Abstract
The translation efficiency of an mRNA molecule is typically determined by its 5′- and/or 3′-untranslated regions (UTRs). Previously, we have found that the 3′-UTR of Turnip yellow mosaic virus (TYMV) RNA enhances translation synergistically with a 5′ cap. Here, we use a luciferase reporter system in cowpea protoplasts to show that the 5′ 217 nucleotides from TYMV genomic RNA enhance expression relative to a vector-derived 17-nucleotide 5′-UTR. Maximum expression was observed from RNAs with a cap and both 5′ and 3′ TYMV sequences. In paired reporter constructs, the 5′ 217 nucleotides harboring the UTR and the first 43 or 41 codons of the two overlapping TYMV open reading frames (ORFs), ORF-69 and ORF-206, respectively, were fused in frame with the luciferase gene. This allowed expression from the initiation codon of each ORF (AUG69 and AUG206) to be monitored separately but from the normal sequence environment. Expression from both AUG codons was heavily dependent on a 5′ cap, with a threefold-higher expression occurring from AUG69 than from AUG206 in the presence of the genomic 3′-UTR. Changes that interrupted the cap/3′-UTR synergy (i.e., removal of the cap or TYMV 3′-UTR) resulted in a higher proportion of initiation from AUG206. Mutation of the 3′-UTR to prevent aminoacylation, as well as deletion of 75% of the 5′-UTR, likewise resulted in a lower ratio of expression from AUG69 relative to AUG206. Mutation of each AUG initiation codon increased initiation from the other. Taken together, these results do not fully conform to the expectations of standard leaky ribosomal scanning and leave open the precise mechanism of ribosome commitment to AUG69 and AUG206. However, our observations do not support a recent proposal based on in vitro studies in which the 3′-UTR is proposed to direct cap-independent initiation specifically at AUG206 and not at AUG69 (S. Barends et al., Cell 112:123-129, 2003).
The 5′- and 3′-untranslated regions (UTRs) play major roles in controlling the translation of cellular and viral RNAs. The 5′ m7GpppN cap structure and the 3′ poly(A) tail, both of which are characteristic features of eukaryotic mRNAs, are bound by the initiation factor eIF4E and poly(A)-binding protein, respectively. Each of these proteins interacts with the scaffolding protein eIF4G, shaping the mRNA into a circularized closed loop that is considered to be the form of actively translated mRNAs (16, 19). Through affinity to initiation factors bound to the above complex, ribosomal small subunits are loaded at the 5′ end of the RNA and then scan in the 3′ direction in search of an initiation codon, which is normally AUG (18). The efficiency of initiation is modulated by the sequence context of the initiation codon, with a purine at −3 and a guanosine at +4 the most important contributors to an optimal context (13, 15).
Among viral RNAs, alternative or additional elements enhancing translation have been identified. For instance, certain 3′-UTRs of nonpolyadenylated viral RNAs enhance translation in a manner similar to that provided by a poly(A) tail, including a synergistic interaction with the 5′ cap. This has been demonstrated for rotavirus mRNAs (23) and for plant viral RNAs that terminate with a tRNA-like structure (TLS): Tobacco mosaic virus (TMV), Brome mosaic virus (6), and Turnip yellow mosaic virus (TYMV) (17). Some 5′-UTRs of capped viral RNAs have been observed to enhance translational expression (7, 11, 21, 22), adding additional control to the minimal elements normally present on mRNAs. The best-studied case is the 5′-UTR enhancer of TMV genomic RNA, the so-called Ω sequence, which has been shown to enhance gene expression via elevated recruitment of the eIF4F complex through the mediation of heat shock protein HS101 (4, 25).
We have recently shown that the 3′-UTR of TYMV acts as a potent translational enhancer in plant cells. The TLS was the main feature responsible for translational enhancement, which was maximal when aminoacylation and eEF1A binding properties were intact (17). Those experiments utilized luciferase (LUC) reporter constructs devoid of viral 5′ sequences. In the present study, we examined the translational role of the TYMV 5′-UTR and its relationship to the enhancing role of the 3′-UTR. This was of particular interest with the recent publication of a proposed scheme by which the 3′-UTR communicates specifically with one of two AUG initiation codons to promote translation by a novel cap-independent mechanism that avoids the quasi-ubiquitous involvement of initiator tRNAMet (1).
The genome of TYMV is expressed from two RNAs, both of which possess a 5′ cap and the 3′-TLS. The genomic RNA is translated to express two extensively overlapping proteins, the movement protein (p69) initiating at nucleotide (nt) 88 (AUG69, the 5′-most AUG triplet) and the replication polyprotein (p206) initiating at nt 95 (AUG206; Fig. 1A). The subgenomic RNA, which is colinear with the 3′-terminal 694 nt of the genomic RNA, is a dedicated mRNA for the translation of the coat protein. With the presence of a 5′ cap and the absence of an extensive upstream untranslated region that could harbor an internal ribosome entry site (IRES) (20), normal cap-dependent translation of the genomic RNA via leaky scanning would seem to adequately explain the expression initiating from the two closely spaced 5′-proximal AUG triplets. Leaky scanning is a proven mechanism explaining expression from more than the first AUG (13), although there is controversy concerning the rules governing ribosome selection of two closely spaced AUG triplets as found in TYMV RNA (24) and an influenza B virus mRNA (12, 26).
FIG. 1.
