Abstract
Foxp3+ regulatory T cells (Tregs) migrating from the skin to the draining lymph node (dLN) have a strong immunosuppressive effect on the cutaneous immune response. However, the subpopulations responsible for their inhibitory function remain unclear. We investigated single-cell gene expression heterogeneity in Tregs from the dLN of inflamed skin in a contact hypersensitivity model. The immunosuppressive genes Ctla4 and Tgfb1 were expressed in the majority of Tregs. Although Il10-expressing Tregs were rare, unexpectedly, the majority of Il10-expressing Tregs co-expressed Gzmb and displayed Th1-skewing. Single-cell profiling revealed that CD43+ CCR5+ Tregs represented the main subset within the Il10/Gzmb-expressing cell population in the dLN. Moreover, CD43+ CCR5+ CXCR3− Tregs expressed skin-tropic chemokine receptors, were preferentially retained in inflamed skin and downregulated the cutaneous immune response. The identification of a rare Treg subset co-expressing multiple immunosuppressive molecules and having tissue-remaining capacity offers a novel strategy for the control of skin inflammatory responses.
CD4+ Foxp3+ regulatory T cells (Tregs) appear to be essential for maintaining immunological self-tolerance and suppressing excessive immune responses during contact hypersensitivity (CHS), a prototype of T-cell mediated delayed-type hypersensitivity1. Using mouse lines expressing the photoconvertible protein Kaede, we have shown previously that Tregs migrating to the draining lymph node (dLN) from inflamed skin in an antigen-specific manner (“migratory Tregs”) more frequently express migratory molecules such as CD103 and display superior suppression of the CHS response compared with Tregs pre-existing within the dLN (“LN-resident Tregs”)1. In this study, a subset of Tregs that expressed exceptionally high levels of CD25 (CD25++ Tregs) became apparent under the CHS condition. CD25++ Tregs were more frequent among migratory Tregs than among LN-resident Tregs. Because of their transient emergence after CHS induction and higher CD25 expression than naturally-occurring Tregs (nTregs)2, CD25++ Tregs might correspond to so-called induced Tregs (iTregs). CD25++ migratory Tregs possess stronger suppressive capacity than CD25int/− migratory Tregs. Thus, CD25++ migratory Tregs are likely to play an important role in the immune regulation of acute skin inflammation.
Recently, various methodologies for single-cell analysis have been developed, including multi-parameter flow cytometry and single-cell RNA sequencing3. Studies using these methods have revealed the heterogeneity of gene and protein expression within various cell populations including tumor cells4 and hematopoietic stem cells5,6. Single-cell analyses have also been used to study T cell biology. Even among CD8+ T cells with the same antigen specificity or bearing the same T cell receptor (TCR) derived from a mouse infected with virus or bacteria, individual T cells have been reported to display heterogeneous expression of effector function- and differentiation-related genes7,8. Single-cell approaches have helped to identify novel T cell subsets and their markers that could not be identified by ordinary bulk analysis.
In the present study, we used single-cell real-time PCR array analyses (scqPCR) to identify novel highly-functional Treg subsets, including CD25++ Tregs in the dLN of CHS-induced mice. Even amongst CD25++ migratory Tregs, we found the rare presence of individual Tregs expressing the Il10 gene, which encodes IL-10, an immunosuppressive cytokine that directly inhibits the CHS response9. Il10-expressing Tregs frequently co-expressed the Treg effector genes, Gzmb, Ctla4, and Tgfb1 in a hierarchical manner. Furthermore, Gzmb-/Il10-expressing Treg subsets expressed the novel markers CD43 and CCR5. CD43+ CCR5+ CXCR3− Tregs highly expressed skin-tropic chemokine receptors CCR4 and CCR8. We also measured the retention of Treg subsets within inflamed skin using mice expressing the photoconvertible protein KikGR, and found that the subset of Tregs that was most highly retained in the skin, CD43+ CCR5+ CXCR3− Tregs, had superior in vivo inhibitory function to other Treg subsets.
Results
CD25++ Tregs form a distinct gene expression cluster
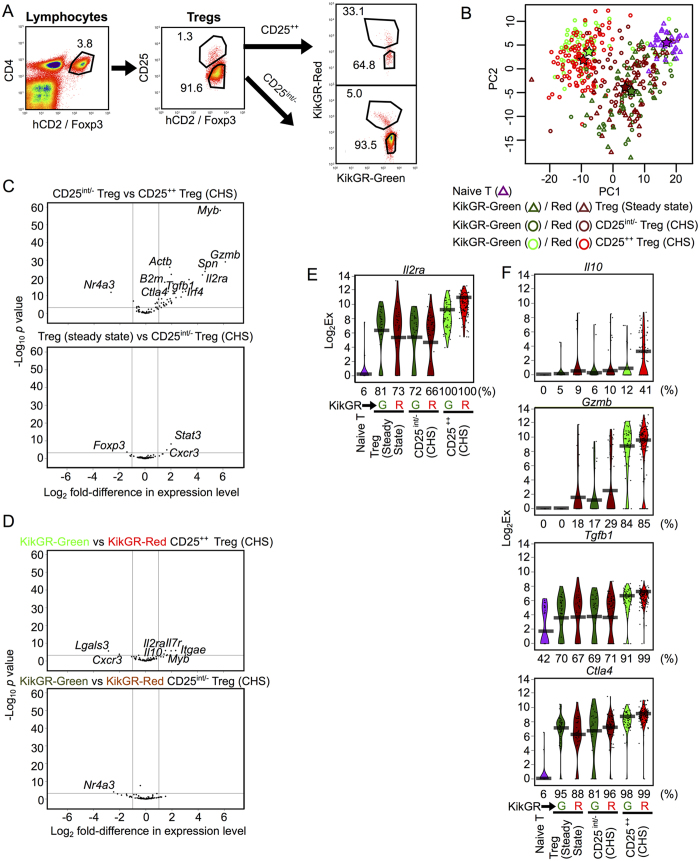
To distinguish LN-resident Tregs from migratory Tregs, we used KikGR/Foxp3hCD2/hCD52 mice. These mice express Kikume Green-Red (KikGR), a photoconvertible fluorescent protein that is irreversibly converted from green (KikGR-Green) to red (KikGR-Red) upon exposure to violet light10,11. These mice also enable detection of Foxp3+ Tregs by surface staining with an anti-human CD2 (hCD2) antibody12. CHS was induced by painting 2,4-dinitro-1-fluoro-benzene (DNFB) onto the abdominal skin of KikGR/Foxp3hCD2/hCD52 mice 5 days after immunization via the dorsal skin (Supplementary Fig. 1A). Photoconversion of the inflamed skin was used to label migratory cells originating from the site of CHS. CD4+ hCD2+ Tregs in the dLN (axillary LN) of CHS mice were sorted into four subpopulations: (1) CD25++ KikGR-Red+ skin-derived migratory Tregs; (2) CD25int/− KikGR-Red+ skin-derived migratory Tregs; (3) CD25++ KikGR-Green+ LN-resident Tregs; and, (4) CD25int/− KikGR-Green+ LN-resident Tregs (Fig. 1A). Lymphocytes from the axillary dLN of non-CHS (steady-state) mice were also isolated as follows: (5) CD25int/− KikGR-Red+ skin-derived migratory Tregs; (6) CD25int/− KikGR-Green+ LN-resident Tregs; and, (7) naive T cells (both KikGR-Red+ and KikGR-Green+). CD25++ Tregs are absent from the dLN in the steady state1.
