Summary
In Arabidopsis thaliana and Oryza sativa, the cytochrome P450 (CYP) 714 protein family represents a unique group of CYP monooxygenase, which functions as a shoot‐specific regulator in plant development through gibberellin deactivation. Here, we report the functional characterizations of PtCYP714A3, an OsCYP714D1/Eui homologue from Populus trichocarpa. PtCYP714A3 was ubiquitously expressed with the highest transcript level in cambium–phloem tissues, and was greatly induced by salt and osmotic stress in poplar. Subcellular localization analyses indicated that PtCYP714A3‐YFP fusion protein was targeted to endoplasmic reticulum (ER). Expression of PtCYP714A3 in the rice eui mutant could rescue its excessive‐shoot‐growth phenotype. Ectopic expression of PtCYP714A3 in rice led to semi‐dwarfed phenotype with promoted tillering and reduced seed size. Transgenic lines which showed significant expression of PtCYP714A3 also accumulated lower GA level than did the wild‐type (WT) plants. The expression of some GA biosynthesis genes was significantly suppressed in these transgenic plants. Furthermore, transgenic rice plants exhibited enhanced tolerance to salt and maintained more Na+ in both shoot and root tissues under salinity stress. All these results not only suggest a crucial role of PtCYP714A3 in shoot responses to salt toxicity in rice, but also provide a molecular basis for genetic engineering of salt‐tolerant crops.
Keywords: cytochrome P450 monooxygenase, development, PtCYP714A3, transgenic plant, salinity
Introduction
As a class of important plant hormones, gibberellins (GAs) play crucial roles in promoting seed germination, stem elongation, leaf expansion and flower development (Eriksson et al., 2006; King et al., 2001; Ogawa et al., 2003; Schwechheimer, 2008). In higher plants, bioactive GA levels can be regulated via controlling the biosynthesis, deactivation and signal transduction of GAs. The biosynthesis of bioactive GAs such as GA1 and GA4 was initiated from geranylgeranyl diphosphate (GGDP), and catalysed by three types of enzymes including plastid‐localized terpene cyclases, membrane‐bound cytochrome P450 monooxygenases (P450s) and soluble 2‐oxoglutarate‐dependent dioxygenases (2ODDs) (Yamaguchi, 2008). The deactivation of GAs also involves several different mechanisms. The major enzymes responsible for the deactivation of bioactive GAs (GA1 and GA4) and their immediate precursors (GA20 and GA9) are GA2‐oxidases (GA2ox) that add a hydroxyl group to the C‐2 position of the substrates (Olszewski et al., 2002; Rieu et al., 2008; Thomas et al., 1999).
In Arabidopsis, AtGA2ox7 and AtGA2ox8 can inactivate the earlier GA biosynthetic intermediates, but cannot deactivate the bioactive GAs lacking C‐20 via 2‐hydroxylation (Lee and Zeevaart, 2005; Schomburg et al., 2003). GA methyltransferase (GAMT1 and GAMT2)‐mediated methylation, which is regulated by developmental stimuli, is another route to inactivate bioactive GAs (Varbanova et al., 2007). In rice, a cytochrome P450 monooxygenase CYP714D1 encoded by Eui gene that was cloned from the recessive tall rice mutant elongated uppermost internode (eui) can deactivate non‐13‐hydroxylated GAs (GA4, GA9 and GA12) by the 16α, 17‐epoxidation (Luo et al., 2006; Zhu et al., 2006). Although the 16α, 17‐[OH]2‐GAs were found in many other plant species, such as Cibotium glaucum (Yamane et al., 1988), Lupinus albus (Gaskin et al., 1992), Malus domestica (Hedden et al., 1993), Pisum sativum (Santes et al., 1995), Prunus avium (Blake et al., 2000) and Populus trichocarpa (Pearce et al., 2002),no any gene as yet was reported having similar functions to Eui gene in these species. Recently, two other CYP714 gene family members, CYP714B1 and CYP714B2, were found to encode GA13‐oxidases that negatively regulate shoot growth and participate in GA homoeostasis in rice (Magome et al., 2013). In Arabidopsis, two Eui‐like genes (ELA1/CYP714A1 and ELA2/CYP714A2) encode Eui homologues that subtly regulate plant growth most likely through catalysing the deactivation of bioactive GAs similar to rice Eui (Zhang et al., 2011). Using a yeast expression system, CYP714A1 was revealed to be a GA‐deactivating enzyme that catalyses the conversion of GA12 to 16‐carboxylated GA12 (16‐carboxy‐16β, 17‐dihydro GA12), while CYP714A2 acts as a 13‐oxidase or 12α‐oxidase of GAs or GA precursors depending on the substrate (Nomura et al., 2013). All these reports suggest that CYP714 gene family members from different plant species might have different functions in GA metabolic pathway.
The endogenous GA levels are also affected by environmental stimuli (Huang et al., 2009a; Nelson et al., 2009). Under salt stress condition, growth inhibition is the primary response of plants and may be the result of positive adaptation mechanisms (Munns, 2002). In Arabidopsis, DELLA proteins were found to integrate responses to independent hormonal and environmental signals of adverse conditions, and confer growth restraint upon salinity treatment (Achard et al., 2006). Recently, DWARF AND DELAYED FLOWERING 1 (DDF1), a salinity‐responsive AP2 transcription factor belonging to the DREB1/CBF subfamily, was reported to bind DRE‐like motifs in GA2ox7 promoter and activate the expression of GA2ox7, leading to growth repression for stress adaptation (Magome et al., 2004, 2008). DDF1‐overexpressing Arabidopsis showed increased tolerance to cold, drought and heat stresses (Kang et al., 2011). In addition, gibberellins were suggested to be involved in Arabidopsis mitochondrial phosphate transporter (AtMPT)‐mediated early response to salt stress (Zhu et al., 2012). However, the potential application of GA‐related genes to improve salt tolerance in other plants including important crops and trees is barely reported (Shan et al., 2014).
Although the functions of some CYP714 gene family members in Arabidopsis and rice have been studied, very limited information is known regarding these enzymes from other plant species. With the finish of genome sequencing and availability of developing genomic tools, Populus has been taken as an ideal model plant for trees (Jansson and Douglas, 2007; Tuskan et al., 2006). Previously, we investigated the function of rice CYP714D1 gene in Populus (Wang et al., 2013). In this work, the role of PtCYP714A3 gene from Populus in plant development and salt stress adaptation was investigated. We found that PtCYP714A3 was a functional homologue of OsCYP714D1/Eui by complementation tests in the rice eui mutant and showed similar but not exactly the same functions in shoot development as did OsCYP714D1/Eui. Further study revealed that PtCYP714A3 regulated Na+/K+ homoeostasis in transgenic rice plants to support their survival under salinity stress. Our results suggest that PtCYP714A3 exerts distinct functions from OsCYP714D1/Eui in plant growth, and plays important roles in plant salt resistance.
Results
PtCYP714A3 encodes a putative cytochrome P450 monooxygenase in Populus
A BLAST search of the cytochrome P450 homepage (http://drnelson.uthsc.edu/ CytochromeP450.html) resulted in the identification of six homologues in the Populus genome, designated as PtCYP714A3 (Potri.019G064600.1), PtCYP714E2 (Potri014G052000.1), PtCYP714E4 (Potri.013G160800.1), PtCYP714E5 (Potri.008G026300.1), PtCYP714E6 (Potri.008G026200.1) and PtCYP714F1 (Potri.010G116300.1). These proteins share 63.33% identity and 69.99% similarity, with PtCYP714E6 as a redundant duplicate of PtCYP714E5 (99% identity). Among these proteins, PtCYP714A3 shares the highest sequence identity with AtCYP714A1 (58.99%), AtCYP714A2 (53.20%) from Arabidopsis and OsCYP714D1 (41.35%) from rice, respectively (Figure 1a,b). Similar to AtCYP714A1, AtCYP714A2 and OsCYP714D1, PtCYP714A3 is characterized by an oxygen binding and activation site, a ERR triad motif and a Haeme binding site, as a common feature of CYP714 subfamily. The fact that Arabidopsis genome encodes only two CYP714 members suggests that Populus could have duplicated this class of gene during its long evolution for better adaptation to the unstable environments over a long lifespan.
