Abstract
The RNA genome of influenza virus is transcribed and replicated by the viral RNA polymerase complex in the cell nucleus. We have generated green fluorescent protein (GFP)-tagged polymerase subunits to study the assembly of the polymerase complex. Our results show that individually expressed polymerase basic protein 1 (PB1) and polymerase acidic protein (PA) subunits were distributed in both the cytoplasm and the nucleus, while the polymerase basic protein 2 (PB2) subunit accumulated in the nucleus. Although it has been reported that PB1 alone accumulates in the nucleus, we demonstrate that PB1 requires the coexpression of PA for efficient nuclear accumulation. Our results support a model which proposes that PB1 and PA are transported into the nucleus as a complex.
The RNA-dependent RNA polymerase complex of influenza A virus is responsible for the transcription (viral RNA [vRNA]→mRNA) and replication (vRNA→cRNA→vRNA) of the eight segments of the negative single-stranded vRNA genome. It is composed of three subunits, polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA) (11, 19, 28). The PB1 subunit plays a central role in the catalytic activities of the RNA polymerase. It contains the conserved motifs characteristic of RNA-dependent RNA polymerases and is directly involved in RNA chain elongation (3, 6). It binds to the promoter sequences of vRNA and cRNA and, depending on its interaction with RNA, performs the endonucleolytic cleavage of capped RNA (21). The PB2 subunit is responsible for recognition and binding of the cap structure of host mRNAs (10, 21). The role of the PA subunit in the transcription and replication of vRNA is less well established. PA can induce generalized proteolysis of both viral and host proteins; however, the biological significance of this activity for viral replication remains obscure (25, 33, 38). More recently, PA was implicated in the endonuclease cleavage of capped RNA primers and as an elongation factor during RNA synthesis (12, 13). It was also reported that PA enhances the 5′-end binding activity of PB1 (20).
The protein-protein interactions among the individual polymerase subunits have been mapped by using a variety of techniques. The PB1 subunit forms the core of the polymerase complex (9). PB1 interacts through its N-terminal region with the C-terminal region of PA, while the C-terminal region of PB1 is involved in an interaction with the N-terminal region of PB2 (14, 31, 34, 41, 43). No direct interaction between PB2 and PA has been demonstrated, but a low-resolution three-dimensional structural model of a recombinant influenza virus RNP generated by electron microscopy suggests that there are extensive contacts among the three polymerase subunits (2).
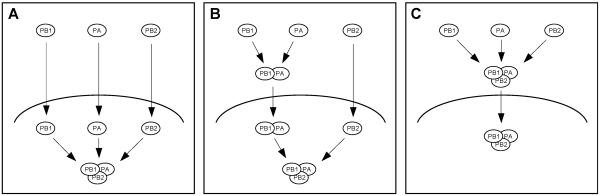
Both the transcription and the replication of vRNA take place in the nucleus of infected cells (15, 17). Therefore, the RNA polymerase has to be transported into the nucleus. Indeed, nuclear localization signals (NLSs) have been identified in all three polymerase subunits, and it has been demonstrated that individually expressed PB1, PB2, and PA can enter the nucleus (1, 18, 24, 27, 29, 30, 39). Although these results could imply that the polymerase complex is assembled in the nucleus (see Fig. 6, model A), the possibility remains that the polymerase assembles in the cytoplasm and enters the nucleus as a heterotrimeric complex (see Fig. 6, model C). Since the RNA polymerase represents a potential target for interfering with viral replication, understanding the assembly of the polymerase complex is of considerable interest.
FIG. 6.
Models for the transport and assembly of the influenza virus RNA polymerase. (A) Subunits PB1, PB2, and PA are transported into the nucleus individually and form a complex in the nucleus. (B) PB1 and PA interact in the cytoplasm and are transported into the nucleus as a dimer. PB2 enters the nucleus as a monomer and binds to PB1-PA in the nucleus. (C) PB1, PB2, and PA interact in the cytoplasm and are transported into the nucleus as a trimeric complex.
In order to study the intracellular localization of the RNA polymerase subunits and the process of polymerase assembly, we generated green fluorescent protein (GFP)-polymerase fusion genes. GFP from the jellyfish Aequorea victoria represents a versatile tool for studying the trafficking of fusion proteins in situ (42). Using a PB1-GFP fusion, we found that individually expressed PB1 does not accumulate efficiently in the cell nucleus, as previously reported (1, 27, 39), but requires the coexpression of PA for the induction of efficient nuclear accumulation.
MATERIALS AND METHODS
Plasmids.