Design of LUC reporter mRNAs investigating the translational regulation of TYMV RNA. (A) Diagram of TYMV genomic RNA and subgenomic RNA with ORF expressing coat protein (CP, gray box). The genomic RNA is translated to produce p69 (hatched box) from initiation at AUG69 and p206 (open box) from initiation at AUG206, but the CP ORF is silent (indicated by dashed outline). The m7GpppN cap of each RNA is indicated by an asterisk. The cruciform structure at the 3′ end represents the TLS. (B) Sequences and proposed conformation (9) of the largely unstructured 5′-UTR of TYMV genomic RNA. The initiation codons at nt 88 and 95 and corresponding translation products are marked. (C) Basic 5′-UTRs of LUC reporter RNA constructs. The sequence of the vector-derived (vec) 5′-UTR is shown, and the 5′ regions of paired constructs reporting initiation from each of the two TYMV AUG initiation codons are diagrammed. The wild-type LUC protein produced from the vec 5′-UTR begins with the dipeptide ME (single-letter amino acid code), as indicated. Constructs with 5′-TYMV sequences produce the LUC fusion proteins 69L or 206L, which have N-terminal fusions of 43 and 41 residues from the N-terminal portions of p69 and p206, respectively. The amino acid sequences derived from p69 and p206 are shown (single-letter code), along with two bridging residues derived from a PstI site used for cloning (LQ, in reverse shading). Reporter RNAs with the 69L or 206L 5′ sequences differ by the addition of a single residue just before the PstI site that functions to match the LUC coding sequences to the reading frame of the N-terminal fragment of p206 (unpublished data). (D) Basic 3′-UTRs of LUC reporter RNA constructs. The sequence of the vector-derived 19-nt 3′-UTR referred to as Bam is shown. This 3′-UTR is produced from DNA templates linearized with BamHI. The two types of TYMV 3′-UTR used are shown: TYg and TYsg, which include the 684-nt UTR of the genomic RNA and the 109-nt UTR of the subgenomic RNA, respectively. Both UTRs are fused as indicated to the generic Bam 3′-UTR sequence. Transcription from plasmid linearized with XmnI produces RNAs with the same 3′ termini as virion RNA.
With these considerations, the proposal based on in vitro studies that the TLS communicates to direct translation initiation of p206 at nt 95 (and not of p69 at nt 88) in a cap-independent manner (1) seemed unnecessarily complex. Our experiments studying expression in cells do not support the proposed scheme. Translation from both AUG initiation codons is strongly cap dependent and is enhanced by the TYMV 3′-UTR, and there is no special functional interaction between the 3′-UTR and initiation at AUG206; the 3′-UTR, including the TLS, is a general enhancer of translation. We further show that the 5′ sequences of TYMV genomic RNA enhance translation in a way that shares some similarities with enhancement by the TMV Ω element.
MATERIALS AND METHODS
Plasmid constructs and in vitro transcription.
To permit ready replacement of 5′ sequences, the firefly LUC reporter plasmid pDCLD (kindly provided by W. W. Chiu) was adapted by replacing the 3′-UTR with the TYMV genomic (g) 3′-UTR from pL-TYg (17) to generate pDCL-TYg. This involved subcloning a fragment spanning the SphI site within the LUC ORF and the polylinker SacI site downstream of the XmnI site used to linearize pL-TYg prior to transcription. TYMV 5′ fragments containing a T7 promoter, 5′-UTR, and the beginning of a LUC fusion ORF were generated with appropriate primers by PCR amplification, followed by insertion into the NotI and PstI sites of pDCL-TYg. This created p69L-TYg, p206L-TYg, and related plasmid constructs. To make p69L-TYsg and p206L-TYsg, the genomic (g) 3′-UTR was replaced by the TYMV subgenomic (sg) 3′-UTR from pL-TYsg (17) (BamHI to SacI fragment). The cloned portions of all plasmid constructs were confirmed by DNA sequencing.
RNA transcripts, in capped (with m7GpppG; Epicentre) or uncapped form as appropriate, were prepared as described previously (17) in the presence of [α-32P]CTP (0.2 μCi per 20-μl reaction) to facilitate assessment of the quality and quantity of the transcripts. Trichloroacetic acid filter precipitation and liquid scintillation counting were used to quantify the transcripts. For examination of RNA quality, transcripts were analyzed by electrophoresis on 1% agarose gels and phosphorimagery.
Protoplast transfection.
Mesophyll protoplasts were prepared from the first leaves of cowpea plants as described previously (17). The washed protoplasts were resuspended in cold EB buffer (0.6 M d-mannitol, 5 mM morpholineethanesulfonic acid, 5 mM CaCl2, and 40 mM KCl; adjusted to pH 5.7 with KOH) at 2 × 106 cells/ml. Aliquots (0.6 × 106 protoplasts) were mixed with RNA transcripts (5 or 10 pmol of capped RNA or 30 pmol of uncapped RNA for a time course experiment) in prechilled electroporation cells (2-mm gap; Bio-Rad) and placed on ice for 10 min. Electroporation was then performed with a GenePulser Xcell device (Bio-Rad) set at 450 V/cm and with a time constant of 50 ms, delivering a capacitance of 900 ± 50 μF. After electroporation, the protoplasts were transferred to a microfuge tube containing 1.13 ml of growth medium (0.44 M d-mannitol, 3% sucrose, 0.01% inositol, 1 mg thiamine/liter, 5 μM 2,4-dichlorophenoxyacetic acid and 0.1 μM kinetin; adjusted to pH 5.7 with NaOH) and held under constant fluorescent illumination at room temperature (21°C).
Analysis of LUC activity.
Protoplasts were collected by pelleting them in a microfuge at 3,000 rpm and then lysed in Passive Lysis Buffer (Promega; 20 μl per 7.5 × 104 cells) for 5 min with constant shaking at room temperature. Portions of each extract (10 μl) were loaded into the wells of black, clear-bottom 96-well plates and mixed with 50 μl of Luciferase Assay Reagent (Promega) before LUC activity was read in a 1450 MicroBeta TriLux counter (Wallac).
Analysis of RNA survival in protoplasts.
At appropriate time points, total RNA was extracted from protoplasts with TRIzol reagent (Gibco-BRL) according to the manufacturer's directions. The resulting RNAs were glyoxalated and analyzed by 1% agarose gel electrophoresis as described previously (24) to assess the physical stability of the RNA. We have used time courses to estimate the functional stability of the RNA by determining the LUC expression half-life, i.e., the time taken for the rate of LUC accumulation to fall from its maximum value to half that. In most experiments, it has not been feasible to produce enough time points for accurate determination of the timing of the 50% rate, but we have found that the time taken to reach half-maximal LUC accumulation (5) produces a similar half-life estimate.
RESULTS
Relative expression of ORF-69 and ORF-206 in protoplasts.
