min on ice in CSK buffer with 0.5% NP-40 and higher salt (400 mM NaCl), followed by centrifugation at 6,000 × g for 2 min. Nuclear and cytoplasmic fractions from FIV IN-expressing cells were mixed with 3 μl of a feline anti-FIV serum (kindly provided by M. Barr; Petaluma) or normal feline serum. Fractions from HIV IN-expressing cells were mixed with 6 μl of a mixture of 3 anti-HIV IN rabbit antisera or normal rabbit serum. These samples were incubated on ice for 20 min, then 30 μl of protein A-Sepharose was added, and samples were incubated at 4°C for 1 h on a rotating platform. Beads were washed three times in CSK buffer containing 0.5% NP-40 and boiled in Laemmli buffer plus β-mercaptoethanol. Samples were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with an anti-LEDGF MAb as described above.
Lentiviral vector and reporter virus production.
Vesicular stomatitis virus glycoprotein G-pseudotyped internally promoted vectors (three plasmid systems) and reporter viruses with env deletions (two plasmid systems) were produced by calcium phosphate cotransfection in 293T cells. FIV vectors have been previously described (46-49) and were produced by cotransfection of 293T cells with 3 μg of FIV packaging plasmid pFP93, 3 μg of enhanced GFP (eGFP) transfer vector pGiNWF or lacZ transfer vector pCT26, and 1 μg of pMD.G. FIV viruses with env deletions were produced by transfecting 293T cells with pCT5efs (35) and 1 μg of pMD.G. For HIV-1 vectors, 3 × 106 293T cells were cotransfected with the three-plasmid system (79): pCMdeltaR8.9 (3 μg), pHR′CMVlacZ (3 μg), and pMD.G (1 μg). To produce HIV-1 reporter viruses, 293T cells were cotransfected with 1 μg of pMD.G and 6 μg of pHIVeGFP, an HIV-1 NL4-3-derived clone with a deletion in HIV-1 env and an insertion of egfp to replace the nef open reading frame. Alternatively, we used 1 μg of pMD.G with 6 μg of pNL43.Luc.R−E− or pNL43.Luc.R+E− (9), which have firefly luciferase (luc) replacing the nef gene, as well as a frameshift in env and vpr or a deletion in env, respectively. Packaging plasmids with the class I integrase mutations (49, 64) were substituted to produce the control preparations described in the preceding section. Eighteen hours after transfection, the culture medium was replaced, and 48 h after transfection, the supernatant was harvested, clarified by low-speed centrifugation, filtered (0.45 μM), and stored at −80°C. Reverse transcriptase (RT) activities and titers on 293T cells were determined for all preparations.
In vitro integration assay and PIC immunoprecipitation.
PICs were isolated as described by Farnet and Haseltine (16) and Lin and Engelman (44) with minor modifications. Briefly, 3 × 106 293T cells were transduced with vesicular stomatitis virus glycoprotein G-pseudotyped vectors or viruses with env deletions at a multiplicity of infection (MOI) of 5. As a control in some experiments, class I integrase mutant HIV and FIV vectors were used which had their RT activities normalized to those of the wild-type preparations. The class I integrase mutant vectors were produced with a packaging plasmid which had mutations (D64N/D116N for HIV-1 and D66V for FIV) as described previously (64). After the addition of virus, cells were left undisturbed for 9 h, at which point they were harvested by trypsinization and washed three times in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Cells were then lysed for 10 min on ice in 800 μl of buffer K (20 mM HEPES [pH 7.6,] 150 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol plus an EDTA-free protease inhibitor mixture [Roche Molecular Biochemicals]) supplemented with 0.25% NP-40. Cytoplasmic fractions were isolated by centrifugation of the cellular lysate at 500 × g for 5 min and used, with and without intervening PIC immunoprecipitation, in integration assays. For PIC immunoprecipitations, 3 μg of anti-LEDGF or control (anti-Myc) MAbs were added to 300 μl of the cytoplasmic fractions and incubated on ice for 20 min. Then 30 μl of magnetic beads coupled to anti-mouse immunoglobulins (Dynabeads) was added and incubated at 4°C for 1 h with rotation. Beads were then washed three times in buffer K containing 0.25% NP-40. In vitro integration assay were done as described previously (16, 44). PICs from 70 μl of total cytoplasmic fractions, or immunoprecipitated PICs, were mixed with 750 ng of PstI-linearized PhiX174 RF I DNA in buffer K and incubated for 45 min at 37°C in a rotary shaker. For a control to establish DNA target dependence, PICs were incubated in the absence of phage DNA. Integration reactions were stopped by treatment with proteinase K (1 mg/ml) in 10 mM EDTA plus 0.5% SDS for 30 min at 55°C. DNA was then recovered by phenol-chloroform extraction and ethanol precipitation. Integration products were analyzed by Southern blotting with random-labeled 32P probes that hybridize within gag-pol of HIV or FIV.
Viral infections.
For infections, cell lines were seeded in six-well plates at 0.45 × 106 cells/well. Twenty-four hours later, cells were treated with aphidicolin (10 μg/ml), which was verified by flow cytometry to produce full G1-S arrest. Cells were transduced 24 h after the aphidicolin block. Aphidicolin was replenished with all medium changes. Dividing cells were seeded at 0.45 × 106 cells/well in a six-well plate and transduced 24 h later. Dividing and nondividing cells were transduced with RT-normalized stocks of HIV-1 or FIV vectors. The transduction supernatant was removed 24 h after transduction. At 48 h after transduction, cells transduced with eGFP-expressing vectors were fixed with 1% formaldehyde and analyzed by flow cytometry (FACScan; Becton Dickinson, Mountain View, Calif.). Jurkat cells were infected overnight with HIV-1 NL4-3 at a 0.1 MOI. After infection, cells were washed three times and cultured for 2 weeks. β-Galactosidase activity was analyzed in lacZ vector-transduced cells with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Luciferase activity was measured in a TopCount apparatus (Packard). HIV-1 p24 was measured by enzyme-linked immunosorbent assay (Coulter).
Quantification of 2-LTR circles.
LEDGF/p75-deficient (si1340 cells, si1340/1428) and control (siScram) cells were treated with 10 μg of aphidicolin/ml 24 h before transduction with eGFP-expressing vectors. Hirt DNA was collected at 3, 6, 12, and 18 h posttransduction. Two-long terminal repeat (2-LTR) circles were quantified by real-time PCR in a Roche LightCycler with primers specific to the 2-LTR circle junctions of FIV and HIV-1 by using the Roche FastStart DNA kit. The HIV-1 primers used were 5′-AACTAGGGAACCCACTGCTTAAG-3′ and 5′-TCCACAGATCAAGGATATCTTGTC-3′; FIV primers are described in reference 64. Five hundred nanograms of HIRT DNA was used for each FIV reaction, and 750 ng was used for each HIV reaction. A 2-LTR circle template was constructed by deleting viral sequences between the two LTRs of pCT5 (59) or pYU-2, a gift of G. Shaw (41). Tenfold dilutions of these templates served as standards. Real-time PCR quantification of human mitochondrial DNA was used to normalize the samples. The identity of PCR products was also confirmed by electrophoresis in agarose gels.
