Abstract
The RNA-dependent RNA polymerase of influenza A virus is responsible for both transcription and replication of negative-sense viral RNA. It is thought that a “switching” mechanism regulates the transition between these activities. We demonstrate that, in the presence of preexisting viral RNA polymerase and nucleoprotein (NP), influenza A virus synthesizes both mRNA (transcription) and cRNA (replication) early in infection. We suggest that there may be no switch regulating the initiation of RNA synthesis and present a model suggesting that nascent cRNA is degraded by host cell nucleases unless it is stabilized by newly synthesized viral RNA polymerase and NP.
As in all negative-sense RNA viruses, the viral genomic RNA (vRNA) of influenza A virus is complexed with an RNA-dependent RNA polymerase (comprising PB1, PB2, and PA subunits) and nucleoprotein (NP) into active viral ribonucleoprotein (vRNP), which serves as a template for both transcription and replication (19). Whereas mRNA transcription requires capped RNA primers snatched from host pre-mRNA and premature poly(A) termination of transcripts, genome replication is primer independent and generates full-length cRNA. cRNA subsequently serves as the template for synthesis of progeny vRNA. Early studies (2, 13, 14, 30) suggested that replication requires de novo protein synthesis, and it was proposed that a “switch” regulates the transition from transcription to replication, but the molecular mechanism for such a switch has remained elusive despite more than two decades of research.
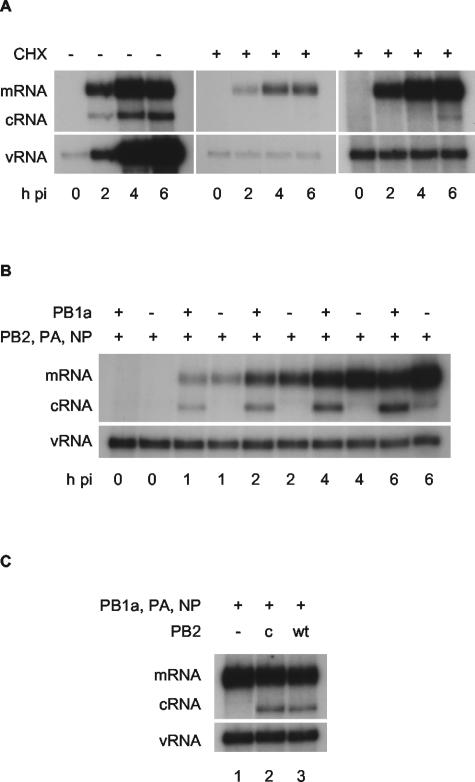
We approached an investigation of this switch in influenza A virus-infected cell cultures by inhibiting the transition from mRNA transcription to RNA replication with cycloheximide (2, 14, 30), an inhibitor of protein synthesis. We confirmed the inhibitory effect of 100 μg of cycloheximide/ml on influenza A/WSN/33 replication in human transformed kidney (293T) cells. Total RNA was isolated by TRIzol (Invitrogen) extraction from 293T cells at various time points after infection with influenza virus A/WSN/33 at a multiplicity of infection (MOI) of 5 in the presence or absence of cycloheximide. The RNA was analyzed by primer extension analysis (8) by using two 32P-labeled primers, one specific for neuraminidase (NA) mRNA and cRNA and the other specific for NA vRNA, generating cDNA with an expected size of 129 nucleotides from vRNA, 160 nucleotides from cRNA, and 169 to 177 nucleotides from mRNA depending on the length of the capped primer (Fig. 1A). Transcription of mRNA from vRNA and replication of vRNA through cRNA intermediates in infected 293T cells in the absence of cycloheximide is clearly demonstrated (Fig. 1A, left panel). RNA detected at zero hours postinfection represents RNA derived from the infecting virus particles. However, whereas vRNA derived from the infecting virus particle is transcribed to mRNA in the presence of 100 μg of cycloheximide/ml, only minimal cRNA synthesis was detected at 6 h postinfection (Fig. 1A, middle and right panels). In addition, the vRNA signal remained constant throughout, representing input vRNA derived from the infecting virus particles. Therefore, synthesis of cRNA and replication of vRNA is severely inhibited in the presence of 100 μg of cycloheximide/ml.
FIG. 1.
