Abstract
Atrial fibrillation (AF) is the most common cardiac rhythm disorder at the clinical setting and accounts for up to 15% of all strokes. Recent genome-wide association studies (GWAS) identified two single nucleotide polymorphisms (SNPs), rs2106261 and rs7193343 in ZFHX3 (zinc finger homeobox 3 gene) and rs13376333 in KCNN3 (encoding a potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3) that showed significant association with AF in multiple populations of European ancestry. Here, we studied a Chinese Han, GeneID cohort consisting of 650 AF patients and 1,447 non-AF controls to test whether the GWAS findings on ZFHX3/KCNN3 and AF can be expanded to a different ethnic population. No significant association was detected for rs7193343 in ZFHX3 and rs13376333 in KCNN3. However, significant association was identified between rs2106261 in ZFHX3 and AF in the GeneID population for both allelic frequencies (P = 0.001 after adjusting for covariates of age, gender, hypertension, coronary artery disease, and diabetes mellitus; OR = 1.32), and genotypic frequencies assuming either an additive or recessive model (OR = 1.29, P = 0.001 and OR = 1.77, P = 0.00018, respectively). When only lone AF cases were analyzed, the association remained significant (OR = 1.50, P = 0.001 for allelic association; OR = 1.45, P = 0.001 for an additive model; OR = 2.24, P = 0.000043 for a recessive model). Our results indicate that rs2106261 in ZFHX3 confers a significant risk of AF in a Chinese Han population. The study expands the association between ZFHX3 and AF to a non-European ancestry population and provides the first evidence of a cross-race susceptibility of the 16q22 AF locus.
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia at the clinical setting, and accounts for approximately one-third of hospitalizations for cardiac rhythm disturbance (Go et al. 2001). AF has an estimated prevalence rate of 0.4–1.0% in the general population, which increases with age (Feinberg et al. 1995; Fuster et al. 2006). Because AF increases susceptibility to develop stroke, especially ischemic stroke, it is an important risk factor for morbidity and mortality in China and the rest of world (Hu and Sun 2008; Shi et al. 2009; Roberts and Gollob 2010).
AF is frequently observed as a complication of hypertension, valvular heart disease, coronary artery disease (CAD), hyperthyroidism, heart failure, and structural heart diseases (Fuster et al. 2006). Some AF cases occur in the absence of clinical or echocardiographic evidence of hypertension, hyperthyroidism, diabetes, CAD, cardiomyopathies, valvulopathies, or other types of cardiac diseases, and are referred to as lone AF (Fuster et al. 2006). AF can be classified into paroxysmal AF (recurrent AF episodes self-terminating in <7 days), persistent AF (>7 days) and permanent AF (ongoing long-term AF episodes) (Marrouche et al. 2003).
Genetic factors play an important role in the pathogenesis of AF. AF has been found to occur in large families (monogenic AF) and can be inherited in either an autosomal dominant model or an autosomal recessive (Oberti et al. 2004). Several genetic loci have been identified for monogenetic AF, including loci on chromosome 1q21, 3p21, 11p15.5, 12p13, 21q22, 17q23–q24, 7q35–36, 5p13, 6q14–16, and 10q22–q24 (Tsai et al. 2008b). At these loci, several AF genes have been identified and they include cardiac sodium channel subunit genes SCN5A, SCN1B, SCN2B, and SCN3B, potassium channel genes KCNQ1, KCNE2, KCNJ2, KCNA5, and KCNH2, as well as NPPA and NUP155 (Chen et al. 2003; Sebillon et al. 2003; Yang et al. 2004; Hong et al. 2005; Olson et al. 2005, 2006; Xia et al. 2005; Hodgson-Zingman et al. 2008; Wang 2008; Zhang et al. 2008; Wang et al. 2010; Ren et al. 2010). Most of these genes encode subunits of ion channels, but interestingly NPPA and NUP155 do not code for ion channels, instead encode natriuretic peptide precursor A and a nuclear pore complex protein, nucleoporin 155 (Hodgson-Zingman et al. 2008; Zhang et al. 2008), indicating that non-ion channel genes are critical to the pathogenesis of AF.