The pcDNA-PB1, pcDNA-PB2, and pcDNA-PA protein expression plasmids for the three polymerase subunits of influenza A/WSN/33 and A/PR/8/34 (Cambridge variant) viruses have been described elsewhere (12, 40). To generate plasmids expressing GFP-tagged polymerase subunits, cloning vectors pcDNA-GFP(EN) and pcDNA-GFP(NX) were constructed. To generate pcDNA-GFP(EN), the GFP open reading frame (ORF) (GFPmut2) (8), without a stop codon, was inserted into the EcoRI/NotI sites of pcDNA3A (12). pcDNA-GFP(NX) was constructed by inserting GFP into the NotI/XbaI sites of pcDNA3A. pcDNA-GFP-PB1, pcDNA-GFP-PB2, and pcDNA-GFP-PA were constructed by inserting the A/WSN/33-derived PB1, PB2, and PA ORFs into the XhoI/XbaI, NotI/XhoI, and NotI/XbaI sites of pcDNA-GFP(EN), respectively. pcDNA-PB1-GFP, pcDNA-PB2-GFP, and pcDNA-PA-GFP were constructed by inserting the A/WSN/33-derived PB1, PB2, and PA ORFs into the HindIII/NotI, BamHI/NotI, and EcoRI/NotI sites of pcDNA-GFP(NX), respectively. The GFP and polymerase ORFs were linked with an Ala-Ala-Ala linker in all constructs, with the exception of pcDNA-GFP-PB1, where an Ala-Ala-Ala-Arg-Ala linker was used.
To generate plasmids pcDNA-PB1-FLAG, pcDNA-PB2-FLAG, and pcDNA-PA-FLAG, plasmids pcDNA-PB1-GFP, pcDNA-PB2-GFP, and pcDNA-PA-GFP, respectively, were digested with restriction enzymes NotI and XbaI to remove the GFP ORF. Two self-annealed deoxyoligonucleotides (5′-GGCCGCCGACTACAAAGACGATGACGACAAGTAGT-3′ and 5′-CTAGACTACTTGTCGTCATCGTCTTTGTAGTCGGC-3′), coding for the FLAG epitope, were inserted into the NotI/XbaI sites. pcDNA-PB1-TAP was generated by inserting the tandem affinity purification (TAP) sequence (37) into the NotI/XbaI sites of pcDNA-PB1-GFP after removal of the GFP ORF with the same restriction enzymes. Plasmids pcDNA-PB1 NLS1, pcDNA-PB1 NLS2, pcDNA-PB1 NLS3, and pcDNA-PB1 NLS12 were generated by site-directed mutagenesis. Plasmids pcDNA-PA(Δ1-256) and pcDNA-PA(Δ694-716), encoding PAs with N-terminal and C-terminal deletions, respectively, were constructed by PCR-based mutagenesis. All sequences generated by PCR were fully sequenced. Sequences of the mutagenic primers are available from us upon request.
RNA isolation and analysis of vRNA, mRNA, and cRNA by primer extension assays.
DNA transfection was performed with 293T cells in suspension in 35-mm dishes (about 106 cells) by using 6 μl of Lipofectamine 2000 (Invitrogen) transfection reagent and 1 μg each of plasmids pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-CAT-RT in 2 ml of minimal essential medium (MEM) containing 10% fetal calf serum (FCS). Cells were harvested 48 h posttransfection, and total RNA was isolated by using TRIzol reagent (Invitrogen). Primer extension assays were performed as described previously (12). Briefly, reverse transcription was performed by using SuperScript II reverse transcriptase (Invitrogen) and two chloramphenicol acetyltransferase (CAT)-specific primers to detect vRNA, mRNA, and cRNA in the same reverse transcription reaction. Transcription products were analyzed on 6% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA buffer and were detected by autoradiography. The expected sizes of the transcription products were 158 nucleotides (nt) (vRNA), 98 to 106 nt (mRNA), and 89 nt (cRNA).
Immunofluorescence.
Vero or COS-7 cells grown on 13-mm coverslips in 24-well plates were transfected as approximately 50% confluent monolayers with 1 μg of expression plasmid(s) by using 2 μl of Lipofectamine 2000 transfection reagent according to the manufacturer's instructions. After 6 h, the transfection mixture was replaced with MEM containing 10% FCS, and the cells were incubated for 18 h. For detecting GFP, the cells were fixed with 4% paraformaldehyde in 250 mM HEPES (pH 7.5). For indirect immunofluorescence, following fixation, the cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 10 min at room temperature, blocked, and incubated with the corresponding antibodies in phosphate-buffered saline containing 1% bovine serum albumin and 0.2% gelatine (pH 8.0) (36) for 1 h at room temperature. Nontagged polymerase subunits were detected by indirect immunofluorescence with the following primary antibodies: rabbit polyclonal anti-PB1 and anti-PA (T. Toyoda, Kurume University, Kurume, Japan) and mouse monoclonal anti-PB2 (4). Cy3-conjugated anti-rabbit and anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Antibodies were used at a 1/500 dilution. Coverslips were mounted in Mowiol containing 4′,6′-diamidino-2-phenylindole (DAPI; Sigma). Cells were viewed by using a Zeiss microscope with a Zeiss ×63/1.25 oil immersion objective, and the images were processed by using Adobe Photoshop 7.0.
Cell fractionation.