Cycloheximide-mediated inhibition of influenza virus A/WSN/33 cRNA synthesis can be rescued by the expression of PB1, PB2, PA, and NP. Viral RNA species were analyzed by NA gene-specific primer extension assays. mRNA is a broad heterogeneous band 9 to 17 nucleotides longer than cRNA due to its 5′ cap-snatched sequence. (A) Time course of viral infection in the absence (−) or presence (+) of 100 μg of cycloheximide (CHX)/ml. The left-hand and middle panels are equally exposed, with a longer exposure of the middle panel at right showing minor breakthrough of cycloheximide inhibition of cRNA synthesis at 6 h postinfection. (B) Time course of viral infection in the presence of 100 μg of cycloheximide/ml after prior (12 to 14 h) transfection of expression plasmids expressing viral proteins as indicated. (C) cRNA rescue with a cap binding mutant of PB2. 293T cells were transfected with plasmids expressing viral proteins (+) or empty plasmid vector (−), as indicated, 12 to 14 h prior to infection by A/WSN/33 virus in the presence of 100 μg of cycloheximide/ml. RNA was harvested at 2 h postinfection. PB1a, PB1-D445A/D446A; PB2c, PB2-F404A; wt, wild type; h pi, hours postinfection.
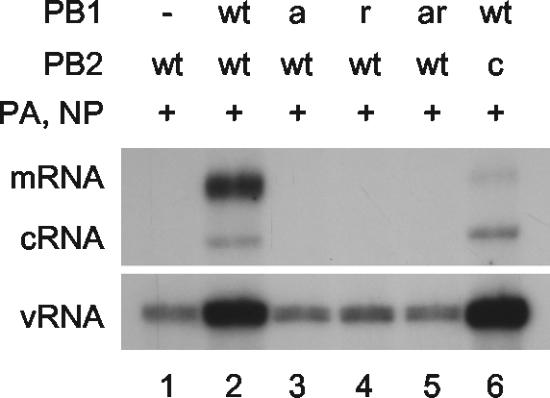
We then investigated whether the switch to cRNA synthesis could be rescued by expressing viral PB1, PB2, PA, and NP in the cells prior to cycloheximide treatment and infection. In order to avoid transcription and replication of viral RNA driven by these expressed proteins, a double mutant in the conserved SDD motif of the active site of the RNA polymerase PB1 subunit (PB1-D445A/D446A [PB1a]) (5) was constructed by site-directed mutagenesis. As expected, PB1a, PB2, and PA expressed from recombinant plasmids in vivo was negative for RNA polymerase activity (i.e., no cRNA or mRNA synthesized above background) on an NA vRNA-like template derived from cotransfected pPOLI-NA-RT (9) (Fig. 2, compare lanes 1 to 3). These data were further confirmed by an in vivo chloramphenicol acetyltransferase (CAT) reporter assay (see Fig. S1 in the supplemental material). 293T cells were then transfected with plasmids expressing PB2, PA, NP, and either PB1a or empty plasmid vector, followed 12 to 14 h later by infection with influenza virus A/WSN/33 at an MOI of 5 in the presence of 100 μg of cycloheximide/ml. Viral RNA species at various time points postinfection were analyzed by primer extension as before. Surprisingly, cRNA was detected, but only in the presence of PB1a, whereas mRNA and vRNA levels were essentially the same in the presence or absence of PB1a (Fig. 1B). Identical results were obtained with nonstructural protein 1 (NS1) gene-specific primers (see Fig. S2 in the supplemental material). Therefore, despite the cycloheximide-mediated inhibition of viral replication, viral cRNA synthesis was evident if viral polymerase and NP were present.
FIG. 2.
Analysis of the in vivo RNA synthesis activity of polymerase mutants used in the study. Plasmids expressing viral proteins (+) or empty plasmid vector (−), as indicated, were transfected into 293T cells together with a plasmid (pPOLI-NA-RT) (9) directing polymerase I-driven transcription of NA vRNA as a template for RNA synthesis. Total RNA was isolated at 15 h posttransfection and analyzed by NA gene-specific primer extension. vRNA detected in the absence of PB1 (lane 1) represents polymerase I-derived template RNA. wt, wild type; PB1a, PB1-D445A/D446A; PB1r, PB1-Y559A; PB1ar, PB1-D445A/D446A/Y559A; PB2c, PB2-F404A.