Building on our previous studies describing the translational control provided by the 3′-UTR, we constructed LUC reporter RNAs that also had 5′ sequences from the genomic TYMV RNA, including the entire 5′-UTR. Paired constructs were designed to monitor expression one at a time from the two closely placed out-of-frame AUG codons, AUG69 at nt 88 and AUG206 at nt 95 (Fig. 1A and B). To preserve the native initiation environments of AUG69 and AUG206, these paired constructs had in common 217 nt from the 5′ end of the TYMV genomic RNA, which is considerably more than the 5′-UTR. Paired constructs differed by only the presence or absence of a single nucleotide needed to fuse either the p69 or p206 reading frame to the LUC ORF (see Fig. 1C). Consequently, ORF-69 expression was monitored by the production of a p69/LUC fusion protein (69L) made up of the first 43 amino acids of p69 fused to the dipeptide Leu-Gln and then to LUC (Fig. 1C). The sister construct monitoring ORF-206 expression encoded a p206/LUC fusion protein (206L) made up of the first 41 amino acids of p206 fused through Leu-Gln to LUC (Fig. 1C).
TYMV UTR function was studied by comparing expression from RNAs with the above TYMV 5′ sequences with expression from RNAs with a 17-nt generic, vector-derived 5′-UTR (referred to as vec in the names of constructs) that directed the synthesis of wild-type LUC (Fig. 1C). To check how the fusion with p69 and p206 sequences affected LUC specific activity (relative light units [RLU] per mole), we produced wild-type LUC (made from RNAs with 5′-vec sequences), 69L, and 206L in [35S]methionine-labeled form by translation in reticulocyte lysates. Bands from sodium dodecyl sulfate-polyacrylamide gel electrophoresis corresponding to each form of LUC were counted to determine the moles of LUC protein made, whereas the LUC activities (RLU) made in those lysates were measured in parallel. No significant differences in specific activity were found (not shown), permitting direct comparisons of LUC activity when translational expression was measured.
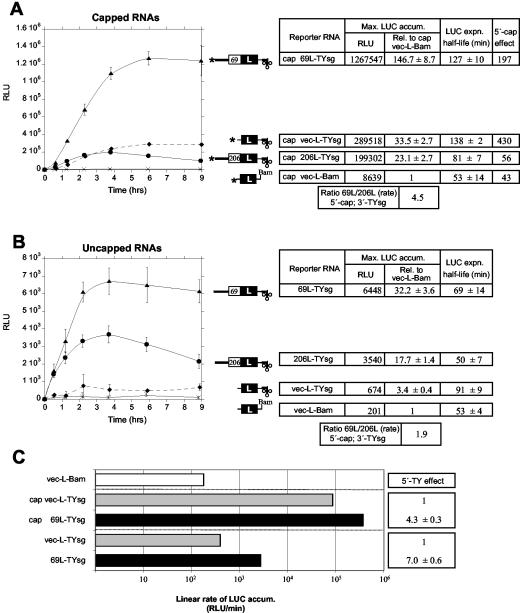
Interactions between the 5′- and 3′-UTRs were studied with RNAs bearing either the 684-nt 3′-UTR of genomic TYMV RNA (TYg), the 109-nt 3′-UTR of the subgenomic RNA (TYsg), or a basal vector-derived 19-nt 3′-UTR (Bam) (Fig. 1D). These same 3′-UTRs were used in our previous study in reporter RNAs with the 5′-vec sequences (17). As in that study, we monitored translation by transfection of cowpea protoplasts with LUC reporter RNAs, followed by the measurement of LUC activities from the protoplast extracts. To enable analysis of LUC expression at early times posttransfection, we delivered RNAs into cowpea protoplasts by electroporation rather than by polyethylene glycol (PEG)-mediated transfection as used previously (17). Expression from capped and uncapped vec-L-TYsg and vec-L-Bam RNAs revealed a 10-fold synergy between the enhancing effect of the 5′ cap and 3′-sgUTR (calculated from the ratio of the 3′-sg effect in the capped [33.5; see Fig. 3A] versus uncapped [3.4; see Fig. 3B] state). The identical synergy value was found previously in our PEG-mediated transfection studies (17), showing that expression patterns are not influenced by the RNA delivery methodology.
FIG. 3.
Combined influences of a 5′ cap and the TYMV 5′- and 3′-terminal sequences. Expression in cowpea protoplasts directed by the indicated capped (A) and uncapped (B) RNAs is presented as in Fig. 2. The 5′-cap effect represents the ratio of maximum LUC expression directed by capped and uncapped forms of the same RNA, with data taken from the time courses in panels A and B. Raw RLU data (after background subtraction) are reported in the tables at right, with the background for these assays being 642 RLU. To improve signals from uncapped RNAs, three times more uncapped than capped RNAs were transfected; expression is linearly dependent on the amount of RNA transfected (up to at least 30 pmol), and all data in this figure are normalized to permit direct comparison to expression data from capped RNAs. All data were derived by pooling two independent transfection experiments with duplicate transcripts per experiment (four datum points). Error bars represent the standard deviation. (C) Comparison of the response to adding 5′ TYMV sequences to reporter RNAs with a TYsg 3′-UTR (data derived from linear rate data from A and B). Note the logarithmic scale.
The 5′-capped reporter RNAs 69L-TYg and 206L-TYg (Fig. 2A) contain the 5′- and 3′-UTRs of the TYMV genomic RNA and thus best reflect the translational behavior of that RNA. The LUC expression from capped 69L-TYg RNA, based on maximum RLU levels reached in time courses after electroporation, was 5.1 times that from capped 206L-TYg RNA (Fig. 2A, 3.5/0.7). We noticed from the time courses, however, that whereas the levels of the 69L protein remained fairly steady after plateau, the levels of the 206L protein reached maximum earlier than 69L and subsequently declined (Fig. 2A). This pattern (also evident in Fig. 3 and 5) indicates a faster inactivation of either the 206L protein or the RNA encoding it, or both. We observed similar stabilities of 69L-TYg and 206L-TYg RNAs as studied by recovery of electroporated radiolabeled RNAs (not shown), a result that was not surprising since these RNAs differ only by the presence or absence of a residue 217 nt from the 5′ end. Further, there was no indication of protoplast death induced by 206L expression (not shown) to explain the decrease in 206L levels. We thus conclude that the 206L protein is less stable in the protoplasts than 69L or wild-type LUC. To minimize the resultant underestimation of 206L expression, we have also estimated relative expression from reporter RNAs in terms of the maximum rate of LUC accumulation (measured in RLU/hour; Fig. 2). On this basis, 69L-TYg RNA supported 3.1 times the expression from 206L-TYg RNA (Fig. 2A). This would correspond to threefold-higher translation of p69 than p206 from the genomic RNA.