The majority of AF cases belong to common complex AF resulting from the interaction among genetic and environmental factors (Wang 2008). Recent genome-wide association studies (GWAS) have revealed several genetic loci for common complex AF. The first GWAS for AF identified a common single nucleotide polymorphism (SNP rs2200733) on chromosome 4q25 that was associated with AF in several populations of European ancestry, a Hong Kong Chinese population, and a mainland Chinese Han population (Gudbjartsson et al. 2007; Viviani Anselmi et al. 2008; Kääb et al. 2009; Shi et al. 2009). Then, two independent GWAS revealed two SNPs in the ZFHX3 gene on chromosome 16q22 that were associated with AF in various populations of European ancestry. Benjamin et al. (2009) found that ZFHX3 SNP rs2106261 was associated with AF, whereas Gudbjartsson et al. (2009) identified a different SNP in ZFHX3, rs7193343 that was associated with AF in the Icelandic population. Gudbjartsson et al. (2009) also assessed the association between ZFHX3 SNP rs7193343 with AF in a small Hong Kong Chinese cohort, but found no association with AF in this population (P = 0.63). These results raise the question whether the association between ZFHX3 SNPs and AF is restricted to the populations of European ancestry, and made further studies on this finding necessary. One very recent GWAS identified significant association between AF and SNP rs13376333 in KCNN3 encoding a potassium intermediate/small conductance calcium-activated channel (subfamily N, member 3) in European ancestry cohorts (Ellinor et al. 2010), but it is unknown whether the same SNP confers risk of AF in a non-European ancestry population. We therefore carried out a large-scale case–control association study with 650 AF patients and 1,447 non-AF controls in a Chinese Han GeneID population to examine whether the two ZFHX3 SNPs (rs2106261 and rs7193343) and rs13376333 in KCNN3 are associated with AF in a non-European ancestry population. SNP rs2106261, but not rs7193343 and rs13376333, was associated with AF in the Chinese Han population.
Methods
Study subjects
The AF patients were selected from the Chinese GeneID database with a large number of study participants with various cardiovascular diseases enrolled from mainland China. This study was approved by the local Ethics Committees and informed consent was obtained from all participants. The Chinese GeneID database has been created for identification of genes for cardiovascular diseases including AF, CAD, stroke and essential hypertension. For AF, study subjects were recruited among individuals who underwent treatment with ablation procedures and electrophysiological examinations from multiple hospitals in Central China and Northern China covering six Provinces. All study participants are of the ethnic Han origin by self-report. Patients with other types of cardiac arrhythmias, hyperthyroidism, cardiomyopathies and valvulopathies were excluded. Evaluation of AF was carried out by expert cardiologists and based on the standard diagnostic criteria according to ACC/AHA/ESC 2006 guidelines for the management of patients with AF (Fuster et al. 2006). Clinical examinations were carried out using rest electrocardiograms (ECG), and sometimes aided with bedside telemetry or ambulatory Holter ECG recordings. Relevant data were also collected by direct interviews or from medical files on age, gender, and history of hypertension, diabetes, CAD, stroke, and hyperthyroidism. AF patients over 75 years old were excluded from this study. Hypertension was defined as a clinical blood pressure of ≥140/90 mmHg or history of medication. Individuals with a coronary stenosis of ≥70%, percutaneous coronary angioplasty, coronary artery bypass graft, or myocardial infarction were classified as CAD patients. Diabetes mellitus was diagnosed by a fasting blood glucose level of ≥7 mmol/L. Diagnosis of stroke was based on medical history, neurological examinations, CT or MRI according to the World Health Organization (WHO) criteria (Goldstein et al. 1989). Echocardiography was preformed for AF patients to exclude cardiomyopathies and valvulopathies. The patients were also examined for thyroid abnormalities by physical examinations, and thyroid function testing was preformed if hyperthyroidism was suspected. AF patients without hypertension, CAD, hyperthyroidism, cardiomyopathies, valvulopathies, or other types of cardiac diseases were classified as lone AF (Fuster et al. 2006).
The controls were unrelated healthy Han individuals who were verified to be free of AF based on ECG or medical files at the time of enrollment. The controls were ascertained from individuals who underwent annual physical exams with or without any disease. Similar to cases, the controls were recruited without consideration of the presence or absence of hypertension, diabetes, CAD, and stroke, but those with other types of cardiac arrhythmias, hyperthyroidism, cardiomyopathies and valvulopathies were excluded. To minimize the effect of population substructure, the cases were matched with controls from the same geographical areas.
SNP genotyping
Blood samples were drawn from study participants and used for isolation of genomic DNA using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s protocol.
SNPs were genotyped using a Rotor-Gene TM 6000 High Resolution Melt system (Corbett Life Science, Concorde, NSW, Australia). Genotyping was performed in 25 µL of standard PCR volume containing 1 µL of LC Green dye, 5 pmol of each primer, 25 ng of genomic DNA, 2.5 µL of 10× PCR buffer with 1.5 mmol/L MgCl2, 5 mmol deoxynucleotide triphosphates and 1 U of Taq polymerase. Two positive controls for each genotype (A/A, A/G and G/G) were included in each run. In order to verify the genotyping results, we randomly selected 50 cases and controls for direct DNA sequence analysis. DNA sequence analysis was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kits on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The sequencing result for the 50 samples perfectly matched that by the HRM method.