Figure 1. Highly expressed genes in CD25++ Tregs and migratory Tregs.
(A) Flow cytometry gating strategy for KikGR-Green+ or KikGR-Red+/CD25++ or CD25int/− Tregs isolated from the dLN of CHS mice. Representative data; values indicate the percentage of the parent population. (B) Principal component analysis plot comparing single-cell expression profiles of seven Treg subsets (Naive T, n = 33; KikGR-Green+ Tregs in the steady state, n = 43; KikGR-Red+ Tregs in the steady state, n = 33; KikGR-Green+ CD25int/− Tregs in CHS, n = 36; KikGR-Red+ CD25int/−Tregs in CHS, n = 73; KikGR-Green+ CD25++ Tregs in CHS, n = 57; KikGR-Red+ CD25++ Tregs in CHS: n = 106). PC1, principal component 1; PC2, principal component 2. Stars indicate the mean value for each subset with the same color. (C,D) Volcano plots indicating genes differentially expressed between CD25int/− Tregs and CD25++ Tregs in CHS (C upper), Tregs in steady state and CD25int/−Tregs in CHS (C lower), KikGR-Green+ and KikGR-Red+ CD25++ Tregs in CHS (D upper), and KikGR-Green+ and KikGR-Red+ CD25int/−Tregs in CHS (D lower). Vertical lines indicate a two-fold change. Horizontal lines indicate p = 0.001. Selected genes that had statistically significant differences in expression between subsets are labeled. (E,F) Violin plots showing expression levels of the gene Il2ra (E) and the genes Il10, Gzmb, Tgfb1, and Ctla4 (F) in individual cells from various Treg subsets. Bars indicate average expression levels. Percentages under each plot indicate the proportion of cells that expressed the gene. The data shown in (E,F) are also shown in Supplementary Fig. 2. Figs 1, 2, 3A,B and Supplementary Figs 2, 3 and 4 show scqPCR data of CD25++ and CD25int/− Tregs and naive T cells.
For each of these seven populations, we measured the single-cell expression levels of 85 transcripts encoding activation markers, regulators of Th and Treg differentiation, cytokines, and molecules associated with migration and adhesion (Supplementary Fig. 1A, Supplementary Table 1). This process generated expression data for a total of 381 individual cells in 7 populations after a batch-wise normalization, in which limit of detection values were adjusted in batches, and after exclusion of single cell data with no expression of Actb, B2m, Cd3e or Cd4. Note that the numbers of single-cells analyzed by scqPCR do not reflect the proportion of actual cell numbers of the Treg subsets in vivo. In particular, the absolute numbers of CD25++ or KikGR-Red populations are much smaller than those for the other populations (Fig. 1A).
Visualization of scqPCR data by principal component analysis (PCA) (Fig. 1B) and hierarchical clustering analysis (HCA) (Supplementary Fig. 1B) revealed two distinctive clusters. Specifically, the CD25++ Treg populations expressed many genes at higher levels than the other populations examined (CD25int/− Treg subsets in CHS, Tregs in the steady state and naive T cells). With the exception of Nr4a3, many genes including Myb, Gzmb, Spn, Il2ra, Irf4, Tgfb1, Ctla4 and reference genes such as Actb and B2m displayed elevated expression in CD25++ Tregs (Fig. 1C, upper panel). These results are consistent with the view that CD25++ Tregs are a highly activated population.
On the other hand, only three genes (Foxp3, Stat3, and Cxcr3) were differentially expressed between CD25int/− Tregs in CHS and CD25int/− Tregs in the steady-state (Fig. 1B,C), suggesting that CHS inflammation influences CD25int/− Tregs only minimally. Similarly, only small differences were detected between skin-derived migratory Tregs (KikGR-Red+) and LN-resident Tregs (KikGR-Green+) (Fig. 1B,D). In CHS mice, only seven genes were differently expressed between CD25++ KikGR-Green+ and KikGR-Red+ Tregs, and only one gene (Nr4a3) was differently expressed between CD25int/− KikGR-Green+ and KikGR-Red+ Tregs (Fig. 1D). These results suggest that the differences between migratory and LN-resident Tregs, including the superior inhibitory function of migratory Tregs1, are largely due to the high proportion of CD25++ cells within the migratory Tregs subset (Fig. 1A).
Gzmb and Il10 expression is more typical of activated Treg subsets than Tgfb1 and Ctla4 expression
In addition to highlighting the inter-population differences described above, scqPCR analysis also revealed a heterogeneity of gene expression levels for each gene between individual Tregs (Supplementary Figs 2 and 3). For example, all CD25++ Tregs expressed Il2ra, which encodes CD25, while CD25int/− Tregs displayed heterogeneous expression of Il2ra (Fig. 1E), consistent with our gating strategy (Fig. 1A).
Compared with CD25int/− Tregs, CD25++ Tregs also highly expressed the Treg effector molecules Tgfb1, Ctla4, and Gzmb (Fig. 1C). While Gzmb was expressed in more than 80% of CD25++ Tregs but in less than 30% of CD25int/− Tregs, Tgfb1 and Ctla4 were expressed in greater than 60% and 80% of cells respectively across all of the Treg populations (Fig. 1F). Although Granzyme B (GzmB), the protein encoded by Gzmb, is best known as a cytotoxic molecule released from killer cells, it has also been shown to act as a Treg effector molecule that suppresses immune responses by killing effector cells or antigen presenting cells13,14,15,16.
Il10 expression was also up-regulated in CD25++ migratory Tregs (Fig. 1D). IL-10 encoded by Il10 is an important immunosuppressive cytokine produced by Tregs13,17 that inhibits the CHS response directly9. However, Il10-expressing cells were rare (less than 12% of total cells) in all Treg populations apart from CD25++ migratory Tregs (41% of total cells) (Fig. 1F).
These observations suggest that Gzmb is a key characteristic of all CD25++ Tregs, whereas Il10 expression is limited to migratory CD25++ Tregs.
