Abstract
The surface glycoprotein (gp95) of the feline immunodeficiency virus (FIV) binds in a strain-specific manner to several cell surface molecules, including CXCR4, heparan sulfate proteoglycans (HSPGs), DC-SIGN, and a 43-kDa cell surface receptor on T cells recently identified as CD134 by M. Shimojima et al. (Science 303:1192-1195, 2004). CXCR4 is the entry receptor in all known cases, and the other molecules act as binding receptors to help facilitate infection. In this report, we confirm and extend the findings regarding CD134 as a primary receptor for FIV. In addition, we show that temperature critically influences the binding properties of FIV gp95 to CXCR4 and HSPGs. The data show that gp95 of the field strain FIV-PPR bound to CXCR4 at 22°C, whereas binding was not detected at 4°C. In contrast, binding of the laboratory adapted FIV-34TF10 gp95 was observed at either 4°C or 22°C, albeit at increased levels at the higher temperature. The level of CXCR4 increased after the temperature was switched from 4 to 22°C, whereas the level of HSPGs decreased, resulting in higher binding of gp95 from both strains to CXCR4 and lower binding of gp95 of FIV-34TF10 to HSPGs (FIV-PPR gp95 does not bind to these molecules). The findings also show that HSPGs facilitate the CXCR4-mediated infectivity of CrFK and G355-5 cells by FIV-34TF10. These two nonlymphoid cell lines express very low levels of CXCR4 and are permissive to FIV-34TF10 but not to productive infection by FIV-PPR. However, overexpression of human CXCR4 in CrFK or G-355-5 cells resulted in extensive cell fusion and infection by FIV-PPR. Taken together, these findings indicate that factors that increase the effective concentration of CXCR4 enhance FIV infectivity and may involve (i) temperature or ligand-induced conformational changes in CXCR4 that enhance SU binding, (ii) coreceptor interactions with gp95 that either alter gp95 conformation to enhance CXCR4 binding and/or raise the localized concentration of receptor or ligand, or (iii) direct increase in CXCR4 concentration via overexpression.
Feline immunodeficiency virus (FIV) is the only nonprimate lentivirus that causes a progressive loss of CD4+ cells and an AIDS-like disease in its natural host, the domestic cat (28). One of the main differences between FIV and the primate lentiviruses is that FIV does not use CD4 as a primary binding receptor and displays a much broader tissue tropism in vivo, infecting CD4+ T cells, as well as CD8+ T cells, B cells, and macrophages (3, 6, 7, 16, 27).
As with the T-cell tropic human immunodeficiency virus type 1 (HIV-1), FIV uses CXCR4 as a common entry receptor (45). In addition, we have shown that the surface glycoprotein of FIV, gp95, could specifically bind other cell surface molecules, including a 43-kDa protein species expressed on peripheral blood mononuclear cells (PBMC) and interleukin-2 (IL-2)-dependent T-cell lines (9), heparan sulfate proteoglycans (HSPGs) that have a ubiquitous tissue expression but are predominantly expressed on epithelial cells and astrocytes (9) and also DC-SIGN (11). Binding of FIV gp95 to each of these receptors was dependent on the origin of gp95. The gp95 from a primary strain (PI) of FIV bound to the 43-kDa receptor and to DC-SIGN but not to HSPGs, whereas gp95 from a tissue culture-adapted (TCA) FIV strain could bind all three receptors (9). Although these differences were observed at the binding level, entry for both PI and TCA FIV was mediated solely via CXCR4 (9, 13, 14, 33). We speculated that the 43-kDa receptor might serve as a primary binding receptor for FIV similar to CD4 for HIV-1 and that entry of FIV would follow the two-step model, followed by the primate lentiviruses (reviewed in reference 12). This model predicts that gp120 initially binds CD4, which induces conformational rearrangement in gp120, exposing the coreceptor binding site on gp120. In a second step, the CD4-gp120 complex binds to its coreceptor.
Recent studies by Shimojima et al. (36) have now demonstrated that the 43-kDa receptor is, in fact, CD134 (OX-40), a molecule specifically upregulated on CD4+ T cells and expressed on a subset of T-cell lines (21). Although structural studies must now be performed to verify the nature of the gp95-CD134 interaction, the overall findings suggest parallels with the CD4-HIV-1 SU interaction and offer an explanation as to why FIV primarily targets CD4+ T cells even though it does not bind CD4 (26).
The purpose of the present studies was to compare the binding properties of FIV adhesins to CXCR4 to results previously reported for HIV SU-chemokine receptor interactions (1, 34, 41, 46). In addition, studies were carried out to formally assess the ability of gp95 adhesins to bind to cells expressing recombinant CD134. The results show that FIV gp95 binds to CD134 and, further, that gp95 of field strain FIVs binds to CXCR4 in a temperature-dependent manner consistent with a conformation dependence for ligand, receptor, or both.
MATERIALS AND METHODS
Cell lines and viruses.
CrFK cells were obtained from the ATCC (Rockville, Md.), and the feline glial cell line (G355-5) was kindly provided by Don Blair (National Institutes of Health [NIH], Bethesda, Md.). Cat PBMC were prepared from heparinized whole feline blood by Ficoll-Paque gradient purification. The IL-2-dependent T-cell line 104-C1 was isolated by limiting dilution cloning of PBMC and was a gift from Chris Grant (Custom Monoclonal Antibodies, Intl.). The IL-2-independent lymphoma cell line 3201 was obtained from William Hardy. The Cf2Th/synCCR5 cells were obtained through the AIDS Research Reagent program, Division of AIDS, National Institute of Allergy and Infectious Disease (NIAID), NIH, from Tajib Mirzabkov and Joseph Sodroski (24). CXCR4high G355-5 cells were obtained by transduction with human CXCR4. Briefly, 293GP cells were transfected with pBABE-CXCR4 and pCMV-VSV-G. Two days later, viral supernatant was used to transduce G355-5 cells. One week after the transduction, CXCR4 positive cells were sorted by fluorescence-activated cell sorting (FACS). pBABE-CXCR4 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Nathaniel Landau (8, 25). Propagation of the different cell lines was previously described (23).
The FIV strains used in the present study were FIV-34TF10, a molecular clone of the FIV Petaluma isolate (39), and FIV-PPR, a molecular clone of the FIV San Diego isolate (29).
Antibodies and reagents.
Anti-CXCR4 antibody (clone 44717.111) was purchased from R&D Systems (Minneapolis, Minn.). The anti-heparan sulfate chain antibody (clone F58-10E4) was purchased from Seikagaku Corp. (Tokyo, Japan). The phycoerythrin- and horseradish peroxidase (HRP)-conjugated anti-human Fc antibodies were purchased from ICN-Cappel (Aurora, Ohio). The bycyclam AMD3100 was kindly provided by Donald Mosier (The Scripps Research Institute). Heparin, heparinase I, bovine serum albumin (BSA), and anti-Fc agarose beads were purchased from Sigma (St. Louis, Mo.). Neutravidin-HRP was purchased from Pierce (Rockford, Ill.). Earle balanced salt solution (EBSS) and Novex Tris-glycine-sodium dodecyl sulfate (SDS)-polyacrylamide gels were purchased from Invitrogen (Carlsbad, Calif.).
SU adhesins.