FIG. 2.
Relative expression of ORF-69 and ORF-206 in protoplasts. LUC expression directed by the indicated capped reporter mRNAs with the TYg 3′-UTR (A) and Bam 3′-UTR (B) is presented. To facilitate comparison, the 3′-Bam datum points also appear in the time course in panel A. In the reporter mRNA diagrams, the asterisk represents a 5′ cap, thick lines represent TYMV 5′- or 3′-terminal sequences, and the cruciform represents the 3′-TLS. Bam indicates the 3′ terminus of the generic Bam 3′-UTR. The graph shows time courses of the accumulation of LUC activity (in RLU) produced from the indicated RNAs after electroporation into cowpea protoplasts. LUC expression is expressed in tabular form based on maximum levels reached and on the maximal linear rate of accumulation. The latter parameter is less sensitive to differences in RNA or protein turnover, which is expressed as the LUC expression half-life. For RNAs encoding the same form of LUC, or equally stable forms of LUC, this parameter equates to RNA functional half-life. Because the half-life of 206L protein expression is shorter than for other forms of LUC, comparisons involving expression of 206L and other forms of LUC (e.g., determination of 69L/206L expression ratio) are best done by using the linear rate data. The time course plots shown are derived from a single experiment with duplicates (the error bars represent the standard deviation), whereas the tabulated data are derived from two such experiments (four datum points). (C) Comparison of the response to adding the TYg 3′-UTR to reporter RNAs with different 5′ sequences (data derived from linear rate data from panels A and B).
FIG. 5.
Differential effect on 69L and 206L expression of a large internal deletion in the TYMV 5′-UTR. Expression in cowpea protoplasts directed by the indicated capped reporter RNAs expressing 69L (A) and 206L (B) is presented as in Fig. 2. The deletion of nt 13 through 79 of the 5′-UTR was adapted from an infectious TYMV variant reported by Hellendoorn et al. (10). The linear rates of LUC accumulation are expressed relative to expression from vec-L-TYg, which is shown in both panels A and B. The data were produced in the same experiments and with the same replication as the data of Fig. 2. (C) Comparison of the response to adding the TYg 3′-UTR to reporter RNAs with different 5′ sequences (data were derived from linear rate data in panels A and B).
Translational enhancement by 5′ TYMV sequences.
Maximal expression from capped 69L-TYg RNA was 3.5-fold that from capped vec-L-TYg RNA (Fig. 2A), suggesting that the 5′ 217 nt of TYMV RNA enhance expression. Because the profiles of LUC expression from these RNAs were similar (similar LUC expression half-lives; Fig. 2A), the expression difference is not complicated by different protein or RNA stabilities and seems to reflect different translational efficiencies. Measurement of the translational efficiency directly as the initial linear rate of LUC increase confirms the response to 5′ TYMV sequences (3.8-fold increase; Fig. 2A). The enhanced expression from the 5′-most AUG (AUG69) occurred despite its weaker context (−3C/+4A) compared to the strong context of the AUG codon after the vec-UTR (−3A/+4G) (Fig. 1). To determine whether this enhancement depended on the presence of 3′ TYMV sequences, we compared expression from capped 69L-Bam and vec-L-Bam RNAs (Fig. 2B). The 5′-TYMV sequences increased expression fivefold (Fig. 2B). Expression from capped 206L-Bam RNA was also fivefold higher than from vec-L-Bam RNA on the basis of initial rates of LUC accumulation (Fig. 2B), indicating similar initiation at AUG69 and AUG206 in the presence of the 3′-Bam UTR. Surprisingly, the 3′-UTR appears to influence the behavior of ribosomes on the TYMV 5′-UTR and the relative initiation at the AUG codons at nt 88 and 95. Barends et al. (1) have postulated a communication between the 3′-UTR and the 5′ end that selectively enhances initiation from AUG206. Our results indicate that stimulated expression derived from the TYMV 3′-UTR in fact benefits initiation from both AUG codons, but more so from AUG69 (Fig. 2C).
Translation of both ORF-69 and ORF-206 is cap dependent.
To better understand the contribution of the TYMV 5′ sequences, we compared expression from capped and uncapped RNAs with or without 5′ TYMV sequences. In these experiments, we also wished to assess the role of the cap in TYMV gene expression. Based on the fact that TYMV RNA is naturally capped and lacks an extended 5′-UTR with upstream AUG codons, it seems most reasonable to expect the RNA to be translated by the standard cap-dependent initiation mechanism, with leaky scanning (13) or random selection (26) explaining the choice between initiation at the closely spaced starts at AUG69 and AUG206. On the other hand, the recent proposal based on in vitro studies (1) calls for ORF-206, though not ORF-69, to be translated by a cap-independent mechanism. Experiments with capped and uncapped RNAs were designed to distinguish between these proposals. Because expression from uncapped RNAs was low, we utilized RNAs with the TYsg 3′-UTR, which is a roughly twofold stronger translational enhancer than TYg (17).
The enhancing effect of the 5′ cap on expression from vec-L-Bam, an RNA lacking a 3′-translational enhancer capable of interacting synergistically with the cap, was 43-fold (Fig. 3A). The cap effect for vec-L-TYsg RNA (Fig. 3A) was 10-fold higher because of synergy between the cap and 3′-TYsg enhancer (17). With TYMV 5′ sequences additionally present, the cap effects were 197- and 56-fold (Fig. 3A), respectively, for 69L and 206L protein synthesis. Clearly, expression from all RNAs tested was strongly cap dependent, especially for each of the RNAs with 3′ TYMV sequences, from which there was additional expression due to synergistic effects with the cap. The cap dependency observed means that expression from uncapped 69L-TYsg and 206L-TYsg RNAs was only 0.5 or 2% that from capped forms of the same RNA (see RLU data, Fig. 3). Indeed, gene expression from uncapped RNAs was so low that even though three times the amount of capped RNAs was transfected (followed by appropriate corrections), LUC signals from some RNAs were close to background (RLU for vec-L-Bam; Fig. 3B).