Figure 1.

Amino acid sequence alignment and phylogenetic tree of different CYP714 protein family members. (a) Multiple alignment of the deduced amino acid sequences of CYP714 proteins from Arabidopsis (AtCYP714A1/A2) and rice (OsCYP714D1). (b) Phylogenetic tree of typical CYP714 proteins in Populus, Arabidopsis and Oryza sativa with bootstrap values conducted with Clustal X and Mega 3 program. The GenBank accession numbers for Populus CYP714 genes are as follows: PtCYP714A3 (XM_00631280.1), PtCYP714E2 (XM_006375124.1), PtCYP714E4 (XM_002319405.2), PtCYP714E5 (XM_002311935.2) and PtCYP714F1 (XM_002314788.1).
Expression pattern of PtCYP714A3 in Populus
To elucidate the possible roles of PtCYP714A3, we first performed quantitative real‐time PCR and investigated its expression pattern in Populus trichocarpa (Torr. & Gray) genotype Nisqually‐1. We observed that PtCYP714A3 was predominantly expressed in the cambium–phloem tissues than in the other tissues such as roots, shoot apexes, leaves, petioles and xylem tissues (Figure 2a). We also generated PtCYP714A3 promoter–GUS reporter vector and introduced it into Shanxin yang. Consistent with the qRT‐PCR results, GUS was ubiquitously expressed in various tissues (Figure 2b), with a strong GUS expression in the cambium zone of stem (Figure 2c).
Figure 2.

Expression pattern and subcellular localization of PtCYP714A3 in populus. (a) Relative expressional levels of PtCYP714A3 gene in different tissues of Populus trichocarpa (Torr. & Gray) genotype Nisqually‐1. R, Root; Ap, apical bud; L, leaf; Pe, petiole; Ca‐Ph, cambium–phloem zone; X, xylem. the expression value of PtCYP714A3 in root was set to 1. Error bars are means ± SD of three biological replicates. The experiment was repeated two times independently. (b, c) Histochemical GUS analysis of a PtCYP714A3 promoter–GUS transgenic plant and a transverse section of the stem. (d) Subcellular localization of PtCYP714A3. Confocal laser scanning microscopy images of poplar mesophyll protoplasts transiently expressing yellow fluorescent protein (YFP), or PtCYP714A3‐YFP alone and cyan fluorescent protein (CFP)‐HDEL together under the control of the 35S promoter were shown.
PtCYP714A3 is targeted to endoplasmic reticulum (ER)
Cellular and subcellular localization of a certain protein may indicate how and/or where it works. Previous studies have shown that OsCYP714D1/Eui, AtCYP714A1 and AtCYP714A2 are all targeted to ER (Zhang et al., 2011; Zhu et al., 2006). To determine the subcellular localization of PtCYP714A3, we transiently expressed PtCYP714A3‐YFP (yellow fluorescent protein) fusion protein in poplar leaf protoplasts. HDEL, an ER‐localized reporter (Dong et al., 2008), was also co‐transformed with PtCYP714A3‐YFP to verify the ER targeting of PtCYP714A3 protein. As we have expected, the fluorescence of PtCYP714A3‐YFP overlapped perfectly with that of CFP‐HDEL, revealing that PtCYP714A3‐YFP fusion protein was indeed targeted to the ER organella (Figure 2d).
PtCYP714A3 can functionally complement the rice eui mutant
The rice eui mutant displays excessive‐shoot‐growth phenotype (Zhu et al., 2006). To understand whether PtCYP714A3 might have similar functions as the rice ortholog, we heterologously expressed PtCYP714A3 driven by the Eui promoter in the eui background (Figure 3a). In parallel, OsCYP714D1/Eui driven by the Eui promoter was separately introduced into eui mutant as well (Figure 3a). At least ten independent transgenic lines were obtained for each gene. RT‐PCR analyses confirmed the expression of OsCYP714D1/Eui or PtCYP714A3 in the eui mutant, respectively (Figure 3b,c). Compared to the wild‐type and eui mutant plants, all transgenic lines (L1 and L4) expressing OsCYP714D1/Eui showed severely stunted growth, and failed to flower and produce seeds (Figure 3b), whereas those expressing PtCYP714A3 only showed semi‐dwarfed growth and set seeds successfully (Figure 3c). Detailed studies with the T4 seeds of two PtCYP714A3 transgenic lines (L7 and L8) showed that the semi‐dwarfed phenotype in PtCYP714A3 transgenic plants was attributed to the shortened upper most (1st), the second and the third internodes (Figure 3d). These results suggest that PtCYP714A3 biologically complemented the excessive‐shoot‐growth phenotype of eui mutant, but did not work exactly the same as did OsCYP714D1.
Figure 3.

Genetic complementation of rice eui mutant by PtCYP714A3. (a) Schematic representation of T‐DNA region transformed into eui mutant. (b) Growth phenotypes of WT, eui and two OsCYP714D1 complementary lines. eui, elongated uppermost internode rice mutant; L1 and L4, independent eui transgenic lines expressing OsCYP714D1/EuicDNA under the control of Eui promoter; WT, wild type. Bar = 10 cm. (c) Growth phenotypes of WT, eui and two PtCYP714A3 complementary lines. L7 and L8, independent eui transgenic lines expressing PtCYP714A3 cDNA under the control of Eui promoter. Bar = 10 cm. (d) Plant heights of eui mutant, wild‐type and the PtCYP714A3 transgenic plants. Values are means ± SD from 30 individual plants with three independent biological replicates. * indicates significant difference in comparison with the WT at P < 0.05 (Student's t‐test).
Expression of PtCYP714A3 inhibits shoot growth and promotes tillering in transgenic rice plants
To investigate the exact function of PtCYP714A3 in plant, we introduced the construct containing the coding sequence of OsCYP714D1/Eui or PtCYP714A3, driven by the Eui promoter, into the genome of rice (ZH11) by Agrobacterium‐mediated transformation (Figure 3a). More than 10 independent transgenic lines were successfully obtained for each gene construct. The integration of PtCYP714A3 into the rice genome was confirmed by PCR analyses (Figure S1a). Further analysis by RT‐PCR indicated the successful expression of PtCYP714A3 in the selected transgenic rice plants (Figure S1b). Overexpression of OsCYP714D1/Eui led to severely dwarfed phenotype in all transgenic lines, just the same as its overexpression in the eui mutant (Figure S1c). However, all transgenic plants ectopically expressing PtCYP714A3 showed consistent semi‐dwarfed phenotype regardless of the expression levels of transgene. Therefore, we selected two independent homozygous transgenic lines which showed high expression of PtCYP714A3 (Z33, Z38) for subsequent phenotypic analyses (Figure S1c). Compared to WT plants, PtCYP714A3 transgenic plants produced shorter shoots, including internodes and panicles (Figure 4a–c,e). In addition, increased tiller number of the transgenic lines was also a prominent difference from the WT (Figure 4d). To determine whether expression of PtCYP714A3 would affect seed development in transgenic plants, wild‐type and homozygous T4 transgenic plants were chosen for field trial in 2013 (transgenic trial permit number: 2013‐T018). Compared to the wild‐type plants, both transgenic lines produced smaller panicles and seeds with decreased seed setting, leading to reduced grain yield per plant (Figure 4e–j; Table S2).