The FIV SU adhesins PPR-Fc and 34-Fc were previously described (9). For JRCSF-Fc, gp120 from the JR-CSF clone of HIV-1 (20) was subcloned (from the threonine residue at position 50 to the alanine residue at position 489) by PCR in the pRSC-Fc-GS vector essentially as described for the FIV SU adhesins. SU adhesins were batch purified from cell supernatants by protein A affinity chromatography. JR-CSF env and Fc plasmid were kindly provided by Dennis Burton (The Scripps Research Institute) and Brian Seed (Massachusetts General Hospital), respectively.
Flow cytometry analyses.
Adhesins and antibodies were incubated with 105 cells for 1 h in 100 μl of EBSS containing 0.1% BSA, washed once in EBSS, and resuspended in 100 μl of EBSS-0.1% BSA supplemented with the fluorescence-conjugated secondary antibody at a dilution of 1:250. Samples were analyzed on a FACScan. The data were acquired with CellQuest software and analyzed with FlowJo software.
Immunoprecipitation.
Cells were cell surface biotinylated as previously described (9). Biotinylated cells were lyzed in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% SDS) and incubated for 30 min on ice. Clear supernatants were incubated with 2 μg of adhesins and 10 μl of anti-Fc agarose beads overnight at 4°C. Beads were then washed three times with the lysis buffer and once in the lysis buffer minus NP-40 and SDS, subjected to SDS-PAGE on a Novex 8 to 16% Tris-glycine gel, transferred to nitrocellulose membrane, and then Western blotted by using neutravidin-HRP.
Receptor overlay assay.
A modified virus overlay protein-binding assay was used. Cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting. Blots were incubated overnight with Fc, PPR-Fc, or 34-Fc at 1 μg/ml in phosphate-buffered saline (PBS) at 4°C. The blots were then washed in PBS and further incubated for one h with anti-Fc-HRP at a 1:1,000 dilution. The blots were again washed, and gp95-binding proteins were revealed by chemiluminescence.
Heparinase treatment.
For the infection assay, G355-5 and CrFK cells, plated the day before at 50 × 104 cells per well in a 12-well plate, were washed once in EBSS, and resuspended in EBSS containing 2 mM CaCl2 and 0.1% BSA in the absence or the presence of heparinase I at 10 U per ml. The cells were incubated for 45 min at room temperature on a rocker platform, washed once in EBSS, and infected for 30 min at room temperature with FIV-34TF10 (27 × 103 cpm reverse transcriptase [RT] activity containing supernatant). The cells were then washed again and resuspended in 4 ml of complete Dulbecco modified Eagle medium. At 7 days postinfection, virus production was assayed on 50 μl of supernatant by an RT assay (9). For the FACS assay, G355-5 cells were detached in EBSS containing 5 mM EDTA, washed once, and resuspended in heparinase buffer (see above) in the absence or presence of 10 U of heparinase per ml. After 45 min of incubation at room temperature, cells were washed twice and resuspended in EBSS-0.1% BSA and proceed for FACS analysis as described above.
CXCR4high cells infectivity assay.
Cells were plated at 1.0 × 105 cells per well in a six-well plate. The next days, cells were infected for 30 min at room temperature with 100 μl of virus stock. The cells were next washed once and incubated in complete Dulbecco modified Eagle medium. At day 5, supernatants were analyzed for virus production by using an RT assay as described above.
PCR assays.
Newly synthesized viral DNA and RNA were analyzed as previously described (10). Briefly, 3201 and 104-C1 cells (5 × 105) were infected with 100 μl of FIV-PPR virus stock (8.5 × 104 cpm RT activity containing supernatant). Total DNA and RNA were extracted at 20 and 72 h postinfection, respectively. Viral DNA was analyzed by PCR with the primer pair GAG1 and GAG2, which amplifies a 325-bp FIV gag (10). Viral RNA was analyzed by RT-PCR with the primer pair LA6 and LA13, which amplifies two viral spliced RNA products of 645 and 710 bp, respectively (10). A CD4 primer pair designed to amplify a short intron between exons 1.2 and 2 was used as a DNA extraction control, and a β-actin primer pair was used as a control for RNA extraction (10).
Cloning of feline CD134.
Poly(A)+ RNA was isolated from 104-C1 cells with an mRNA messenger kit (Gibco-BRL) and examined for its integrity by Northern blot analysis with a radiolabeled GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA as probe. cDNA with a 5′ EcoRI restriction end and a 3′ XhoI end was generated from 5 μg of poly(A)+ RNA by using the Superscript cDNA Synthesis kit (Invitrogen) but according to the manufacturer's protocol outlined by Stratagene. A total of 500 ng of cDNA and 0.5 μg of hygroMaRXII vector (38) digested with EcoRI and XhoI were ligated and transformed into ElectroMAX DH10B cells (Invitrogen) to generate the cDNA library. We estimated that the primary library included 5 × 106 independent clones. To determine the quality of the library, 32 randomly chosen clones were subjected to restriction analysis. The results indicated that 90% of these clones contained unique cDNA inserts with an average size of 1 kb. Feline CD134 was next cloned by rapid amplification of cDNA ends (RACE) from the 104-C1 cDNA library. Briefly, two CD134 regions of high homology at the amino acid level, QACK and PIQE, were identified by alignment of human mouse and rat CD134. Degenerative primers were synthesized. One, sense and corresponding to QACK, had the sequence 5′-CARGCCTGCAAGCCCTGGACCAA-3′, and the other, antisense and corresponding to PIQE, had the sequence 5′-GGCTAGATCTTGGCCAGAGTGGAGTKKGCGGTC-3′. After a first round of PCR with these two primers, a 350-bp QACK/PIQE amplicon was obtained and confirmed to be homologous to CD134 by sequencing. Next, the 5′ end and the 3′ end of feline CD134 was obtained by RACE. Finally, amplification of the entire cat CD134 was performed and cloned into the MIGR1-green fluorescent protein (GFP) vector (11). Both MIGR1-GFP and MIGR1-GFP/CD134 vectors were transduced in CrFK cells for SU binding studies.
RESULTS
Cell surface receptors interacting with FIV SU.
We previously reported an experimental approach to scan for cell surface receptors involved in interactions with FIV surface glycoprotein gp95 (SU) by using adhesins of gp95 fused in-frame with the Fc domain of human immunoglobulin G1 (9). Two SU adhesins were constructed: one using SU from the primary isolate (PI) FIV-PPR (PPR-Fc) and the other using SU from the TCA isolate FIV-34TF10 (34-Fc). Using PPR-Fc and 34-Fc in flow cytometry analyses, blocking assays with specific chemokines and heparin, and immunoprecipitation assays, we identified three cell surface receptors interacting with either one or both adhesins (9) (the results are summarized in Table 1). Under the conditions used, PPR-Fc bound to a 43-kDa protein expressed on the surface of feline PBMC and primary feline T cells, but direct binding to CXCR4 was not detected. The 43-kDa receptor is distinct from CXCR4 in that SDF1α and AMD3100, while efficiently inhibiting infection by primary isolates, failed to block binding of PPR-Fc to the 43-kDa receptor (9). The much broader tropism of the TCA strain was reflected in the ability of the 34-Fc adhesin to bind not only the 43-kDa receptor but also to bind directly to CXCR4 and to HSPGs in a cell-specific manner that directly reflected the TCA infectivity pattern (Table 1). Based on competition studies with AMD3100, the sole entry receptor for both PI and TCA FIVs is CXCR4 (9) (Table 1). We speculated that (i) the 43-kDa receptor might serve as a primary binding receptor for PI FIVs and that (ii) FIV entry followed a two-step model similar to primate lentiviruses in that binding to the 43-kDa receptor would induce conformational rearrangement in gp95 to facilitate binding to CXCR4. In this scenario, the TCA isolate would be the FIV equivalent of a CD4-independent HIV isolate and thus already possess a conformation compatible with high-affinity binding to chemokine receptor.