Vero cells in 35-mm-diameter dishes (about 60% confluent) were transfected with pcDNA-PB1 or pcDNA-PB1, pcDNA-PB2, and pcDNA-PA by using Lipofectamine 2000 transfection reagent according to the manufacturer's instructions. At about 24 h posttransfection, the cells were released by trypsin treatment, resuspended in MEM containing 10% FCS, and pelleted by centrifugation. Cells were washed with 1 ml of MEM containing 10% FCS, and cell pellets were resuspended in 100 μl of MEM containing 50 μg of digitonin (Sigma)/ml and then incubated on ice for 10 min. Following centrifugation at 2,000 × g at 4°C, supernatants (cytoplasmic fraction) were transferred to a fresh tube, and pellets (nuclear fraction) were resuspended in 100 μl of MEM. In order to lyse nuclei, an equal volume of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer was added, and the nuclei were sonicated for 30 s. Equivalent proportions of cytoplasmic and nuclear fractions were analyzed by SDS-PAGE and Western blotting with a rabbit polyclonal anti-PB1 antibody (9). Under the conditions used, PB2 was predominantly present in the nuclear fraction, indicating that no significant leakage of nuclear proteins into the cytoplasmic fraction had occurred (results not shown).
Assays for polymerase subunit interactions.
The TAP method (37) was used to purify PB1 and interacting PA. 293T cells in 35-mm dishes were transfected with 2 μg each of pcDNA-PB1-TAP and pcDNA-PA (wild type or mutants) by using 8 μl of Lipofectamine 2000 transfection reagent. Cells were harvested at 30 h posttransfection, and cell lysates were prepared by resuspending cells in 100 μl of Tris lysis buffer (50 mM Tris-HCl [pH 8.0]; 200 mM NaCl; 25% glycerol; 0.5% Igepal CA-630 [Sigma]; 1 mM dithiothreitol; 1 mM phenylmethylsulfonyl fluoride; 1 Complete Mini, EDTA-free protease inhibitor cocktail tablet [Roche]/10 ml) on ice. PB1-TAP and interacting proteins (PA) were purified by incubating 80 μl of cell lysates with 10 μl (packed volume) of immunoglobulin G (IgG)-Sepharose (Amersham) at 4°C for 3 h in a final volume of 320 μl. IgG-Sepharose was washed four times with 1 ml of wash buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Igepal CA-630, 1 mM phenylmethylsulfonyl fluoride) and two times with 1 ml of cleavage buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Igepal CA-630, 1 mM dithiothreitol, 0.5 mM EDTA). PB1 and interacting proteins were released from IgG-Sepharose by cleavage with 20 U of tobacco etch virus protease (Invitrogen) at 16°C for 2 h. Proteins were analyzed by Western blotting with a polyclonal anti-PA antibody.
RESULTS AND DISCUSSION
Characterization of GFP-tagged influenza virus RNA polymerase subunits.
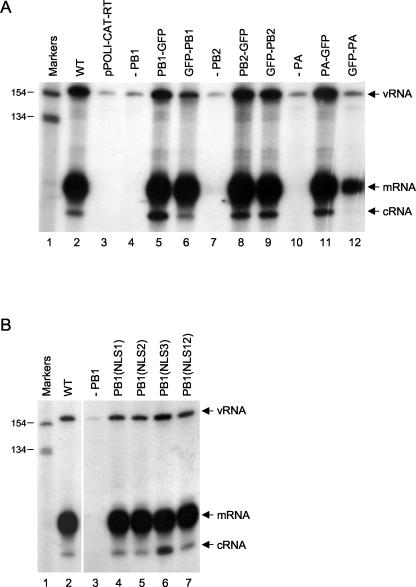
In order to study the subcellular localization of RNA polymerase and the process of complex formation, we generated expression plasmids to express RNA polymerase subunits derived from influenza A/WSN/33 virus tagged with GFP at the N or C terminus. The levels of expression of the GFP-tagged polymerase subunits were similar to those of the nontagged subunits, indicating no significant levels of degradation (data not shown). The activities of the polymerase-GFP fusions were tested by expressing RNP complexes containing GFP-tagged polymerase subunits and assaying for transcriptional and replicative activities by analyzing vRNA, mRNA, and cRNA levels in transfected cells with primer extension assays (12, 35) (Fig. 1A). The GFP tag did not inhibit activity, except when fused to the N terminus of PB1 or PA. GFP-PB1 had reduced replicative activity (three- to fivefold reduction) (Fig. 1A, lane 6), while GFP-PA had some residual transcriptional activity but minimal replicative activity (lane 12). Therefore, the polymerase subunits tagged at the C terminus were selected for further work.
FIG. 1.