Statistical analysis
We used PLINK1.06 to assess allelic and genotypic associations as well as Hardy–Weinberg equilibrium (HWE). Odds ratios (ORs) and 95% confidence intervals (CIs) were estimated using the χ2 test (PLINK1.06 or SPSS, version 13.0). When the samples were stratified, Breslow–Day tests were performed to analyze the homogeneity between ORs from each sub-group (SPSS, version 13.0). Multivariable logistic regression analysis was carried out using SAS version 9.0 (SAS Institute, Inc). Empirical P values were determined using the PLINK v1.06 program with 100,000 Monte Carlo simulations.
Power analysis was carried out using Power and Sample Size Program 3.0 or nQuery Advisor 7.0 assuming an alpha value of 0.05, a two-sided test, ORs from published reports, and minor allele frequencies for the Chinese population from the HapMap database (http://www.hapmap.org, phase 2, HapMap-HCB). The T allele of SNP rs7193343 was associated with risk of AF in populations of European ancestry (Gudbjartsson et al. 2009). Based on a frequency of 68.2% for the T allele in the Chinese population from the HapMap database (http://www.hapmap.org, phase 2, Hap-Map-HCB) and ORs of 1.21 from the Gudbjartsson et al. (2009) studies, the power to detect a significant association between rs7193343 and AF in our GeneID population was 78%. Benjamin et al. (2009) revealed significant association between the minor allele A of SNP rs2106261 and AF in populations of European ancestry. Assuming an OR of 1.44 from Benjamin et al. (2009) study and a risk allele frequency of 36.7% for the Chinese population from the HapMap database, the power to detect a significant association between rs2106261 and AF in our GeneID population was 99%. For SNP rs13376333 in KCNN3, our population has a power of 74% to detect the association with AF assuming a previously reported OR of 1.56 and a minor allele frequency of 0.033 in the Chinese population (http://www.hapmap.org, phase 2, HapMap-HCB).
Results
In order to test whether SNPs in the ZFHX3 gene are associated with AF in a non-European ancestry population, we carried out a case–control association study with a large cohort of samples from 650 Chinese Han AF patients and 1,447 non-AF controls. The general and clinical characteristics of the study population are summarized in Table 1. The average age for AF cases and controls was 58.4 ± 15.9 and 59.7 ± 12.2 years, respectively, but the difference was not statistically significant (Table 1). In the AF group, 27.7% of cases qualified to be lone AF cases. A majority of AF cases (64%) were classified as paroxysmal AF, and 32.5 and 3.5% were persistent AF and permanent AF, respectively (Table 1). As expected, known risk factors for AF, including hypertension, CAD, and diabetes, were more prevalent in the AF group than in the control group (Table 1). The prevalence rate of hypertension, CAD, and diabetes in controls was low. The rate of ischemic stroke in the AF group (14.2%) was much higher than that in the control group (0.3%) (P < 0.01).
Table 1.
General clinical characteristics of the GeneID AF case control cohort
| Characteristic | Cases (n = 650) |
Controls (n = 1,447) |
P* |
|---|---|---|---|
| Gender (male/female) | 398/252 | 902/545 | 0.63 |
| Age (mean ± SD, years) | 58.4 ± 15.9 | 59.7 ± 12.2 | 0.84 |
| Lone AF (%) | 180 (27.7) | N/A | – |
| Paroxysmal AF (%) | 416 (64.0) | N/A | – |
| Persistent AF (%) | 211 (32.5) | N/A | – |
| Permanent AF (%) | 23 (3.5) | N/A | – |
| Hypertension (%) | 273 (42.0) | 39 (2.7) | <0.01 |
| Ischemic stroke (%) | 92 (14.2) | 4 (0.3) | <0.01 |
| CAD (%) | 181 (27.8) | 2 (0.01) | <0.01 |
| T2DM (%) | 49 (7.5) | 14 (0.9) | <0.01 |
Age age-at-onset of AF for cases and age at the enrollment for controls, P* derived from an unpaired Student’s t test or Pearson’s chi-square test, SD standard deviation, CAD coronary artery disease, T2DM type 2 diabetes mellitus
The distribution of three genotypes for SNPs rs2106261, rs7193343, and rs13376333 was in Hardy–Weinberg equilibrium in the control group (P > 0.05) (Table 2). Highly significant allelic association was identified between SNP rs2106261 and AF (Pobs = 6.67 × 10−5, OR = 1.32, 95% CI of 1.15–1.51) (Table 2). The association remained significant after adjusting for covariates of age, gender, CAD, hypertension and diabetes mellitus (Padj = 0.001, Table 2). Similarly, highly significant genotypic association was also found between SNP rs2106261 and AF assuming either an additive or recessive model (Pobs = 0.0001 and 4.53 × 10−5, respectively) (Table 3). The genotypic association remained significant after adjusting for covariates of age, gender, CAD, hypertension and diabetes mellitus (Padj = 0.001 and 1.81 × 10−4, respectively; Table 3).