TABLE 1.
Summary of the receptors used by FIV
| Cell type | SU adhesin bounda | Binding receptorb | Entry receptorc |
|---|---|---|---|
| Feline | |||
| PBMC | PPRFc and 34Fc | 43-kDa receptor | CXCR4 |
| IL-2-dependent T cells | PPRFc and 34Fc | 43-kDa receptor | CXCR4 |
| IL-2-independent T cells | 34Fc only | CXCR4 | CXCR4 |
| Epithelial and glial cells | 34Fc only | HSPGs | CXCR4 |
| Human | |||
| Jurkat | 34Fc only | CXCR4d | CXCR4d |
| HeLa | 34Fc only | HSPGs | CXCR4d |
| U87 | 34Fc only | HSPGs | Nonee |
| Hamster | |||
| CHO-K1 | 34Fc only | HSPGs | Nonee |
| CHO-pgsA745 | Nonef |
SU adhesins from FIV-PPR and FIV-34TF10 strains are denoted PPRFc and 34Fc, respectively.
The binding receptor involved was identified by FACS, neutralization, and immunoprecipitation assays (9).
The entry receptor implicated was determined by inhibition of infection by SDF1-α and AMD3100.
FIV uses human CXCR4 as well for binding and entry. A block of infection occurs at a postentry level (30, 45).
These cell lines are deficient in CXCR4 expression.
CHO-pgsA745 cells are deficient in HSPGs, as well as in CXCR4 expression.
Binding of FIV SU to CXCR4 is temperature dependent.
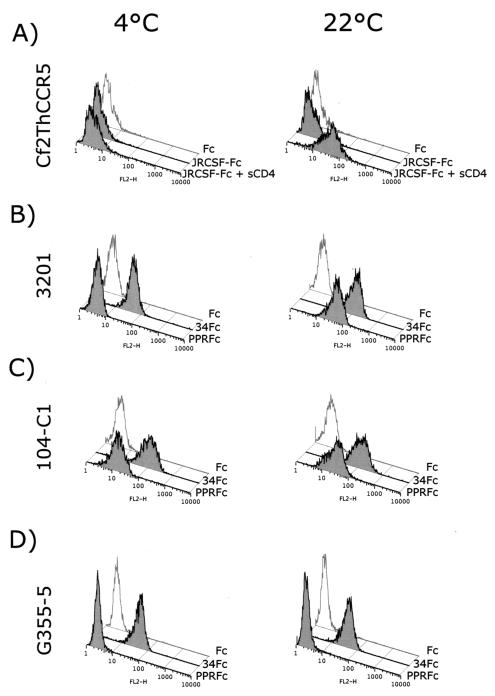
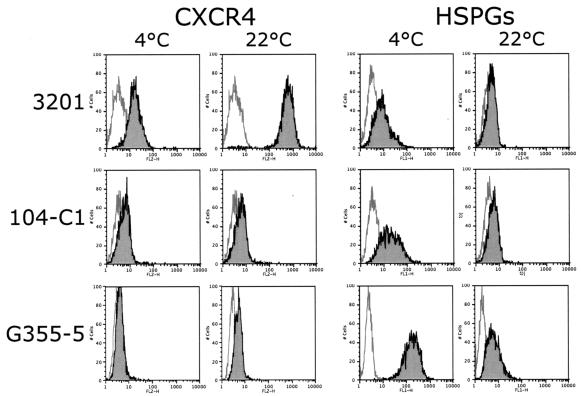
In order to test the hypotheses outlined above, additional assays were carried out to further assess the nature of binding by the FIV SU adhesins. All of our previous binding studies had been performed at 4°C (9). Since binding studies with HIV have revealed a temperature dependence for SU-receptor interactions, we compared binding properties of our adhesins at 4 and 22°C (Fig. 1). As a control, an HIV-1 gp120 adhesin was prepared by using SU of the CCR5-dependent JR-CSF strain of HIV-1 (20), and binding at 4 and 22°C was assessed on Cf2ThsynCCR5 cells (CD4− CCR5+) (24) in the absence or presence of sCD4 (Fig. 1A). Binding of JRCSF-Fc to CCR5 was observed only in the presence of sCD4 and, most importantly, only at 22°C, a finding consistent with a temperature-dependent conformational rearrangement of the gp120-sCD4 complex to facilitate binding to the chemokine receptor at 22°C but not at 4°C. Binding of the two FIV SU adhesins to feline cell lines, including 3201 (Fig. 1B; CXCR4+), 104-C1 (Fig. 1C; 43-kDa receptor+ CXCR4+), and G355-5 cells (Fig. 1D; CXCR4+ HSPG+) was then assessed at 4 and 22°C. As previously observed, binding of PPR-Fc adhesin was not detected on 3201 cells at 4°C (expressing only CXCR4), but the 34-Fc adhesin bound strongly at this temperature (Fig. 1B). In contrast, when the assay was performed at 22°C, substantial binding was noted with the PPR-Fc adhesin, and some increase in binding was also noted for 34-Fc (Fig. 1B). Binding to the 104-C1 T-cell line (expressing both CXCR4 and the 43-kDa receptor) was substantial with both adhesins at either temperature, a finding consistent with a lack of temperature dependence for interaction with the 43-kDa receptor (Fig. 1C). Consistent with past findings (9), the PI adhesin failed to bind to the adherent G355-5 cell line (as well as CrFK cells [not shown]), but the TCA adhesin bound strongly at both temperatures (Fig. 1D).
FIG. 1.
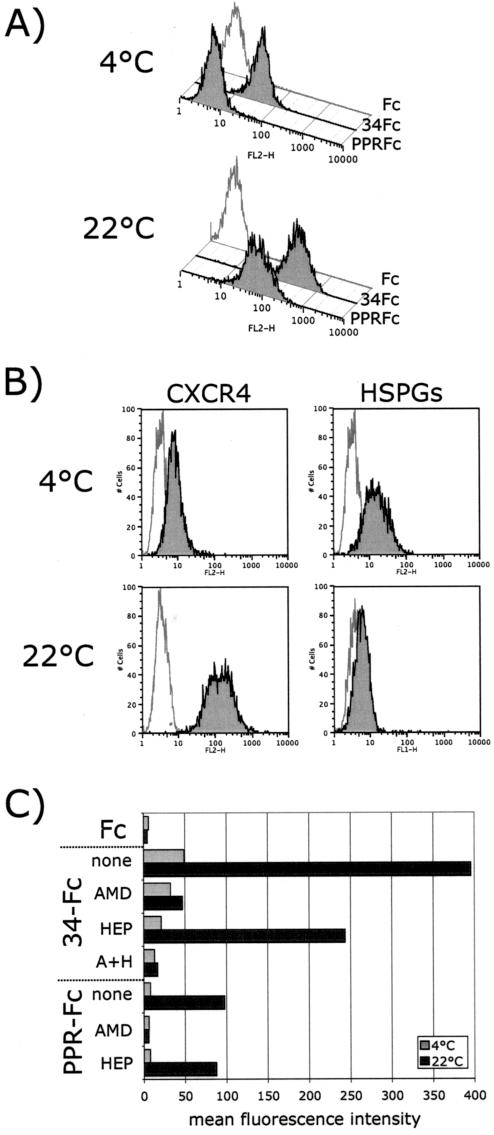
Binding profile of FIV SU adhesins: conformation-dependent binding of FIV SU to CXCR4. (A) FACS analysis of the binding of HIV-1 JRCSF-Fc to the CD4− CCR5+ Cf2Th synCCR5 cells requires sCD4 and a temperature shift from 4 to 22°C. (B) FACS analysis of the binding of FIV PPR-Fc and 34-Fc to the CXCR4+ 3201 feline lymphoma cells. Binding of PPR-Fc to CXCR4 requires only a shift in temperature; likewise, binding of 34-Fc is increased after the temperature shift. (C and D) FACS analysis of the binding of PPR-Fc and 34-Fc at 4 and 22°C on 104-C1 and G355-5, respectively. Fc alone was used as a negative control.