Primer extension assays of vRNA, mRNA, and cRNA. (A) Effects of GFP tags on transcription and replication of influenza virus RNA. Primer extension assays of vRNA, mRNA, and cRNA isolated from 293T cells expressing influenza virus polymerase proteins (not tagged, i.e., wild type [WT]; tagged with GFP at the C terminus [PB1-GFP, PB2-GFP, and PA-GFP]; or tagged with GFP at the N terminus [GFP-PB1, GFP-PB2, and GFP-PA]), NP, and vRNA-like CAT RNA were performed as described previously (12). Negative controls include the analysis of RNA isolated from cells transfected with pPOLI-CAT-RT alone (lane 3) or with the omission of pcDNA-PB1 (lane 4), pcDNA-PB2 (lane 7), or pcDNA-PA (lane 10). (B) Effects of mutations in the potential NLSs of PB1 on the transcription and replication of influenza virus RNA. Primer extension assays of vRNA, mRNA, and cRNA isolated from 293T cells expressing influenza virus PB1 (wild type [WT] or the indicated mutants), PB2, PA, NP, and vRNA-like CAT RNA were performed as described previously (12). Positions of vRNA, mRNA, and cRNA signals are indicated on the right. Sizes of markers (32P-labeled 1-kb ladder [Invitrogen]) are indicated on the left.
Cellular localization of influenza virus RNA polymerase subunits.
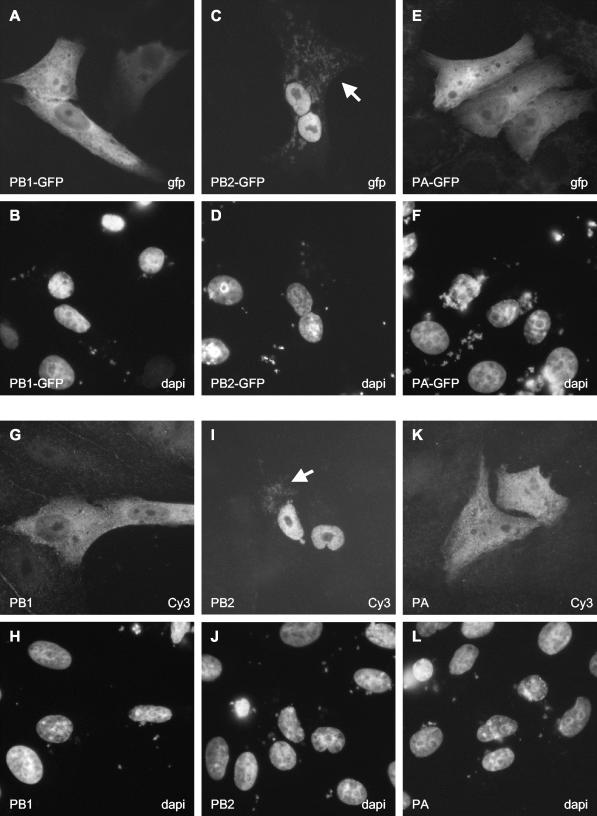
Next, we studied the pattern of localization of individually expressed GFP-tagged polymerase subunits in Vero cells (Fig. 2A to F). PB2 localized to the nucleus (Fig. 2C), while PA was observed in both the cytoplasm and the nucleus (Fig. 2E), as reported previously (18, 24, 30, 39). Surprisingly, we found that PB1 was also distributed in both the cytoplasm and the nucleus (Fig. 2A) rather than accumulating in the nucleus, as observed previously (1, 27, 39). In order to rule out the possibility that the GFP tag interfered with the localization of PB1, we used indirect immunofluorescence for the detection of nontagged polymerase subunits; the results obtained were similar to those obtained for the GFP-tagged subunits (Fig. 2G to L). Significantly, the nontagged PB1 subunit did not accumulate efficiently in the nucleus (Fig. 2G). Similar results were obtained with N-terminally GFP-tagged PB1 or C-terminally FLAG-tagged PB1 (results not shown). To exclude the possibility that the cell type affected the localization, we tested PB1 and PB1-GFP localization in both COS-7 and 293T cells; the results obtained were similar to those obtained in Vero cells (results not shown). In addition, to exclude the possibility that this localization pattern was unique to our clone of A/WSN/33-derived PB1, we tested the localization of A/PR/8/34 (Cambridge variant)-derived PB1 in both Vero and COS-7 cells (data not shown). Efficient nuclear accumulation was observed only for PB2 and not for PB1 or PA. Interestingly, in addition to its nuclear localization, low levels of PB2 were also observed to colocalize with mitochondria (data not shown), as determined by costaining with MitoTracker (Molecular Probes), a stain specific for mitochondria (S. M. Carr and E. Fodor, unpublished data) (Fig. 2C and I). Overall, the localization pattern for PB1 was not significantly affected by the amount of transfected plasmid or the time posttransfection at which the cells were analyzed (results not shown).
FIG. 2.
Intracellular distribution of GFP-tagged and nontagged polymerase subunits in transfected Vero cells. Cells expressing GFP-tagged polymerase subunits (A, C, and E) were analyzed directly for GFP expression by fluorescence microscopy. Cells expressing nontagged polymerase subunits (G, I, and K) were examined after staining with the corresponding subunit-specific primary and Cy3-conjugated secondary antibodies as described in Materials and Methods. Panels B, D, F, H, J, and L show the same fields of cells as panels A, C, E, G, I, and K, respectively, stained for DNA to indicate the locations of nuclei. The expressed protein is indicated in the bottom left corner of each panel. The method of detection (GFP, Cy3, or DAPI) is indicated in the bottom right corner of each panel. The white arrows in panel C and I indicate the mitochondrial localizations of PB2-GFP and PB2, respectively.