Gzmb and Il10 are dominantly expressed in Tregs expressing multiple genes related to Treg effector molecules
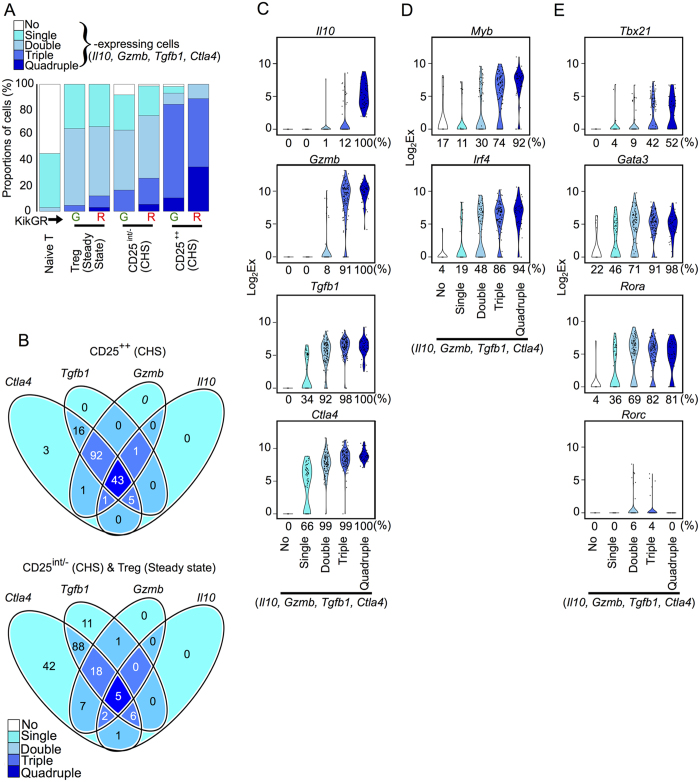
The heterogeneous single-cell expression profiles of genes encoding Treg effector molecules prompted us to categorize individual cells depending on their expression of none, one, two, three, or four of the genes Il10, Gzmb, Tgfb1, and Ctla4 (“single”, “double”, “triple” or “quadruple” expressing cells, respectively). More than 75% of CD25++ Tregs were quadruple- or triple-expressing cells, while more than 75% of CD25int/− Tregs were double- or single-expressing cells (Fig. 2A). Migratory Tregs (KikGR-Red+) included more quadruple-expressing cells than LN-resident Tregs (KikGR-Green+) across all Treg subsets (Fig. 2A). These data are consistent with Tregs migrating from the skin having superior inhibitory activity to LN-resident Tregs, as we have reported previously1.
Figure 2. Gzmb and Il10 are dominantly expressed in Tregs expressing multiple genes associated with Treg effector molecules.
(A) The proportions of individual Tregs or naive T cells co-expressing the genes Il10, Gzmb, Tgfb1, and Ctla4. (B) Venn diagrams showing the numbers of individual cells with co-expression of the genes Il10, Gzmb, Tgfb1 and Ctla4 among CD25++ Tregs in CHS (upper) or CD25int/− Tregs in CHS and Tregs in steady state (lower). (C,D,E) Violin plots showing expression levels of the genes Il10, Gzmb, Tgfb1 and Ctla4 (C), Myb and Irf4 (D), and Tbx21, Gata3, Rora and Rorc genes (E) in individual no-, single-, double-, triple- and quadruple-expressing cells (n = 23, 70, 115, 125 and 48, respectively). Percentages under each plot indicate the proportion of cells that expressed the gene.
Furthermore, the genes Il10, Gzmb, Tgfb1, and Ctla4 were expressed in a hierarchical manner (Fig. 2B). Most double-expressing Tregs expressed Tgfb1 and Ctla4 in both CD25++ and CD25int/− Tregs, while almost all triple-expressing CD25++ Tregs expressed Gzmb, Tgfb1, and Ctla4, but not Il10. Violin plots created by pooling data from the six Treg subsets shown in Fig. 2B highlighted the hierarchical expression of these four genes and the restriction of Il10 expression mainly to quadruple-expressing Tregs (Fig. 2C).
Tregs expressing Gzmb and Il10 are Th1-polarized
To understand how Treg effector molecule expression is regulated, we first compared the expression of the Treg differentiation regulators Myb and Irf4, which were highly up-regulated in CD25++ Tregs (Fig. 1C upper panel). The expression of both Myb and Irf4 on an individual cell was correlated closely with the number of Treg effector molecules expressed by that cell (Fig. 2D), unlike the expression of other Treg differentiation regulators (Supplementary Fig. 4). Myb is also associated with CD4+ T cell development via its target gene Gata318, which is known as a master regulator of CD4+ helper T cell type 2 (Th2) differentiation. Irf4 is also a crucial factor in the differentiation of Th1/2/17 lymphocytes and Tregs19.
These observations motivated us to further investigate the expression profiles of other master regulators of Th differentiation such as Tbx21 (Th1), Gata3 (Th2), and Rora and Rorc (Th17)20,21. Rorc was expressed by only a small number of double or triple-expressing Tregs, whereas Gata3 and Rora were expressed in a large proportion of double-, triple- and quadruple-expressing Tregs (Fig. 2E). Similarly, the proportion of Tbx21-expressing cells was markedly higher in triple- and quadruple-expressing Tregs (42 and 52%, respectively) compared with non-expressing or single-/double-expressing Tregs (0, 4, and 9%). Given that the expression of the respective Th master genes in Tregs causes polarization towards other CD4+ Th subsets22, our Tbx21 data suggest that activation of Gzmb/Il10-expressing Tregs involves polarization towards a Th1 phenotype.
CD43, CCR5, CXCR3 are markers for Gzmb + Tregs
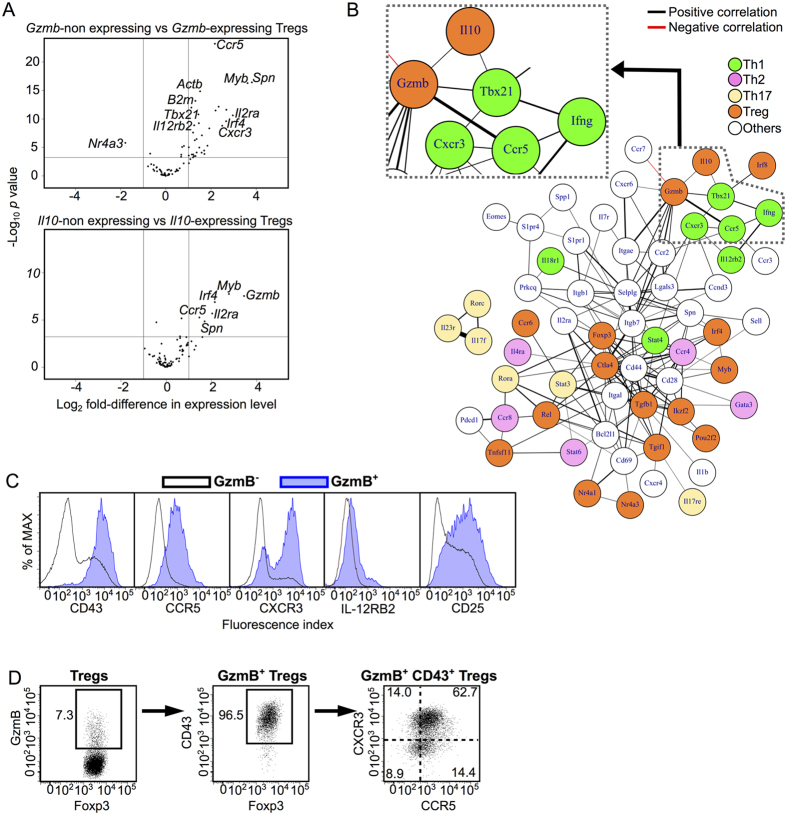
Th1-polarized Gzmb/Il10-expressing Tregs are likely to be the primary population responsible for the immunosuppressive function of CD25++ Tregs. However, since CD25int/− Tregs were present in much greater numbers than CD25++ Tregs (about 70-fold more frequent; Fig. 1A), the CD25int/− Treg population also contains a significant number of Gzmb/Il10-expressing Tregs (Fig. 2A). We thus compared gene expression levels between individual Tregs with and without expression of the genes Gzmb and Il10 (Fig. 3A) in order to determine surface markers other than CD25 that identify Gzmb-/Il10-expressing Tregs. A number of surface molecules including Spn, Ccr5, and Il2ra were commonly and highly expressed in both Gzmb- and Il10-expressing cells, and Gzmb-expressing cells highly expressed the Th1-related genes Cxcr3, Il12rb2, and Tbx21 (Fig. 3A). Furthermore, a subcluster of Th1-related genes including Ccr5 and Tbx21 positively correlated with Gzmb and Il10 in a correlation network derived from CD25int/− scqPCR data (Fig. 3B). This result suggested that it may be possible to use the expression of these genes to distinguish Gzmb/Il10-expressing Tregs from other Tregs.