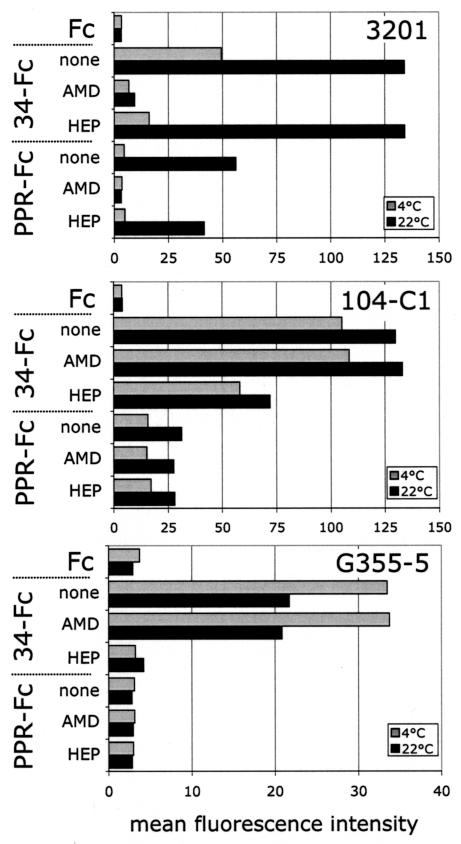
We previously reported that 34-Fc binding on 3201 cells was mediated through CXCR4 and that binding could be blocked by the CXCR4 antagonist, AMD3100 (9). To investigate whether the increased binding of PPR-Fc that we observed at 22°C was also mediated through CXCR4, we tested the inhibitory activity of AMD3100 (Fig. 2, top panel). The binding of PPR-Fc and 34-Fc to 3201 cells at 22°C was CXCR4 dependent, since AMD3100 inhibited the binding by both SU adhesins by >90%. In parallel, we also tested the inhibitory activity of heparin, since 34-Fc has the ability to bind HSPGs. The results show that heparin partially inhibited the binding of 34-Fc to 3201 cells at 4°C but had no significant inhibitory activity at 22°C (Fig. 2, top panel). Neither AMD3100 nor heparin had a significant effect on binding of either adhesin to 104-C1 cells at either temperature (Fig. 2, middle panel). Likewise, AMD3100 did not significantly block binding of 34-Fc to G355-5 cells at either temperature. However, binding was substantially blocked at both temperatures by heparin (Fig. 2, bottom panel). The overall results are consistent with a temperature-dependent increase in binding of FIV SU to CXCR4 but not to either the 43-kDa receptor or HSPGs.
FIG. 2.
Inhibition profile of FIV SU adhesins. Inhibition of PPR-Fc and 34-Fc binding to 3201, 104-C1, and G355-5 cells at 4 and 22°C by AMD3100 and heparin. Cells were incubated with the indicated SU adhesins in the absence (none) or presence of AMD3100 (AMD), heparin (HEP), or both (A+H). SU binding, expressed as the mean fluorescence intensity, was analyzed by FACS analysis as described in Materials and Methods. Heparin and AMD3100 were used at 1 μg/ml. Fc alone was used as a negative control.
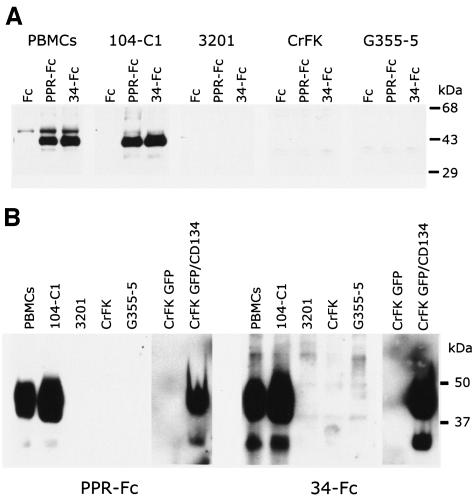
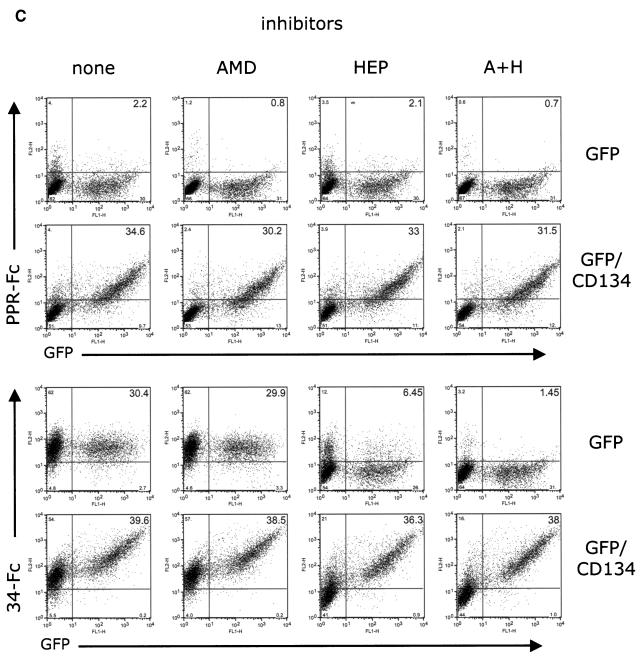
The strong binding of both adhesins to the 104-C1 cell line and to PBMC is facilitated by a 43-kDa protein species unrelated to either CXCR4 or HSPGs (9). This is reiterated in the data shown in Fig. 3A, in which the PI and TCA adhesins were able to drag down biotinylated receptor from PBMC and 104-C1 cells in immune precipitations but not from 3201, CrFK, or G355-5 cells. While the present study was in progress, Shimojima et al. (36) published findings consistent with the notion that the 43-kDa receptor was the T-cell marker CD134 (OX-40). We subsequently cloned and expressed feline CD134 on CrFK cells and then assayed for specific binding by the adhesins (Fig. 3B). The findings verify that both the PI and TCA adhesins bind strongly to CD134 and that the size of the expressed protein and binding pattern is identical to that observed on PBMC and the 104-C1 T-cell line (Fig. 3, compare panels A and B). Binding of either adhesin to the CD134 expressing cells was not blocked by AMD3100, heparin, or a combination of the two inhibitors (Fig. 3C).
FIG. 3.