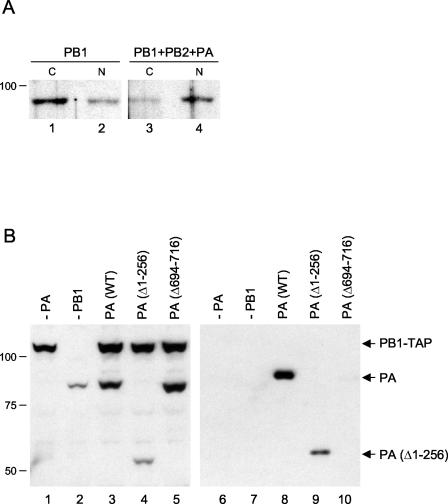
Our observation that PB1 cannot accumulate efficiently in the cell nucleus when expressed alone contradicts previous results (1, 27, 39). We do not understand the reason for this discrepancy. Therefore, to obtain further evidence for the intracellular distribution of PB1, we fractionated cells expressing PB1 alone or PB1 in the presence of PB2 and PA, followed by Western blot analysis of PB1 in the cytosolic and nuclear fractions (Fig. 3A). We observed that the proportion of individually expressed PB1 in the cytosolic fraction was greater than that in the nuclear fraction (Fig. 3A, compare lanes 1 and 2). This finding is consistent with the imaging data, which showed that individually expressed PB1 does not accumulate efficiently in the cell nucleus (see above). However, we observed a shift of PB1 from the cytosolic fraction to the nuclear fraction in the presence of PB2 and PA (Fig. 3A, compare lanes 3 and 4), suggesting that PB2 and/or PA are required for the nuclear accumulation of PB1.
FIG. 3.
(A) Intracellular distribution of PB1 in the absence or presence of PB2 and PA. 293T cells expressing PB1 (lanes 1 and 2) or PB1, PB2, and PA (lanes 3 and 4) were fractionated into cytoplasmic (C) and nuclear (N) fractions, and equivalent amounts were subjected to SDS-PAGE and Western blotting with a polyclonal anti-PB1 antibody. (B) Expression and binding to PB1 of mutant PA proteins. Extracts from 293T cells expressing PA (wild type [WT] or the indicated deletion mutants) and TAP-tagged PB1 (37) were analyzed by SDS-PAGE and Western blotting with a polyclonal anti-PA antibody before (left panel) and after (right panel) purification. Note the cross-reaction of PB1-TAP in the left panel due to the presence of the TAP tag. Sizes (in kilodaltons) are indicated on the left.
Effect of PA on the nuclear accumulation of PB1.
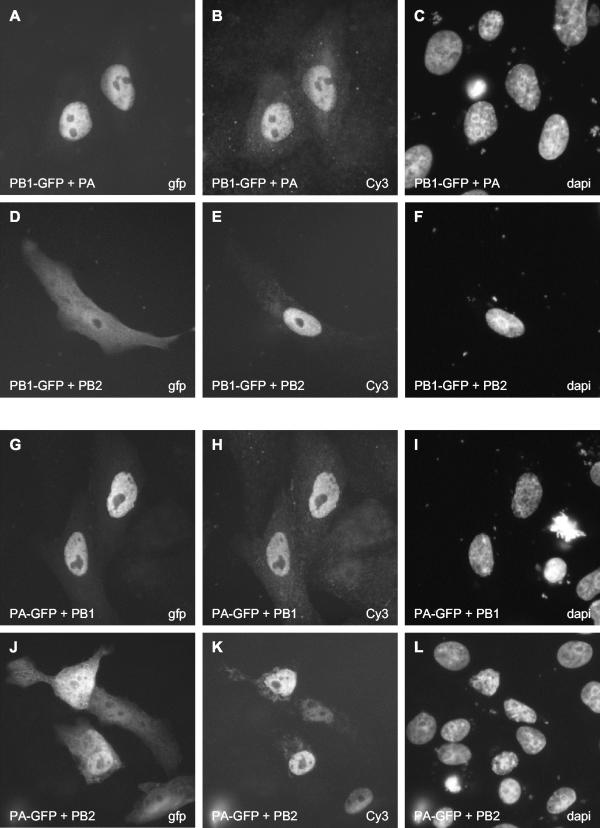
In infected cells, PB1 is detected predominantly in the cell nucleus (1, 18, 30). In order to identify the factor(s) required for the efficient accumulation of PB1 in the nucleus, we coexpressed PB1-GFP with PB2 or PA (Fig. 4A to F). The coexpression of PB2 did not affect the localization of PB1-GFP (Fig. 4D). In contrast, in the presence of PA, PB1-GFP efficiently accumulated in the cell nucleus (Fig. 4A). Moreover, when PA-GFP was coexpressed with PB1, we observed efficient accumulation of PA-GFP in the cell nucleus (Fig. 4G). PB2 had no effect on PA-GFP localization (Fig. 4J). We obtained similar results when we used COS-7 and 293T cells and either WSN- or PR8-derived polymerase subunits (data not shown). The coexpression of nucleoprotein (NP), which is known to bind PB1 (5, 22, 23), did not affect the localization of PB1 (results not shown).
