Abstract
Optogenetic gene expression systems enable spatial-temporal modulation of gene transcription and cell behavior. Although applications in biomedicine are emerging, the utility of optogenetic gene switches remains elusive in cancer research due to the relative low gene activation efficiency. Here, we present an optimized CRISPR-Cas9-based light-inducible gene expression device that controls gene transcription in a dose-dependent manner. To prove the potential utility of this device, P53 was tested as a functional target in the bladder cancer cell models. It was illustrated that the light-induced P53 inhibited proliferation of 5637 and UMUC-3 cell effectively. The “light-on” gene expression system may demonstrate a novel therapeutic strategy for bladder cancer intervention.
Keywords: light-inducible gene expression, CRISPR-Cas9, gene switch, bladder cancer.
Introduction
Chemical-regulated gene expression systems have been widely applied in biomedical research and biotechnology 1, 2. However, molecular-inherent drawbacks such as diffusion-based transportation and difficulties in removing the inducer have limited their utility when precisely timed or localized induction is required. Compared with chemicals, light is an ideal gene expression inducer due to its unprecedented tunability and spatiotemporal resolution 3-6. Therefore, the development of light-inducible gene expression device is highly desired and has received much attention 7. Undoubtedly, it will be exciting to witness the development of bio-production processes and biomedical interventions with light, such as cancer interventions.
Recently, a variety of light-inducible gene expression devices in mammalian cells have been reported 7. Such devices mainly fall into two categories including: (I) those using the light-triggered homo-dimerization of a photoreceptor for reconstitution a DNA-binding domain to guide a transcriptional activation domain (AD) (VP64, P65, etc.) to the target gene 8, 9; (II) those employing the light-regulated recruitment of an AD to a DNA-binding protein (ZFN, TALE, etc.) following two-hybrid principle (such as interaction between cryptochrome 2, CRY2 and its binding partner, CIB1, etc.) 10-13. Generally, the first class system can activate a stronger gene transcription (>100-fold). And thus, first applications in biomedicine field are starting to emerge including treating diabetes mouse model by light-induced production of insulin 9. The second strategy is more widely interrogated with the development of gene targeting technology, as it can be customized to target many DNA sites including endogenous genes. Up to date, CRISPR-Cas9-based light-inducible transcription systems have been reported, affording greater flexibility and simplicity in activating any gene 11, 13. However, the relative lower gene activation efficiency still limited their potential applications.
Here, we described an optimized light-inducible gene expression device based on dCas9 and the light-sensitive CRY2-CIB1 interaction. The transcription activation components were optimized by testing different combinations of dCas9-CIBN and CRY2-AD infusions in bladder cancer cell models. Furthermore, to prove the potential utility of this device, tumor suppressor p53 gene, frequently mutated in bladder cancer cell 14, was used as a therapeutic target. Our results demonstrated that light-induced P53 expression can effectively inhibit proliferation of bladder cancer cell 5637 and UMUC-3 in vitro, which might become a new promising strategy for bladder cancer therapy.
Results and discussion
Optimization of CRISPR-Cas9-based light-inducible gene expression system in bladder cancer cell
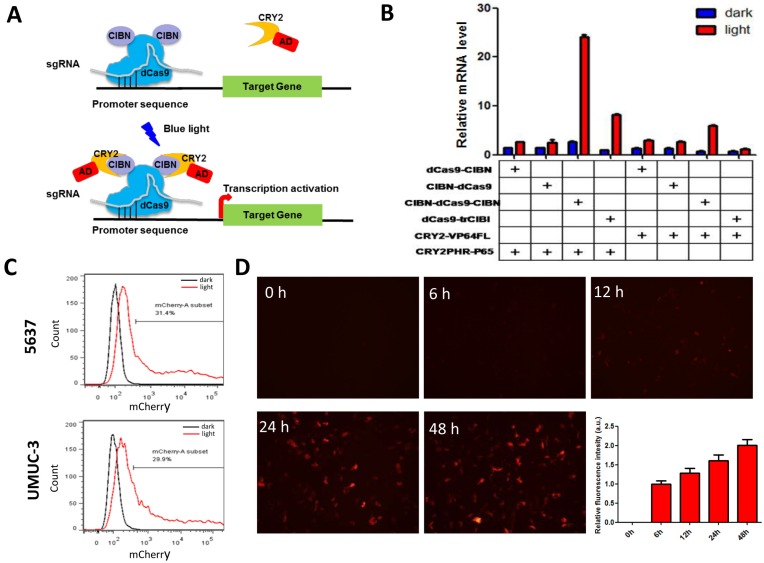
The CRISPR-Cas9-based light-inducible gene expression system consists of genomic anchor (dCas9-CIB1 fusion protein), transcriptional activator (CRY2-AD fusion protein), sgRNA and reporter/effector gene (Fig.1A). In the absence of blue light, the genomic anchor binds to the targeted promoter sequence, guided by the sgRNA, while the activator is freely diffused within nucleus. Upon blue light illumination, CRY2 and CIBI are heterodimerized, recruiting transcriptional ADs to the target locus to activate gene transcription.