A 43-kDa binding receptor for gp95 on PBMC and the IL-2-dependent T-cell line 104-C1 corresponds to the T-cell marker, CD134. (A) Immunoprecipitation studies identified a 43-kDa protein species on PBMC and 104-C1 cells that specifically interacts with FIV SU adhesins. Biotinylated cell lysates were incubated with the indicated adhesins. Complexes were resolved by SDS-PAGE, and gp95-binding proteins were revealed by Western blotting with a neutravidin-HRP antibody. This receptor was not immunoprecipitated from 3201, CrFK, or G355-5 cells. (B) A modified virus overlay assay shows that FIV SU adhesins interact with a 43-kDa protein species on PBMC and 104-C1 cells but not 3201, CrFK, and G355-5 cells. A similarly sized molecule was bound by both PPR and 34TF10 SU adhesins on CrFK transduced with MIGR1-GFP/CD134 (CrFK GFP/CD134) but not on CrFK transduced with MIGR1 vector expressing only GFP (CrFK GFP). The indicated cell lysates were resolved by SDS-PAGE. Blots were then overlaid with the indicated adhesins. Binding of gp95 was revealed by an anti-Fc-HRP antibody. (C) FACS analysis revealing the specific interaction of FIV SU with CD134. CrFK cells were transduced with MIGR1-GFP or MIGR1-GFP/CD134. Binding with PPR-Fc and 34-Fc was assessed by FACS analysis in the absence (none) or presence of AMD3100 (AMD), heparin (HEP), or both (H+P). Note that neither AMD3100, heparin, or a combination of both inhibitors inhibited the binding of either SU adhesin to CrFK expressing CD134. Heparin and AMD3100 were used at 1 μg/ml.
Detection of CXCR4 and HSPGs is distinct at 4 and 22°C.
It has previously been shown that CXCR4 can adopt different conformations from one cell type to another (2). The results observed here with 3201 cells (Fig. 1B) suggest that the shift in the temperature from 4 to 22°C increases the affinity of FIV SU for CXCR4, probably through conformational rearrangement of SU. However, the increase in affinity could also be the result of a change in the conformation of CXCR4 between the two temperatures. We also observed a difference in the sensitivity of 34-Fc binding to inhibition by heparin at 4 and 22°C (Fig. 2), which might indicate a conformational change that influences HSPG exposure and/or presentation. We therefore analyzed the relative detection of CXCR4 and HSPGs at 4 and 22°C on the different cell lines (Fig. 4) by using an anti-human CXCR4 monoclonal antibody that cross-reacts with feline CXCR4 (clone 44717.111; R&D Systems) and another monoclonal antibody that specifically recognizes heparan sulfate chains present on cell surface proteoglycans (clone F58-10E40; Seikagaku Corp.). CXCR4 detection, particularly on 3201 cells, is increased markedly upon switching the temperature from 4 to 22°C (Fig. 4, top panel) but less pronounced on 104-C1 or G355-5 cell lines that express lower levels of CXCR4. Pretreatment of 3201 cells with cycloheximide (30 μg/ml) or sodium azide (0.005%) did not abolish the increase in CXCR4 detection (data not shown), a finding consistent with the concept that a change in conformation rather than an increase in CXCR4 expression (either via protein synthesis or a change in membrane turnover rate) was responsible for increased antibody binding. In contrast, we observed a consistent decrease in the detection of HSPGs when the temperature was switched from 4 to 22°C (Fig. 4). The decrease was substantial in the case of G355-5 cells and detectable with the other cells (Fig. 4). The results offer an explanation as to why heparin has reduced inhibitory activity on the binding of 34-Fc to 3201 cells at 22°C (Fig. 2), since binding under these conditions is almost exclusively via CXCR4 interactions.
FIG. 4.
Temperature-dependent detection of CXCR4 and HSPGs. Cells were labeled at the indicated temperatures, and the detection of CXCR4 and HSPGs was carried out by FACS analysis. The switch from 4 to 22°C induced an increase in CXCR4 detection that was most noticeable on 3201 cells. In contrast, HSPG detection was higher at 4°C and markedly decreased when the temperature was raised to 22°C.
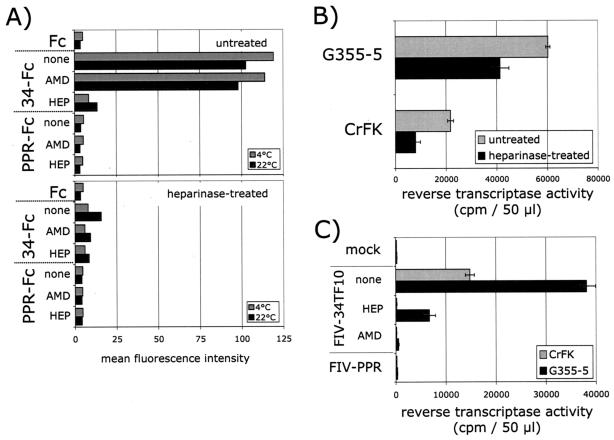
Heparan sulfate chains facilitate infection of CrFK and G355-5 cells by FIV-34TF10.
The entry receptor, CXCR4, is expressed at relatively low levels on G355-5 cells and CD134 is not expressed on these cells (Fig. 3). Thus, the presence of a binding receptor such as HSPGs might improve efficiency of FIV-34TF10 infection. To address this issue, binding studies were carried out on G355-5 cells in the presence or absence of heparinase treatment (Fig. 5A). Treating the cells with heparinase reduced the binding of 34-Fc to G355-5 cells by >90% and was more pronounced at 4°C than at 22°C. We next investigated the role of HSPGs in FIV-34TF10 infection of CrFK and G355-5 cells (Fig. 5B and C). In the first experiment (Fig. 5B), cells were left untreated or treated with heparinase and then washed and infected with FIV-34TF10, and virus production was assessed at 7 days by RT assay. As shown on Fig. 5B, heparinase treatment reduced FIV infectivity by ca. 35% on both cell lines, a finding consistent with a facilitating role of HSPGs in the infectivity by FIV-34TF10 in these adherent cells. In another set of experiments, infections were carried out with FIV-34TF10 in the absence or presence of heparin (10 μg/ml) or AMD3100 (1 μg/ml), and virus production was measured at 7 days postinfection (Fig. 5C). The results show that heparin neutralizes FIV-34TF10 infection of CrFK and G355-5 cells (Fig. 5C), with reductions of 90 and 80%, respectively. Complete inhibition of infection of both cell lines by AMD3100 confirmed that the entry receptor is CXCR4. The overall data suggest that HSPGs play a significant role in the permissivity of CrFK and G355-5 cells to FIV-34TF10 infection, presumably by acting as a primary binding receptor, in the absence of CD134.
FIG. 5.
HSPGs facilitate TCA FIV binding and infection of G355-5 and CRFK cells. (A) Binding of FIV SU adhesins was analyzed at 4 and 22°C before and after heparinase treatment of G355-5 cells. Heparinase treatment significantly reduced the binding of 34-Fc. Inhibition of binding by AMD3100 (AMD) and heparin (HEP) showed that residual binding after heparinase treatment is mediated by both CXCR4 and HSPGs. (B) Heparinase treatment reduced FIV-34TF10 infectivity. CrFK and G355-5 Cells were treated in the absence or presence of heparinase, washed, and then infected with FIV-34TF10. Virus production was monitored by an RT assay at 7 days postinfection. (C) Heparin efficiently neutralized FIV infection. Cells were mock infected or infected with FIV-34TF10 and FIV-PPR. Cells infected with FIV-34TF10 were treated in the absence (none) or presence of heparin (10 μg/ml, HEP) or AMD3100 (1 μg/ml, AMD). Virus production was monitored by an RT assay at 7 days postinfection.