Joint contribution of 5′ and 3′ TYMV sequences to enhanced expression.
As previously observed with capped RNAs (17), enhancement by the TYsg 3′-UTR (33.5-fold; Fig. 3A) was about twice (1.6 times) that from TYg (21.0-fold; from data in the Fig. 2 experiment). In comparison with 3′-TYg, 3′-TYsg displayed a similar relationship with the capped TYMV 5′ sequences: similar ratios of 69L to 206L expression and relationship to the levels of LUC produced from the vec 5′-UTR and strong translation from the combined effects of 5′ and 3′ TYMV sequences (compare the time courses of Fig. 2A and 3A).
Comparing expression from 69L-TYsg and vec-L-TYsg RNAs showed that the TYMV 5′ sequences enhanced expression from AUG69 in both the capped (Fig. 3A) and uncapped (Fig. 3B) states. This occurred despite a minor decrease in RNA stability (shorter LUC expression half-life; Fig. 3A and B). Note also that functional RNA half-lives (see Materials and Methods) of uncapped RNAs are no less than 50% relative to the capped form of the same RNA, as previously observed (17).
The 5′ TYMV sequences also enhanced expression initiating from AUG206 of uncapped 206L-TYsg RNA relative to that from vec-L-TYsg (Fig. 3B), but the LUC activities derived from the capped forms of the two RNAs were similar (Fig. 3A). Likewise, a lower TYMV 5′-UTR-dependent enhancement from capped than from uncapped RNAs was evident with 69L-TYsg RNA (Fig. 3C). Interestingly, the response of 69L and 206L expression to the addition of 5′ TYMV sequences is different, so that the ratio of initiation from AUG69 relative to AUG206 was 1.9 from the uncapped 3′-sg RNAs (Fig. 3B) but 4.5 from the capped RNAs (Fig. 3A). This is reminiscent of a ratio of 0.9 from capped RNAs with 3′-Bam UTR (Fig. 2B) but 3.1 for capped RNAs with the TYg 3′-UTR (Fig. 2A). In both cases, the presence of two terminal translation enhancers (5′ cap and TYMV 3′-UTR) resulted in a more skewed initiation profile in favor of ORF-69. Also in both cases, this resulted from a greater stimulation of ORF-69 than ORF-206 expression when compared to related reporter constructs lacking one of the terminal enhancers. In Fig. 2C, this is seen by comparing the response from adding a TYg 3′-UTR to the basal 3′-Bam in the presence of 5′-69L (9.9-fold increase) compared to 5′-206L sequences (2.9-fold increase). Interestingly, the greatest stimulation due to 3′-TYg occurred in the presence of the vec 5′-UTR (13-fold increase). The parallel trend is evident in Fig. 3A in the gradation of cap effects for RNA with 3′-TYsg and differing 5′-UTRs (5′-vec > 69L > 206L). Note, however, that whereas the response to the addition of a 5′ cap or TYMV 3′-UTR is smaller in the presence of 5′ TYMV sequences, absolute expression levels are highest for RNAs with all enhancing elements, cap and 5′ and 3′ TYMV sequences (Fig. 3C).
Another way to view these results is in terms of the combined response of adding a capped 5′-UTR and TYsg 3′-UTR to the basal reporter vec-L-Bam RNA. For RNAs with the vec 5′-UTR, we observed a 10-fold-higher response to the addition of both 5′ cap and 3′-TYsg than expected from the individual effects of these enhancers (derived from data in Fig. 3). This corresponds to previous estimates of 5′ cap/3′-TYsg synergy (17). When the TYMV 5′-UTR was added together with the cap, the “excess” response was only 4.6-fold when 5′-69L sequences were added and 1.3-fold when 206L sequences were added. These results suggest some functional overlap between the TYMV 5′-UTR and the other enhancers, and an influence of the terminal enhancers on the ratio of 69L and 206L expression (see also later).
Role of valylation in 3′-UTR translational enhancement.
In previous studies, we have shown that maximal translational enhancement by the TYsg 3′-UTR requires aminoacylation (17). The valylation of TYsg can be completely destroyed by a point mutation that changes the anticodon from CAC to CGC, a mutation whose effect we studied with the TYsg(CGC) 3′-UTR against different capped 5′-UTRs (Fig. 4). Because comparisons between 206L, 69L, and wild-type LUC expression (from vec-L RNAs) are best made on the basis of rates of LUC accumulation, we focused in these experiments on early times postelectroporation.
FIG. 4.
Influence of 3′ aminoacylation on expression of 69L and 206L. Time courses of LUC accumulation in cowpea protoplasts directed by the indicated capped RNAs with 5′-vec (A), 5′-69L (B), and 5′-206L (C) sequences are shown. The CGC mutation refers to mutation of the CAC anticodon of the 3′-TLS to CGC, resulting in abolition of valylation. The time course plots shown are derived from a single experiment with duplicates (error bars represent standard deviation), while the tabulated data are derived from two experiments (four datum points). (D) Tabulation of results from the time courses.
With a vec 5′-UTR, the CGC mutation decreased LUC expression by a factor of 5.2 (Fig. 4D, column b). 69L and 206L expression were decreased by factors of 2.8 and 1.7, respectively, in response to the anticodon mutation (Fig. 4D, column b). This means that the ratio of expression from AUG69 and AUG206 was 3.6 from RNA with the wild-type sg 3′-UTR (Fig. 4D, column c) and 2.2 in the presence of the CGC mutation (Fig. 4D, column d).