LEDGF/p75 RT-PCR.
cDNA was prepared from 500 ng of total RNA from control or LEDGF/p75-deficient cell lines by random hexamer priming. Minus RT controls omitted RT. One-seventeenth of the total cDNA was used in a PCR with LEDGF/p75-specific sense (5′-GAAGCCAGAAGTTAAGAAAGTGG-3′) and antisense (5′-GCTTTTTGTTTGGCCCTTTCTTCC-3′) primers. β-Actin was amplified as a loading control.
RESULTS
To facilitate study of native FIV and HIV-1 integrase protein properties and interactions with endogenous cellular proteins in the absence of other viral proteins or any fusion partners, we first addressed two problems that are recognized to complicate integrase localization studies: Rev dependence (7, 58, 65) and the tendency of integrase to artifactually localize or precipitate after transient transfection-mediated overexpression (7).
Intron A confers Rev independence to lentiviral integrases.
Integrase is normally expressed as a C-terminal proprotein within the Rev-dependent Gag/Pol precursor, from which it is cleaved by the viral protease. The integrase-encoding portion of the pol mRNA contains particularly strong instability (INS or CRS) elements that complicate expression from a subcloned cDNA (8, 65). The use of other viral RNA transport elements and placement of the Rev response element in the 3′ untranslated region of the integrase mRNA and supplying Rev in trans (ordinarily quite successful for Rev-dependent genes) also have not reliably enabled expression (58; data not shown), and in any case, we wished to express integrase in the absence of any other viral proteins, including Rev. Another option for HIV-1 pol-encoded proteins, a codon-optimized cDNA (7, 65), has not been synthesized yet for FIV.
After testing a variety of cellular and viral promoter arrangements in expression plasmids, we found that incorporating intron A of the human CMV immediate early gene enabled Rev-independent expression of full-length FIV and HIV-1 integrase proteins (Fig. 1). Using the same CMV promoter without intron A consistently failed to enable Rev-independent expression, as did a variety of other commercially available expression plasmids (Fig. 1B). In contrast to the lentiviral integrases, MoMLV integrase expression did not require the presence of intron A but was considerably enhanced by it (Fig. 1B).
FIG. 1.
Intron A is necessary and sufficient for Rev-independent HIV-1 and FIV integrase expression. (A) Expression plasmids; (B) intron A dependence. 293T cells transfected with the indicated plasmids were analyzed by Western blotting (WB) with an anti-Myc MAb. +, present; −, absent.
Stable expression of integrase proteins.
As previously observed (7), we found that transient transfection in HeLa or 293T cells of integrase-expressing constructs could produce heterogeneous artifactual inclusions in cytoplasmic or perinuclear locations (data not shown). These bore no resemblance to the intracellular distributions ultimately observed with stable, lower expression. This source of artifact was not a problem in our hands if the integrase was part of a larger fusion with a nonviral protein such as GFP or PK, but we deemed it critical to study the properties of native, unfused integrase proteins as well. Because stable expression would enable lower, more physiologically relevant and more consistent integrase expression levels and improve analysis of interactions with endogenously expressed cellular proteins, it is the principal method used here for both FIV and HIV-1 integrase. To accomplish this, we inserted poliovirus IRES-linked selectable marker genes downstream of the integrase cDNAs in the intron A-containing plasmids described above (Fig. 1A). In addition, to exclude artifactual localizations that have been shown to result from cryptic splicing (14, 42) or that could result from protein degradation, the expression of single proteins of predicted size was confirmed by immunoblotting in all experiments described in subsequent sections.
Intracellular distribution of retroviral integrases.
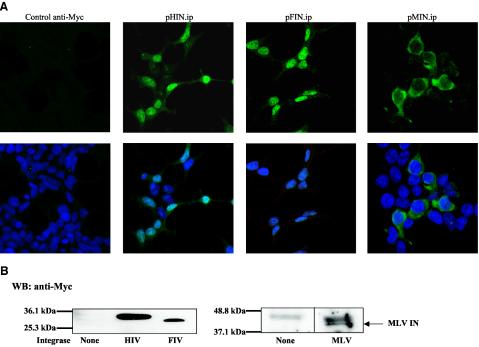
The stably expressed HIV-1 and FIV integrases were consistently located in the nucleus. Integrase expression was clearly identified by immunostaining in more than 90% of cells selected in puromycin, with some variation in intensity of expression from cell to cell, and was always nuclear (Fig. 2A). Myc epitope-tagged integrases detected with anti-Myc antibodies and untagged integrase detected by polyclonal antibodies to HIV-1 and FIV integrase had identical appearances (data not shown). Immunoblotting showed single, predicted-size proteins when performed with either anti-Myc antibodies (Fig. 2B) or polyclonal antisera against FIV, HIV-1, and MoMLV integrases (data not shown). In clear contrast to the lentiviral integrases, stably expressed MoMLV integrase was found exclusively in the cytoplasmic compartment. This subcellular localization was observed whether the cells were stained with an anti-MoMLV polyclonal antibody (Fig. 2A) or with an anti-Myc MAb (data not shown). An interesting finding was that when stable cell lines employing the same plasmid background (including intron A) were evaluated on Western blots with the same anti-Myc antibody, MoMLV integrase was consistently seen to accumulate to lower levels than the lentiviral integrases but was nevertheless readily detected.
FIG. 2.
Karyophilic properties of retroviral integrase proteins in stable cell lines. (A) Confocal immunofluorescence microscopy of puromycin-selected 293T cells stably transfected with pHIN.ip, pFIN.ip, and pMIN.ip. The primary antibody used in the left three panels was an anti-Myc MAb. In the rightmost panel, an anti-MoMLV IN antibody was used. The bottom panels are overlays with nuclear DAPI staining. Photographic exposure times were equal. (B) Immunoblotting of the same cell lines. HIV-1, FIV, and MoMLV integrase are 34, 32, and 46 kDa, respectively.
Lack of transferable NLSs in lentiviral integrases.