CXCR4 expression level is a limiting factor in permissivity of G355-5 cells to productive infection by FIV-PPR.
As indicated above, primary strains such as FIV-PPR do not productively infect CrFK and G355-5 cells. However, after 3 to 4 weeks in culture, adaptation occurs and viruses emerge that are then able to productively infect these cells with kinetics of infection similar to that of FIV-34TF10 (22). The low level of CXCR4 expression on CrFK and G355-5 cells and/or the inability of the PI strain to bind to HSPGs may explain why these cells are not permissive to FIV-PPR infection. To address this issue, human CXCR4, known to bind FIV and facilitate infection (5, 43, 45), was transfected into G355-5 cells, and CXCR4high cells were sorted by flow cytometry. Binding experiments with the PPR-Fc and 34-Fc adhesins on CXCR4high cells (Fig. 6A) revealed a pattern of binding similar to that observed with 3201 T cells (Fig. 1) in that PPR-Fc efficiently bound CXCR4high cells at 22°C but not at 4°C. Binding of 34-Fc also increased with G355-5 cells at 22°C (Fig. 6A). Also, as observed with 3201 cells (Fig. 4), we noted significant increases in the detection of CXCR4 on the transduced cells at 22°C (Fig. 6B). The profile of HSPG detection at both temperatures was similar to that of the parental cells, although the levels of expression were lower (compare Fig. 6B and Fig. 4, bottom right panel). We next analyzed the inhibitory activity of AMD3100 and heparin on the binding of both SU adhesins (Fig. 6C). A difference in the inhibitory activity pattern of both compounds was observed between 4 and 22°C. As with binding to 3201 cells (Fig. 1 and 2), only 34-Fc bound to the CXCR4 transduced cells at 4°C, and the inhibition of binding was most pronounced when both heparin and AMD3100 were added together (Fig. 6C). PPR-Fc binding observed at 22°C was not influenced by heparin but was completely inhibited by AMD3100, indicating that the sole binding receptor for PPR-Fc is CXCR4 (Fig. 6C).
FIG. 6.
Overexpression of CXCR4 results in efficient binding of PPR-Fc on G355-5 cells. (A) Binding of FIV SU adhesins at 4 and 22°C on CXCR4high G355-5 cells. At 22°C, PPR-Fc efficiently bound the CXCR4high cells, whereas no binding was observed at 4°C. (B) Detection profile of CXCR4 and HSPGs at 4 and 22°C on CXCR4high G355-5 cells. As with the 3201 cells (Fig. 4), CXCR4 detection was temperature dependent and increased after the temperature was raised to 22°C. The detection profile of HSPGs was similar to parental cells (Fig. 4). (C) Inhibition of binding of FIV SU adhesins to CXCR4high cells. Cells were incubated at 4 or 22°C with the indicated SU adhesins in the absence (none) or presence of AMD3100 (AMD), heparin (HEP), or both (A+H). SU binding, expressed as the mean fluorescence intensity, was analyzed by FACS analysis as described in Materials and Methods. Heparin and AMD3100 were used at 1 μg/ml. Fc alone was used as a negative control.
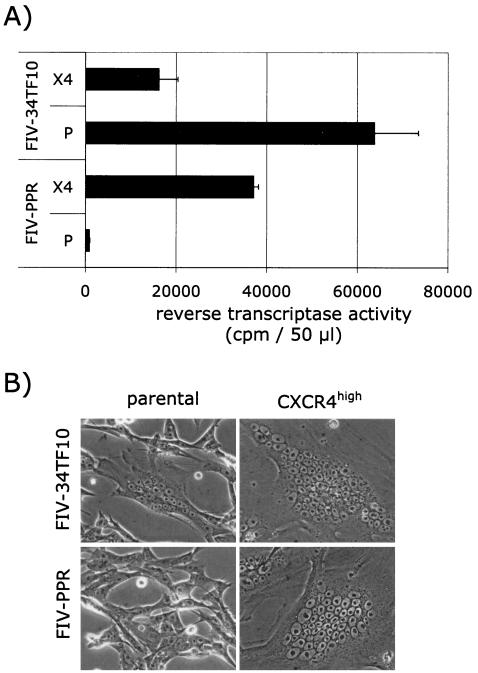
Next, parental and CXCR4high cells were infected with FIV-PPR and FIV-34TF10, and virus production was monitored at 5 days postinfection (Fig. 7A). Nontransduced G355-5 cells were permissive only to FIV-34TF10 infection, whereas CXCR4high cells were permissive to both FIV strains. Massive cell death started to occur at 5 days after the initiation of the infection in the CXCR4high cell population and was more significant for the 34TF10 infection, resulting in lower RT levels detected in the CXCR4high infected cell cultures than in the infected parental cell cultures (Fig. 7A). No syncytia were observed with the parental cells infected with the PPR strain, whereas a few syncytia with fewer than 15 nuclei were observed with parental cells infected with the 34TF10 strain (Fig. 7B). In contrast, giant syncytia with more than 40 to 50 nuclei were commonly observed in CXCR4high cells infected with either FIV strain, a finding consistent with a massive cell fusion activity (Fig. 7B).
FIG. 7.
Overexpression of CXCR4 renders G355-5 cells permissive to FIV-PPR infection. (A) Parental (P) and CXCR4high (X4) G355-5 cells were infected with FIV-34TF10 and FIV-PPR, and virus production was monitored at 5 days postinfection. (B) Massive cell fusion was observed in the CXCR4high cell population infected with either FIV strain at 3 days postinfection. Small syncytia with fewer nuclei were observed only with the parental cells infected with FIV-34TF10.
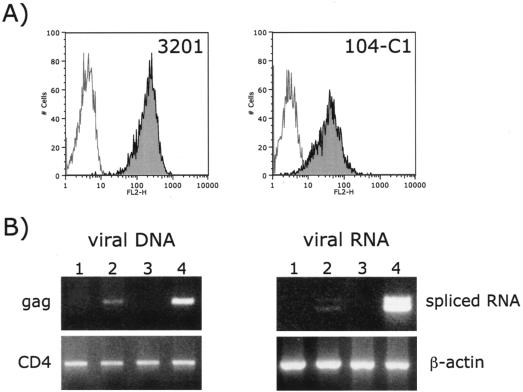
We have previously shown that 3201 cells were permissive to FIV-34TF10 infection and that productive infection with FIV-PPR occurred after 2 to 3 weeks of adaptation (23). Adapted viruses could then productively infect the 3201 cells with a peak in virus production usually observed 5 to 7 days after the initiation of infection. We surmised that since 3201 cells lacked the 43-kDa receptor, the binding to CXCR4 was inefficient (as noted at 4°C, Fig. 1A) and thus failed to facilitate virus entry and productive replication. However, in the present study, we observed substantial binding of the PPR-Fc adhesin at 22°C (Fig. 1B and 8A) but no productive infection, indicating that more than simple binding was involved in producing an infection that could generating significant levels of detectable virus. Kinetic analyses were performed, comparing viral DNA and RNA expression in 104-C1 and 3201 cells infected with FIV-PPR. The findings show that a degree of infection of 3201 cells does occur (Fig. 8B, lanes 2) but does not amplify as in 104-C1 cells (Fig. 8B, lanes 4), a finding consistent with either inefficient entry or a postentry block.
FIG. 8.