FIG. 4.
Effect of PA or PB2 on the localization of PB1-GFP and effect of PB1 or PB2 on the localization of PA-GFP in transfected Vero cells. Cells expressing the indicated combinations of proteins (see bottom left of each panel) were analyzed directly for the GFP-tagged polymerase subunits (A, D, G, and J) and stained for the nontagged subunits with the corresponding subunit-specific primary and Cy3-conjugated secondary antibodies (B, E, H, and K). The three panels in each row show the same field of cells. The method of detection (GFP, Cy3, or DAPI) is indicated in the bottom right corner of each panel.
These data suggest that an interaction between PB1 and PA is required for their efficient nuclear accumulation. The following scenarios can be envisaged. Individually expressed PB1 or PA could shuttle between the nucleus and the cytoplasm, but they could be retained in the nucleus if they were coexpressed and formed a dimeric complex. Dimer formation between PB1 and PA could lead to the exposure of a nuclear retention signal or masking of a nuclear export signal. However, treatment of cells with leptomycin B, an inhibitor of CRM1-dependent nuclear export, did not affect the localization of PB1-GFP or PA-GFP (results not shown). Although we cannot rule out the possibility that PB1 and PA use another, CRM1-independent pathway for nuclear export, these results suggest that PB1 and PA are not shuttling proteins. We favor an alternative scenario in which PB1 and PA are imported into the nucleus as a heterodimer. Interactions between PB1 and PA in the cytoplasm might induce a conformational change resulting in the exposure of a strong NLS on either PB1 or PA or both. Recognition of this signal by the cellular nuclear transport system could promote efficient transport of the dimeric PB1-PA complex into the nucleus.
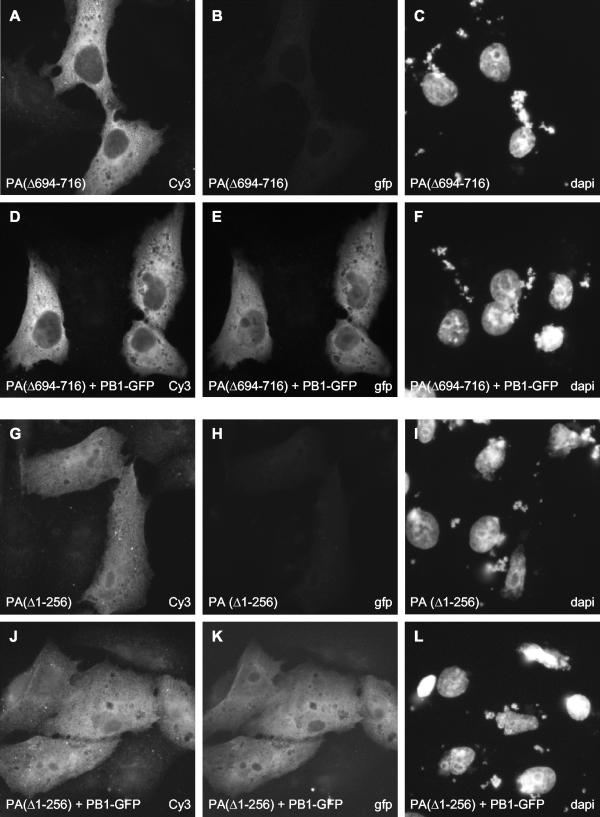
To address this latter possibility, we studied the effect of PA deletion mutants on the localization of PB1 (Fig. 5A to L). A deletion mutant with a C-terminal deletion of 24 amino acids [PA(Δ694-716)] and with no detectable transcriptional and replicative activities (12) was unable to induce the nuclear accumulation of PB1 (Fig. 5E). In agreement with previous results (41, 43), this PA mutant had a greatly reduced ability to bind to PB1, as determined by purification of TAP-tagged PB1 with the TAP method (37) and copurification of PA from transfected 293T cells (Fig. 3B, lane 10). These results show that the interaction of PA with PB1 is essential for induction of the nuclear accumulation of PB1. Moreover, a deletion mutant of PA lacking the N-terminal 256 amino acids [PA(Δ1-256)] failed to induce the nuclear accumulation of PB1 (Fig. 5K), although it was able to bind to PB1 (Fig. 3B, lane 9). Since the N-terminal one third of PA is reported to contain an NLS (29), it is possible that the transport of the PB1-PA dimer is preferentially driven by the NLS present in the N terminus of PA. Alternatively, we cannot exclude the possibility that in the absence of the N terminus, PA(Δ1-256) could not induce a conformational change in PB1 that might be required for its nuclear accumulation.
FIG. 5.