Figure 3. CD43, CCR5 and CXCR3 are markers for Gzmb+ Tregs.
(A) Volcano plots indicating genes differentially expressed between non-Gzmb-expressing and Gzmb-expressing Tregs (upper) and between non-Il10-expressing and Il10-expressing Tregs (lower) under CHS condition. Vertical lines indicate a two-fold change in expression. Horizontal lines indicate p = 0.001. Selected genes that had statistically significant differences in expression between subsets are labeled. (B) A correlation network constructed using scqPCR data from CD25int/− Tregs in CHS (Spearman’s rank correlation coefficient, ρ > 0.3). Genes are colored according to their function. Line colors indicate positive (black) or negative (red) correlations. Line thickness indicates correlation coefficient (a thicker line indicates a higher correlation coefficient). (C) CD43, CCR5, CXCR3, IL-12RB2 and CD25 protein expression in GzmB+ and GzmB– Treg subsets isolated from the dLN of CHS mice. (D) CD43, CCR5, and CXCR3 protein expression in GzmB+ Treg subsets in the dLN. Flow cytometry data are representative of three independent experiments; values on the plots indicate the percentage of the parent population.
To confirm whether the molecules encoded by these genes are appropriate markers for Gzmb-/Il10-expressing Tregs, we examined cell surface marker expression on intracellular GzmB-stained freshly-isolated Tregs without in vitro stimulation. GzmB+ Tregs were mainly observed in the dLN rather than in the non-dLN of CHS mice (Supplementary Fig. 5A). The molecules CD43 (an activation marker encoded by Spn23), CCR5, CXCR3, IL12RB2, and CD25 were expressed to a greater extent by GzmB+ Tregs than by GzmB− Tregs (Fig. 3C). However, a large proportion of GzmB+ Tregs did not express IL-12RB2, and the majority of both GzmB– and GzmB+ Tregs expressed CD25. Thus, CD43, CCR5 and CXCR3, but not IL-12RB2 or CD25, appear to be good candidate markers for Gzmb-/Il10-expressing Tregs.
More than 95% of GzmB+ Tregs expressed CD43, and the majority of CD43+ GzmB+ Tregs expressed CXCR3 and/or CCR5 (Fig. 3D). Fifty-one percent of CD43+ CCR5+ CXCR3+ Tregs expressed GzmB, whereas no CD43− Tregs expressed GzmB (Supplementary Fig. 5B). Thus, use of the markers CD43, CCR5, and CXCR3 offers a way to detect GzmB+ Tregs without using the marker CD25.
CD43+ CCR5+ Tregs are a major subset of Gzmb-/IL10-expressing Tregs
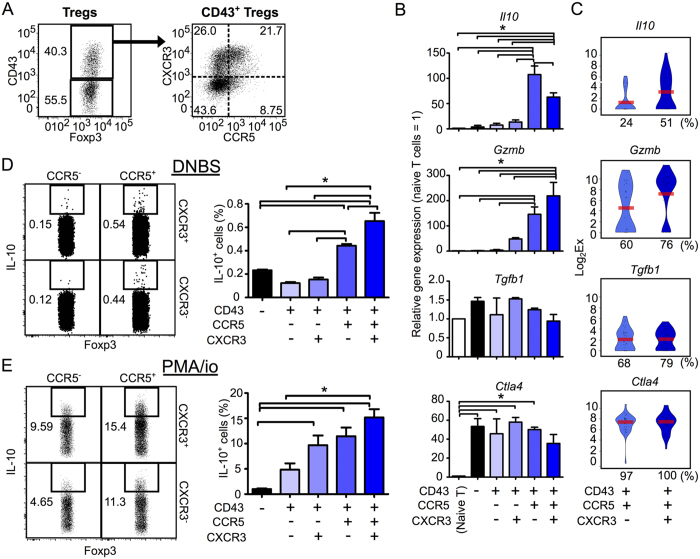
We next analyzed the expression of genes for Treg effector molecules in five Treg subsets defined by the markers CD43−, CD43+ CCR5− CXCR3−, CD43+ CCR5+ CXCR3−, CD43+ CCR5− CXCR3+ and CD43+ CCR5+ CXCR3+ (Fig. 4A). CD43+ CCR5+ (CXCR3+/−) Tregs expressed both the Gzmb and Il10 genes more highly than other Treg subsets (Fig. 4B). Moreover, an extra scqPCR analysis revealed that CD43+ CCR5+ CXCR3+ Tregs expressed Il10 at higher levels and with higher frequency (51%) than CD43+ CCR5+ CXCR3− Tregs (24%) (Fig. 4C) and KikGR-Red+ CD25++ Tregs (41%) (Fig. 1F). The pattern of Gzmb expression was consistent with that of GzmB protein expression (Fig. 4B vs. Supplementary Fig. 5B). Tgfb1 and Ctla4 were expressed at similar levels across each of the five Treg subsets.
Figure 4. CD43+ CCR5+ Tregs are a major subset of Gzmb-/IL10-expressing Tregs.