Figure 4.

Expression of PtCYP714A3 inhibits shoot growth and promotes tillering in transgenic rice plants. (a) Field‐grown plants. (b, c) Internode lengths. (d) Tiller numbers. (e) Mature panicles. (f) Seed‐setting rates. (g) Seed numbers per panicle. (h) Grain yields per plant. (I) One‐hundred‐grain weights. (j) Seeds with and without seed coat. WT, wild type; Z33 and Z38, independent transgenic lines. Values are means ± SD from 30 individual plants with three independent biological replicates. * indicates significant difference in comparison with the WT at P < 0.05 (Student's t‐test).
Expression of PtCYP714A3 reduces the contents of bioactive GAs in transgenic rice plants
To understand whether PtCYP714A3 indeed functions in GA deactivation, endogenous levels of GA precursors and bioactive GAs in the internodes of WT and PtCYP714A3 transgenic plants were examined. Compared to the WT, the levels of both bioactive GAs (GA1 and GA4) and GA12, the common precursor in GAs biosynthesis pathway, were extremely lower or even undetectable in transgenic plants (Figure 5). On the contrary, the level of the other important precursor, such as GA53, was significantly higher in both transgenic lines, especially in line Z33 (Figure 5). We also determined several other immediate precursors of bioactive GAs. Both GA19 and GA20 were measurable in transgenic rice plants with the concentrations waving from 0.79 to 6.99 (ng/g), but they were undetectable in WT plants (Figure 5). GA9 was too low to be detected in both WT and transgenic plants (data not shown).
Figure 5.

Endogenous GA levels in the stems of 2.5‐month‐old wild‐type and PtCYP714A3 transgenic plants. WT, wild type; Z33 and Z38, independent transgenic lines. Values are means ± SD of three biological replicates of ten individual plants from the WT or the transgenic lines. ND, not detected.
To explain why GA accumulation was affected in transgenic plants, we examined the expression levels of genes in GA biosynthesis, deactivation and signal pathways by quantitative real‐time PCR. Among these genes, the transcription levels of those involved in the synthesis of the common precursor GA12, such as CPS, KS1, KO2 and KAO, all decreased significantly (Figure 6). GA20ox and GA3ox are the main enzymes that catalyse the syntheses of bioactive GAs (GA1 and GA4) (Itoh et al., 2001; Sasaki et al., 2002). We found that the expression level of GA20ox2 decreased whereas that of GA3ox2 increased, implying that they showed opposite variation in these transgenic plants. In addition, the transcription levels of the bioactive GA deactivation gene GA2ox3 (Sakai et al., 2003), the GA receptor gene GID1 (Ueguchi‐Tanaka et al., 2005) and the F‐box protein gene GID2 (Sasaki et al., 2003) also decreased in transgenic rice compared with that in the WT plants (Figure 6). These results indicate that expression of PtCYP714A3 influences GA accumulation and GA metabolic gene expression.
Figure 6.

Relative expressional levels of GA‐related genes in wild‐type and PtCYP714A3 transgenic plants. WT, wild type; Z33 and Z38, independent transgenic lines. The name and the accession numbers of the genes are as the following: CPS1, ent‐copalyl diphosphate synthase (AP004572); KS1, ent‐kaurene synthase (OSJN00255); KO2, eht‐kaurene oxidase (AP005471); KAO, ent‐kaurenoic acid oxidase (AP000616); GA20ox2, GA 20‐oxidase (AB077025); GA3ox2, GA 3‐oxidase (AB056519); GA2ox3, GA 2‐oxidase (NM_001050827); GID1, soluble GA receptor (AB211399); GID2, F‐box protein (AB100246). Values are means ± SD of three biological replicates from the WT or the transgenic lines. Significant differences were analysed with Student's t‐test. *, P < 0.05; **, 0.05 < P < 0.01; ***, 0.01 < P < 0.001.
Expression of PtCYP714A3 confers salt tolerance on transgenic rice plants
Previous studies have shown that GAs also function in plant response to abiotic stress (Magome et al., 2004, 2008; Shan et al., 2014). To investigate the functions of PtCYP714A3 and its potential value in improving salt tolerance in plants, we first extracted total RNA from the leaves of Shanxin yang treated with 150 mm NaCl for increasing periods of time and performed qRT‐PCR. We found that expression of PtCYP714A3 was steadily induced by salt from 0 to 24 h, and reached a maximal 87‐fold induction at 18 h (Figure 7a). We then examined the effects of salt on the growth of transgenic plants at whole‐plant scale. Under normal growth condition, both WT and transgenic lines all grew well (Figure 7b). However, after the plants were treated with 150 mm NaCl for 12 days, obvious differences were observed between WT and transgenic plants. While WT plants became wilted, transgenic lines appeared to be less impaired by salt stress (Figure 7b). Although salt stress considerably affected the growth of all the plants in general, transgenic plants were remarkably more vigorous. Compared to WT, most PtCYP714A3‐expressing plants exhibited near‐to‐normal leaf colour and showed a superior survival rate at the end of treatment: more than 70% of PtCYP714A3 transgenic, whereas less than 30% of WT plants survived (Figure 7c). All these results indicate that expression of PtCYP714A3 in rice enhanced salt tolerance in transgenic plants.
Figure 7.
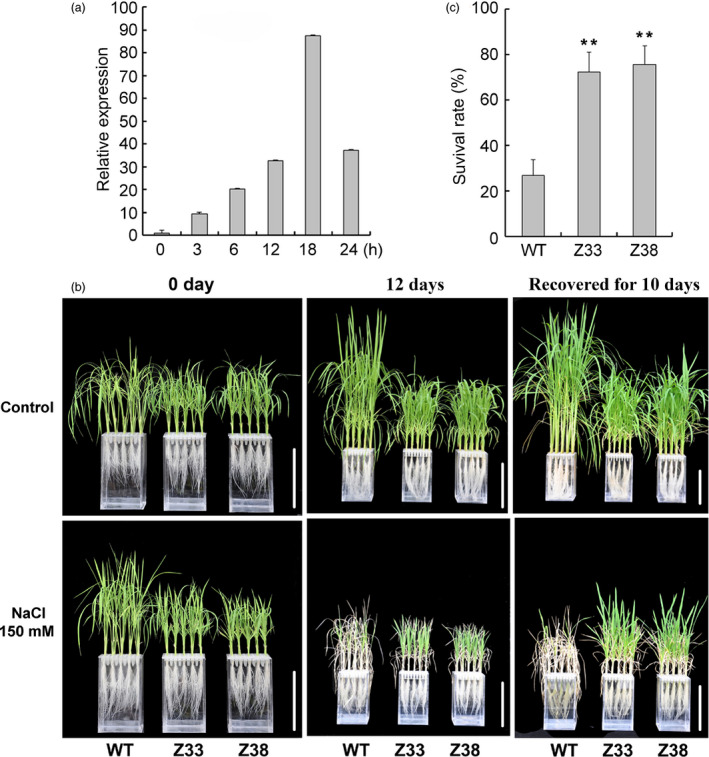
Expression PtCYP714A3 in response to high‐salinity and salt stress tolerance analyses of transgenic rice plants. (a) Expression of PtCYP714A3 is responsive to NaCl stress. Poplar leaves were cut into sections and divided into several groups that were treated separately with 200 mm NaCl for different time points. Samples were collected at 0, 3, 6, 12, 18 and 24 h after the initiation of treatment. Data represent the average of three independent experiments. (b, c) Phenotypes (b) and survival rates (c) of three‐week‐old seedlings of wild‐type and PtCYP714A3 transgenic plants treated with 150 mm NaCl for 12 d followed by 10 days of recovery without NaCl. WT, wild type; Z33 and Z38, independent transgenic lines. Values are means ± SD (n = 4) from three independent experiments. Asterisks indicate statistically significant difference in comparison with the WT (Student's t‐test, **, P < 0.01).