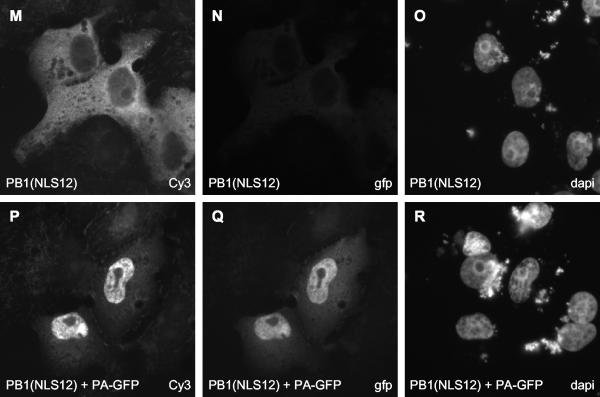
Effect of mutations in PA and PB1 on the nuclear accumulation of PB1. PA deletion mutants (A, D, G, and J) and the PB1 NLS12 mutant (M and P) were detected with rabbit polyclonal anti-PA and anti-PB1 antibodies, respectively, followed by staining with a Cy3-conjugated anti-rabbit secondary antibody. PB1-GFP (E and K) and PA-GFP (Q) were detected directly by fluorescence microscopy. The three panels in each row show the same field of cells. The method of detection (GFP, Cy3, or DAPI) is indicated in the bottom right corner of each panel.
Effect of NLS mutations on the nuclear accumulation of PB1.
We also attempted to generate mutants of PB1 with an exclusively cytoplasmic localization by introducing mutations into the proposed NLS sequences and then studying whether the nuclear localization of the resulting mutants could be rescued by the coexpression of PA. Nath and Nayak (27) proposed that two discrete regions containing two stretches of positively charged amino acid residues, 187-RKRR-190 and 207-KRKQR-211, are essential for the nuclear localization of PB1. We mutated these sequences to 187-RAAR-190 and 207-KAAQR-211 (mutated residues are underlined), either individually (mutants NLS1 and NLS2, respectively) or together (mutant NLS12). We also mutated 668-PKRNRSI-674, a stretch of basic amino acid residues predicted to be a potential NLS in a computer search (PSORT II) of PB1, to 668-PAANRSI-674 (mutant NLS3). None of these mutants, however, showed an exclusively cytoplasmic localization (Fig. 5M and results not shown). Although mutant NLS12 appeared to be more cytoplasmic than wild-type PB1 (compare Fig. 5M and Fig. 2A), NLS12, as well as NLS1, NLS2, and NLS3, could accumulate efficiently in the cell nucleus when coexpressed with PA (Fig. 5P and results not shown). Moreover, all of these PB1 mutants were active in transcription and replication (Fig. 1B).
Conclusions.
We conclude that, in contrast to previously published data (1, 27, 39), PB1 requires the coexpression of PA for its efficient nuclear accumulation. This conclusion is supported by the observation of Nieto and colleagues (30) that PB1 stimulates the nuclear accumulation of PA. Taken together, these results are consistent with a model in which PB1 and PA interact in the cytoplasm and are transported into the nucleus as a complex (Fig. 6, model B). However, our results cannot exclude the possibility that in infected cells, PB2 is also assembled into the complex in the cytoplasm and the trimeric PB1-PB2-PA complex is transported into the nucleus (Fig. 6, model C). Moreover, the presence of PA or of PA and PB2 in the nucleus might stabilize PB1 and thereby contribute to its nuclear accumulation. We observed that all three polymerase subunits accumulated efficiently in the nuclei of MDBK cells infected with influenza A/WSN/33 virus as early as 2 to 3 h postinfection (results not shown), in agreement with our model that PB1 and PA are transported into the nucleus as a complex. We found no evidence for the previously reported delayed transport of PA compared to PB1 (1, 30).
Our results show that the N-terminal 256 amino acids of PA are critical for the induction of the nuclear accumulation of PB1. Since this region is proposed to contain an NLS (29), we suggest that the transport of the PB1-PA dimer into the nucleus might be preferentially driven by the NLS present in the N-terminal one third of PA. This NLS might be sterically hindered in PA, explaining the inefficient nuclear accumulation of individually expressed PA. However, after a conformational change induced by PB1 binding, it might become exposed. It is not clear, however, what role the previously identified NLSs in PB1 (27) play, since their mutation had no obvious effect on PB1-PA localization.
The observation that a dimeric PB1-PA complex can accumulate efficiently in the nucleus in the absence of PB2 raises the question of whether the PB1-PA dimer could perform any of the activities proposed for RNA polymerase. Indeed, several studies have concluded that PB1 and PA synthesize cRNA in the absence of PB2 (16, 26). However, these findings have not yet been confirmed by others (7, 20, 32).
Acknowledgments
We thank J. Robinson and A. Taylor for DNA sequencing; M. Crow for plasmids; and S. Inglis, A. Portela, and T. Toyoda for antibodies. We also thank G. G. Brownlee, O. Engelhardt, F. Vreede, and S. Carr for helpful discussions.
This work was supported by the MRC (senior nonclinical fellowship G117/457 to E.F.).