These results once again echo the observation above on the influence of the 5′ TYMV sequences in tempering the stimulatory effects of the terminal translational enhancers. Thus, the enhancement derived from the 5′ TYMV sequences was stronger in RNAs with the weakened 3′ enhancer TYsg(CGC) than with the wild-type TYsg (Fig. 4D, column d versus column c). Another expression of these relationships is that for capped RNAs with the 5′ TYMV sequences, 206L expression benefits less from the 3′ translation enhancement effect than 69L (Fig. 4D, column b: 1.7- versus 2.8-fold). This is opposite to the proposed influence of the 3′-UTR specifically on ORF-206 expression (1).
Differential effect on ORF-69 and ORF-206 translation after internal deletion of the 5′-UTR.
In order to gain more insight into coordination of ORF-69 and ORF-206 translation by the TYMV genomic 5′- and 3′-UTRs, we tested expression from RNAs with three-quarters of the 5′-UTR missing due to deletion of nt 13 through 79. This deletion leaves a 21-nt UTR with unchanged context around the initiation codons. TYMV RNA with this mutation was viable and genetically stable in plants, although somewhat defective for systemic spread (10).
Introduction of the 5′-UTR deletion (5′Δ) into the 5′ TYMV sequences of capped RNAs decreased the rate of 69L accumulation 2-fold (Fig. 5A), while increasing 206L accumulation 3.5-fold (Fig. 5B). Consequently, in the presence of the 5′Δ-UTR, the rate of 206L expression was higher than 69L (also observed for RNAs with 3′-Bam; Fig. 5C), meaning that many ribosomes were bypassing the first AUG without initiating. These changes in 69L and 206L expression were made without significantly altering the functional stability of the RNA (Fig. 5A and B).
Another reason the 5′Δ mutation was of interest to us was in relation to the 5′-UTR function in the scheme proposed by Barends et al. (1). If the 3′-TLS were to direct translation specifically of ORF-206 as proposed, there should be some collaborative cis elements near the AUG, perhaps in the 5′-UTR. If removed by the 5′Δ deletion, this should result in specific negative effects in the relationship between ORF-206 expression and the 3′-UTR. We observed that replacement of the Bam with the TYg 3′-UTR had the same effect (5.4-fold increase; Fig. 5C) on the rate of expression of both 69L and 206L. In contrast, with the wild-type 5′ TYMV sequences, this same 3′ replacement increased expression of 69L 9.9-fold and of 206L 2.9-fold (Fig. 2C). Thus, the initiation behavior at AUG69 and AUG206 was changed by the 5′Δ mutation, with ORF-206 expression favored. This suggests that the deleted portion of the TYMV 5′-UTR does not collaborate with the 3′-UTR in promoting initiation of ORF-206.
Knockout of either start codon increases gene expression from the reciprocal start codon.
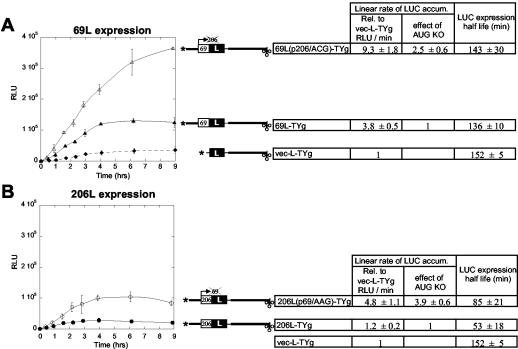
The previously discussed experiments indicated an influence of the terminal enhancer elements (the 5′ cap and 3′-UTR) on the selection by ribosomes between initiation at AUG69 and AUG206. An understanding of this surprising effect necessitates knowledge of the normal selection mechanisms between these AUGs. We have previously used rabbit reticulocyte lysates to study this issue, with mutations to inactivate each initiation codon (24). Mutation of the AUG69 to AAG eliminated in vitro expression of ORF-69, while slightly increasing ORF-206 expression. The reciprocal mutation of AUG206 to ACG, while retaining the native coding sequence of ORF-69, substantially increased the expression of ORF-69 (24), behavior that is not expected of simple leaky scanning (12). To examine the situation in vivo, we revisited this issue with the appropriate LUC reporter RNAs.
Knockout of AUG69 increased expression from AUG206 (downstream) by 3.9-fold [206L(p69/AAG)-TYg RNA; Fig. 6B ], while knockout of AUG206 increased expression from AUG69 (upstream) by 2.5-fold [69L(p206/ACG)-TYg RNA; Fig. 6A]. The functional stabilities of the RNAs were only slightly increased by the mutations (Fig. 6), indicating substantial differences in translational behavior. The effect of the p206/ACG mutation in increasing expression from the upstream AUG69 is similar to that observed in vitro (24) and is abnormal behavior for leaky scanning as the process determining ribosome selection between AUG69 and AUG206. On the other hand, we observed a greater increase of ORF-206 expression upon removal of the upstream AUG in these results than in reticulocyte lysates, a result expected of leaky scanning.
FIG. 6.
Mutation of either AUG69 or AUG206 increases expression from the remaining AUG initiation codon. Time courses of LUC accumulation in cowpea protoplasts directed by the indicated capped RNAs expressing 69L (A) and 206L (B) is presented as in Fig. 2. In 69L(p206/ACG)-TYg RNA (A), the ORF-206 initiation codon has been mutated to ACG, and in 206L(p69/AAG)-TYg RNA (B), the ORF-69 initiation codon has been mutated to AAG. No mutations alter the encoded amino acid sequence. Both AUG mutations resulted in abolished expression of the respective gene in vitro (24). The data were produced in the same experiments and with the same replication as the data of Fig. 2.
DISCUSSION
5′ TYMV sequences enhance translation. In comparison to a 17-nt generic (vec) 5′-UTR, 5′ genomic TYMV sequences increased expression from the 5′-most AUG without significantly affecting the stability of the RNA (Fig. 2 and 3). In transfected protoplasts, the expression was enhanced 4.3-fold from capped RNAs and 7.0-fold in the absence of a cap (Fig. 3C). Future studies will need to refine the properties of this enhancement, since the mRNAs used in the present study varied in 5′-UTR length and in the context surrounding the initiation codons (Fig. 1B). Since the context of the TYMV AUG69 codon is considerably weaker than that of the AUG codon in the vec 5′-UTR (−3C/+4A versus −3A/+4G), we may well have underestimated the enhancement. Even with three-quarters of the TYMV 5′-UTR deleted, producing a UTR similar in length to the vec UTR, expression from AUG69 (the 5′-most AUG) was stronger than from the vec UTR (Fig. 5A). This finding supports the conclusion that the TYMV 5′-UTR does provide for efficient translation.