PtCYP714A3 expression regulates tissue Na+ and K+ distribution
To understand how the introduced PtCYP714A3 gene improved salt‐tolerant capacity in transgenic plants, we examined the distribution of Na+ and K+ contents in different tissues of WT and transgenic rice plants after treated with or without 150 mm NaCl for 12 days. Under normal growth condition, WT showed less Na+ and more K+ content in the roots, although both WT and transgenic Z33, Z38 lines contained approximately equal Na+ and K+ content in the shoots (Figure 8a,c). Under salt stress condition, Na+ content increased in all plant tissues accompanied by a decrease in K+ content. However, Na+ content in Z33 and Z38 was significantly higher than that in WT in both shoots and roots (Figure 8b). Generally, WT and transgenic plants accumulated equivalent levels of K+ in both shoots and roots (Figure 8d). Taken together, our data suggest that PtCYP714A3 regulates Na+ and K+ homoeostasis under salt stress condition in transgenic rice plants.
Figure 8.

Na+ and K+ contents in roots and shoots of wild‐type (WT) and transgenic rice plants (Z33 and Z38). At the end of 0 or 150 mm NaCl treatment, plant materials were harvested and pooled into roots and shoots for the measurement of Na+ and K+ contents. (a) Na+ contents of different tissues at 0 mm NaCl. (b) Na+ contents of different tissues at 150 mm NaCl. (c) K+ contents of different tissues at 0 mm NaCl. (d) K+ contents of different tissues at 150 mm NaCl. Values are means ± SD (n = 4) from two independent experiments. Asterisks indicate statistically significant difference in comparison with the WT (Student's t‐test, *, P < 0.05).
Expression of salt stress‐related genes is affected in transgenic plants
To further investigate the role of PtCYP714A3 in salt stress adaptation of plants, the relative transcript levels of a series of salt stress‐related marker genes before or after NaCl stress treatment were analysed (Figure 9). NHX1 and SOS1 encode Na+/H+ the antiporters that compartment Na+ from cytoplasm into the vacuole (NHX1) or out of the plant cell (SOS1). The expression levels of NHX1 and SOS1 in transgenic rice (except SOS1 in Z33) were slightly higher than in WT under nonstress condition, and increased significantly after salt stress in both WT and transgenic lines. Compared to NHX1, the expression increase of SOS1 in transgenic plants was more significant than that in WT, indicating that in transgenic rice, more sodium ions were pumped out of the plant cells than were compartmented into the vacuole under salt stress. The enzyme of P5CS (Δ1‐pyrroline‐5‐carboxylate synthase) controls the rate‐limiting step of glutamate‐derived proline biosynthesis and is induced by high salt, dehydration, cold and ABA treatment (Igarashi et al., 1997). The transcription of P5CS was also induced by NaCl, and an almost fourfold increase was observed in Z33 upon NaCl treatment, much higher than that in WT (about 2 folds). The SPY gene encodes an O‐linked N‐acetylglucosamine transferase, a negative regulator of plant GA signalling, and is inducible by drought stress and slightly responsive to salt stress in Arabidopsis (Qin et al., 2011; Steiner et al., 2012). The expression of a homologous SPY in rice named SPY‐like was analysed in this study. The expression level of SPY‐like was slightly lower in Z33, but higher in Z38 than in WT before NaCl treatment, although no obvious difference was seen among these three lines after NaCl stress. DREBs (the dehydration‐responsive element‐binding proteins) are a kind of transcript factors in response to drought, high‐salt and cold stresses (Dubouzet et al., 2003; Wang et al., 2008; Mao and Chen 2012). So the expressions of DREB1 genes in rice were analysed. All of these genes were induced to varying degrees by salt stress in the wild‐type rice. All these genes were expressed at higher levels in Z33 and Z38 than in WT under normal situations (except DREB1F in Z33). Under NaCl stress, the expression levels of DREB1A, DREB1C and DREB1F were decreased in Z38. All these results indicate that expression of PtCYP714A3 altered the expression of salt stress‐related genes in transgenic plants.
Figure 9.

Quantitative real‐time PCR analyses of salt tolerance‐related marker genes. Three‐week‐old seedlings treated with 150 mm NaCl for 12 h or 0 h (Control) were harvested for total RNA extraction, transverse transcription and real‐time PCR analyses. WT, wild type; Z33 and Z38, independent transgenic lines. Values are means ± SD of three biological replicates from the WT or the transgenic lines. Asterisks indicate statistically significant difference in comparison with the WT (Student's t‐test, *, P < 0.05). The TIGR loci of these genes are as follows: NHX1 (Os07g0600900); SOS1 (Os12g0641100); P5CS (Os05g0455500); SPY ‐like (Os08g0559300); DREB1A (Os09g35030); DREB1F (Os01g73770); DREB2A (Os01g07120).
Discussion
Previous reports that CYP714D1/Eui functions as a GA 16α, 17‐epoxidase to inactivate GA12, GA9 and GA4 in rice suggest that 16α, 17‐epoxidation might be an important process of GA deactivation existed in a variety of plant species. Indeed, 16α, 17‐[OH]2‐GAs have been detected in many plant species including Populus trichocarpa (Blake et al., 2000; Gaskin et al., 1992; Hedden et al., 1993; Pearce et al., 2002; Santes et al., 1995; Yamane et al., 1988; Zhu et al., 2006). In our previous study, heterologous expression of CYP714D1/Eui gene led to improved growth rate and biomass of transgenic Populus, just opposite to the phenotypes of CYP714D1/Eui‐overexpressing rice (Wang et al., 2013). To understand these unexpected results, it is necessary to verify whether Populus CYP714 gene family members have similar functions to CYP714D1/Eui. As PtCYP714A3 has the maximum sequence similarity with CYP714D1/Eui, we cloned this gene and verified its possible biological function in transgenic rice plants. We show here that PtCYP714A3, a member of the P450 (CYP) 714 gene family, is involved in GA deactivation and salt resistance in rice.
PtCYP714A3 shares very high amino acid sequence identity with CYP714D1/Eui (OsCYP714D1), AtCYP714A1 and AtCYP714A2 that contain all the highly conserved domains (Figure 1a,b; Zhang et al., 2011; Zhu et al., 2006), indicating its possible role as a putative cytochrome P450 monooxygenase in Populus. As the genus Populus has become an ideal model plant for forest trees (Jansson and Douglas, 2007; Tuskan et al., 2006), a comparative study on the genetic functions of CYP714 gene family between rice and Populus could shoot inspective light into the difference in the mechanisms of plant growth and development under different environmental conditions between herbaceous and woody plants. PtCYP714A3 was predominantly expressed in cambium–phloem cells, and its expression was strongly induced by salt stress in poplar (Figures 2a–c and 7a). Therefore, the high expression of PtCYP714A3 in cambium–phloem tissues of wild‐type poplar may imply a crucial role of PtCYP714A3 in GA metabolism and abiotic stress response in trees.
With a view to the high sequence homology of CYP714D1/Eui and PtCYP714A3, the biological role of PtCYP714A3 was investigated by eui mutant analyses (Figure 3a–d). The most dramatic phenotypic change in eui mutant was the elongated shoot growth, especially the uppermost internode. PtCYP714A3 restored the growth phenotype of eui to the wild type, indicating that PtCYP714A3 can be employed as a functional allele of CYP714D1/Eui in plants. However, a severely dwarfed phenotype was observed in the transgenic eui plants complemented with CYP714D1/Eui (Figure 3b–d). One explanation of this observation is that CYP714D1/Eui and PtCYP714A3 may have overlapped but not exactly the same function in rice and poplar. Another possibility is that CYP714D1/Eui is more powerful than PtCYP713A3 in controlling plant height. This postulation is also supported by the observation in transgenic poplar plants overexpressing PtCYP714A3, which showed no significant changes in the growth rate and biomass production (Figure S2a–e). More detailed molecular and biochemical studies will help to dissect the exact functions of both proteins in plants.