Table 2.
Analysis of association of allelic frequencies of SNPs rs2106261, rs7193343 and rs13376333 with AF
| Cases/controls (n) | Risk allele frequency in cases/controls |
Phwe | Pobs | Padj | OR (95% CI) | Pemp | |
|---|---|---|---|---|---|---|---|
| SNP rs2106261 | |||||||
| Total AF | 650/1447 | 0.39/0.33 | 0.191 | 6.67 × 10−5 | 0.001 | 1.32 (1.15–1.51) | 1.97 × 10−4 |
| Lone AF | 180/1447 | 0.43/0.33 | 0.191 | 3.52 × 10−4 | 0.001 | 1.50 (1.20–1.87) | 5.68 × 10−4 |
| Other AF | 470/1447 | 0.38/0.33 | 0.191 | 0.004 | 0.048 | 1.22 (1.00–1.47) | 0.005 |
| SNP rs7193343 | |||||||
| Total AF | 650/1447 | 0.32/0.32 | 0.856 | 0.850 | 0.932 | 0.99 (0.87–1.16) | 0.857 |
| Lone AF | 180/1447 | 0.34/0.32 | 0.856 | 0.421 | 0.423 | 1.10 (0.87–1.39) | 0.364 |
| Other AF | 470/1447 | 0.31/0.32 | 0.856 | 0.723 | 0.681 | 0.96 (0.78–1.18) | 0.636 |
| SNP rs13376333 | |||||||
| Total AF | 650/1447 | 0.04/0.03 | 1.000 | 0.220 | 0.225 | 1.24 (0.88–1.75) | 0.333 |
| Lone AF | 180/1447 | 0.05/0.03 | 1.000 | 0.067 | 0.062 | 1.66 (1.00–2.75) | 0.066 |
| Other AF | 470/1447 | 0.04/0.03 | 1.000 | 0.696 | 0.698 | 1.08 (0.72–1.62) | 0.857 |
Phwe P values for Hardy–Weinberg equilibrium tests; Pobs unadjusted P values; Padj adjusted P values using multivariable logistic regression analysis by including covariates of sex, age, T2DM, hypertension, and CAD; Pemp empirical P values obtained by performing 100,000 Monte–Carlo simulations; OR odds ratio; CI confidence interval
A P value of 0.0055 was considered significant after Bonferroni correction for 9 multiple tests (3 SNPs × 3 subgroups of AF)
Table 3.
Analysis of association for genotypic frequencies of SNPs rs2106261, rs7193343 and rs13376333 with AF under different genetic models
| Model | Pobs | Padj | OR (95% CI) | Pemp |
|---|---|---|---|---|
| SNP rs2106261 | ||||
| Total AF | ||||
| Dominant | 9.52 × 10−3 | 0.048 | 1.25 (1.01–1.56) | 0.006 |
| Recessive | 4.53 × 10−5 | 1.81 × 10−4 | 1.77 (1.31–2.39) | 7.00 × 10−5 |
| Additive | 1.00 × 10−4 | 0.001 | 1.29 (1.11–1.51) | 9.64 × 10−5 |
| Lone AF | ||||
| Dominant | 0.064 | 0.085 | 1.34 (0.96–1.86) | 0.100 |
| Recessive | 1.13 × 10−5 | 4.34 × 10−5 | 2.24 (1.53–3.34) | 3.57 × 10−5 |
| Additive | 6.17 × 10−5 | 0.001 | 1.45 (1.16–1.82) | 6.57 × 10−5 |
| Other AF | ||||
| Dominant | 0.033 | 0.002 | 1.53 (1.17–2.02) | 0.036 |
| Recessive | 0.008 | 1 × 10−4 | 2.40 (1.61–3.57) | 0.014 |
| Additive | 0.013 | 1 × 10−4 | 1.55 (1.27–1.89) | 0.022 |
| SNP rs7193343 | ||||
| Total AF | ||||
| Dominant | 0.834 | 0.901 | 1.02 (0.81–1.28) | 1.000 |
| Recessive | 0.873 | 0.581 | 1.12 (0.76–1.65) | 0.750 |
| Additive | 0.951 | 0.731 | 1.03 (0.87–1.23) | 1.000 |
| Lone AF | ||||
| Dominant | 0.206 | 0.425 | 1.14 (0.82–1.59) | 0.209 |
| Recessive | 0.743 | 0.786 | 1.08 (0.62–1.88) | 0.75 |
| Additive | 0.337 | 0.460 | 1.10 (0.86–1.41) | 0.400 |
| Other AF | ||||
| Dominant | 0.644 | 0.268 | 0.86 (0.67–1.12) | 0.750 |
| Recessive | 0.992 | 0.979 | 0.99 (0.63–1.58) | 1.000 |
| Additive | 0.889 | 0.386 | 0.91 (0.74–1.12) | 0.857 |
| SNP rs13376333 | ||||
| Total AF | ||||
| Dominant | 0.288 | 0.085 | 0.70 (0.47–1.05) | 0.371 |
| Recessive | 0.181 | 0.113 | 17.02 (0.51–566.10) | 0.476 |