The block in infection of 3201 cells occurs at a postbinding level. (A) FACS analysis comparing binding of PPR-Fc at 22°C to 3201 (not permissive) and 104-C1 (permissive) cells. (B) PCR-based assay showing an apparent block of replication in 3201 cells. 3201 (lanes 1 and 2) and 104-C1 (lanes 3 and 4) cells were mock infected (lanes 1 and 3) or FIV-PPR infected (lanes 2 and 4), and newly synthesized viral DNA and viral RNA were analyzed at 20 and 72 h postinfection, respectively. PCR with primer pairs specific for a CD4 intron and β-actin were used as controls of DNA and RNA extractions, respectively.
DISCUSSION
FIV and HIV-1 share multiple similarities in the pathologies they induce in their respective hosts. One interesting paradox that emerges between both viruses is that the CD4 molecule is used as a primary binding receptor only by HIV-1, whereas both viruses induce a specific depletion of the CD4+-T-cell subset. The reason this cell population is selectively depleted in the case of FIV infection is interesting when one considers that FIV has a much broader tropism and also infects B cells, CD8+ cells, monocytes, and macrophages (3, 6, 7, 16, 27). Uncoupling of the use of CD4 and the specific depletion of CD4+ T cells observed with the cat model clearly indicates that this common mechanism in the pathogenesis of both infections is independent of the strict usage of CD4+ as a binding receptor. This notion is supported by the natural CD4-independent HIV-2 strains that have recently been described (15, 19, 31, 32).
One common point that exists between both viruses is that they use chemokine receptors for entry into target cells. Two main chemokine receptors, CCR5 and CXCR4, have been described for HIV, whereas only CXCR4 has been identified thus far for FIV (8, 17, 45). The presence of CD4 and an appropriate coreceptor define the tropism of HIV-1 at the level of virus entry. In the case of FIV, tropism is apparently governed by the tissue distribution of CXCR4. However, virus tropism may be influenced by several factors, including the affinity of the viral envelope for the receptor, the level of expression of the receptor, the conformational heterogeneity of the viral envelope as well as the receptor, the presence of a coreceptor, and/or the presence of attachment cofactors such as DC-SIGN (18) and HSPGs.
To date, three different receptors have been identified that interact with FIV gp95: CXCR4, HSPGs, and a 43-kDa gp95-binding protein (9, 45). Although CXCR4 interaction with FIV has been well documented (reviewed in reference 44), the role played by the two other attachment receptors is less clear. The recent findings of Shimojima et al. (36) that indicate that the primary binding receptor on T cells is CD134 is an extremely important observation and fits well with the binding data shown previously (9) and extended in the present studies. The molecular size and SU binding properties of CD134 cloned and expressed on CrFK cells parallels precisely with the size and binding properties of the 43-kDa receptor on primary T cells (9) (Fig. 3). The present study extends the Shimojima study in demonstrating that CD134 binds directly to FIV SU, thus confirming its role as a binding receptor rather than a facilitating role through some indirect means.
In addition, the present study demonstrates that the binding of FIV gp95 to CXCR4 is influenced by temperature and is presumed to be a function of the conformational heterogeneity of CXCR4, gp95, or both molecules. Furthermore, the data show that the level of CXCR4 expression is critical to the relative ability of a given SU to facilitate virus infection. Finally, data are presented that support a facilitator role for HSPGs and presumably for CD134 to enhance infection by TCA and PI FIVs, respectively. CD134 is expressed on PBMC and IL-2-dependent T-cell lines, targets for productive infection by PI and TCA FIVs. Since permissivity to FIV-PPR infection is correlated to cells expressing CD134 (9, 36; the present study), binding of gp95 to this receptor may increase exposure of the CXCR4 binding site on gp95 similar to what is observed upon binding of gp120 by CD4.
A switch in temperature from 4 to 22°C induced a dramatic increased in the extent of binding of gp95 to CXCR4, a finding consistent with a temperature-dependent conformational change for ligand, receptor, or both proteins. Although the temperatures used in the assays reported here are not physiological, this temperature differential facilitated discrimination of binding properties of the viral glycoproteins under in vitro conditions that maintain cellular integrity during FACS analyses. Similar results were observed at 22 and 37°C but with improved cell viability at the former temperature. The results suggest that CXCR4 may undergo a conformational rearrangement dependent on the temperature of the labeling assay. Indeed, results showing a marked specific increase in anti-CXCR4 antibody binding at 22°C versus at 4°C (Fig. 4) supports the notion of a temperature-dependent conformational change in the chemokine receptor. It is also known that different conformations of CXCR4 exist from one cell type to another (2). Whether gp95 also undergoes a conformational change to facilitate better CXCR4-binding remains to be formally verified.
HSPGs are well known in virus research since they have often been implicated as adsorption receptors for many viruses, including HIV and FIV (4, 9, 35, 40). Here we showed that HSPGs can facilitate FIV infection, at least with laboratory-adapted strains such as FIV-34TF10, a virus that has been adapted to grow on CrFK cells (39). CrFK cells are usually not permissive to FIV infection by primary FIV strains. Adaptation to productively grow in CrFK cells requires several weeks of passage and is associated mainly with mutations of amino acid residues at positions 407 and 409 to positively charged lysine residues in the V3 loop of SU (37, 42), resulting in an increase in the electrostatic charge in that region of the surface glycoprotein. Whether these mutations induced an increase in the affinity of SU for CXCR4 or HSPGs remains to be determined, but our data support the second hypothesis. First, CrFK cells express very low levels of CXCR4 and very high levels of HSPGs (Fig. 4). Second, heparin treatment had a profound inhibitory and neutralizing effect on FIV 34TF10 SU binding and virus infectivity (Fig. 5). Finally, infection of G355-5 cells by FIV-34TF10 was less sensitive to heparin neutralization, and we attributed this to a slightly higher level of CXCR4 that compensated for the loss in binding to HSPGs.
The overall data clearly indicated (i) that CXCR4 expression is the limiting factor in the productive infection of these cells by FIV and (ii) that the level of CXCR4 expression and the relative affinity of SU for CXCR4 control the degree of virus spread and cytopathogenicity; i.e., actions that increase the effective concentration of CXCR4 result in substantial increases in SU binding, which, in the majority of cases, translates into a productive infection. Additional structural study will be required to define the precise relationship between SU and the CD134 coreceptor. The binding of PI PPR SU to CXCR4 did not require the presence of a soluble form of CD134, in contrast to binding by HIV SU, where sCD4 was strictly required for gp120 binding to its coreceptor. However, CD134 likely compensates for the relatively low CXCR4 expression we observed on primary T cells by facilitating SU/CXCR4 association. A similar role may be played by HSPGs on CD134− CrFK and G355-5 cells, where binding to SU of the TCA strain increases the effective concentration of the ligand or receptor. The experiments showing that overexpression of CXCR4 on CrFK and G355-5 cells apparently over-rode the need for a secondary binding receptor support this model.
Acknowledgments
This study was supported by grant R01 AI25825 from the National Institutes of Health.
We are grateful to Dennis Burton, Donald Mosier, Joseph Sodroski, Tajib Mirzabkov, Nathaniel Landau, Brian Seed, and Chris Grant for the contribution of certain cell lines and plasmids used in this study and to Sohela de Rozières for careful reading of the manuscript. We thank the AIDS Research and Reference Reagent Program for providing the Cf2Th/synCCR5 cells and plasmid pBABE.CXCR4. We also thank Jackie Wold for assistance in preparation of the manuscript.