To clarify the exact biological functions of PtCYP714A3, we further ectopically expressed PtCYP714A3 in ZH11 (Figures 4a–i and S1a–c), and examined the endogenous GA contents and the transcript levels of GA pathway‐related genes. We found that PtCYP714A3 transgenic rice showed the most comparability with transgenic plants expressing AtCYP714A2 (Nomura et al., 2013; Zhang et al., 2011). First, both PtCYP714A3 and AtCYP714A2 transgenic rice plants were semi‐dwarfed, with increased tillers (Figure 4a–d; Zhang et al., 2011). Second, the variation tendencies of most endogenous GA levels in these two kinds of transgenic plants were consistent. Based on the previous report, in AtCYP714A2 transgenic plants, the levels of GAs in the non‐13‐hydroxylation pathway, including GA12, GA15, GA24 and GA4, all decreased, whereas those of 13‐hydroxy GAs, including GA44, GA19, GA20 and GA1, all increased with the exception of GA53 which was unaffected (Nomura et al., 2013). In our study, most of detected GAs (GA12, GA4, GA53, GA19 and GA20) showed similar tendencies in PtCYP714A3 transgenic rice, except for bioactive GA1, which decreased rather than increased (Figure 5). In addition, the expression level variations of GA pathway‐related genes in PtCYP714A3‐expressing plants sustained that PtCYP714A3 most likely possesses the function of AtCYP714A2. The expression of GA receptors (GID1), F‐box protein (GID2) as well as bioactive GA‐deactivating enzyme (GA2ox3) was all reduced along with decreased bioactive GAs (GA1 and GA4) (Figure 6). The transcript level of GA3ox which catalyses the conversion of GA20 and GA9 to bioactive GAs (GA1 and GA4) also increased. However, the transcript level of GA20ox2 which encode the important GA‐oxidase involved in most steps of the bioactive GA biosynthesis pathway decreased, opposite to that of the GA3ox2 gene (Figure 6). As the contents of GA19 and GA20, the intermediates produced in the 13‐hydroxylation (13‐H) pathway of GA biosynthesis, dramatically increased (Figure 5), we presumed that the inconformity between the expressions of GA20ox2 and GA3ox2 was attributed to the regulatory mechanism of intermediate substrates. Among the detected GA pathway genes, the most intriguing ones are those genes (CPS, KS, KO and KAO) participating in the biosynthesis of GA12, the common precursor of both 13‐H and non‐13‐H GA pathways. Expressions of all these genes reduced significantly, although the content of GA12 was extremely low or even undetectable in transgenic rice (Figures 5 and 6). In Arabidopsis, AtCYP714A2 was reported as a bifunctional enzyme that preferentially catalyses C‐12 hydroxylation of the ent‐gibberellane carbon skeleton, leading to the conversion of GA12 to 12α‐hydroxy GA12, and C‐13 hydroxylation of the ent‐kaurane carbon skeleton, by which steviol (ent‐13‐hydroxy asurenoic acid) was detected as the sole product when ent‐kaurenoic acid was added as a substrate (Nomura et al., 2013). As the content of GA53 was not affected along with the decrease of its immediate precursor GA12 in PtCYP714A3 transgenic rice (Figure 5), and GA53 was also produced from steviol in AtCYP714A2 transgenic plants (Nomura et al., 2013), we postulate that PtCYP714A3 might have similar function as AtCYP714A2 and could also catalyse the production of steviol‐like substance, and thus, the transcripts of CPS, KS, KO and KAO were inhibited by feedback regulation. Therefore, we deduced that, just similar to AtCYP714A2, PtCYP714A3 also functions directly in deactivating GA12 and converting ent‐kaurenoic acid into steviol, and thus plays an important role in regulating plant growth and development through fine‐tuning GA homoeostasis. However, due to the tremendous species differences between perennial woody Populus and annual herbaceous Arabidopsis or rice, delicate differences could exist among the CYP714 families members, including their expression patterns (Figure 2; Zhang et al., 2011), their functions on the metabolism of GA1 (Figure 5; Nomura et al., 2013; Zhang et al., 2011), and phenotypes in transgenic Populus and rice overexpressing PtCYP714A3 (Figures 4, S1c and S2d).
Previous studies have suggested that GA2ox genes including GA2ox7 were up‐regulated by high‐salinity stress in Arabidopsis (Magome et al., 2008). Overexpression of OsGA2ox5 or DDF1, an AP2 transcription factor of the DREB/CBF subfamily, activated the expression of GA2ox7, and enhanced the salt tolerance in transgenic plants (Magome et al., 2004, 2008; Shan et al., 2014). We found that expression of PtCYP714A3 was also responsive to high‐salinity and osmotic stress (Figures 7a and S3a), and transgenic rice seedlings expressing PtCYP714A3 showed improved tolerance to salt and osmotic stress, with higher survival rates and less growth inhibition, than did the wild‐type seedlings (Figures 7b,c and S3b,c).
Maintaining low levels of sodium ions in the cell cytosol is critical for plant growth and development. Salt‐tolerant plants usually had lower Na+ contents (Huang et al., 2009b; Tang et al., 2014). Under high‐salt stress condition, Na+ content in both shoots and roots of the PtCYP714A3 transgenic rice was higher than that of in WT (Figure 8b). This could be due to the different functional mechanisms of PtCYP714A3 and other salt‐resistant genes (Tang et al., 2014). Similar results were also observed in transgenic poplar plants overexpressing PtCLB10s which accumulated lower Na+ in leaves but higher Na+ in stems than did the WT plants (Tang et al., 2014). In addition, the semi‐dwarfed transgenic rice plants might have higher concentration ratio of sodium ion than the wild type.
To further understand the salt‐tolerant mechanism in PtCYP714A3‐overexpressing plants, we compared the transcript levels of marker genes related to abiotic stress before or after high‐salinity stress (Figure 9). Three kinds of genes have been selected: antiporters (NHX1 and SOS1) or enzyme genes (P5CS), negative regulator gene (SPY‐like), and DREB transcript factor genes (DDF1‐like and DREB1A‐G). In transgenic rice plants (Z33 and Z38), almost all selected genes were expressed at higher levels than that in the WT plants under normal condition. After treated with NaCl stress, all of them were expressed highly in both WT and transgenic plants, indicating that these genes are high‐salinity‐responsive. Among them, the expression of SOS1 gene, which encodes an Na+/H+ antiporter pumping Na+ out of the plant cell from cytosol, was significantly enhanced in NaCl‐treated transgenic lines compared with the WT plants, suggesting that one of functions of PtCYP714A3 is promoting the Na+ efflux in transgenic rice, although the exact mechanism that how PtCYP714A3 affects the expression of SOS1 gene still remains to be clarified. Another intriguing observation is the expression patterns of dehydration‐responsive element‐binding protein 1 (DREB1s). In Arabidopsis, DREB1A, DREB1B, DREB1C, DREB2A, and DREB2B proteins are probably the major transcription factors that function in cold‐, high‐salt‐ and drought‐inducible gene expression (Dubouzet et al., 2003). In rice, ten putative DREB1 homologues (OsDREB1A to OsDREB1J) have been identified and several of them were induced by cold, drought or salinity stress (Dubouzet et al., 2003; Mao and Chen, 2012; Wang et al., 2008). As DREB genes were responsive to drought, we also examined the dehydration tolerance of PtCYP714A3 transgenic plants (Figure S3b,c). After PEG treatment for 20 days, growth inhibitions were observed in both WT and transgenic rice. Following the recovery without PEG for 2 weeks, transgenic line Z38 grew quite better than did Z33 (Figure S3b). The percentage of biomass with and without PEG treatment (relative biomass) showed that, under osmotic stress condition, the relative biomass ratio in shoots of line Z38 was about 70% of the control, obviously higher than that of the WT plants (49.05%) and transgenic line Z33 (36.88%). These results suggest that although PtCYP714A3 has function on enhancing salt stress tolerance in transgenic rice, it might have no relevant to dehydration resistance of plants.