| Additive | 0.276 | 0.152 | 0.75 (0.51–1.11) | 0.265 |
| Lone AF | ||||
| Dominant | 0.128 | 0.760 | 0.92 (0.52–1.62) | 0.148 |
| Recessive | 0.002 | 0.018 | 23.5 (1.72–320.49) | 0.050 |
| Additive | 0.005 | 0.869 | 1.05 (0.62–1.78) | 0.013 |
| Other AF | ||||
| Dominant | 0.651 | 0.905 | 1.03 (0.66–1.60) | 0.750 |
| Recessive | 0.568 | 0.105 | 66.09 (0.42–10520.15) | 1.000 |
| Additive | 0.749 | 0.663 | 1.10 (0.72–1.68) | 0.857 |
/ data not available because there were only two genotypes
A P value of 0.0055 was considered significant after Bonferroni correction for 9 multiple tests (3 SNPs × 3 subgroups of AF)
In the AF group, a total of 180 patients were classified as lone AF. Significant allelic and genotypic association was detected for SNP rs2106261 and lone AF (Pobs = 3.5 × 10−4, OR = 1.50 for allelic frequencies, 1.1 × 10−5 for a recessive model, and 6.2 × 10−5 for an additive model) (Tables 2, 3). Both allelic and genotypic associations remained significant after adjusting for covariates of age, gender, CAD, hypertension and diabetes mellitus (Padj = 0.001, 4.34 × 10−5, and 0.001, respectively; Tables 2, 3). No significant allelic association was identified for other AF (total AF − lone AF), but significant genotypic association was identified under an additive or recessive model (Tables 2, 3). Breslow–Day tests did not find a significant difference between the OR for lone AF (1.50) and the other AF (1.22) (P = 0.19).
There was no significant allelic or genotypic association between SNP rs7193343 and AF (Tables 2, 3), which is consistent with the result in the Hong Kong AF cohort reported by Gudbjartsson et al. (2009). SNP rs13376333 in KCNN3 did not show significant allelic or genotypic association with AF either (Tables 2, 3).
Discussion
In this study, we show that intronic SNP rs2106261 in the ZFHX3 gene on chromosome 16q22 is associated with AF in a non-European ancestry population. We carried out a case–control association study involving 650 Chinese AF patients and 1,447 non-AF controls, all from a large Chinese GeneID population of Han ethnic descent. Highly significant allelic association was identified with an OR of 1.32 (Padj = 0.001) (Table 2). Similarly, genotypic association was also significant with an additive model (Padj = 0.001) and a recessive model (Padj = 1.81 × 10−4) (Table 3). These results expand the association between ZFHX3 variant and AF beyond European ancestry populations.
In a 2009 GWAS, Gudbjartsson et al. (2009) identified the association between ZFHX3 SNP rs7193343 and AF in several European ancestry populations, and further assessed its association with AF in a Hong Kong-based Chinese population. No significant association was identified (Gudbjartsson et al. 2009). In this study, we also failed to find any association between rs7193343 and AF. On the other hand, although both SNPs rs7193343 and rs2106261 are located in the same intron, intron 1 of ZFHX3, association of rs2106261 with AF was highly significant in this Chinese GeneID study. These results highlight the importance of analyzing multiple SNPs in the same gene or genomic region in replication studies, in particular in a different ethnic population.