Figure 1.
Optimization of CRISPR-Cas9-based light-inducible gene expression system in bladder cancer cell. A) Schematic of the CRISPR-Cas9-based light-inducible gene expression system; B) Reporter mRNA expression induced by each combination of genomic anchor and activator components; C) Flow cytometry analysis of mCherry induction in bladder cancer cell under the optimized combination; D) Time course of light-induced reporter (mCherry) fluorescence intensity. Data are shown as mean ±SD.
To assemble an effective light-inducible gene expression system for bladder cancer cell, we assessed the induction efficiency of different combinations of genomic anchors and activators after 24 hours' illumination. It was found that the fusion of CIBN to both end (N- and C- termini) of dCas9 led to a significant increase in reporter gene expression in bladder cell, compared with those inductions by dCas9 fusions to a single CIBN domain (Fig.1B). Besides, the P65 transcription AD is much more efficient to induce reporter gene expression (>10-fold) than VP64 in bladder cancer cell (Fig.1B). Thus, the combination of CIBN-dCas9-CIBN and CRY2PHR-P65 was sufficient to induce gene expression in bladder cancer cell 5637 and UMUC-3 (Fig.1C, Fig.S1A), with an activation efficiency of 3 times as high as that previously described 11.
Besides, we also investigated the time course of light-induced reporter gene expression in 5637 cell using the optimized combination. It was shown that the fluorescence intensity of reporter gene increased as the illumination time prolonged (Fig.1D). Although mcherry mRNA level reduced after 12 hours' illumination, it remained at a high level after 48 hours' illumination (30-fold) (Fig.S1B).
The effects of blue light illumination dose on reporter and effector gene expression
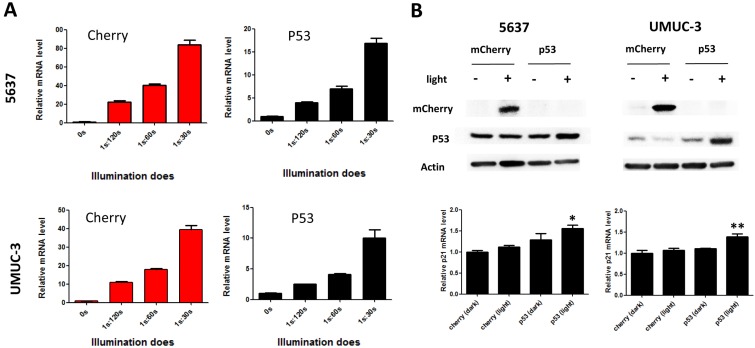
To demonstrate the potential utility of the system in therapeutic application, the best known tumor suppressor used in gene therapy 15, 16, p53 was tested as an effector gene. Bladder cancer cells (5637 and UMUC-3) transfected with light-inducible mcherry and p53 gene expression systems were treated with different illumination dose for 12h (Fig.2A). Similar to mcherry gene, p53 mRNA level increased with the illumination dose in both 5637 and UMUC-3 cells. Notably, the calculated induction efficiency of p53 was much lower than that of mcherry. This is partially caused by the endogenous expression of p53 which increases the background, while mcherry is absent in both cells. Besides, p53 mRNA may also be subjected to a faster degradation rate than mcherry mRNA in cell, which deserve further investigation.
Figure 2.