Semi‐dwarfism has been described as ‘Green Revolution’ morphological phenotype for more resistant to wind and rain damage (Sakamoto, 2006; Sakamoto et al., 2003). Previous report has shown that AtCYP714A2‐expressing rice produced more yielding tillers and resulted in higher grain productivity than did the wild‐type rice, suggesting a favourable approach for molecular designing of crops with higher grain yield (Zhang et al., 2011). To explore the potential of PtCYP714A3 gene, we compared the yield of wild‐type and transgenic plants grown in the field with or without high‐salinity stress (Table S2). Compared with the wild‐type control, the trait of increased tiller numbers for transgenic plants was not affected by high‐salt stress. In addition, although under both normal and salt stress conditions, the 100‐grain weight and plot yield of the transgenic plants were lower than the wild‐type control, the reductions of 100‐grain weight caused by high‐salinity stress in the transgenic plants were 1.04% (Z33) and 1.92% (Z38), much lower than that of in the wild‐type control (6.23%). All these results suggest that PtCYP714A3 could be used as an effective gene for engineering transgenic rice with improved salt resistance. The undesirable affects in grain size and plot yield could be due to the unsuitable OsCYP714D1/Eui promoter used in this study, as OsCYP714D1/Eui was strongly expressed in young panicle and flowering panicle during the heading stage (Zhu et al., 2006). Therefore, more suitable promoters such as the OsGA3ox2 gene promoter could be tried in the future work to avoid the influence of PtCYP714A3 expression on flower and grain development in transgenic plants (Sakamoto et al., 2003; Zhang et al., 2011). Taken together, our data suggest that PtCYP714A3 could function in one or several pathway(s) by affecting GA biosynthesis and metabolism. Although the precise mode of the action of PtCYP714A3 in plant growth and response to abiotic stress is still intangible, the results of our study provide direct evidence that altered expression of PtCYP714A3 can significantly modify GA biosynthesis, growth and salt resistance in transgenic plants.
Experimental procedures
Plant materials and growth conditions
Populus trichocarpa (Torr. & Gray) genotype Nisqually‐1 and a commercial hybrid clone Shanxin yang (P. davidiana Dode × P. bolleana Lauche) were used in this study. Plants were subcultured on MS medium (Murashige and Skoog, 1962) supplemented with 0.1 mg/L naphthalene acetic acid (NAA). Plants were also grown in the greenhouse under a 12‐h light/12‐h dark photoperiod at 20–25 °C.
Wild‐type rice (Oryza sativa) Zhonghua 11 (ZH11) and the eui mutant were grown under field conditions or in a greenhouse with a 16‐h/8‐h light and dark photoperiod at 28–30 °C.
Gene isolation, vector construction and plant transformation
PtCYP714A3 was cloned into the Sma I site in pBlueScript II KS (pKS, Stratagene, La Jolla, CA) for sequence confirmation. For PtCYP714A3 promoter–GUS construction, the 5'‐flanking DNA (2046 bp) of the PtCYP714A3 coding region was cloned into the pCAMBIA1300 + pBI101 vector (Liu et al., 2003).
For rice transformation, a 2.5‐kb fragment containing the promoter region of Eui was cloned into the EcoR V site in pKS for sequence confirmation, then digested with Pst I and Bgl II, and cloned into pCAMBIA1301 to replace the original CaMV 35S promoter. To construct the expression vector of ProEui::PtCYP714A3, PtCYP714A3 was placed downstream of the Eui promoter in p1301‐ProEui. For the construction of the control expression vector ProEui::OsCYP714D1, CYP714D1/Eui cDNA was also cut off with BamH I and Kpn I from the vector 35S‐C1301 (Zhu et al., 2006) and put downstream of the Eui promoter. ProEui::PtCYP714A3 and ProEui::OsCYP714D1 were separately transformed into wild‐type ZH11 and eui mutant to generate independent transgenic and complementary plants, respectively, as described previously (Hiei et al., 1994). T3 or T4 generations of PtCYP714A3‐expressing plants were used for phenotypic analyses. Plant height and other agronomic traits were compared upon maturing with 30 plants for each line.
For poplar transformation, the relative vector was transformed into Shanxin yang as described previously (Wang et al., 2011).
β‐galactosidase (GUS) expression analyses
For histochemical GUS activity assays, the whole plantlet or hand‐cut sections of the stem from three‐week‐old wild‐type and transgenic plants were stained as described previously (Gallagher, 1992).
Subcellular localization analyses
To determine the subcellular localization of PtCYP714A3‐YFP fusion protein, the encoding region without the stop codon of PtCYP714A3 was fused in‐frame to the N‐terminal of yellow fluorescent protein (YFP) via the Xho I/Spe I sites in the pA7‐YFP vector and transfected into the mesophyll protoplasts of Shanxin yang by polyethylene glycol (PEG)‐mediated transfection as described previously (Yoo et al., 2007). pA7‐YFP was used as a positive control. For colocalization, the encoding region without the start codon of HDEL, an endoplasmic reticulum (ER)‐localized marker protein (Dong et al., 2008), was amplified from Arabidopsis cDNA, digested with BamH I/Spe I and cloned into the pA7‐CFP vector. After transfected with plasmid DNA, the protoplasts were incubated at 23 °C for 16 h and then examined using a confocal laser scanning microscope (Zeiss LSM 510, Oberkochen, Germany). The excitation wavelengths for YFP and CFP were 514 and 433 nm, respectively.
PCR and reverse transcriptase (RT)‐PCR analyses
For PCR analyses, genomic DNA was isolated from fresh leaves (about 500 mg for each sample) of WT and transgenic plants cultured in greenhouse as described previously (Kang et al., 2010). Gene‐specific primers (Table S1) and GC buffer (TaKaRa, Dalian, China) were used to amplify a 530‐bp PCR product.
For RT‐PCR analyses, total RNA was isolated with the RNAiso Reagent (TaKaRa, Japan) from leaves (for Populus) or stems (for rice) of WT and transgenic plants cultured in the greenhouse for 1 month (Populus) or 2.5 months (rice). After treated with DNase I (Promega, Madison, USA), 2 μg of total RNA was subjected to reverse transcription reaction using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Burlington, Canada) at 42 °C for 1 h. GC buffer and gene‐specific primers were the same as used for PCR analysis (30 cycles). The elongation factor gene PtEF1β (for Populus) and ubiquitin gene Ubi_1 (for rice) were employed as internal controls with the primers shown in Table S1.
Quantitative real‐time (qRT)‐PCR
For qRT‐PCR analyses, total RNA was extracted from different organs and tissues of Populus or rice as needed, and subjected to reverse transcriptions using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) at 42 °C for 1 h. qRT‐PCR was performed on an AceQ qPCR SYBR Green Master Mix (Vazyme Biotech, Nanjing, China) using a CFX Connect Real‐Time System (Bio‐Rad, Hercules, California) with gene‐specific primers (Table S1). The log2 fold change in value was calculated based on 2−▵Ct method. The relative expression of each target gene was normalized using the housekeeping gene OsUbi‐1(for rice) or PtEF1β (for Populus). The error bars were calculated from three biological replicates, and the experiment was repeated at least two times. For expression pattern analysis, the expression value of PtCYP714A3 in root was set to 1. For the expression level analyses of the selected marker genes, the expression value of wild type was set to 1. For the response analyses of PtCYP714A3 to high salinity, the expression of the gene under nonsalt stress condition was set to 1.