(A) Flow cytometry gating strategy for five Treg subsets (CD43− and CD43+ CCR5+/− CXCR3+/− Tregs) isolated from the dLN of CHS mice. (B) The population-wide expression levels of the genes Il10, Gzmb, Tgfb1 and Ctla4 genes within CD43− and CD43+ CXCR3+/− CCR5+/− Treg subsets, relative to expression levels in naive T cells (n = 3). (C) Violin plots showing expression levels of the genes Il10, Gzmb, Tgfb1 and Ctla4 genes in individual Tregs in the CD43+ CCR5+ CXCR3− or CXCR3+ subsets (n = 37 each). Bars indicate average expression levels. Percentages under each plot indicate the proportion of cells that expressed the gene. Figures 4C and 5A show scqPCR data for CD43+ CCR5+ CXCR3− or CXCR3+ Tregs. (D,E) The proportions of IL-10-producing cells in Treg subsets (n = 3) stimulated with DNBS (D) or PMA/ionomycin (E). Flow cytometry dot plots indicate IL-10 expression in the CD43+ CXCR3+/− CCR5+/− Treg subsets. Flow cytometry data are representative of three independent experiments; values on the plots indicate the percentage of the parent population. Data in bar graphs represent means ± SEM. Statistical comparisons were performed by one-way ANOVA with Tukey’s post-hoc test (*p < 0.05).
The IL-10 protein producing ability of CD43+ CCR5+ Tregs compared to CD43− and CD43+ CCR5− Tregs was further confirmed by antigen-specific stimulation in vitro using 2,4-dinitrobenzene sulfonic acid (DNBS, which has the same antigenicity as DNFB) (Fig. 4D) and phorbol myristate acetate (PMA) / ionomycin stimulation (Fig. 4E).
Within the overall Treg population in the dLN, the proportion of CD43+ CCR5+ Tregs (Fig. 4A; 12.3% = 40.3% × (21.7% + 8.75%)) was almost 10-times larger than the proportion of CD25++ Tregs (Fig. 1A; 1.3%). These results indicate that CD43+ CCR5+ Tregs are a major subset of the Gzmb-/Il10-expressing Treg population, while CD25++ Tregs represent only a minor subset. Thus, although rare, CD43+ CCR5+ Tregs are likely to be the primary Treg subset responsible for CHS inhibition.
CD43+ CCR5+ CXCR3− Tregs are preferentially retained in inflamed skin
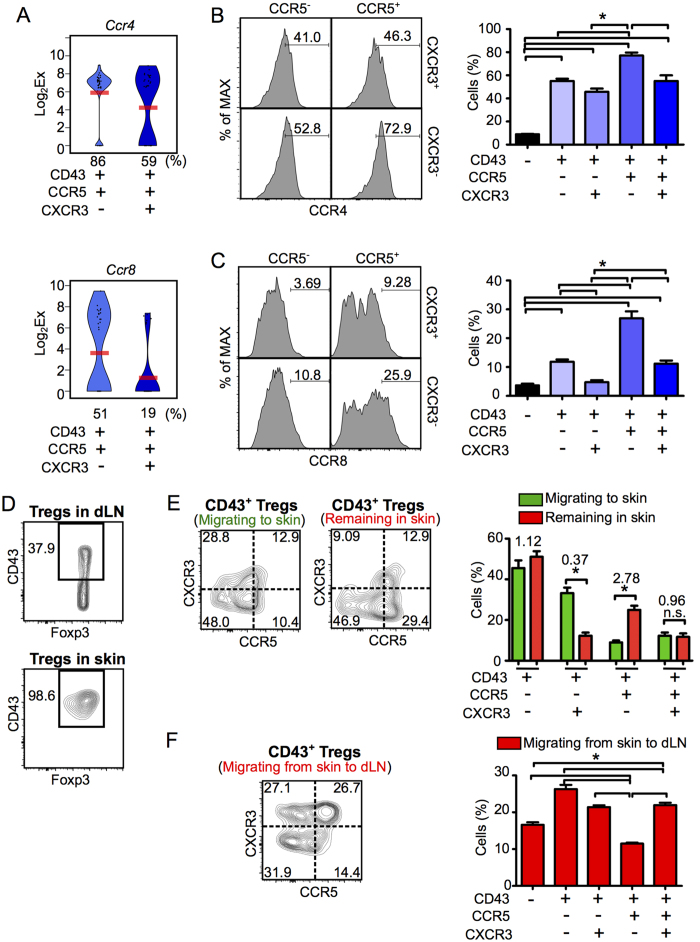
Single-cell expression profiles of two Gzmb-/Il10-expressing Treg populations (CD43+ CCR5+ CXCR3− and CD43+ CCR5+ CXCR3+ Tregs) revealed differential expression of Tbx21, Ccr5 and Cxcr3 (data not shown) and the skin-tropic chemokine receptors Ccr4 and Ccr824,25 (Fig. 5A). Moreover, flow cytometric analysis showed that both CCR4 and CCR8 expression was highest in CD43+ CCR5+ CXCR3− Tregs (Fig. 5B,C). These results suggest that movement between the dLN and inflamed skin differs between Treg subsets, and that the CD43+ CCR5+ CXCR3− Treg subset is most highly retained in inflamed skin. To further investigate the migration of Treg subsets, we photoconverted cells in the inflamed skin tissue and then measured the influx of new Tregs to the skin (KikGR-Green+ cells) and the retention of Tregs in the skin (KikGR-Red+ cells) (Supplementary Fig. 6A).
Figure 5. CD43+ CCR5+ CXCR3− Tregs are preferentially retained in inflammatory skin.
(A) Violin plots showing expression levels of the genes Ccr4 (upper panel) and Ccr8 (lower panel) genes in individual Tregs within the CD43+ CCR5+ CXCR3− or CXCR3+ subsets (n = 37 each). Bars indicate average expression levels. Percentages under each plot indicate the proportion of cells that expressed the gene. (B,C) The proportions of CCR4+ (B) and CCR8+ cells (C) in each Treg subset (n = 3). Flow cytometry histograms (left) indicate CCR4 and CCR8 expression in the CD43+ CXCR3+/− CCR5+/− Treg subsets. (D) CD43 expression in Tregs isolated from the dLN and the skin of CHS mice (n = 3). (E,F) KikGR/Foxp3hCD2/hCD52 mice were treated as shown in Supplementary Fig. 6, and flow cytometric analyses were performed on day 3 following DNFB challenge. Proportions of CD43– and CD43+ CXCR3+/– CCR5+/– Treg subsets in Tregs migrating from the circulation to the skin (KikGR-Green+) and remaining in the skin (KikGR-Red+) (E; n = 9), and in Tregs migrating from skin to dLN (F; n = 6). Values above the bars in the graph in E indicate ratios (for each subset) of the proportion of KikGR-Red+ Tregs remaining in skin to the proportion of KikGR-Green+ Tregs migrating to skin. Flow cytometry data are representative of three independent experiments; values on the plots indicate the percentage of the parent population. Data in bar graphs represent means ± SEM. Statistical comparisons were performed by one-way ANOVA with Tukey’s post-hoc test (*p < 0.05).