Dose-dependent reporter (mCherry) and effector gene (P53) expression induced by light in bladder cancer cell. A) mcherry and p53 mRNA level induced by increasing illumination dose in 5637 and UMUC-3 cell. 1s:30-120s stands for 1 second illumination (0.84 W/m2) every 30-120 seconds; B) mCherry, p53 protein expression induced by light (0.84 W/m2, 1s:30s) and downstream p21 mRNA level. Data are shown as mean ±SD; *, p<0.05; **, p<0.01.
At the illumination frequency of 1s (light): 30s (dark) with an average irradiance of 0.84 W/m2, 16- and 9- fold inductions of p53 mRNA were achieved in 5637 and UMUC-3 cell (Fig.2A), respectively. Correspondently, induction of mCherry and P53 protein were detected under the illumination condition in both cell lines (Fig.2B). In addition, the downstream p21 mRNA was significantly up-regulated in both cell lines with induced p53 expression by light (Fig.2B). Those results suggested that the system may be applied to bladder cancer interventions effectively.
The effects of light-induced P53 expression on bladder cancer cell proliferation
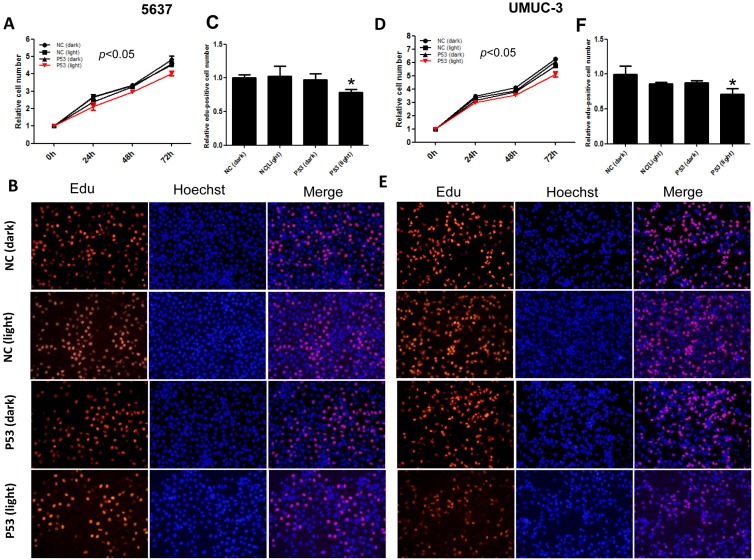
Furthermore, we investigated the function of light-induced P53 expression on bladder cancer cell proliferation under the same illumination condition (0.84 W/m2; 1s:30s). By CCK-8 assay, it was indicated that light-induced p53 expression inhibited the growth of both 5637 (Fig.3A) and UMUC-3 (Fig.3D) cell significantly (p<0.05) during 72 hours' exposure to the blue light, compared with the negative controls. Edu incorporation assay was carried out to interrogate the effect of light-induced P53 on the proliferation of bladder cancer cell in more detail (Fig.3B, E). Relative number of Edu-positive cell reduced significantly (p<0.05) in 5637 and UMUC-3 cells, by ~22% (Fig.3C) and 29% (Fig.3F) respectively. These results demonstrated that light-induced tumor suppressor gene expression can inhibit proliferation of bladder cancer cell remarkably, providing new strategy for cancer therapy.
Figure 3.
Inhibition of bladder cancer cell proliferation by light-induced P53 expression. The growth curves of 5637 (A) and UMUC-3 cell (D) treated with induced P53 under continuous illumination (0.84 W/m2; 1s:30s) were monitored by CCK-8 assay; The proliferation of 5637 (B) and UMUC-3 cell (E) were monitored by Edu staining assay; Relative Edu-positive cell number in 5637 (C) and UMUC-3 cell (F) under different treatment were statistically analyzed. Data were shown as mean ±SD; *, p<0.05.
However, it's worth noting that the suppression efficiency here (20-30%) was lower than that (37.5-59.7%) reported by a P53 over-expression Adeno-associated virus vector 16. This may partly ascribe to the relative lower transfection rate compared with virus transduction. Furthermore, it has been most recently demonstrated that the tandem fusion of multiple ADs (VP64-p65-Rta, VPR) increases CRISPR-dCa9-based transcriptional activation efficiently 17. Therefore, there is still more room for optimizing the transcription activation modules in our system.