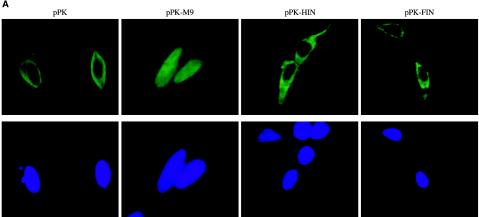
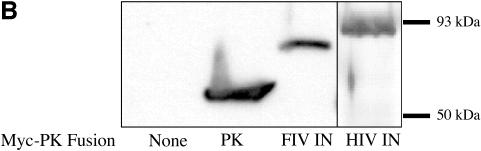
To evaluate whether FIV and HIV-1 integrases have transferable NLSs that determine their observed karyophilia, we constructed fusion proteins in which each was fused in frame to the carboxyl terminus of N-terminally Myc-tagged chicken PK (Fig. 1A). PK is commonly used in this way as an indicator to identify NLSs because it is a large (ca. 55 to 60 kDa), normally strictly cytoplasmic and monomeric protein that exceeds the diffusion limit of nucleopores (1, 20, 69). As shown in Fig. 3A, PK localizes, as expected, to the cytoplasm. As a positive control, PK was transported into the nucleus when fused in frame with M9 (60), a 38-amino-acid NLS from the hnRNP A1 protein (Fig. 3A). In contrast, the fusions of PK with FIV and HIV-1 integrase remained cytoplasmic. Single bands corresponding to full-length fusion proteins were verified in immunoblots (Fig. 3B). The same results were obtained when HeLa cells were used instead of 293T cells (data not shown). These data established that neither lentiviral integrase contains a transferable NLS as assayed by fusion to PK. By comparison, we found that smaller integrase fusion proteins that do not clearly exceed the nucleopore diffusion limit (e.g., when GFP was fused to either integrase), produced inconsistent localizations with variable distribution between the nucleus and cytoplasm, but nuclei were favored on average (data not shown). In addition, we constructed the PK-integrase fusion proteins in a pcDNA3.1 background. Other experiments (data not shown) confirmed the recent report (14) that aberrant proteins generated by cryptic splicing caused the apparent nuclear localization previously observed with pcDNA1 constructs encoding HIV-1 integrase fused to PK.
FIG. 3.
Lentiviral integrases lack transferable NLSs. (A) Cellular distribution of PK fusion proteins. 293T cells were transfected with pPK, pPK-M9, pPKHIN, or pPKFIN and analyzed by confocal immunofluorescence microscopy with anti-Myc antibody. (B) Immunoblotting of cells shown in panel A with an anti-Myc MAb.
FIV integrase interacts with endogenous LEDGF/p75.
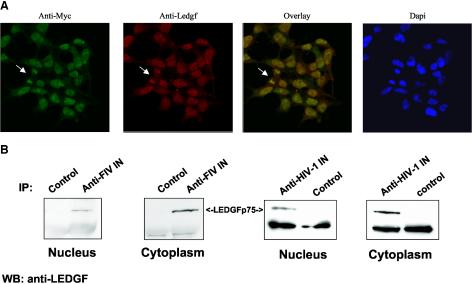
FIV and HIV-1 integrase share three conserved amino acids involved in catalysis (D64, D116, and E152) (64), but the level of homology between the two proteins (37% overall identity) precluded assuming from the known interaction with HIV-1 integrase (6) that FIV integrase would interact with LEDGF/p75. We found, however, in confocal immunofluorescence analyses of the stably expressing cells that this nonprimate lentiviral integrase does colocalize with endogenous human LEDGF/p75, both of which were found to be prominently nuclear (Fig. 4A). Colocalization of FIV integrase with LEDGF/p75 was identical to the pattern seen when HIV-1 integrase was stably expressed (data not shown). Colocalization to condensed chromosomes in mitotic figures was also observed, which is consistent with a strong interaction with chromatin (Fig. 4A).
FIG. 4.
Interaction of lentiviral integrases with LEDGF/p75. (A) FIV integrase colocalizes with endogenous LEDGF/p75. Cells stably expressing FIV integrase were double stained with an anti-Myc polyclonal antibody and an anti-LEDGF MAb and analyzed by confocal immunofluorescence microscopy. The arrow indicates a mitotic figure. (B) FIV and HIV-1 integrases coimmunoprecipitate with endogenous LEDGF/p75. Samples were immunoprecipitated (IP) with a feline anti-FIV antiserum or rabbit anti-HIV integrase antiserum and then probed with an anti-LEDGF MAb.
The stable cell lines also facilitated direct evaluation of interactions of endogenous LEDGF/p75 with the endogenously expressed lentiviral integrases. We immunoprecipitated FIV and HIV-1 integrase from nuclear and cytoplasmic fractions of the respective stable integrase-expressing cells by using anti-integrase-specific antisera. Immunoprecipitated proteins were immunoblotted for LEDGF/p75 with a specific MAb. As shown in Fig. 4B, LEDGF/p75 coimmunoprecipitated with both HIV-1 integrase and FIV integrase. It is noteworthy that the lentiviral integrase-LEDGF interactions are detected by coimmunoprecipitation from both the cytoplasmic and nuclear compartments despite the strongly nuclear colocalization detected by immunofluorescence. LEDGF/p75 may be a shuttle protein or the proteins may interact before either is imported into the nucleus. Secondly, nuclear LEDGF is tightly bound to chromatin (Fig. 4A), which may reduce the amount recovered from nuclei and result in an apparently larger amount of LEDGF in cytoplasm by sensitive immunoprecipitation assays.
Generation of LEDGF/p75-deficient cell lines.
To further evaluate the role of LEDGF/p75, we stably expressed siRNAs to generate LEDGF/p75-deficient 293T and Jurkat cell lines. To avoid introducing viral sequences that could subsequently confound analyses of retroviral integrations, all knockdown lines were made by stable plasmid transfection rather than transduction. siRNA target sites downstream of the p52/p75-differentiating splice junction were used to achieve p75-specific targeting. Two 21-nt sequences, one at nt 1340 and the other at nt 1428 of the coding sequence, were targeted by using siRNA constructs in which an intervening 9-nt loop connects the sense and antisense portions (2). These two siRNAs were selected for use after their potencies were determined by transient cotransfections of the expression plasmids with or without a LEDGF/p75 expression plasmid. Each proved potent, eliminating not only detectable endogenous LEDGF/p75 but also exogenously cotransfected protein (data not shown). The 1340 and 1428 hairpin siRNA sequences are under the transcriptional control of Pol III promoters (H1 and U6, respectively), with separate Pol II-promoted transcription units encoding puromycin (1340) or hygromycin (1428) resistance.