Irrespective of their migratory status, CD43− Tregs in CHS-induced skin were extremely scarce (Fig. 5D), suggesting that the CD43− subset does not migrate from the dLN to inflamed skin, unless they differentiate into CD43+ Tregs during migration to the skin. CD43+ KikGR-Green+ Tregs in the skin (Fig. 5E left contour plot) contained the four Treg subsets present within CD43+ Tregs in the dLN (Fig. 4A), suggesting that CD43+ Tregs migrate from the dLN to the site of inflammation. The proportion of CD43+ CCR5+ CXCR3+ Tregs was equivalent within the population of Tregs retained in the skin and the population of Tregs newly migrating to the skin (Fig. 5E). However, the proportion of CD43+ CCR5+ CXCR3− Tregs (which highly expressed both CCR4 and CCR8) was almost three-fold higher in the population retained in the skin (KikGR-Red+; Fig. 5E right contour plot) than in the population that had newly migrated to the skin (KikGR-Green+; Fig. 5E left contour plot). Moreover, the proportion of CD43+ CCR5+ CXCR3− Tregs within KikGR-Red+ Tregs migrating from photoconverted skin to the dLN was the lowest of the four CD43+ subsets (Supplementary Fig. 6B and Fig. 5F). These data suggest that CD43+ CCR5+ CXCR3− Tregs are preferentially retained at the site of inflammation.
To exclude the possibility that the high proportion of CD43+ CCR5+ CXCR3− cells retained in the skin was due to in situ proliferation, we used FucciG1-#639 FucciS/G2/M-#474 double transgenic mice (Fucci-Tg mice) (Supplementary Fig. 6C). In Fucci-Tg mice, cells in the S/G2/M phase of the cell cycle appear as Fucci green+ Fucci orange− cells26. In the skin, only a small proportion of cells in each Treg subset were in S/G2/M phase, while in the dLN a much greater proportion of Tregs were proliferating, particularly CD43+ CCR5+ and CD43+ CXCR3+ Tregs.
CD43+ CCR5+ CXCR3− Tregs, but not CD43+ CCR5+ CXCR3+ Tregs, exhibit immunosuppressive activity in inflamed skin in vivo
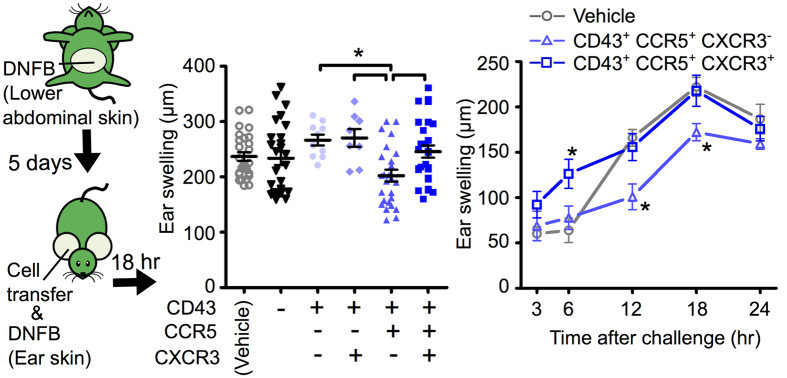
To determine the immunosuppressive activity of the newly identified Treg subsets in vivo, Tregs isolated from the dLN of CHS-induced mice were adoptively transferred into ear skin immediately prior to CHS induction. Of the Gzmb-/Il10-expressing Treg subsets that were transferred, CD43+ CCR5+ CXCR3− Tregs but not CD43+ CCR5+ CXCR3+ Tregs suppressed ear swelling (Fig. 6). This result suggests that high expression of Gzmb and Il10 with co-expression of Tgfb1 and Ctla4 is insufficient, and that retention in inflamed skin is required, for Tregs to suppress the CHS response.
Figure 6. CD43+ CCR5+ CXCR3− Tregs, but not CD43+ CCR5+ CXCR3+ Tregs, exhibit in vivo immunosuppressive activity in inflammatory skin in vivo.
Ear swelling in B6 WT CHS mice at 18 h (left) and over a 24 h time course (right). Mice were adoptively transferred with a no-cell vehicle or CD43− and CD43+ CXCR3+/− CCR5+/− Treg subsets immediately prior to CHS induction. Data represent means ± SEM. Statistical comparisons were performed by one-way ANOVA with Tukey’s post-hoc test (left) or two-way ANOVA with Bonferroni post-test (right, compared with vehicle control) (* p < 0.05). Illustration was created by R.I. using Power Point software.
Discussion
The Treg subsets that regulate the CHS response remain poorly defined. In the present study, using KikGR mice, scqPCR and flow cytometry, we identified a rare subset of CD43+ CCR5+ CXCR3− Tregs that co-expressed Gzmb and Il10 genes at high levels. These Tregs also expressed the skin-tropic chemokine receptors CCR4 and CCR8, were preferentially retained in inflamed skin, and were equipped with in vivo immunosuppressive function. We also characterized the degree to which different Treg subsets infiltrate and are retained in the skin and express the genes Gzmb and Il10 (Table 1). Even if only present in small numbers, highly activated Tregs that co-express Gzmb and Il10 and that have the capacity to remain in inflamed tissue are likely to be clinically relevant due to the role of these molecules in the control of excessive immune responses27,28.
Table 1. Characterization of the five Treg subsets defined by CD43, CCR5, and CXCR3.
| CD43 | CCR5 | CXCR3 | % in dLNa | Gzmb expression | Il10 expression | Skin infiltration | Skin remaining | Inhibition of CHS response |
|---|---|---|---|---|---|---|---|---|
| − | 54.5 ± 1.1 | Low | Low | − | − | − | ||
| + | − | − | 19.9 ± 1.0 | Low | Low | + | Low | − |
| + | 9.8 ± 0.4 | High | Low | + | Low | − | ||
| + | − | 4.6 ± 0.5 | High | High | + | High | + | |
| + | 8.4 ± 0.8 | High | High | + | Low | − | ||
aProportion of each Treg subset among total Tregs isolated from the dLN of CHS mice (mean ± SEM). The data is also presented in Fig. 4A.
The flexibility and plasticity of CD4+ Th cell types are thought to be regulated by the expression of the Th1/2/17 differentiation-related genes Tbx21, Gata3, Rora and Rorc22. Polarization may play a role in the induction of Treg inhibitory function. For example, GzmB expression in CD4+ T cells is induced by antigen stimulation together with exposure to a Th1-polarized environment29,30, and Il10 gene expression is downregulated in T-bet-deficient Tregs during Th1-type inflammation31. Our scqPCR data also revealed Th1 polarization in highly activated Tregs with Il10 and Gzmb expression. Furthermore, Th1 differentiation is induced by TCR signaling of proper intensity without relation to the extrinsic environment32. The rapid cell cycling that we observed in CD43+ CCR5+ and CD43+ CXCR3+ Tregs in the dLN (Fig. 6D) and the differential TCR Vβ repertoire between CD43+ CCR5+ CXCR3− Tregs and CD43− Tregs (data not shown) suggested that the Tregs in our experiments received proper TCR stimulation. Thus, it seems likely that Tbx21 expression induced by proper TCR stimulation could trigger Gzmb expression and subsequent Il10 expression.