Conclusion
In conclusion, we have developed an optimized CRISPR-Cas9 based light-inducible gene expression system in bladder cancer cell. The device controls gene expression in a dose-dependent manner and can inhibit proliferation of cancer cell by modulating expression of p53 gene. This work may provide a new strategy for quantitatively controlling specific targets in human cancer.
Materials and methods
Plasmids construction
Genomic anchor (CIBN-dCas9, dCas9-CIBN, CIBN-dCas9-CIBN and dCas9-trCIB1), activator components (CRY2-VP64FL and CRY2PHR-P65), Tet promoter targeting sgRNA and mCherry reporter vector (Tet-Cherry) were purchased from Addgene (http://www.addgene.org/). The target sequence of sgRNA (CTCCCTATCAGTGATAGAGA) was located in the 13×tetO (Tet operator) region in Tet-mCherry vector 11. The corresponding plasmid IDs were listed in Table 1. P53 effector vector (Tet-P53) was constructed by replacing mcherry with synthetic wide type p53 gene using EcoR I and Bgl II sites.
Table 1.
Plasmids from Addgene.
| Plasmid ID | Plasmid name |
|---|---|
| 60551 | pcDNA3.1-CIBN-dCas9 |
| 60552 | pcDNA3.1-dCas9-CIBN |
| 60553 | pcDNA3.1-CIBN-dCas9-CIBN |
| 60554 | pcDNA3.2-Cry2FL-VP64 |
| 64119 | NLS-dCas9-trCIB1 |
| 64124 | NLS*3-CRY2PHR-P65 |
| 64128 | Tet-mCherry |
Cell culture
Human bladder cancer cells (5637 and UMUC-3) were purchased from the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, China. The 5637 cell was maintained in RPMI-1640 media supplemented with 10% FBS and 1% antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin sulfates). The UMUC-3 cell was maintained in DMEM media supplemented with the same concentration of FBS and antibiotics with that in DMEM media. Both cells were grown at 37 °C in an atmosphere of 5 % CO2.
Cell transfection and Illumination
Cells were transfected with Lipofactamine 3000 (Invitrogen) following the manufacture's protocols. Typically, genomic anchor, activator, sgRNA, and reporter/effector were transfected at a ratio of 1:1:1:1, meaning 0.25 μg for each plasmid in a 12-well. After transfection in dark for 12h, cells were illuminated by a blue light LED lamp (460 nm peak, 0.84 W/m2 of average irradiance) or remained in dark for 0-72h. The LED lamps were controlled by a timer to adjust the illumination dose.
Quantitative Real-Time PCR Analysis
Total RNAs from cells under different illumination conditions were isolated using the Trizol reagent (Invitrogen, USA) following the manufacture's instructions. cDNA was then synthesized using SuperScript III® (Invitrogene, USA) kit. Quantitative real-time PCR (qRT-PCR) was performed in triplicate on the ABI PRISM 7000 Fluorescent Quantitative PCR system (Applied Biosystems, USA) with SYBR Green Premix (Takara, Japan). All samples were normalized to gapdh (the internal control) and the relative mRNA levels were calculated using the △△Ct method. The primers using here were shown as below in 5' to 3' direction:
gapdhF: TCATCCCTGCCTCTACTG;
gapdhR: TGCTTCACCACCTTCTTG;
mcherryF: CTGAAGGGCGAGATCAAGCA;
mcherryR: TAGTCCTCGTTGTGGGAGGT;
p53F: GAGGTTGGCTCTGACTGTACC;
p53R: TCCGTCCCAGTAGATTACCAC;
p21F: CGATGGAACTTCGACTTTGTCA;
p21R: GCACAAGGGTACAAGACAGTG.
Flow cytometry
Transfected cells were harvested after incubation under light/dark for 24h. Reporter gene expression in bladder cell was determined by flow cytometry on a FACSCanto SORP device (BD Biosciences, USA). The gates were set up in the way that >99.5% of cells without illumination were counted as mCherry-negative cells.