GA content determination
Sampled rice stems from 2.5‐month‐old plants (mainly including nodes and internodes) were homogenized in liquid nitrogen using a mortar and pestle, and then lyophilized. An amount of 0.5 g dry weight (DW) of each sample was purified and analysed as described previously (Chen et al., 2011).
Salt stress treatment
To analyse whether the expression of PtCYP714A3 gene is inducible by salt and other abiotic stresses, leaves of Shanxin yang were cut into pieces (about 1 cm2 each piece) and treated with 150 mm NaCl for 0, 3, 6, 12, 18 and 24 h.
For salt tolerance tests, rice seeds were sown in a 96‐well plate (bottom removed). The plate was floated on water and placed to a growth chamber with a 13‐h light (28 °C)/11‐h dark (26 °C) photoperiod. Five days later, the seedlings were cultured with Yoshida's culture solution (Yoshida et al., 1976) and the solution was refreshed every 3 days. For salt treatment, three‐week‐old seedlings were transferred to Yoshida's culture solution supplemented with 150 mm NaCl. The solutions were changed every 3 days. The survival rates were counted 10 days after recovering. Plants were also grown in Binhai (Jiangsu Province, China) for field trial in 2013 (transgenic trial permit number: 2013‐T018).
Na+ and K+ content assays
After salt treatments, plant materials were harvested and pooled as roots and shoots. The samples were dried for 48 h at 80 °C, milled to fine powder, weighed and digested with concentrated HNO3 at 90 °C for 1–2 h. The concentrations of Na+ and K+ were determined in the digested liquid using an atomic absorption spectrophotometer as described previously (Hitachi Z‐8000, Tokyo, Japan; Wang and Zhao, 1995).
Statistical analysis
For statistical analyses, the Student's t‐test was used to generate every P value. The tests were one‐tailed. All data in this work were obtained from at least three independent experiments with three replicates each.
Supporting information
Figure S1 Molecular identification and phenotypes of transgenic plants.
Figure S2 Molecular confirmation and phenotype analyses of transgenic Shanxin yang overexpression of PtCYP714A3.
Figure S3 Expression of PtCYP714A3 gene in response to PEG treatment and osmotic stress analyses of wild‐type and transgenic plants.
Table S1 Primers used in this study.
Table S2 Mean comparisons for tillers and yields of field‐grown wild‐type and PtCYP714A3 transgenic rice plants with or without high‐salinity stress.
Acknowledgements
We are very grateful to Prof. Zhuhua He for providing us the eui mutant seeds, Prof. Hongxuan Lin for ion measurement. This work was jointly supported by the following grants: the National Science Foundation of China (NSFC) 31370670, 31371228 and 31171169; the National Mega Project of GMO Crops 2014ZX08001003‐007, 2014ZX08004002‐006 and 2014ZX0800942B; The comprehensive Survey of Lithium, Boron and Biological Resources in Salt Lake on the Tibetan Plateau [2015]02‐04‐04‐001; and the Strategic Priority Research Program of the Chinese Academy of Sciences XDA08030108.
References
- Achard, P. , Cheng, H. , De Grauwe, L. , Decat, J. , Schoutteten, H. , Moritz, T. , Van Der Straeten, D. et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science, 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Blake, P.S. , Browning, G. , Benjamin, L.J. and Mander, L.N. (2000) Gibberellins in seedlings and flowering trees of Prunus avium L. Phytochemistry, 53, 519–528. [DOI] [PubMed] [Google Scholar]
- Chen, M.L. , Huang, Y.Q. , Liu, J.Q. , Yuan, B.F. and Feng, Y.Q. (2011) Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis‐time of flight‐mass spectrometry. J. Chromatogr. B, 879, 938–944. [DOI] [PubMed] [Google Scholar]
- Dong, C.H. , Rivarola, M. , Resnick, J.S. , Maggin, B.D. and Chang, C. (2008) Sub‐cellular co‐localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 53, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet, J.G. , Sakuma, Y. , Ito, Y. , Kasuga, M. , Dubouzet, E.G. , Miura, S. , Seli, M. et al. (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought‐, high‐salt‐ and cold‐responsive gene expression. Plant J. 33, 751–763. [DOI] [PubMed] [Google Scholar]
- Eriksson, S. , Bohlenius, H. , Moritz, T. and Nilsson, O. (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell, 18, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992) GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. San Diego, CA: Academic Press. [Google Scholar]
- Gaskin, P. , Hoad, G.V. , Macmillan, J. , Makinson, I.K. and Readman, J.E. (1992) Gibberellins A82 and A83 in seed of Lupinus albus . Phytochemistry, 31, 1869–1877. [Google Scholar]
- Hedden, P. , Hoad, G.V. , Gaskin, P. , Lewis, M.J. , Green, J.R. , Furber, M. and Mander, L.N. (1993) Kaurenoids and gibberellins, including the newly characterized gibberellin A88, in developing apple seeds. Phytochemistry, 32, 231–237. [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Huang, J.G. , Yang, M. , Liu, P. , Yang, G.D. , Wu, C.A. and Zheng, C.C. (2009a) GhDREB1 enhances abiotic stress tolerance, delays GA‐mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant, Cell Environ. 32, 1132–1145. [DOI] [PubMed] [Google Scholar]
- Huang, X.Y. , Chao, D.Y. , Gao, J.P. , Zhu, M.Z. , Shi, M. and Lin, H.X. (2009b) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi, Y. , Yoshiba, Y. , Sanada, Y. , Yamaguchi‐Shinozaki, K. , Wada, K. and Shinozaki, K. (1997) Characterization of the gene for Δ1‐pyrroline‐5‐carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol. Biol. 33, 857–865. [DOI] [PubMed] [Google Scholar]
- Itoh, H. , Ueguchi‐Tanaka, M. , Sentoku, N. , Kitano, H. , Matsuoka, M. and Kobayashi, M. (2001) Cloning and functional analysis of two gibberellin 3β‐hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl Acad. Sci. USA, 98, 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S. and Douglas, C.J. (2007) Populus, a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. [DOI] [PubMed] [Google Scholar]
- Kang, B.G. , Ye, X. , Osburn, L.D. , Stewart, C.N. Jr and Cheng, Z.M. (2010) Transgenic hybrid aspen overexpressing the Atwbc19 gene encoding an ATP‐binding cassette transporter confers resistance to four aminoglycoside antibiotics. Plant Cell Rep. 29, 643–650. [DOI] [PubMed] [Google Scholar]
- Kang, H. , Kim, J. , Kim, B. , Jeong, H. , Choi, S.H. , Kim, E.K. , Lee, H. et al. (2011) Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana . Plant Sci. 180, 634–641. [DOI] [PubMed] [Google Scholar]
- King, E.K. , Moritz, T. and Harberd, N.P. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics, 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.J. and Zeevaart, J.A.D. (2005) Molecular cloning of GA 2‐oxidase 3 from spinach and its ectopic expression in Nicotiana sylvestris . Plant Physiol. 138, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Xu, Z.H. , Luo, D. and Xue, H.W. (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36, 189–202. [DOI] [PubMed] [Google Scholar]
- Luo, A. , Qian, Q. , Yin, H. , Liu, X. , Yin, C. , Lan, Y. , Tang, J. et al. (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47, 181–191. [DOI] [PubMed] [Google Scholar]
- Magome, H. , Yamaguchi, S. , Hanada, A. , Kamiya, Y. and Oda, K. (2004) dwarf and delayed‐flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37, 720–729. [DOI] [PubMed] [Google Scholar]
- Magome, H. , Yamaguchi, S. , Hanada, A. , Kamiya, Y. and Oda, K. (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin‐deactivating gene, GA2ox7, under high‐salinity stress in Arabidopsis . Plant J. 56, 613–626. [DOI] [PubMed] [Google Scholar]