Although well-known as an effector molecule of cytotoxic T lymphocytes, in Tregs GzmB also plays a role as an immunosuppressive molecule that works by killing effector cells or antigen presenting cells13,14,15,16. GzmB also promotes transmigration of CD8+ T cells in postcapillary venules by cleaving basement membranes33. On the other hand, injection of IL-10 into the ear inhibited ear swelling in a model of CHS9, suggesting that IL-10 secreted from skin-infiltrating Tregs or other immune cells can directly inhibit inflammation. Accordingly, co-expression of Gzmb and Il10 by CD43+ CCR5+ Tregs is likely to result in synergistic effects on migration and immunosuppression in the skin.
Hierarchical and rare expression of Gzmb and Il10 in Tregs could limit the numbers of Tregs with hyper-immunosuppressive activity and regulate their tissue-homing capacity, thus representing a mechanism to limit “global” excessive immunosuppression. Gzmb expression was positively correlated with the expression of Th1-asocciated trafficking receptors (Ccr5, Cxcr6 and Ccr2) and negatively did with the expression of LN homing receptor, Ccr7 (Fig. 3B). This finding suggests that these migration-related genes could influence tissue tropism of Gzmb- (and Il10-) expressing Tregs. Moreover, there were a number of other correlations between the expression of genes related to Treg effector function and migration, such as Ctla4 and Itgb7 or Tgfb1 and Ccr4. This possibility is consistent with the report that CTLA-4 and IL-10 produced by Tregs mainly act in lymphoid and peripheral tissues, respectively34. In our CHS model, retention in the skin and the co-expression of Gzmb and Il10 were sufficient to elicit “local” immunosuppression of the CHS response.
CCL3, CCL4 and CCL5 (CCR5 ligands) and CXCL9 and CXCL10 (CXCR3 ligands) are expressed in the skin following induction of CHS35,36. CCR5 expression by Tregs is required for the migration of Tregs to Leishmania major-infected skin and subsequent downregulation of the effector immune response37. In addition, T-bet+ Tregs migrate to sites of Th1-type inflammation by a CXCR3-dependent mechanism21,38. CD43 also mediates the migration of Th1 cells to inflammatory skin39. Accordingly, it is consistent with these reports that that CD43+ CCR5+ and CD43+ CXCR3+ Tregs migrate from the dLN to the skin in CHS. Moreover, high expression of CCR4 and CCR8 in CD43+ CCR5+ CXCR3− Tregs might enable retention within the inflamed skin. This hypothesis is supported by the findings that skin-tropic Tregs express CCR4 and CCR824,25 and that the ligands for CCR4 and CCR8 are expressed in the inflamed skin35,36,40. It remains to be elucidated which activation pathways induce the expression of CD43, CCR5, CXCR3, CCR4, and CCR8 in Tregs.
In this study, we used a combination of scqPCR high-dimensional gene expression profiling data with spatiotemporal information about cellular movement from KikGR mice to elucidate the role and characteristics of Treg subsets in CHS. An understanding of the hierarchical expression of effector molecules and migratory status of T-cell subsets will support the development of novel strategies for immune regulation by T cell-mediated therapy.
Methods
Mice
To generate KikGR/Foxp3hCD2/hCD52 and Fucci/Foxp3 hCD2/hCD52 mice, KikGR-transgenic mice and Fucci-Tg mice10,26 were crossed with Foxp3hCD2/hCD52 mice (kindly provided by Dr. Shohei Hori, IMS, RIKEN, Japan), in which Foxp3+ cells express human CD2 and human CD52 fusion protein on the cell surface12. Mice were housed in specific pathogen-free facilities at Kyoto University or Osaka Ohtani University. All animal experiments were conducted in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan), and all protocols were approved by the Institutional Animal Care and Use Committee of Kyoto University Faculty of Medicine and the Animal Research Committee of Osaka Ohtani University.
Photoconversion
Skin photoconversion was performed on 1.5, 2 or 2.5 days following DNFB challenge, as described previously1,41. Briefly, KikGR/Foxp3hCD2/hCD52 mice were anesthetized and shaved skin was subjected to two 2.5 min exposures to violet light (436 nm, 100 mW/cm2) using SP500 spot UV curing equipment with a 436 nm bandpass filter (USHIO). Based on assessment of concanavalin A-induced T cell proliferation in vitro and Il1b and Tnfa mRNA expression in the skin in vivo, this photoconversion protocol does not induce significant inflammation in the skin1,42.
Flow cytometry
Antibody-stained samples (see Supplementary Methods) were analyzed using a LSRFortessa (BD Biosciences) or SP6800 (SONY) flow cytometer. Data were analyzed using FlowJo software (Tree Star). Tregs were identified as CD4+ hCD2+ cells falling within a lymphocyte forward scatter and side scatter gate.
To detect IL-10-producing cells, cells from the dLN were incubated at 37° C for 6 h with DNBS (50 μg/mL, Alfa Aesar) or PMA and ionomycin (50 ng/mL and 1 μg/mL), with the addition of brefeldin A (5 μg/mL, BioLegend) for the last 4 h. Intracellular staining for GzmB and IL-10 was performed using Fixation Buffer and 10× Permeabilization Wash Buffer (BioLegend), as described in the manufacturer’s protocol.
Measurement of single-cell gene expression
The experimental scheme for the measurement of single-cell gene expression is depicted in Supplementary Fig. 1 and described in Supplementary Methods. Briefly, the isolation of individual Tregs, cell lysis, RT-PCR, and pre-amplification specific for 85 genes were performed using a C1 Single-Cell Auto Prep system (Fluidigm) with a Single Cell-to-CT kit (Life Technologies) and a C1 Single-Cell AutoPrep Reagent kit (Fluidigm), as described in the manufacturer’s protocol. scqPCR was performed using 48.48 or 96.96 Dynamic Arrays (Fluidigm) with SsoFast EvaGreen Supermix with Low ROX (Bio-Rad Laboratories) and a BioMark system (Fluidigm). The primers used to amplify the 85 genes in the C1 system were designed and synthesized by Fluidigm and Life technologies (see Supplementary Methods).
Adoptive cell transfer experiments
Donor cells were isolated from Foxp3hCD2/hCD52 mice in which CHS had been induced in the upper abdominal skin as described above. CD43−, CD43+ CCR5– CXCR3−, CD43+ CCR5– CXCR3+, CD43+ CCR5+ CXCR3–, and CD43+ CCR5+ CXCR3+ Tregs were sorted from the dLNs (axillary LNs) using MoFlo Astrios (Beckman Coulter), FACSAria III (BD Biosciences) or SH800 (SONY) flow cytometer. B6 WT recipient mice were sensitized with 25 μL of 0.6% DNFB to the abdominal skin and challenged with 20 μL of 0.36% DNFB to each ear. Immediately prior to challenge, 4,000 cells from each sorted Treg subset in 20 μL PBS were injected subcutaneously into the ventral surface of the ears of recipient mice. Ear thickness was measured using a micrometer (TECLOCK) before and after challenge. The extent of ear swelling was expressed as the change in ear thickness before and after challenge.