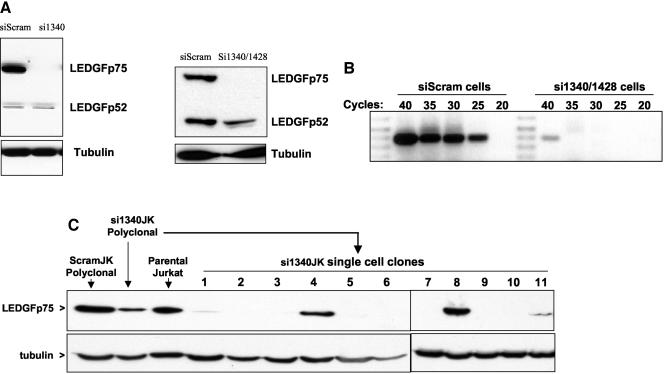
Stable cell lines expressing these siRNAs and control cells stably expressing a scrambled siRNA were then generated. The resulting drug-resistant cell lines were highly polyclonal, representing stable outgrowth in puromycin or hygromycin from at least 1,000 separate stable transfection events. As shown in Fig. 5A, polyclonal 293T cells stably expressing the 1340 siRNA (si1340 cells) or both the 1340 and 1428 siRNAs (si1340/1428 cells) lack detectable expression of LEDGF/p75 protein. LEDGF/p75 was never detectable on Western blots in these cell lines, even with maximal film exposure (data not shown), while the control siRNA line (designated siScram cells) and the parental cell line express large and equivalent amounts of LEDGF/p75. Moreover, the siRNAs had no effect on the expression of the p52 variant of LEDGF, providing a rigorous intrasample control for loading and blotting sensitivity with the N terminus reactive antibody and proving that the siRNA specifically targets the spliced p75 mRNA as planned (Fig. 5A). No differences in morphology, growth rate, or adherence were discernible between si1340, si1340/1428, siScram, and parental cell lines, and continued propagation under selection for multiple passages over several months did not result in the emergence of increased LEDGF/p75 expression in the silenced cells. In agreement with the lack of detectable protein by immunoblotting, LEDGF/p75 was also not detectable by immunofluorescence microscopy in either the si1340 cell line or the si1340/1428 cells but was prominently seen under the same conditions in the nuclei of siScram cells (data not shown). The drastic extent of the siRNA knockdown was confirmed at the RNA level by RT-PCR (Fig. 5B). mRNA could be detected in si1340/1428 cells only intermittently and was at the limit of detection, i.e., only with 40 cycles of PCR in some assays (Fig. 5B) but not others (data not shown); from the comparisons with siScram cells, we estimate an approximate 4 to 5 log knockdown.
FIG. 5.
Generation of LEDGF/p75-deficient cell lines. (A) Immunoblotting. LEDGF/p75 protein expression was evaluated by Western blotting with anti-LEDGF MAb in siScram cells and si1340 cells (left) and si1340/1428 cells (right). (B) mRNA levels. LEDGF/p75 mRNA was quantitated by RT-PCR in the same cell lines. (C) LEDGF/p75 protein expression in Jurkat cell lines.
These cell lines derived from 293T cells displayed the most complete knockdown of cell lines we have so far tested. The stable knockdown achieved in siRNA-expressing Jurkat cell lines (designated si1340JK and si1340/1428JK cells) was only partial at the polyclonal level (Fig. 5C). The addition of the 1428 siRNA to si1340JK cells did not result in enhanced knockdown (data not shown). Immunoblotting of single cell clones derived from the si1340JK cells by limiting dilution revealed that the low levels of LEDGF/p75 seen in the polyclonal si1340JK cell population result from the contributions of high-expressing clones amidst clones that expressed no detectable LEDGF/p75 protein (Fig. 5C). Immunoblot-negative si1340JK clones showed a drastic decrease in LEDGF/p75 mRNA, although at 40 cycles of RT-PCR, mRNA was detectable (data not shown). Because the 293T cell-derived si1340 and si1340/1428 cells were more stringently defective for LEDGF/p75 expression than the Jurkat-derived lines, and because an adherent cell line was more suitable for nuclear localization studies, we initially concentrated on them for analyses.
Endogenous LEDGF/p75 is required for nuclear localization and chromatin tethering of two lentiviral integrase proteins.
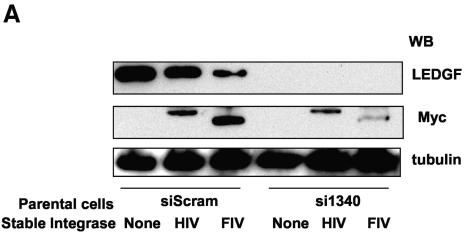
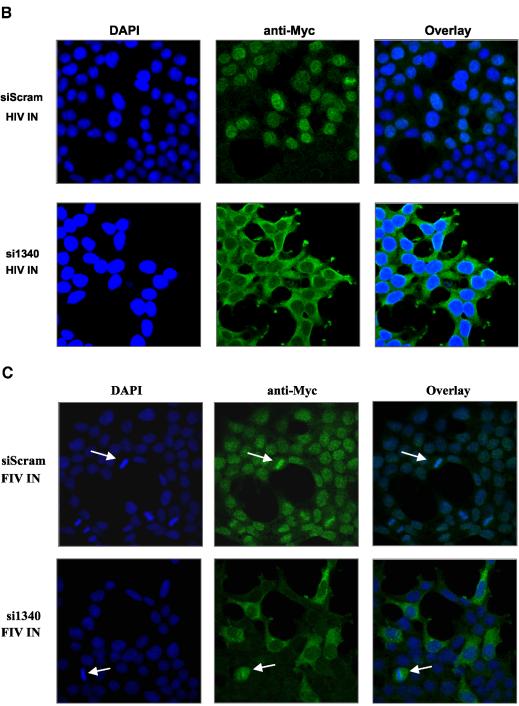
We proceeded to introduce Myc-tagged HIV-1 and FIV integrase constructs into the hygromycin-selected stable siScram and si1340 cell lines. Puromycin selection was used. The expression of both LEDGF/p75 and integrase proteins was evaluated by immunoblotting (Fig. 6A) and immunofluorescence (Fig. 6B and C). HIV-1 and FIV integrases localized in nuclei of the siScram cells as expected. In striking contrast, both lentiviral integrases were restricted to the cytoplasm in cells specifically deficient in LEDGF/p75 (Fig. 6B and C). In clear contrast to the striking relocalization of the lentiviral integrases, the strictly cytoplasmic distribution of MoMLV integrase was the same in both si1340 cells and siScram cells (data not shown).
FIG. 6.
LEDGF is required for nuclear localization of lentiviral integrase proteins. (A) Stable HIV-1 and FIV integrase expression in si1340 cells and siScram cells; (B and C) subcellular distributions of lentiviral integrase proteins in the same cells were analyzed by confocal immunofluorescence microscopy with anti-Myc antibody.