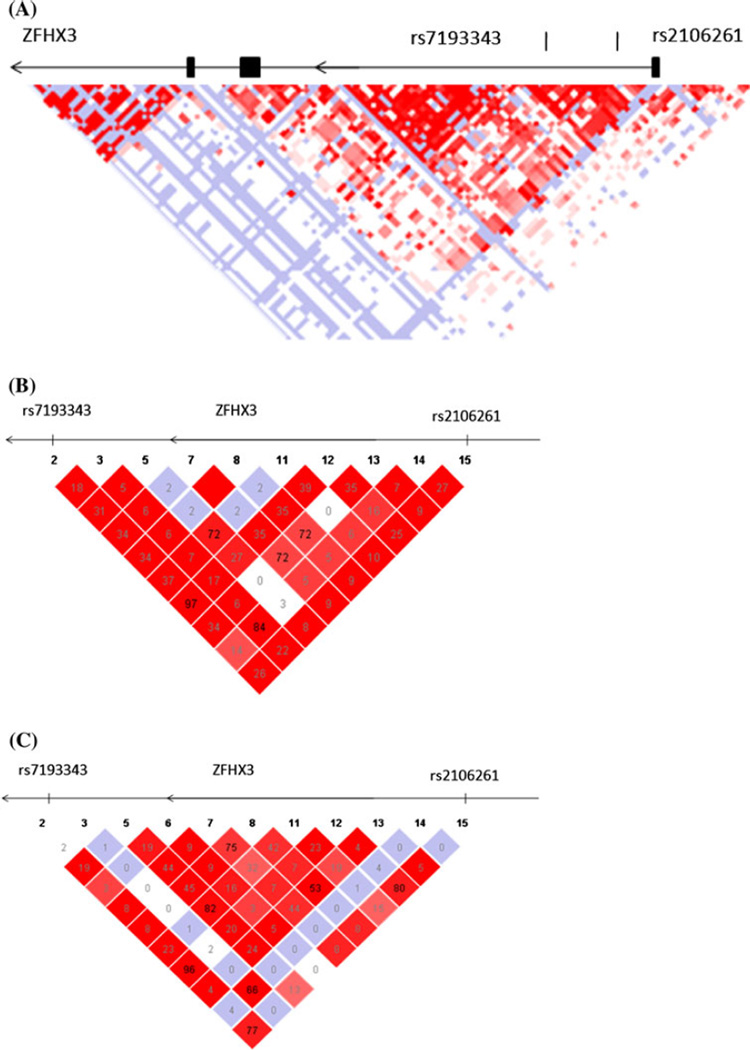
We constructed a LD map for a 160 kb genomic region involving intron 1 of the ZFHX3 gene and two SNPs rs2106261 and rs7193343 using the genotyping data for the Chinese samples from the HapMap database (http://www.hapmap.org, phase 2, HapMap-HCB) (Fig. 1). Although rs2106261 and rs7193343 were 22 kb apart, the LD correlation between two SNPs was weak, and r2 was 0.26 (Fig. 1). The r2 predicted from the HapMap data on individuals of European ancestry (http://www.hapmap.org, phase 2, HapMap-CEU) was 0.77. Thus, it is reasonable to predict that the causative SNP(s) for AF at the 16q22 locus may be tagged with both rs2106261 and rs7193343 in populations of European ancestry, but with rs2106261 only in the Chinese Han population. In the populations of European ancestry, the minor allele T of rs7193343 was the risk allele for AF and had a frequency of about 15%. In contrast, the frequency of rs7193343 T allele in the Chinese GeneID population was 68%, representing the common allele. The dramatic difference of allele frequencies between the populations of European ancestry and Chinese populations may be a cause for lack of association between rs7193343 and AF in the Chinese cohorts.
Fig. 1.
a Overview of linkage disequilibrium (LD) of the 160 kb region around intron 1 of ZFHX3. The LD structure around SNPs rs7193343 and rs2106261 was derived from the genotyping data of SNPs from the HAPMAP database (http://www.hapmap.org, phase 2, HapMap-CEU). The pairwise correlation between SNPs was measured as D′ and a black diamond without a number refers to D′ = 100. Exons are marked with black boxes. The direction of ZFHX3 transcription is to the left as marked by the arrows. b LD structure between SNPs rs7193343 and rs2106261 predicted based on the HapMap data for the Chinese population (http://www.hapmap.org, phase 2, HapMap-HCB). c LD structure between SNPs rs7193343 and rs2106261 predicted based on the HapMap data for the population of European ancestry (http://www.hapmap.org, phase 2, HapMap-CEU)
In this study, we also tested the association between ZFHX3 SNP rs2106261 and lone AF. Significant allelic association was identified for lone AF with an OR of 1.50 (Padj = 0.001). The OR for lone AF was higher than that for other AF, 1.50 versus 1.22, but the difference was not statistically significant. This finding is in drastic contrast to the results we recently reported for SNP rs2200733 on chromosome 4q25 (Shi et al. 2009). The chromosome 4q25 was the first locus identified by GWAS for AF, and we recently showed that the OR for association of rs2200733 with lone AF was significantly higher than the OR for other AF, 2.40 versus 1.59 (P = 0.022) (Shi et al. 2009). These results suggest that some AF loci (e.g., 4q25 locus) play a more important role in lone AF than other AF, whereas other AF loci (e.g., 16q22 locus in this study) play an equally important role in both lone AF and other AF.