REFERENCES
- 1.Akkina, R. K., T. M. Chambers, D. R. Londo, and D. P. Nayak. 1987. Intracellular localization of the viral polymerase proteins in cells infected with influenza virus and cells expressing PB1 protein from cloned cDNA. J. Virol. 61:2217-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Area, E., J. Martín-Benito, P. Gastaminza, E. Torreira, J. M. Valpuesta, J. L. Carrascosa, and J. Ortín. 2004. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl. Acad. Sci. USA 101:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos, P. 1988. A sequence motif in many polymerases. Nucleic Acids Res. 16:9909-9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bárcena, J., M. Ochoa, S. de la Luna, J. A. Melero, A. Nieto, J. Ortín, and A. Portela. 1994. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 68:6900-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownlee, G. G., and J. L. Sharps. 2002. The RNA polymerase of influenza A virus is stabilized by interaction with its viral RNA promoter. J. Virol. 76:7103-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Digard, P., V. C. Blok, and S. C. Inglis. 1989. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fechter, P., L. Mingay, J. Sharps, A. Chambers, E. Fodor, and G. G. Brownlee. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 11.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, p. 1-29. In C. W. Potter (ed.), Influenza. Elsevier, Amsterdam, The Netherlands.
- 12.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., L. J. Mingay, M. Crow, T. Deng, and G. G. Brownlee. 2003. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J. Virol. 77:5017-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, S., T. Zürcher, and J. Ortín. 1996. Identification of two separate domains in the influenza virus PB1 protein invloved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herz, C., E. Stavnezer, R. M. Krug, and T. Gurney. 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell 26:391-400. [DOI] [PubMed] [Google Scholar]
- 16.Honda, A., K. Mizumoto, and A. Ishihama. 2002. Minimum molecular architectures for transcription and replication of the influenza virus. Proc. Natl. Acad. Sci. USA 99:13166-13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, D. A., A. J. Caton, S. J. McCready, and P. R. Cook. 1982. Influenza virus RNA is synthesized at a fixed site in the nucleus. Nature 296:366-368. [DOI] [PubMed] [Google Scholar]
- 18.Jones, I. M., P. A. Reay, and K. L. Philpott. 1986. Nuclear location of all three influenza polymerase proteins and a nuclear signal in polymerase PB2. EMBO J. 5:2371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1530. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Lee, M. T. M., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medcalf, L., E. Poole, D. Elton, and P. Digard. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mena, I., E. Jambrina, C. Albo, B. Perales, J. Ortín, M. Arrese, D. Vallejo, and A. Portela. 1999. Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J. Virol. 73:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukaigawa, J., and D. P. Nayak. 1991. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J. Virol. 65:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naffakh, N., P. Massin, and S. van der Werf. 2001. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the PA subunit. Virology 285:244-252. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa, Y., K. Oda, and S. Nakada. 1996. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J. Virol. 70:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nath, S. T., and D. P. Nayak. 1990. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1N1). Mol. Cell. Biol. 10:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann, G., G. G. Brownlee, E. Fodor, and Y. Kawaoka. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 283:121-143. [DOI] [PubMed] [Google Scholar]
- 29.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, and J. Ortín. 1994. Complex structure of the nuclear translocation signal of influenza virus polymerase PA subunit. J. Gen. Virol. 75:29-36. [DOI] [PubMed] [Google Scholar]
- 30.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, J. Valcárcel, J. A. Melero, and J. Ortín. 1992. Nuclear transport of influenza virus polymerase PA protein. Virus Res. 24:65-75. [DOI] [PubMed] [Google Scholar]
- 31.Ohtsu, Y., Y. Honda, Y. Sakata, H. Kato, and T. Toyoda. 2002. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol. Immunol. 46:167-175. [DOI] [PubMed] [Google Scholar]
- 32.Perales, B., and J. Ortín. 1997. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol. 71:1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. F. Santarén, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez, D. R., and R. O. Donis. 2001. Functional analysis of PA binding by influenza A virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75:8127-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleschka, S., S. R. Jaskunas, O. G. Engelhardt, T. Zürcher, P. Palese, and A. García-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pombo, A., P. Cuello, W. Schul, J.-B. Yoon, R. G. Roeder, P. R. Cook, and S. Murphy. 1998. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 17:1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Séraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 38.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortín, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, G. L., J. Z. Levin, P. Palese, and B. Moss. 1987. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology 160:336-345. [DOI] [PubMed] [Google Scholar]
- 40.Subbarao, K., H. Chen, D. Swayne, L. Mingay, E. Fodor, G. G. Brownlee, X. Xu, X. Lu, J. Katz, N. Cox, and Y. Matsuoka. 2002. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology 305:192-200. [DOI] [PubMed] [Google Scholar]
- 41.Toyoda, T., D. M. Adyshev, M. Kobayashi, A. Iwata, and A. Ishihama. 1996. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 77:2149-2157. [DOI] [PubMed] [Google Scholar]
- 42.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 43.Zürcher, T., S. de la Luna, J. J. Sanz-Ezquerro, A. Nieto, and J. Ortín. 1996. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77:1745-1749. [DOI] [PubMed] [Google Scholar]