- Magome, H. , Nomura, T. , Hanada, A. , Takeda‐Kamiya, N. , Ohnishi, T. , Shinma, Y. , Katsumata, T. et al. (2013) CYP714B1 and CYP714B2 encode gibberellin 13‐oxidases that reduce gibberellin activity in rice. Proc. Natl Acad. Sci. USA, 110, 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, D. and Chen, C. (2012) Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold‐responsive pathway. PLoS ONE, 7, e47275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns, R. (2002) Comparative physiology of salt and water stress. Plant, Cell Environ. 25, 239–250. [DOI] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–495. [Google Scholar]
- Nelson, D.C. , Riseborough, J.A. , Flematti, G.R. , Stevens, J. , Ghisalberti, E.L. , Dixon, K.W. and Smith, S.M. (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T. , Magome, H. , Hanada, A. , Takeda‐Kamiya, N. , Mander, L.N. , Kamiya, Y. and Yamaguchi, S. (2013) Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 54, 1837–1851. [DOI] [PubMed] [Google Scholar]
- Ogawa, M. , Hanada, A. , Yamauchi, Y. , Kuwahara, A. , Kamiya, Y. and Yamaguchi, S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell, 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N. , Sun, T.P. and Gubler, F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell, 14(Suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, D.W. , Hutt, O.E. , Rood, S.B. and Mander, L.N. (2002) Gibberellins in shoots and developing capsules of Populus species . Phytochemistry, 59, 679–687. [DOI] [PubMed] [Google Scholar]
- Qin, F. , Kodaira, K.S. , Maruyama, K. , Mizoi, J. , Tran, L.S. , Fujita, Y. , Morimoto, K. et al. (2011) SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu, I. , Eriksson, S. , Powers, S.J. , Gong, F. , Griffiths, J. , Woolley, L. , Benlloch, R. et al. (2008) Genetic analysis reveals that C19‐GA 2‐oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell, 20, 2420–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, M. , Sakamoto, T. , Saito, T. , Matsuoka, M. , Tanaka, H. and Kobayashi, M. (2003) Expression of novel rice gibberellin 2‐oxidase gene is under homeostatic regulation by biologically active gibberellins. J. Plant. Res. 116, 161–164. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T. (2006) Phytohormones and rice crop yield: strategies and opportunities for genetic improvement. Transgenic Res. 15, 399–404. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T. , Morinaka, Y. , Ishiyama, K. , Kobayashi, M. , Itoh, H. , Kayano, T. , Iwahori, S. et al. (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat. Biotechnol. 21, 909–913. [DOI] [PubMed] [Google Scholar]
- Santes, C.M. , Hedden, P. , Gaskin, P. and Garcia‐Martinez, J. (1995) Gibberellins and related compounds in young fruits of pea and their relationship to fruit‐set. Phytochemistry, 40, 1347–1355. [Google Scholar]
- Sasaki, A. , Ashikari, M. , Ueguchi‐Tanaka, M. , Itoh, H. , Nishimura, A. , Swapan, D. , Ishiyama, K. et al. (2002) Green revolution: a mutant gibberellin‐synthesis gene in rice. Nature, 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Sasaki, A. , Itoh, H. , Gomi, K. , Ueguchi‐Tanaka, M. , Ishiyama, K. , Kobayashi, M. , Jeong, D.H. et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F‐box mutant. Science, 299, 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M. , Bizzell, C.M. , Lee, D.J. , Zeevaart, J.A.D. and Amasino, R.M. (2003) Overexpression of a novel class of gibberellin 2‐oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell, 15, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. (2008) Understanding gibberellic acid signaling – are we there yet? Curr. Opin. Plant Biol. 11, 9–15. [DOI] [PubMed] [Google Scholar]
- Shan, C. , Mei, Z. , Duan, J. , Chen, H. , Feng, H. and Cai, W. (2014) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE, 9, e87110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, E. , Efroni, I. , Gopalraj, M. , Saathoff, K. , Tseng, T.S. , Kieffer, M. , Eshed, Y. et al. (2012) The Arabidopsis O‐linked N‐acetylglucosamine transferase SPIDLY interacts with classI TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell, 24, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, R.J. , Yang, Y. , Yang, L. , Liu, H. , Wang, C.T. , Yu, M.M. , Gao, X.S. et al. (2014) Poplar calcineurin B‐like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant, Cell Environ. 37, 573–588. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G. , Phillips, A.L. and Hedden, P. (1999) Molecular cloning and functional expression of gibberellin 2‐oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl Acad. Sci. USA, 96, 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan, G.A. , DiFazio, S. , Jansson, S. , Bohlmann, J. , Grigoriev, I. , Hellsten, U. and Bokhsar, D. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Ueguchi‐Tanaka, M. , Ashikari, M. , Nakajima, M. , Itoh, H. , Katoh, E. , Kobayashi, M. , Chow, T.Y. et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature, 437, 693–698. [DOI] [PubMed] [Google Scholar]
- Varbanova, M. , Yamaguchi, S. , Yang, Y. , McKelevy, K. , Hanada, A. , Borochov, R. , Yu, F. et al. (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell, 19, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B.S. and Zhao, K.F. (1995) Comparison of extractive methods of Na+, K+ in wheat leaves. Plant Physiol. Commun. 31, 50–52 (in Chinese). [Google Scholar]
- Wang, Q. , Guan, Y. , Wu, Y. , Chen, H. , Chen, F. and Chu, C. (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67, 589–602. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Wang, C. , Liu, H. , Tang, R. and Zhang, H. (2011) An efficient Agrobacterium‐mediated transformation and regeneration system for leaf explants of two elite aspen hybrid clones Populus alba × P. Berolinensis and Populus Davidiana×P. Bolleana . Plant Cell Rep. 30, 2037–2044. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Bao, Y. , Wang, Q. and Zhang, H. (2013) Introduction of rice CYP714D1 gene into Populus inhibits expression of its homologous genes and promotes growth, biomass production and xylem fiber length in transgenic trees. J. Exp. Bot. 64, 2847–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008) Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Yamane, H. , Fujioka, S. , Spray, C.R. , Phinney, B.O. , Macmillan, J. , Gaskin, P. and Takahashi, N. (1988) Endogenous gibberellins from sporophytes of two tree ferns, Cibotiumglaucum and Dicksonia antarctica . Plant Physiol. 86, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Forno, D.A. , Cock, J.H. and Gomez, K.A. (1976) Laboratory Manual for Physiological Studies of Rice. Manila, Philippines: International Rice Research Institute. [Google Scholar]
- Zhang, Y. , Zhang, B. , Yan, D. , Dong, W. , Yang, W. , Li, Q. , Zeng, L. et al. (2011) Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 67, 342–353. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Nomura, T. , Xu, Y. , Zhang, Y. , Peng, Y. , Mao, B. , Hanada, A. et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell, 18, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Miao, Q. , Sun, D. , Yang, G. , Wu, C. , Huang, J. and Zheng, C. (2012) The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana . PLoS ONE, 7, e43530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Molecular identification and phenotypes of transgenic plants.
Figure S2 Molecular confirmation and phenotype analyses of transgenic Shanxin yang overexpression of PtCYP714A3.
Figure S3 Expression of PtCYP714A3 gene in response to PEG treatment and osmotic stress analyses of wild‐type and transgenic plants.
Table S1 Primers used in this study.
Table S2 Mean comparisons for tillers and yields of field‐grown wild‐type and PtCYP714A3 transgenic rice plants with or without high‐salinity stress.