The molecular mechanism by which intronic SNP rs2106261 of ZFHX3 confers risk of AF is unknown. Because rs2106261 is located in intron 1, one possibility is that it affects the expression level of ZFHX3 by modulating regulation of ZFHX3 transcription or pre-mRNA splicing because such regulatory roles have been found for intronic sequences (Greenwood and Kelsoe 2003). The ZFHX3 gene encodes a widely expressed transcription factor. ZFHX3 was found to be a regulatory factor for STAT3-mediated signal transduction (Nojiri et al. 2004). Tsai et al. suggested that activation of the angiotensin II/Rac1/STAT signaling transduction pathway may contribute to the structural and inflammatory changes associated with AF (Tsai et al. 2008a). Therefore, we hypothesize that ZFHX3 variant may increase risk of AF by affecting the JAK/STAT signaling, but future investigations are needed to test this hypothesis.
There are several limitations for this study. One limitation is that this is the first time that SNP rs2106261 was found to be associated with AF in the Chinese Han population, therefore, the finding needs to be further replicated in additional independent Chinese Han populations. Second, the rate of lone AF cases in the population under this study has reached 27.7%, which may reflect the selection bias of ascertaining AF from individuals who underwent treatment with ablation procedures and electrophysiological examinations. Third, the mean age of the AF patients was 58.4 ± 15.9 years, which was younger than the mean age of typical AF in the community. Fourth, the control group was selected from individuals undergoing annual physical exams, and appeared to be very healthy as the prevalence of hypertension, CAD, stroke, and diabetes mellitus was low. This may cause selection bias for the control group. Fifth, the findings in this study may not be generalized to other Chinese ethnicities or to the AF in the general community due to selection or referral bias. Moreover, the significant SNP rs2106261 associated with AF may not be causal, and the functional relation between the genomic region and AF is unknown.
A very recent GWAS identified association between SNP rs13376333 in KCNN3 on chromosome 1q21 and lone AF in populations of European ancestry (Ellinor et al. 2010). We assessed this association using the same GeneID samples under this study, but did not find significant association between SNP rs13376333 and lone AF or other AF (Tables 2, 3). The lack of association between SNP rs13376333 in KCNN3 and AF may be due to the very low, 3% minor allele frequency of the SNP in the Chinese population and the small sample size, although the cohort under this study has a power of 74%.
In conclusion, we found that the A allele of SNP rs2106261 of the ZFHX3 gene on chromosome 16q22 confers a highly significant risk of AF in the Chinese GeneID population. The results expand the association of ZFHX3 SNPs with AF previously identified in multiple cohorts of European descent to a non-European ancestry population and provide the first piece of evidence of a cross-race susceptibility of the 16q22 locus to AF.
Acknowledgments
We thank the study participants for their important support of this research. We thank Dr Baofeng Yang for help and discussion. This study was supported by a Hubei Province Natural Science Key Program (2008CDA047), the China National Basic Research Program (973 Program 2007CB512002), a China National 863 Scientific Program (2006AA02Z476), a National Natural Science Foundation grant of China (30670857 and 30800457), a Key Academic Program Leader Award of Wuhan City (200951830560), and in part by an NIH grant (R01 HL094498). This study has no relationship with any industry.
Contributor Information
Cong Li, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Fan Wang, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Yanzong Yang, First Affiliated Hospital of Dalian Medical University, Dalian, China.
Fenfen Fu, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Chengqi Xu, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Lisong Shi, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Sisi Li, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Yunlong Xia, First Affiliated Hospital of Dalian Medical University, Dalian, China.
Gang Wu, Renmin Hospital of Wuhan University, Wuhan, China.
Xiang Cheng, Institute of Cardiology, Union Hospital, Cardio-X Institute, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, China.
Hui Liu, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Chuchu Wang, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Pengyun Wang, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Jianjun Hao, Wuhan No.1 Hospital, Wuhan, China.
Yuhe Ke, Wuhan No.1 Hospital, Wuhan, China.
Yuanyuan Zhao, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Mugen Liu, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Rongfeng Zhang, First Affiliated Hospital of Dalian Medical University, Dalian, China.
Lianjun Gao, First Affiliated Hospital of Dalian Medical University, Dalian, China.
Bo Yu, The 2nd Affiliated Hospital of Harbin Medical University, Harbin, China.