As a control to demonstrate that the rescued cRNA signal does not derive from “decapped” mRNA, we investigated whether the cRNA could be rescued by influenza virus polymerase containing a previously described cap binding mutant of the PB2 subunit (7) (PB2-F404A [PB2c]). The replication-positive but transcription-negative phenotype of polymerase containing PB2c was confirmed on a polymerase I-transcribed vRNA template by an in vivo RNA synthesis assay as described above (Fig. 2, compare lanes 2 and 6). However, expression of polymerase containing PA, PB1a, and wild-type or mutant PB2, together with NP, prior to infection yielded identical cRNA signals in primer extension analyses (Fig. 1C, compare lanes 2 and 3) demonstrating that, as expected, rescue of cRNA was independent of the cap binding activity of the preexpressed RNA polymerase.
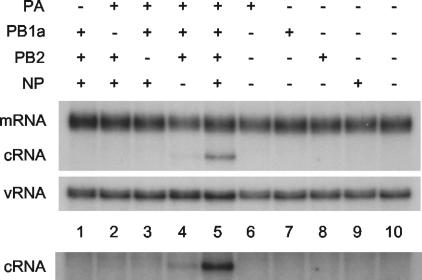
The determinants of the switch to cRNA synthesis were analyzed in more detail by replacing individual viral protein-expression plasmids in the transfection mixture with empty plasmid vector or by transfecting the plasmids separately. Expression of proteins (PB1a, PB2, PA, and NP) from transfected plasmids was confirmed by Western blot analyses (see, for example, Fig. S5 in the supplemental material). Omission of any one of the three polymerase subunits (PB1a, PB2, or PA) resulted in elimination of the cRNA signal (Fig. 3, compare lanes 1 to 3 with lane 5). However, in the absence of NP (lane 4), the cRNA signal was reduced but nevertheless remained clearly detectable (on a longer exposure) above background (lane 10). It follows that expression of any individual protein component alone (lanes 6 to 9) would yield no detectable cRNA signal. Overall, therefore, these data suggest that the polymerase, comprising subunits PB1, PB2, and PA, is necessary and sufficient for switching to cRNA synthesis, but it is strongly reinforced by NP. However, NP alone is insufficient for switching.
FIG. 3.
RNA polymerase and NP are essential for rescuing cRNA. 293T cells were transfected with plasmids expressing viral proteins (+) or empty plasmid vector (−), as indicated, 12 to 14 h prior to infection with A/WSN/33 virus in the presence of 100 μg of cycloheximide/ml. RNA was harvested at 2 h postinfection and the viral RNA species were analyzed by NA gene-specific primer extension assays. A longer exposure of the cRNA-specific bands is shown below to emphasise the detection of cRNA in lane 4. PB1a, PB1-D445A/D446A.
The dose dependence and stoichiometry of the polymerase-NP complex required to rescue cRNA was investigated by titrating the amount of polymerase and/or NP-expressing plasmids in the transfections. The effect of increasing plasmid transfection levels on protein expression 12 to 14 h posttransfection could be confirmed by Western blotting (see, for example, Fig. S5 in the supplemental material). Altering the transfection levels of plasmids expressing the polymerase subunits by 0.5- to 3-fold over that used in Fig. 3 had no significant effect on cRNA signals (see Fig. S3A in the supplemental material), suggesting that the concentration of expressed polymerase is not limiting. However, cRNA signals were found to increase with increasing NP (tested up to 4 μg of transfected plasmid; see Fig. S3B in the supplemental material). Although this showed that suboptimal levels of NP were used in the experiments described in the present study, practical considerations prevented optimization. Nevertheless, no cRNA could be detected if plasmids expressing the polymerase subunits were omitted, even at the maximum NP concentration tested (see Fig. S3B, lane 2, in the supplemental material).