A previous study using β-glucuronidase reporter genes failed to detect translational enhancement by the TYMV 5′-UTR in tobacco protoplasts (8). This was evidently due to the design of the reporter constructs used, with the β-glucuronidase initiation codon placed downstream of the AUG69 codon. With a similar reporter, we observed no significant enhancement in expression from the downstream AUG206 codon upon addition of 5′ TYMV sequences (Fig. 2A, 206L-TYg versus vec-L-TYg; Fig. 3A, 206L-TYsg versus vec-L-TYsg).
The 5′ TYMV sequences diminish the enhancing effects of the cap and 3′-UTR.
The highest expression observed in these studies came from RNAs containing all three enhancing elements: the 5′ cap, the 5′ TYMV sequences, and the TYMV 3′-UTR (Fig. 3, capped 69L-TYsg RNA). These elements are clearly important in optimizing expression from TYMV RNA. However, stimulatory effects (enhancement ratios) provided by the cap (Fig. 3A) and the TYMV 3′-UTR (Fig. 2C) were lower when 5′-TYMV sequences were present. Vice versa, the enhancing effects of the 5′ TYMV sequences in capped RNAs were greater when the TYMV 3′-UTR was absent or weakened by mutation of the anticodon (Fig. 4D, column c versus column d).
The behavior of the 5′ TYMV sequences shares similarities with that of the TMV Ω enhancer. The presence of the Ω element in an RNA decreases the stimulatory effect of a cap, particularly in the presence of a poly(A) tail (with which it can synergistically interact) and vice versa (4). Among capped RNAs, enhancement by Ω was greatest in the absence of a poly(A) tail. It was concluded from those studies that Ω and the cap provide overlapping or partially redundant functions that can be in competition, which can also affect the ability of the cap to interact synergistically with the poly(A) tail. Thus, whereas the presence of both cap and Ω provides the highest levels of translation, the enhancing potential of each of these elements is blunted in the presence of the other, perhaps because of overlapping action such as the recruitment of eIF4F, which is typically limiting (4).
A further similarity between the TYMV and TMV systems is in the influence of the viral 5′ sequences on the earliest stages of translation. Inspection of the earliest parts of the LUC expression time courses revealed a longer lag before a maximal linear rate of LUC accumulation was reached for RNAs lacking 5′ TYMV sequences (Fig. 2A, 3A, and 4). This finding suggests that, as hypothesized for the TMV Ω element (4), the TYMV 5′ sequences facilitate the earliest rounds of translation in a way that the cap is not capable of.
Expression of the overlapping ORFs of TYMV RNA.
Using capped reporter RNAs with the same 5′- and 3′-terminal sequences as genomic TYMV RNA (69L-TYg and 206L-TYg RNAs), we estimated the ratio of ORF-69 to ORF-206 expression in cowpea protoplasts to be about 3 (Table 1). This estimate relied on the comparison of the maximum rates of LUC accumulation early in the expression profile (Fig. 2A) in order to avoid bias due to a short half-life of 206L expression compared to 69L. Neither protoplast death induced by 206L expression nor RNA lability (RNAs with 5′ 69L and 206L sequences differ only by the presence or absence of 1 nt that matches LUC to ORF-69 or ORF-206) appear to be responsible for the faster turnover of 206L expression. We therefore deduce that 206L is a less stable protein. It will be interesting to determine whether this is true of the native TYMV proteins with the same N-terminal sequence, p206 and its proteolytically processed daughter, p141. Ironically, accelerated turnover of p69, not p206, has been reported, although residues implicated in ubiquitin-dependent degradation (2) were not present in our 69L constructs.
TABLE 1.
Ratio of the rate of expression of 69L to 206L proteinsa
The likeliest expression strategy for ORF-69 and ORF-206 involves the conventional process of ribosome association at the cap followed by scanning through the 5′-UTR. Leaky scanning, or some variant of it, could explain the choice by ribosomes between AUG69 and AUG206 for initiation. The upstream AUG69 context is rather weak and should permit sufficient ribosomes to continue scanning for initiation at AUG206. We observed that initiation at both AUGs was strongly dependent on a 5′ cap (Fig. 3A), a finding consistent with expression by ribosomes that were initially recruited to the 5′ terminus by the cap. Mutation of the upstream AUG69 increased the expression rate from AUG206 3.9-fold (Fig. 6B), an observation that is consistent with leaky scanning. However, contrary to the behavior expected of leaky scanning, mutation of AUG206 led to an increase (2.5-fold; Fig. 6A) of expression from AUG69, which lies upstream. This response might be explained by the release of a restriction on ORF-69 expression at the elongation phase of translation by a relatively slow progress of ribosomes translating the overlapping ORF-206. Slow elongation on ORF-206 might occur because of slow decoding at certain codons, and ribosomes translating ORF-69 would be limited to the same elongation rate because ribosomes are unable to pass on an mRNA. Expression limitation of this sort has been suggested for reovirus S1 RNA, which also has overlapping ORFs (3). Perhaps ORF-206 expression is normally limited by both slow elongation and limited initiation due to interference by the upstream AUG69.
A proposal by Williams and Lamb (26) provides another model for ribosome behavior on TYMV RNA. These authors studied the synthesis of influenza B virus NB and NA proteins from overlapping ORFs that are separated by 4 nt as in TYMV RNA. Based on a mutagenic study and expression in cells, they suggested that ribosomes reach the paired AUG initiation codons by conventional scanning but then select between the closely spaced AUGs by random choice. This scheme has been disputed by Kozak (12) based on studies with the same closely spaced initiation codons. However, those studies were performed in vitro and with non-influenza virus sequences flanking the initiation sites and so may not have dealt with the same phenomenon as that described by Williams and Lamb (26). A reciprocal increase in initiation from the remaining AUG in response to mutation of one AUG triplet, as we observed, is an expected property of a random selection process.