These data involving specific, stable, and highly effective removal of LEDGF/p75 from cells with siRNAs show that both lentiviral integrases depend on LEDGF/p75 for their nuclear localization. It is also clear that in the absence of LEDGF/p75, LEDGF/p52 does not supply this role. In addition, MoMLV integrase is not affected by the siRNA targeting and lacks nuclear targeting whether LEDGF/p75 is present or absent. Thus, LEDGF/p75 direction of integrases to nuclei and tethering to chromatin appears to be a lentiviral integrase-specific phenomenon.
Endogenous LEDGF/p75 is a component of functional HIV-1 and FIV PICs.
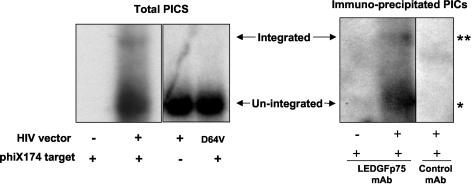
Having characterized the interactions with free integrase proteins, we asked whether endogenous LEDGF/p75 associates with HIV-1 and FIV PICs in infected cells. 293T cells were infected with single-round HIV-1 and FIV vectors. As a control to confirm the detection of functional PIC integrase activity, vectors of two types were used: those having wild-type integrase and those having a class I integrase mutation, i.e., a single amino acid change that selectively disables only integrase catalytic function while leaving intact other aspects of the viral life cycle (38, 49, 64). PICs were isolated from the cytoplasmic fraction of infected cells and evaluated in a previously described in vitro integration assay that uses linearized, double-stranded phiX174 phage DNA as an integration target, followed by vector-specific Southern blotting to detect a high-molecular-weight integration-specific band as well as a more prominent lower band representing total PICs (44). PICs of each vector were evaluated in this assay in two ways: directly and after immunoprecipitation with an anti-LEDGF/p75 MAb or a control MAb (anti-Myc) (Fig. 7). Only PICs produced by virions having wild-type integrase integrated into the target DNA, establishing that the generation of this band requires a catalytically functional integrase (Fig. 7, left panel). Formation of an integration band also depended on inclusion of the phiX174 target DNA in the reaction mixture. Three important results were noted. First, as seen in the right panel of Fig. 7, HIV-1 PICs were prominently immunoprecipitated with the anti-LEDGF/p75 MAb but not by the control MAb. Second, integrated HIV-1 vector genomes were detected in reactions with the anti-LEDGF/p75 MAb-immunoprecipitated samples. Third, the ratio of integrated to total PICs was similar to that in nonimmunoprecipitated PICs shown in the left panel of Fig. 7. The same results were found when FIV vector PICs were used (data not shown). These data show that LEDGF/p75 is a constituent of integration-competent PICs of both lentiviruses.
FIG. 7.
LEDGF/p75 is a component of functional PICs. HIV-1 vector PICs were isolated and used in an in vitro DNA integration assay before (left) and after (right) immunoprecipitation with an MAb to LEDGF/p75 or a control (anti-Myc) MAb. Integration products are detected as a larger band that is a fraction of the nonintegrated PICs. The reaction is shown to require both a functional integrase (IN mutant tested is D64V) and the presence of the target DNA. Single asterisk, HIV-1 PICs prominently immunoprecipitated with anti-LEDGF/p75 MAb; double asterisk, integrated HIV-1 vector genomes detected in reactions with anti-LEDGF/p75 MAb-immunoprecipitated samples. The same results were obtained with FIV PICs (data not shown).
Normal PIC nuclear import and single-round infection by HIV-1 and FIV in drastically LEDGF/p75-deficient cells.
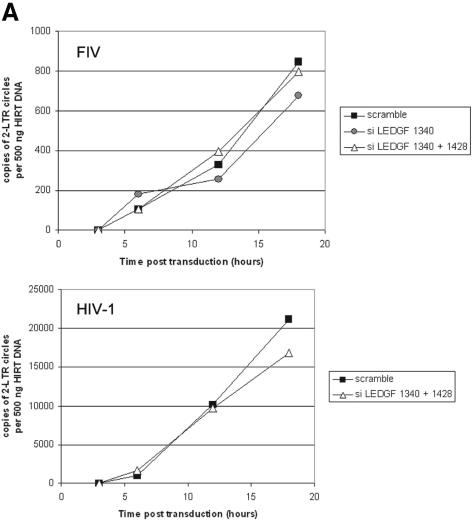
Having demonstrated that this protein that targets lentiviral integrases to nuclei is also a component of functional lentiviral PICs, it was of strong interest to determine the influence of LEDGF/p75 on PIC nuclear import. siScram, si1340, and si1340/1428 cells were growth arrested with aphidicolin treatment and infected with HIV-1 and FIV vectors encoding a reporter transgene. At 3, 6, 12, and 18 h postinfection, low-molecular-weight DNA was extracted and 2-LTR circles were quantified by real-time PCR. As shown in Fig. 8A, the kinetics of 2-LTR circle formation were identical in both the siScram and LEDGF/p75-deficient cell lines. These data indicated that the knockout of LEDGF/p75 in these cell lines does not affect the nuclear import of HIV-1 and FIV PICs.
FIG. 8.
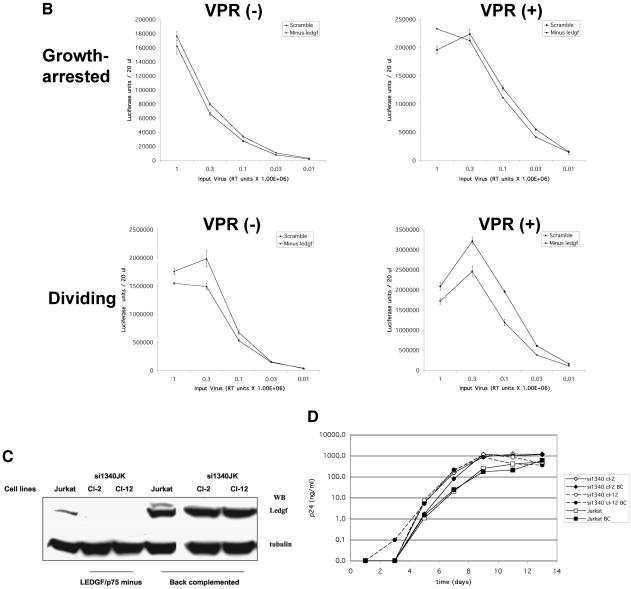
Lentiviral infection of LEDGF/p75-deficient cells. (A) Two-LTR circle formation in growth-arrested control and LEDGF/p75-deficient cells infected with HIV eGFP or an FIV lacZ vector (CT26). (B) Infection of LEDGF/p75-deficient si1340/1428 cells (squares) and siScram (diamonds) with pNL43.Luc.R−E− or pNL43.Luc.R+E−. Luminescence units are shown on the y axes. (C) Back complementation of si1340JK clones with LEDGF/p75. (D) HIV-1 NL4-3 replication in si1340JK clones with or without back complementation with LEDGF/p75. Jurkat BC cells are wild-type Jurkat E6 cells which were stably transfected with the same LEDGF/p75 expression construct.