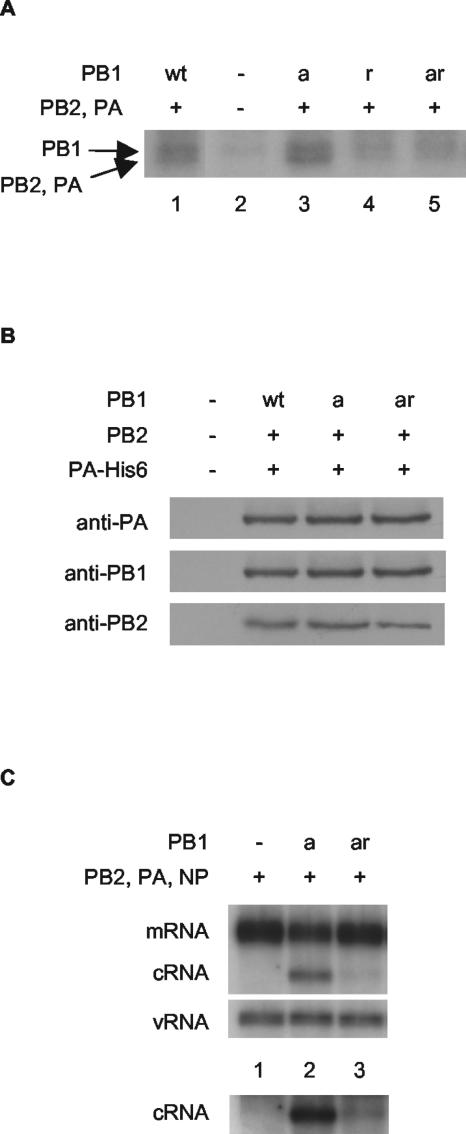
We then examined whether the RNA-binding activity of the preexpressed polymerase was required for cRNA rescue. With this aim in mind, we identified a mutation in PB1 which inhibits cRNA promoter binding. Substituting tyrosine with alanine at residue 559, immediately adjacent to the proposed binding site for the 5′ end of the vRNA promoter (20), in either PB1 (to yield PB1-Y559A [PB1r]) or in PB1a (to yield PB1-D445A/D446A/Y559A [PB1ar]), was found to result in significantly reduced but still detectable cRNA promoter binding in an in vitro cross-linking assay (Fig. 4A, compare lane 1 with lane 4 and lane 3 with lane 5). Polymerase possessing the Y559A mutation in PB1 was found to have no detectable transcriptional or replicative activity, both in vivo by an RNA synthesis (Fig. 2, compare lanes 2 and 4) or a CAT reporter (see Fig. S4A in the supplemental material) assay and in vitro by an ApG-primed transcription assay (see Fig. S4B in the supplemental material). The amino acid substitution was shown by Western blot analyses not to inhibit expression levels of PB1ar compared to PB1a in transfected cells (see Fig. S5 in the supplemental material). Furthermore, PB1a and PB1ar were shown to be complexed with the other polymerase subunits, since PB1 and PB2 could be readily detected by Western blotting after nickel-affinity purification of His-tagged PA (Fig. 4B). Strikingly, expression of PB1ar in the cRNA rescue assay yielded significantly reduced, but still detectable, cRNA signals compared to PB1a (Fig. 4C, compare lanes 2 and 3; lane 1 is background). This suggests that rescue of the cRNA signal is dependent on binding of the cRNA promoter by the polymerase.
FIG. 4.
cRNA rescue is reduced by a mutation inhibiting the cRNA promoter-binding activity of PB1. (A) Substitution of tyrosine at residue 559 in PB1 or PB1a with alanine inhibits photochemical cross-linking of the polymerase complex to labeled cRNA. Partially purified His-tagged polymerase from cells transfected with plasmids expressing wild-type or mutant PB1, PB2, and PA-His6 (8), or empty vectors, as indicated, were cross-linked with a 32P-labeled RNA probe corresponding to the 3′-end cRNA in the presence of an excess unlabeled 5′-end cRNA by UV irradiation (8). The products were analyzed by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The positions of the PB1 and comigrating PB2 and PA cross-linked bands are shown. (B) The Y559A mutation in PB1 does not inhibit polymerase complex formation. Partially purified His-tagged polymerase from cells transfected with plasmids expressing wild-type or mutant PB1, PB2, and PA-His6 (8), or empty vectors, as indicated, was subjected to Western blotting with antibodies raised against the individual subunits as shown. (C) PB1-Y559A severely restricts cRNA rescue. 293T cells were transfected with plasmids expressing viral proteins (+) or empty plasmid vector (−), as indicated, 12 to 14 h prior to infection by A/WSN/33 virus in the presence of 100 μg of cycloheximide/ml. RNA was harvested at 2 h postinfection, and viral RNA species were analyzed by NA gene-specific primer extension assays. A longer exposure of the cRNA-specific bands is shown below to emphasize the differences in cRNA levels. wt, wild type; PB1a, PB1-D445A/D446A; PB1r, PB1-Y559A; PB1ar, PB1-D445A/D446A/Y559A.