Future studies will further test the applicability of the random choice model to TYMV gene expression. Such studies will have to take into account the surprising observations summarized in Table 1. We observed the expression from AUG69 relative to AUG206 to vary depending on the presence or absence of translational enhancers at the 5′ and 3′ termini, i.e., the cap and TYg or TYsg 3′-UTRs. The expression ratio was also different after deletion of much of the 5′-UTR (5′Δ). The presence of translational enhancers—cap, 5′ TYMV sequences, or TYMV 3′-UTR—favored ORF-69 over ORF-206 expression. The effect of the 5′-UTR deletion may be explained by the positioning of AUG69 close enough to the 5′ end to promote ribosome bypassing, a phenomenon described by Kozak (14). The other observations suggest a mechanism whereby translation that is occurring on an RNA for which there is synergy between 5′ and 3′ enhancers is carried out principally by ribosomes that are recycled from the 3′ end and that carry with them some attribute that favors initiation at the first AUG codon.
Is ORF-206 translation facilitated by “tRNA mimicry as a molecular Trojan horse?”
Our studies have provided a test of some aspects of the TYMV gene expression model recently proposed by Barends et al. (1) and described in terms of a molecular Trojan horse. Unlike those studies, which were conducted in wheat germ extracts, our experiments used protoplasts capable of supporting a TYMV infection. Although we used LUC reporter RNAs, gene expression was assayed in the same milieu as that of a natural infection, and the reporter RNAs had considerable TYMV sequence at both ends. The tested aspects of the proposed scheme were: (i) is ORF-206 translation cap independent and (ii) does the TYMV 3′-UTR containing the TLS enhance, perhaps in an obligatory way, expression of ORF-206 but not of ORF-69?
Our experiments make it clear that translation from both AUG69 and AUG206 is strongly cap dependent. We have also observed that the TYMV 3′-UTR is a general enhancer, with a principal mode of action being synergy with the cap (17). The 3′-UTR-enhanced translation from RNAs with 5′ vec or 5′ TYMV sequences and from AUG69, as well as from AUG206 (Fig. 2C). We did observe a difference in the influence of the 3′-UTR on initiation at AUG69 compared to AUG206, but ORF-206 expression was less dependent on the 3′-UTR than expression of ORF-69 (Fig. 2C). This is opposite to the Trojan horse proposal (1).
Our experiments thus do not support the findings of Barends et al. (1) model and, in fact, contradict them. That model may describe an in vitro phenomenon or the result of studies that attempted to identify TYMV proteins without immunological verification.
Acknowledgments
This study was supported by NSF grant MCB0235563. L. Bauer was the beneficiary of an REU supplement from NSF.
REFERENCES
- 1.Barends, S., H. H. Bink, S. H. van den Worm, C. W. Pleij, and B. Kraal. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112:123-129. [DOI] [PubMed] [Google Scholar]
- 2.Drugeon, G., and I. Jupin. 2002. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83:3187-3197. [DOI] [PubMed] [Google Scholar]
- 3.Fajardo, J. E., and A. J. Shatkin. 1990. Translation of bicistronic viral mRNA in transfected cells: regulation at the level of elongation. Proc. Natl. Acad. Sci. USA 87:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallie, D. R. 2002. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30:3401-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 6.Gallie, D. R., and M. Kobayashi. 1994. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 142:159-165. [DOI] [PubMed] [Google Scholar]
- 7.Gallie, D. R., D. E. Sleat, J. W. Watts, P. C. Turner, and T. M. Wilson. 1987. The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15:3257-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallie, D. R., D. E. Sleat, J. W. Watts, P. C. Turner, and T. M. Wilson. 1987. A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 15:8693-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellendoorn, K., P. J. Michiels, R. Buitenhuis, and C. W. Pleij. 1996. Protonatable hairpins are conserved in the 5′-untranslated region of tymovirus RNAs. Nucleic Acids Res. 24:4910-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellendoorn, K., P. W. Verlaan, and C. W. Pleij. 1997. A functional role for the conserved protonatable hairpins in the 5′ untranslated region of turnip yellow mosaic virus RNA. J. Virol. 71:8774-8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jobling, S. A., and L. Gehrke. 1987. Enhanced translation of chimaeric messenger RNAs containing a plant viral untranslated leader sequence. Nature 325:622-625. [DOI] [PubMed] [Google Scholar]
- 12.Kozak, M. 1995. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl. Acad. Sci. USA 92:2662-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 14.Kozak, M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1:111-115. [PMC free article] [PubMed] [Google Scholar]
- 15.Lukaszewicz, M., M. Feuermann, B. Jerouville, A. Stas, and M. Boutry. 2000. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 154:89-98. [DOI] [PubMed] [Google Scholar]
- 16.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the posttranscriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda, D., and T. W. Dreher. 2004. The tRNA-like structure of Turnip yellow mosaic virus RNA in a 3′-translational enhancer. Virology 321:36-46. [DOI] [PubMed] [Google Scholar]
- 18.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 20.Sarnow, P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomashevskaya, O., A. Solovyev, O. Karpova, O. Fedorkin, N. Rodionova, S. Morozov, and J. Atabekov. 1993. Effects of sequence elements in the potato virus X RNA 5′ non-translated alpha beta-leader on its translation enhancing activity. J. Gen. Virol. 74:2717-2724. [DOI] [PubMed] [Google Scholar]
- 22.Turner, R. L., M. Glynn, S. C. Taylor, M. K. Cheung, C. Spurr, D. Twell, and G. D. Foster. 1999. Analysis of a translational enhancer present within the 5′-terminal sequence of the genomic RNA of potato virus S. Arch. Virol. 144:1451-1461. [DOI] [PubMed] [Google Scholar]
- 23.Vende, P., M. Piron, N. Castagne, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiland, J. J., and T. W. Dreher. 1989. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 17:4675-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells, D. R., R. L. Tanguay, H. Le, and D. R. Gallie. 1998. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 12:3236-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams, M. A., and R. A. Lamb. 1989. Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J. Virol. 63:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]