We then assessed the ability of LEDGF/p75-deficient cells to support infection with HIV-1 and FIV. siScram, si1340, and si1340/1428 cells were first infected with pNL43.Luc.R−E− or pNL43.Luc.R+E− (9). Infections were done over a wide dynamic range of viral input and under conditions of proliferation and growth arrest. The expression of luciferase was analyzed 48 h after infection. As shown in Fig. 8B, control and LEDGF/p75-deficient cells displayed equivalent luciferase expression, whether dividing or growth arrested. Multiple other experiments with various combinations of HIV-1 or FIV vectors and viruses under different conditions were performed and corroborated these and the 2-LTR circle results in showing a lack of a simple antiviral effect from drastic LEDGF/p75 knockdown (supplemental material is available at http://mayoresearch.mayo.edu/mayo/research/poeschla/index.cfm). Briefly, no differences in titer or in mean fluorescence intensity occurred between siScram and si1340/1428 cells after either HIV-1 or FIV eGFP vector transduction. Growth-arrested siScram and si1340/1428 cells transduced with HIV and FIV lacZ vectors having wild-type integrase or class I integrase mutations expressed substantial but equivalent amounts of β-galactosidase at 48 h, which has previously been shown to be proportional to unintegrated nuclear cDNA accumulation, predominantly 2-LTR circles (64). This result suggests again that PIC nuclear import was not affected. For both lentiviral vectors as well, β-galactosidase expression was identical in control and LEDGF/p75-deficient cell lines after 14 days of passage, consistent with equivalent levels of integration and stable transgene maintenance in the presence and absence of LEDGF/p75. Similarly, in multiple LEDGF/p75-deficient si1340JK cell clones, an HIV-1 GFP reporter virus produced a percent transduction and mean fluorescence intensity equivalent to those of siScramJK cells or parental Jurkat cells. The use of viruses that lack Vpr and vectors that lack the central DNA flap and Vpr rules out any LEDGF/p75-overriding contribution by these potentially karyophilic elements.
Taken together, these results show that, despite its profound effect on free integrase trafficking, removal of LEDGF/p75 does not alter susceptibility to infection of the cell lines tested here with HIV-1 or FIV, whether the cells are dividing or nondividing. In addition to indicating that a simple antiretroviral effect does not result from knockdown, they also suggest that despite the strong association of LEDGF/p75 with chromatin, knockdown does not alter integration site distributions to a degree that is sufficient to strongly influence aggregate short-term proviral expression after integration.
To determine whether the presence of LEDGF/p75 in the producer cell is important (e.g., similar to APOBEC3G/CEM15), we produced HIV-1 NL43.Luc.R−E− in si1340/1428 and siScram cells, normalized for RT activity, and determined infectivity on si1340/1428, siScram, and 293T cells. The LEDGF/p75 expression status of the producer cells did not affect luciferase expression, whether target cells were LEDGF/p75 positive or LEDGF/p75 negative (data not shown). The same equivalence was observed when cells were evaluated 1 week after transduction.
HIV-1 replication in a LEDGF/p75-deficient T-cell line.
We next evaluated whether HIV-1 replicates normally when CD4+ T cells that are permissive for productive replication are made LEDGF/p75 deficient. As noted above, Western blotting revealed a substantial but still partial knockdown in the polyclonal si134JK cells, which was shown by single-cell cloning to represent variable, clonally distinct LEDGF/p75 expression levels, which range from wild type to undetectable (Fig. 5C). Two clones in which LEDGF/p75 was not detected, Cl-2 and Cl-12, were selected (Fig. 8C). Cl-2 and Cl-12 were verified by flow cytometry to have substantial cell surface CD4 and CXCR4, equivalent to that of parental Jurkat cells (data not shown). RT-PCR confirmed severe LEDGF/p75 knockdown (data not shown). These two si1340JK single cell clones were then back complemented by stable transfection of a LEDGF/p75 cDNA mutated in the siRNA target region (and linked by an IRES to a puromycin resistance gene), thus generating clone pairs positive and negative for LEDGF/p75. The seven introduced nucleotide changes in the siRNA target site were synonymous, preserving the protein sequence. This is the most rigorous strategy for such experiments, since it eliminates the potential for the oligoclonal selection artifacts that could eventuate from simply comparing populations of Jurkat cells selected to express the active and scrambled siRNAs; it also ensures that any observed phenotype is due specifically to lack of the protein in question (25). Back complementation in each si1340JK cell clone was confirmed by immunoblotting (Fig. 8C). Both LEDGF/p75-deficient and back-complemented cells were then infected at a 0.1 MOI, as were parental Jurkat cells. Viral replication was not affected by the absence of LEDGF/p75 (Fig. 8D). Engineering of supranormal levels of LEDGF/p75 in parental Jurkat cells (Jurkat BC cells; Fig. 8C) also had no significant effect on HIV-1 replication (Fig. 8D).
DISCUSSION
Here we identify specific interactions of the transcriptional coactivator LEDGF/p75 with the PICs and integrase proteins of two different lentiviruses. These interactions are evolutionarily conserved between primate and nonprimate lentiviruses and are not observed with the integrase of a murine oncoretrovirus, MoMLV, suggesting a role specific to the biology of lentiviruses. Further extension of this comparative approach to nonlentiviruses and nongammaretroviruses will be informative.
It is clear that the interaction of LEDGF/p75 with FIV and HIV-1 integrases fully explains each protein's karyophilia. The striking permanent redistribution to the cytoplasm in cells made stably deficient in LEDGF/p75 by siRNA expression from integrated Pol III transcription units provides direct, definitive evidence that it is LEDGF/p75 that determines lentiviral integrase protein trafficking rather than long-sought, classically acting NLSs within these proteins. The failure of LEDGF/p75 to colocalize with MoMLV integrase is similarly consistent with the observed lack of nuclear targeting of this integrase. It is unlikely that HIV-1 integrase has a weak NLS, as has been suggested by some studies. Our siRNA and PK fusion data indicate that such a hypothesis would require postulating either an unknown cytoplasmic binding partner that sequesters integrase in the cytosol in the absence of LEDGF/p75 or a nuclear export signal that is dominant only in the absence of LEDGF/p75.