We have, therefore, demonstrated that infecting influenza A virions can initiate both capped RNA-dependent mRNA transcription and primer-independent RNA replication early in infection. We have shown that the detection of significant levels of cRNA is dependent on the expression of viral polymerase and NP and that the cRNA signal is significantly reduced if the RNA-binding capacity of the PB1 subunit of the polymerase is impaired. Therefore, we postulate that viral polymerase and NP expressed prior to virus infection bind nascent cRNA synthesized by the incoming vRNPs.
Various models of switching from transcription to replication in influenza A virus infection have been proposed (27). NP was identified as a prime candidate for a switching molecule based on several temperature-sensitive NP mutants defective in replication and RNA binding (18, 22, 29) and biochemical studies that suggested that free NP is required for the synthesis of full-length transcripts (3, 28). It was proposed that interaction of NP with the polymerase (4, 23) or with the promoter element of the template RNA (10, 17) alters the mode of transcriptional initiation. More recently, the PB2 and PA subunits of the polymerase have also been implicated from studies of mutations affecting viral replication but not transcription (11, 16). However, all of these models were based on the assumption that cRNA is not synthesized early in infection, an assumption that is now challenged. In fact, low levels of cRNA have previously been detected early in infection in the presence of cycloheximide (2), but these data were discounted at the time.
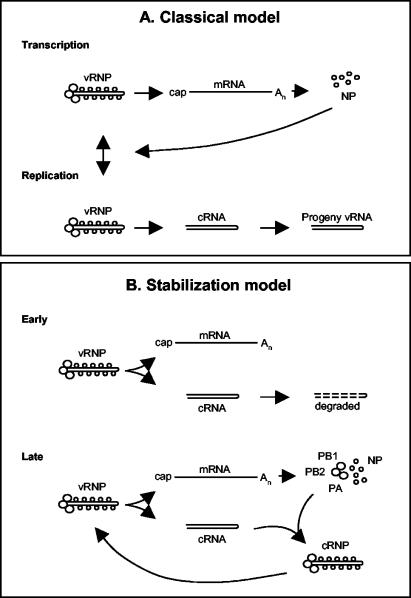
Based on our results, we propose a new “stabilization” model for influenza viral infection (Fig. 5) in which vRNPs derived from the infecting virus synthesize both mRNA and cRNA. mRNA is protected from normal cellular degradative processes by the presence of a 5′ cap and a 3′ poly(A) tail, whereas nascent cRNA is presumably rapidly degraded by host cell nucleases. The transition to a replicative phase occurs when cRNA is protected by the specific binding of RNA polymerase (assembled from newly synthesized PB1, PB2, and PA) to the cRNA promoter (12). The fact that polymerase alone (i.e., in the absence of NP), which is known to bind both the 5′ and 3′ ends of cRNA (6), can rescue cRNA suggests that host 5′ and 3′ exonucleases may be involved. The cRNA-polymerase promoter complex then serves as a nucleation point for binding of free newly synthesized NP, leading to the formation of active and stable cRNPs suitable for replicative vRNA synthesis. Crucially, this model proposes that there is no switch regulating initiation of mRNA or cRNA synthesis but that newly synthesized polymerase and NP stabilize cRNA transcripts later in infection. We suggest that it explains why mutations in NP (4, 22, 23, 28), PB2 (16), or PA (11), which may interfere with cRNP complex formation, can affect the transition between mRNA transcription and RNA replication phases.
FIG. 5.
Models for switching between influenza A viral transcription and replication. (A) Classical model. Early in infection, vRNA is transcribed to mRNA (primary transcription). Expression of NP switches primary transcription to replication (synthesis of cRNA and vRNA) and subsequent secondary transcription. (B) Proposed “stabilization” model. Early in infection, mRNA and cRNA are synthesized, but cRNA is degraded. Later in infection, once PB1, PB2, PA, and NP are synthesized, cRNA is stabilized as a cRNP complex that can be replicated to vRNA. See the text for further details.