Statistics
The one-way ANOVA with Tukey’s post-hoc test, two-way ANOVA with Bonferroni post-test and Wilcoxon’s matched pair test were performed using GraphPad Prism version 5.0 (GraphPad Software). Data in bar graphs represent means ± SEM. P-values of less than 0.05 were considered to be statistically significant.
Additional Information
How to cite this article: Ikebuchi, R. et al. A rare subset of skin-tropic regulatory T cells expressing Il10/Gzmb inhibits the cutaneous immune response. Sci. Rep. 6, 35002; doi: 10.1038/srep35002 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by JSPS Grants-in-Aid for Scientific Research in Innovative Areas “Analysis and Synthesis of Multidimensional Immune Organ Network” (JP24111007); JSPS Grants-in-Aid for Scientific Research (C) (JP22590442); Special Coordination Funds for Promoting Science and Technology of the Japanese Government; Astellas Pharma Inc. through the Formation of Innovation Centers for the Fusion of Advanced Technologies Program; JSPS Grant-in-Aid for Young Scientists (B) (JP25870396); JSPS Grant-in-Aid for Young Scientists (B) (JP15K19687); and by grants from the Sumitomo Science Foundation. Materials provided by Dr. Shohei Hori from the RIKEN Research Center were of great assistance in this study. We would like to thank Dr. Naganari Ohkura (Osaka University) for providing useful advice, and Dr. Shiki Takamura (Kinki University) for technical guidance.
Footnotes
Author Contributions R.I., S.T and M.T. designed experiments. R.I. and M.T. performed experiments. T.H. supervised adoptive cell transfer and skin cell flow cytometric experiments. R.I., S.T. and A.V. performed statistical analysis of scqPCR data. R.I., S.T., A.V., T.H., Y.N. and M.T. analyzed data. R.I., S.T., A.V., F.H.W.S., T.W. and M.T. wrote the manuscript.
References
- Tomura M. et al. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J. Clin. Invest. 120, 883–893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum M. D. et al. Response to self antigen imprints regulatory memory in tissues. Nature 480, 538–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P. K., Gierahn T. M., Roederer M. & Love J. C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 15, 128–135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham C. E. & Morrison S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G. et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C. A. et al. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2, e159 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenio J. et al. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat. Immunol. 15, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatz L. & Roychoudhuri R. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc. Natl. Acad. Sci. USA 108, 5724–5729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Mckenzie R. C. & Sauder D. N. Interleukin-10 inhibits the elicitation phase of allergic contact hypersensitivity. J. Invest. Dermatol. 103, 811–814 (1994). [DOI] [PubMed] [Google Scholar]
- Tomura M. et al. Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci. Rep. 4, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Karasawa S., Shimizu H., Nukina N. & Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 6, 233–238 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N. et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. USA 106, 1903–1908 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach E. M. Mechanisms of Foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009). [DOI] [PubMed] [Google Scholar]
- Cao X. et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 27, 635–646 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao D.-M., Thornton A. M., DiPaolo R. J. & Shevach E. M. Activated CD4+ CD25+ T cells selectively kill B lymphocytes. Blood 107, 3925–3932 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonnas A. et al. Foxp3+ T cells induce perforin-dependent dendritic cell death in tumor-draining lymph nodes. Immunity 32, 266–278 (2010). [DOI] [PubMed] [Google Scholar]
- Tran D. Q. TGF-β : the sword. the wand. and the shield of FOXP3+ regulatory T cells. J. Mol. Cell Biol. 4, 29–37 (2012). [DOI] [PubMed] [Google Scholar]
- Maurice D., Hooper J., Lang G. & Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 26, 3629–3640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M. & Lohoff M. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44, 1886–1895 (2014). [DOI] [PubMed] [Google Scholar]
- Dong S. et al. Multiparameter single-cell profiling of human CD4+ FOXP3+ regulatory T-cell populations in homeostatic conditions and during graft-versus-host disease. Blood 122, 1802–1812 (2013). [DOI] [PubMed] [Google Scholar]
- Koch M. A. et al. T-bet+ Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 37, 501–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S., Takahashi H., Kanno Y. & O’Shea J. J. Helper T cell diversity and plasticity. Curr. Opin. Immunol. 24, 297–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R. B., Meyerholz D. K. & Varga S. M. Foxp3+ CD4 regulatory T cells limit pulmonary immunopathology by modulating the CD8 T cell response during respiratory syncytial virus infection. J. Immunol. 185, 2382–2392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. A. & Luster A. D. T cell homing to epithelial barriers in allergic disease. Nat. Med. 18, 705–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iellem A. et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 194, 847–53 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomura M. et al. Contrasting quiescent G0 phase with mitotic cell cycling in the mouse immune system. PLoS One 8, e73801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebbermann J. et al. Regulatory T cells expressing granzyme B play a critical role in controlling lung inflammation during acute viral infection. Mucosal Immunol. 5, 161–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C., Mauze S., Leach M. W., Coffman R. L. & Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J. Exp. Med. 190, 995–1004 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher L. H., Townsend M. J., Sullivan B. M. & Lord G. M. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 4, 900–911 (2004). [DOI] [PubMed] [Google Scholar]
- Lazarevic V. & Glimcher L. H. T-bet in disease. Nat. Immunol. 12, 597–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M. A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo N. J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash M. D. et al. Granzyme B promotes cytotoxic lymphocyte transmigration via basement membrane remodeling. Immunity 41, 960–972 (2014). [DOI] [PubMed] [Google Scholar]
- Campbell D. J. & Koch M. A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebeler M. et al. Differential and sequential expression of multiple chemokines during elicitation of allergic contact hypersensitivity. Am. J. Pathol. 158, 431–440 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui G. et al. Effect of a topical steroid on gene expressions for chemokines in mice with contact hypersensitivity. Int. Immunopharmacol. 4, 57–69 (2004). [DOI] [PubMed] [Google Scholar]
- Yurchenko E. et al. CCR5-dependent homing of naturally occurring CD4+ regulatory T cells to sites of Leishmania major infection favors pathogen persistence. J. Exp. Med. 203, 2451–2460 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. O. H. et al. The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity 37, 511–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M. et al. CD43 collaborates with P-selectin glycoprotein ligand-1 to mediate E-selectin-dependent T cell migration into inflamed skin. J. Immunol. 178, 2499–2506 (2007). [DOI] [PubMed] [Google Scholar]
- Islam S. A. et al. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ TH2 cells. Nat. Immunol. 12, 167–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomura M., Itoh K., Kanagawa O. & Alerts E. Naive CD4+ T lymphocytes circulate through lymphoid organs to interact with endogenous antigens and upregulate their function. J. Immunol. 184, 4646–4653 (2010). [DOI] [PubMed] [Google Scholar]
- Tomura M. et al. Monitoring cellular movement in vivo with photoconvertible fluorescence protein ‘Kaede’ transgenic mice. Proc. Natl. Acad. Sci. USA 105, 10871–10876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.