In addition to using stable expression and knockdowns, and its comparative virology, our study differs from others in its focus on the analysis of native integrase proteins having no fusion partner at all or only a minimal epitope tag. Our data show that in the absence of LEDGF/p75, HIV-1 integrase is entirely excluded from the nucleus, a result that differs from the diffuse cellular location of the GFP-HIV integrase fusion protein in a study by Maertens et al. (see Fig. 9 in reference 50). Our data support a model in which LEDGF/p75 is the principal actor mediating nuclear import and chromatin tethering of lentiviral integrase proteins. While the immunofluorescence analyses detected LEDGF/p75 and the associated lentiviral integrases overwhelmingly in the nucleus, we also found that LEDGF/p75 could be coimmunoprecipitated from both cytoplasmic and nuclear compartments with antibodies to HIV-1 or FIV integrase. One explanation is that tight association with chromatin (Fig. 4A and 6C) is likely to reduce the relative recovery of nuclear versus preimport cytoplasmic LEDGF-integrase complexes in coimmunoprecipitations. These data are also consistent with a model in which LEDGF/p75 may be a shuttle protein.
Technical aspects of lentiviral integrase analyses apparent in these experiments deserve comment. We have established that intron A of human CMV permits Rev-independent integrase expression. The nuclear degradation pathway triggered by INS elements is not yet known, making unclear the mechanism by which this maneuver prevents the otherwise rapid INS-mediated decay of the integrase RNA. However, intron A has also been shown to enable expression of a different Rev-dependent cDNA (the envelope glycoprotein) at levels sufficient for DNA-based vaccines (29). Thus, this approach can be a convenient solution to Rev-independent expression of lentiviral proteins.
Our data also explain the contradictory findings previously observed with various integrase fusion proteins. Fusions to either a short epitope tag or eGFP are found entirely or predominantly in the nucleus, but GFP fusions are less clearly informative because their smaller sizes do not exclude diffusion. An important point to emerge from this study is that conclusions drawn from analyses of integrase fusion proteins are necessarily qualified and may produce contradictory findings that are difficult to interpret in the absence of structural information for the particular fusion protein. For example, interaction with LEDGF/p75 should in principle direct transport of PK-integrase fusions to the nucleus. Their observed confinements to the cytoplasm, and failure to interact with LEDGF/p75, may reflect steric interference by the PK moiety. An alternative possibility is interference with proper folding and/or dimerization of the lentiviral integrases. Like our PK-integrase fusions, Kukolj and colleagues observed that a fusion of a larger partner (β-galactosidase) to HIV-1 integrase resulted in exclusion from nuclei (37), whereas in another study of GFP and glutathione S-transferase fusions to FIV integrase, nuclear targeting was preserved in the fusions and the N-terminal zinc-binding domain, which is thought to be involved in multimerization, was required (75). Our results are also consistent with the data of Cherepanov et al. (6), who identified the interaction of LEDGF/p75 with multimeric forms of the HIV-1 integrase, and Maertens et al. (50), who found that transient siRNA knockdown of LEDGF/p75 caused a GFP-integrase fusion protein to be located diffusely in both the nucleus and cytoplasm. The relatively low identity in the protein sequences of FIV and HIV-1 integrases suggests that the conserved interaction of these proteins with LEDGF/p75 is less likely to occur via shared, discrete peptide determinants but rather with discontinuous structures common to these proteins in their multimerized states. Also in support of this concept is a report that nuclear localization of HIV-1 integrase is prevented by a point mutation that disrupts its ability to oligomerize (57).
While LEDGF/p75 targets integrase proteins to nuclei and mediates chromatin association, the role it plays in the PICs of lentiviruses is not certain and is a priority for further study. Lentiviral PICS were immunoprecipitated specifically by antibodies to LEDGF/p75, suggesting that PIC-associated integrase could recruit cytoplasmic LEDGF/p75. The immunoprecipitated PICs were also shown to be functional in that they are competent for integration in vitro. Because the lentiviral specificity of the LEDGF/p75-integrase interaction correlates with the differential capacity of lentiviruses and oncoretroviruses to infect nondividing cells, and because of its presence in functional PICs, we initially hypothesized that LEDGF/p75 could mediate PIC nuclear import in a nonredundant manner, i.e., the protein would be required for or augment lentiviral infection of nondividing cells. However, our results establish clearly that this is not the case, at least in the cell lines studied here. LEDGF/p75-deficient Jurkat and 293T cells were as susceptible to HIV-1 and FIV infection as wild-type or control siRNA-expressing cells, whether or not these cells were dividing or growth arrested. Also, cells analyzed several weeks after infection with lentiviral vectors had equivalent transgene expression, indicating an absence of a gross defect in viral integration. Since several other PIC components have been proposed to mediate HIV-1 PIC nuclear import, we took care to extend our analyses to subgenomic vectors lacking both Vpr and the central DNA flap. The vector and reporter virus data were additionally corroborated when the kinetics of HIV-1 and FIV cDNA nuclear import were found to be unaffected in nondividing LEDGF/p75-deficient cells. Two markers of PIC nuclear import, the traditional 2-LTR circle quantification (31) and quantification of reporter gene expression from nonintegrated viral genomes in growth-arrested cells (64), support this conclusion. Again, these experiments were also designed with accessory gene and flap-minus HIV vectors. Thus, LEDGF/p75 cannot be considered a required cofactor for HIV-1 PIC nuclear import or for HIV-1 integration. An unresolved question is whether LEDGF/p75 is incorporated in the target cell after infection or in the producer cell, e.g., in the manner of APOBEC3G (30). However, we have established that a deficiency of LEDGF/p75 in the producer cell does not lead to an infectivity defect. It remains possible that, in other cell types or in primary cells, LEDGF/p75 will have a critical role.
We also conclude from the present data that a cellular environment in which virion-unassociated lentiviral integrase proteins are karyophilic is dispensable for infection of nondividing cells, since such infections proceed normally in cells in which this is abolished. More generally, our data are consistent with those of Petit et al. (57), who concluded that the karyophilic properties of HIV-1 integrase are not required for nuclear import of the proviral DNA. It is possible, however, that a virion-derived integrase resident in the PIC may have different properties than free integrase protein in this regard and still modulate PIC karyophilia in the absence of LEDGF/p75. Redundant mechanisms may regulate PIC import, and cell-type-specific variation may occur. Mapping NLSs in LEDGF/p75 and clarifying its interactions with nuclear import proteins (17) will be of interest.
Determining the function of LEDGF/p75 interaction in the viral life cycle thus remains an important goal. A yet unexplored but plausible role could be to modulate the distribution of integration loci, which are now known to differ between retroviruses (66, 76). Because LEDGF/p75 associates with HIV-1 but not MoMLV integrase, it is possible that LEDGF/p75 participates in modulating these biases or the local status of chromatin after integration.