Although we cannot at present exclude other models, we believe that the data best fit our stabilization model. For instance, it may be argued that the preexisting inactive polymerase used in our experiments induces a host factor (24) or forms a higher-order polymerase complex or lattice (21), switching the polymerase in the infecting virus to replicative activity. However, the demonstrated requirement for RNA-binding activity of the preexisting polymerase would argue against these alternative models.
Our stabilization model suggests that the initiation of mRNA or cRNA synthesis by influenza A virus polymerase may be regulated stochastically; that is, random primer-dependent or independent initiation dictating the synthesis of mRNA or cRNA, possibly involving subtle conformational differences in the structure of the polymerase (15, 26). Stochastic regulation at the level of initiation of cRNA synthesis, while being “wasteful” by generating cRNA destined for degradation, would reflect the limited genetic resources possessed by RNA viruses for complex regulation. It will be interesting to use our new model and our methodological approach to determine whether a similar mechanism of gene regulation also occurs in other negative- and positive-sense RNA viruses in which it has proved difficult to define the precise mechanism regulating transcription and replication (1, 19, 25).
Supplementary Material
Acknowledgments
We thank Ervin Fodor for helpful discussions and Julian Robinson for DNA sequencing.
This study was supported by MRC grants G9523972 and G9901312 to G.G.B. and an EPA Fellowship to T.E.J.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 2002. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim. Biophys. Acta 1577:337-353. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, T., A. J. Wolstenholme, and B. W. J. Mahy. 1979. Transcription and replication of influenza virus RNA. Virology 98:211-225. [DOI] [PubMed] [Google Scholar]
- 3.Beaton, A. R., and R. M. Krug. 1986. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl. Acad. Sci. USA 83:6282-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crow, M., T. Deng, M. Addley, and G. G. Brownlee. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fechter, P., L. Mingay, J. Sharps, A. Chambers, E. Fodor, and G. G. Brownlee. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 8.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gastaminza, P., B. Perales, A. M. Falcón, and J. Ortín. 2003. Mutations in the N-terminal region of influenza virus PB2 protein affect virus RNA replication but not transcription. J. Virol. 77:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatada, E., M. Hasegawa, J. Mukaigawa, K. Shimizu, and R. Fukuda. 1989. Control of influenza virus gene expression: quantitative analysis of each viral RNA species in infected cells. J. Biochem. 105:537-546. [DOI] [PubMed] [Google Scholar]
- 14.Hay, A. J., B. Lomniczi, A. R. Bellamy, and J. J. Skehel. 1977. Transcription of the influenza virus genome. Virology 83:337-355. [DOI] [PubMed] [Google Scholar]
- 15.Honda, A., K. Mizumoto, and A. Ishihama. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells. 4:475-485. [DOI] [PubMed] [Google Scholar]
- 16.Huarte, M., A. Falcón, Y. Nakaya, J. Ortín, A. García-Sastre, and A. Nieto. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klumpp, K., R. W. H. Ruigrok, and F. Baudin. 1997. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 16:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krug, R. M., M. Ueda, and P. Palese. 1975. Temperature-sensitive mutants of influenza WSN virus defective in virus-specific RNA synthesis. J. Virol. 16:790-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1530. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 20.Li, M.-L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 22.Medcalf, L., E. Poole, D. Elton, and P. Digard. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mena, I., E. Jambrina, C. Albo, B. Perales, J. Ortín, M. Arrese, D. Vallejo, and A. Portela. 1999. Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J. Virol. 73:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, G., M. A. Whitt, and Y. Kawaoka. 2002. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J. Gen. Virol. 83:2635-2662. [DOI] [PubMed] [Google Scholar]
- 26.Penn, C. R., and B. W. Mahy. 1984. Capped mRNAs may stimulate the influenza virion polymerase by allosteric modulation. Virus Res. 1:1-13. [DOI] [PubMed] [Google Scholar]
- 27.Portela, A., and P. Digard. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723-734. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro, G. I., and R. M. Krug. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 62:2285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiura, A., M. Ueda, K. Tobita, and C. Enomoto. 1975. Further isolation and characterization of temperature-sensitive mutants of influenza virus. Virology 65:363-373. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, J. M., R. Illmensee, S. Litwin, L. Herring, B. Broni, and R. M. Krug. 1977. Use of specific radioactive probes to study transcription and replication of the influenza virus genome. J. Virol. 21:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.