Qiutang Zeng, Institute of Cardiology, Union Hospital, Cardio-X Institute, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, China.
Yuhua Liao, Institute of Cardiology, Union Hospital, Cardio-X Institute, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, China.
Bo Yang, Renmin Hospital of Wuhan University, Wuhan, China.
Xin Tu, Email: xtu@mail.hust.edu.cn, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China.
Qing K. Wang, Email: qkwang@mail.hust.edu.cn, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Cardio-X Institute, Huazhong University of Science and Technology, Wuhan, China; Center for Cardiovascular Genetics, Cleveland Clinic, Cleveland, OH, USA.
References
- Benjamin E, Rice K, Arking D, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xu S, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- Ellinor P, Lunetta K, Glazer N, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg W, Blackshear J, Laupacis A, et al. Prevalence, age distribution, and gender of patients with atrial fibrillation: analysis and implications. Arch Intern Med. 1995;155:469. [PubMed] [Google Scholar]
- Fuster V, Rydén L, Cannom D, et al. ACC/AHA/ESC 2006 guidelines for the management of patients with atrial fibrillation: full text. Europace. 2006;8:651. doi: 10.1093/europace/eul097. [DOI] [PubMed] [Google Scholar]
- Go A, Hylek E, Phillips K, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA. 2001;285:2370. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- Goldstein M, Barnett H, Orgogozo J, et al. Recommendations on stroke prevention, diagnosis and therapy: report of the WHO task force on stroke and other cerebrovascular disorders. Stroke. 1989;20:1407–1431. doi: 10.1161/01.str.20.10.1407. [DOI] [PubMed] [Google Scholar]
- Greenwood T, Kelsoe J. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D, Arnar D, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson-Zingman D, Karst M, Zingman L, et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Bjerregaard P, Gussak I, et al. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16:394–396. doi: 10.1046/j.1540-8167.2005.40621.x. [DOI] [PubMed] [Google Scholar]
- Hu D, Sun Y. Epidemiology, risk factors for stroke, and management of atrial fibrillation in China. J Am Coll Cardiol. 2008;52:865. doi: 10.1016/j.jacc.2008.05.042. [DOI] [PubMed] [Google Scholar]
- Kääb S, Darbar D, Van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrouche N, Martin D, Wazni O, et al. Phased-array intracardiac echocardiography monitoring during pulmonary vein isolation in patients with atrial fibrillation: impact on outcome and complications. Circulation. 2003;107:2710. doi: 10.1161/01.CIR.0000070541.83326.15. [DOI] [PubMed] [Google Scholar]
- Nojiri S, Joh T, Miura Y, et al. ATBF1 enhances the suppression of STAT3 signaling by interaction with PIAS3. Biochem Biophys Res Commun. 2004;314:97–103. doi: 10.1016/j.bbrc.2003.12.054. [DOI] [PubMed] [Google Scholar]
- Oberti C, Wang L, Li L, et al. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation. 2004;110:3753. doi: 10.1161/01.CIR.0000150333.87176.C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson T, Michels V, Ballew J, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson T, Alekseev A, Liu X, et al. Kv1.5 channelopathy due to KCNA 5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- Ren X, Xu C, Zhan C, et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin Chim Acta. 2010;411:481–485. doi: 10.1016/j.cca.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Roberts J, Gollob M. Impact of genetic discoveries on the classification of lone atrial fibrillation. J Am Coll Cardiol. 2010;55:705–712. doi: 10.1016/j.jacc.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Sebillon P, Bouchier C, Bidot L, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Li C, Wang C, et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum Genet. 2009;126:843–849. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- Tsai C, Lin J, Lai L, et al. Membrane translocation of small GTPase Rac1 and activation of STAT1 and STAT3 in pacinginduced sustained atrial fibrillation. Heart Rhythm. 2008a;5:1285–1293. doi: 10.1016/j.hrthm.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Tsai CT, Lai LP, Lin JL, et al. Molecular genetics of atrial fibrillation. Acta Cardiol Sin. 2008b;24:177–190. [Google Scholar]
- Viviani Anselmi C, Novelli V, Roncarati R, et al. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart. 2008;94:1394. doi: 10.1136/hrt.2008.148544. [DOI] [PubMed] [Google Scholar]
- Wang Q. Atrial fibrillation from bench to bedside. New Jersey: Humana Press; 2008. Atrial fibrillation: genetic consideration; pp. 131–141. [Google Scholar]
- Wang P, Yang Q, Wu X, et al. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98–104. doi: 10.1016/j.bbrc.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Jin Q, Bendahhou S, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xia M, Jin Q, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genetics. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]