Acknowledgments
We thank Daniah Thompson for assisting with cell line generation, Sharon Delgado for diverse technical assistance, Michael Malim, Zeger Debyser, Monica Roth, and Gideon Dreyfuss for sharing expression plasmids, Didier Trono for HIV-1 vector constructs, Stephen Goff for antibodies to MoMLV integrase, and Margaret Barr for FIV antisera.
NIH grant AI47536 and a Pfizer Scholars Grant for New Faculty provided funding to E.M.P.
REFERENCES
- 1.Bouyac-Bertoia, M., J. Dvorin, R. Fouchier, Y. Jenkins, B. Meyer, L. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinskaya, A. G., A. Ghorpade, N. K. Heinzinger, T. E. Smithgall, R. E. Lewis, and M. Stevenson. 1996. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc. Natl. Acad. Sci. USA 93:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman, F. D. 1999. Host proteins in retroviral cDNA integration. Adv. Virus Res. 52:301-317. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P., W. Pluymers, A. Claeys, P. Proost, E. De Clercq, and Z. Debyser. 2000. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 14:1389-1399. [DOI] [PubMed] [Google Scholar]
- 8.Cochrane, A. W., K. S. Jones, S. Beidas, P. J. Dillon, A. M. Skalka, and C. A. Rosen. 1991. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65:5305-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 12.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 13.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 14.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 16.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 19.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, A. V. Albright, F. Gonzalez-Scarano, and M. H. Malim. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed, E. O., G. Englund, F. Maldarelli, and M. A. Martin. 1997. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell 88:171-174. [DOI] [PubMed] [Google Scholar]
- 22.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 25.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 26.Ge, H., Y. Si, and R. G. Roeder. 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17:6723-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 28.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 29.Haigwood, N. L., C. C. Pierce, M. N. Robertson, A. J. Watson, D. C. Montefiori, M. Rabin, J. B. Lynch, L. Kuller, J. Thompson, W. R. Morton, R. E. Benveniste, S. L. Hu, P. Greenberg, and S. P. Mossman. 1999. Protection from pathogenic SIV challenge using multigenic DNA vaccines. Immunol. Lett. 66:183-188. [DOI] [PubMed] [Google Scholar]
- 30.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 31.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 34.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemler, I., R. Barraza, and E. M. Poeschla. 2002. Mapping of the encapsidation determinants of feline immunodeficiency virus. J. Virol. 76:11889-11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenna, M. A., C. B. Brachmann, S. E. Devine, and J. D. Boeke. 1998. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 18:1115-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leavitt, A. D., L. Shiue, and H. E. Varmus. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 268:2113-2119. [PubMed] [Google Scholar]
- 39.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limon, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llano, M., T. Kelly, M. Vanegas, M. Peretz, T. E. Peterson, R. D. Simari, and E. M. Poeschla. 2002. Blockade of human immunodeficiency virus type 1 expression by caveolin-1. J. Virol. 76:9152-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loewen, N., C. Bahler, W. Teo, T. Whitwam, M. Peretz, R. Xu, M. Fautsch, D. H. Johnson, and E. M. Poeschla. 2002. Preservation of aqueous outflow facility after second-generation FIV vector-mediated expression of marker genes in anterior segments of human eyes. Investig. Ophthalmol. Vis. Sci. 43:3686-3690. [PubMed] [Google Scholar]
- 47.Loewen, N., R. Barraza, T. Whitwam, D. Saenz, I. Kemler, and E. Poeschla. 2003. FIV vectors. Methods Mol. Biol. 229:251-271. [DOI] [PubMed] [Google Scholar]
- 48.Loewen, N., M. Fautsch, M. Peretz, C. Bahler, J. D. Cameron, D. H. Johnson, and E. M. Poeschla. 2001. Genetic modification of human trabecular meshwork with lentiviral vectors. Hum. Gene Ther. 12:2109-2119. [DOI] [PubMed] [Google Scholar]
- 49.Loewen, N., D. Leske, Y. Chen, W. Teo, D. Saenz, M. Peretz, J. Holmes, and E. M. Poeschla. 2003. Comparison of wild-type and class I integrase mutant-FIV vectors in retina demonstrates sustained expression of integrated transgenes in retinal pigment epithelium. J. Gene Med. 5:1009-1017. [DOI] [PubMed] [Google Scholar]
- 50.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 51.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore, S. P., L. A. Rinckel, and D. J. Garfinkel. 1998. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulder, L. C., L. A. Chakrabarti, and M. A. Muesing. 2002. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277:27489-27493. [DOI] [PubMed] [Google Scholar]
- 55.Mumm, S. R., P. J. Hippenmeyer, and D. P. Grandgenett. 1992. Characterization of a stable eukaryotic cell line expressing the Rous sarcoma virus integrase. Virology 189:500-510. [DOI] [PubMed] [Google Scholar]
- 56.Nishizawa, Y., J. Usukura, D. P. Singh, L. T. Chylack, Jr., and T. Shinohara. 2001. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 305:107-114. [DOI] [PubMed] [Google Scholar]
- 57.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 59.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 60.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 61.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saenz, D., N. Loewen, M. Peretz, T. Whitwam, R. Barraza, K. Howell, J. H. Holmes, M. Good, and E. M. Poeschla. 2004. Unintegrated lentiviral DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 67.Singh, D. P., N. Fatma, A. Kimura, L. T. Chylack, Jr., and T. Shinohara. 2001. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Commun. 283:943-955. [DOI] [PubMed] [Google Scholar]
- 68.Singh, D. P., N. Ohguro, T. Kikuchi, T. Sueno, V. N. Reddy, K. Yuge, L. T. Chylack, Jr., and T. Shinohara. 2000. Lens epithelium-derived growth factor: effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem. Biophys. Res. Commun. 267:373-381. [DOI] [PubMed] [Google Scholar]
- 69.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vodicka, M. A. 2001. Determinants for lentiviral infection of non-dividing cells. Somat. Cell Mol. Genet. 26:35-49. [DOI] [PubMed] [Google Scholar]
- 72.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Schwedler, U., R. S. Kornbluth, and D. Trono. 1994. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. USA 91:6992-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitwam, T., M. Peretz, and E. M. Poeschla. 2001. Identification of a central DNA flap in feline immunodeficiency virus. J. Virol. 75:9407-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woodward, C. L., Y. Wang, W. J. Dixon, H. Htun, and S. A. Chow. 2003. Subcellular localization of feline immunodeficiency virus integrase and mapping of its karyophilic determinant. J. Virol. 77:4516-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 77.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 7:920-926. [DOI] [PubMed] [Google Scholar]
- 78.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 79.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]