Western blotting
After incubation under light/dark for 48h, transfected cells were harvested and lysed in RIPA buffer (Beyotime, China). Equal amounts of protein were subjected to SDS-PAGE and transferred to PVDF membranes (Bio-Rad, USA). After blocked with 5% skim milk, the membranes were incubated with appropriate primary antibodies (P53 antibody, Cell Signaling Technology, USA; mCherry antibody, Abcam, UK; Actin antibody, Transgene, China) overnight at 4 ℃. After washing for three times, blots were incubated with horseradish peroxidase-labeled secondary antibodies (Transgene, China) for 1h at RT. Detection was carried out using Pierce ECL Westerning Blotting substrates (Thermofisher, USA). Chemiluminescence signal was captured and analyzed by the ChemiDoc XRS software (Bio-Rad, USA).
CCK-8 assay
The effects of light-induced p53 expression on cell growth were determined by Cell Counting Kit-8 assay (Transgene, China). Briefly, 5×103 cells/well were seeded in a 96-well plate for 12h and then transfected as mentioned above (each vector was adjust to 25 ng/well). At 0, 24, 48, 72 hours after blue light illumination, 100 μl fresh medium containing 10% of CCK-8 was replaced to each well and the cells were cultured for 1 hour. The absorbance at 450 nm was determined using an ELISA microplate reader (Bio-Rad, Hercules, CA, USA). Experiments were repeated for three times.
Edu incorporation assay
The effects of light-induced P53 expression on cell proliferation were determined by Ethynyl-2-deoxyuridine incorporation assay using Cell-Light™ EdU Apollo®567 In Vitro Imaging Kit (Ribobio, China). Briefly, after illumination for 24h, the Edu was added to each well with a finial concentration of 50 μM. After 2 hours' culture, cells were fixed with 4% paraformaldehyde in PBS. After washed in PBST (PBS containing 0.1% Triton X-100) for 3 times, cells were incubated with 1 X Apollo solution for 30 min at room temperature in the dark. Finally, cells were subjected to nuclear staining with 1X DAPI for 30 min and then observed using fluorescence microscopy.
Statistical analysis
All data were presented as mean ± standard deviation (SD). Statistical analysis was conducted by Student's t-test or ANOVA using SPSS version 19.0 software (SPSS Inc. Chicago, IL, USA). p < 0.05 was considered statistically significant.
Supplementary Material
Supplementary figure S1.
Acknowledgments
This work was supported by the National Key Basic Research Program of China (973 Program) (2014CB745201), China Postdoctoral Foundation (2015M582463), Shenzhen Municipal Government of China (ZDSYS201504301722174, JCYJ20150330102720130, ZDSY20130531165409949, GJHZ20150316154912494), Special Support Funds of Shenzhen for Introduced High-Level Medical Team, and Shenzhen High-Level Medical Discipline Development Program (2016031638).
References
- 1.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 2.Tigges M, Fussenegger M. Recent advances in mammalian synthetic biology-design of synthetic transgene control networks. Current opinion in biotechnology. 2009;20:449–60. doi: 10.1016/j.copbio.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–8. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 4.Moglich A, Hegemann P. Biotechnology: Programming genomes with light. Nature. 2013;500:406–8. doi: 10.1038/500406a. [DOI] [PubMed] [Google Scholar]
- 5.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M. et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–2. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 6.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nature methods. 2011;8:35–8. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller K, Naumann S, Weber W, Zurbriggen MD. Optogenetics for gene expression in mammalian cells. Biological chemistry. 2015;396:145–52. doi: 10.1515/hsz-2014-0199. [DOI] [PubMed] [Google Scholar]
- 8.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature chemical biology. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature methods. 2012;9:266–9. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 10.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–6. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chemistry & biology. 2015;22:169–74. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. Journal of the American Chemical Society. 2012;134:16480–3. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nature chemical biology. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nature reviews Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 15.Fodor I, Timiryasova T, Denes B, Yoshida J, Ruckle H, Lilly M. Vaccinia virus mediated p53 gene therapy for bladder cancer in an orthotopic murine model. The Journal of urology. 2005;173:604–9. doi: 10.1097/01.ju.0000143196.37008.2c. [DOI] [PubMed] [Google Scholar]
- 16.Ruifa H, Liwei L, Binxin L, Ximing L. Additional gene therapy with rAAV-wt-p53 enhanced the efficacy of cisplatin in human bladder cancer cells. Urologia internationalis. 2006;77:355–61. doi: 10.1159/000096341. [DOI] [PubMed] [Google Scholar]
- 17.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI. et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015;12:326–8. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1.