REFERENCES
- 1.Babcock, G. J., T. Mirzabekov, W. Wojtowicz, and J. Sodroski. 2001. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 276:38433-38440. [DOI] [PubMed] [Google Scholar]
- 2.Baribaud, F., T. G. Edwards, M. Sharron, A. Brelot, N. Heveker, K. Price, F. Mortari, M. Alizon, M. Tsang, and R. W. Doms. 2001. Antigenically distinct conformations of CXCR4. J. Virol. 75:8957-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beebe, A. M., N. Dua, T. G. Faith, P. F. Moore, N. C. Pedersen, and S. Dandekar. 1994. Primary stage of feline immunodeficiency virus infection: viral dissemination and cellular targets. J. Virol. 68:3080-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobardt, M. D., A. C. Saphire, H. C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. A. Gallay. 2003. Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 18:27-39. [DOI] [PubMed] [Google Scholar]
- 5.Brelot, A., N. Heveker, K. Adema, M. J. Hosie, B. Willett, and M. Alizon. 1999. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 73:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, W. C., L. Bissey, K. S. Logan, N. C. Pedersen, J. H. Elder, and E. W. Collisson. 1991. Feline immunodeficiency virus infects both CD4+ and CD8+ T lymphocytes. J. Virol. 65:3359-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunner, D., and N. C. Pedersen. 1989. Infection of peritoneal macrophages in vitro and in vivo with feline immunodeficiency virus. J. Virol. 63:5483-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major coreceptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 9.de Parseval, A., and J. H. Elder. 2001. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J. Virol. 75:4528-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Parseval, A., D. L. Lerner, P. Borrow, B. J. Willett, and J. H. Elder. 1997. Blocking of feline immunodeficiency virus infection by a monoclonal antibody to CD9 is via inhibition of virus release rather than interference with receptor binding. J. Virol. 71:5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Parseval, A., S. V. Su, J. H. Elder, and B. Lee. 2004. Specific interaction of feline immunodeficiency virus surface glycoprotein with human DC-SIGN. J. Virol. 78:2597-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 13.Egberink, H. F., E. De Clercq, A. L. Van Vliet, J. Balzarini, G. J. Bridger, G. Henson, M. C. Horzinek, and D. Schols. 1999. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J. Virol. 73:6346-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo, Y., Y. Goto, Y. Nishimura, T. Mizuno, T. Watari, A. Hasegawa, T. Hohdatsu, H. Koyama, and H. Tsujimoto. 2000. Inhibitory effect of stromal cell derived factor-1 on the replication of divergent strains of feline immunodeficiency virus in a feline T-lymphoid cell line. Vet. Immunol. Immunopathol. 74:303-314. [DOI] [PubMed] [Google Scholar]
- 15.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 16.English, R. V., C. M. Johnson, D. H. Gebhard, and M. B. Tompkins. 1993. In vivo lymphocyte tropism of feline immunodeficiency virus. J. Virol. 67:5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 18.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 19.Hoxie, J. A., C. C. LaBranche, M. J. Endres, J. D. Turner, J. F. Berson, R. W. Doms, and T. J. Matthews. 1998. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J. Reprod. Immunol. 41:197-211. [DOI] [PubMed] [Google Scholar]
- 20.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 21.Latza, U., H. Durkop, S. Schnittger, J. Ringeling, F. Eitelbach, M. Hummel, C. Fonatsch, and H. Stein. 1994. The human OX-40 homolog: cDNA structure, expression and chromosomal assignment of the ACT35 antigen. Eur. J. Immunol. 24:677-683. [DOI] [PubMed] [Google Scholar]
- 22.Lerner, D. L., and J. H. Elder. 2000. Expanded host cell tropism and cytopathic properties of feline immunodeficiency virus strain PPR subsequent to passage through interleukin-2-independent T cells. J. Virol. 74:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerner, D. L., C. K. Grant, A. de Parseval, and J. H. Elder. 1998. FIV infection of IL-2-dependent and -independent feline lymphocyte lines: host cells range distinctions and specific cytokine upregulation. Vet. Immunol. Immunopathol. 65:277-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirzabekov, T., N. Bannert, M. Farzan, W. Hofmann, P. Kolchinsky, L. Wu, R. Wyatt, and J. Sodroski. 1999. Enhanced expression, native purification, and characterization of CCR5, a principal HIV-1 coreceptor. J. Biol. Chem. 274:28745-28750. [DOI] [PubMed] [Google Scholar]
- 25.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norimine, J., T. Miyazawa, Y. Kawaguchi, K. Tomonaga, Y. S. Shin, T. Toyosaki, M. Kohmoto, M. Niikura, Y. Tohya, and T. Mikami. 1993. Feline CD4 molecules expressed on feline non-lymphoid cell lines are not enough for productive infection of highly lymphotropic feline immunodeficiency virus isolates. Arch. Virol. 130:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novotney, C., R. V. English, J. Housman, M. G. Davidson, M. P. Nasisse, C. R. Jeng, W. C. Davis, and M. B. Tompkins. 1990. Lymphocyte population changes in cats naturally infected with feline immunodeficiency virus. AIDS 4:1213-1218. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, T. R., R. Talbott, C. Lamont, S. Muir, K. Lovelace, and J. H. Elder. 1990. Comparison of two host cell range variants of feline immunodeficiency virus. J. Virol. 64:4605-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poeschla, E. M., and D. J. Looney. 1998. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J. Virol. 72:6858-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potempa, S., L. Picard, J. D. Reeves, D. Wilkinson, R. A. Weiss, and S. J. Talbot. 1997. CD4-independent infection by human immunodeficiency virus type 2 strain ROD/B: the role of the N-terminal domain of CXCR-4 in fusion and entry. J. Virol. 71:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves, J. D., A. McKnight, S. Potempa, G. Simmons, P. W. Gray, C. A. Power, T. Wells, R. A. Weiss, and S. J. Talbot. 1997. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology 231:130-134. [DOI] [PubMed] [Google Scholar]
- 33.Richardson, J., G. Pancino, R. Merat, T. Leste-Lasserre, A. Moraillon, J. Schneider-Mergener, M. Alizon, P. Sonigo, and N. Heveker. 1999. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J. Virol. 73:3661-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saphire, A. C., M. D. Bobardt, Z. Zhang, G. David, and P. A. Gallay. 2001. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 75:9187-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimojima, M., T. Miyazawa, Y. Ikeda, E. L. McMonagle, H. Haining, H. Akashi, Y. Takeuchi, M. J. Hosie, and B. J. Willett. 2004. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 303:1192-1195. [DOI] [PubMed] [Google Scholar]
- 37.Siebelink, K. H., J. A. Karlas, G. F. Rimmelzwaan, A. D. Osterhaus, and M. L. Bosch. 1995. A determinant of feline immunodeficiency virus involved in Crandell feline kidney cell tropism. Vet. Immunol. Immunopathol. 46:61-69. [DOI] [PubMed] [Google Scholar]
- 38.Sun, P., P. Dong, K. Dai, G. J. Hannon, and D. Beach. 1998. p53-independent role of MDM2 in TGF-β1 resistance. Science 282:2270-2272. [DOI] [PubMed] [Google Scholar]
- 39.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanabe-Tochikura, A., T. S. Tochikura, J. R. Blakeslee, Jr., R. G. Olsen, and L. E. Mathes. 1992. Anti-human immunodeficiency virus (HIV) agents are also potent and selective inhibitors of feline immunodeficiency virus (FIV)-induced cytopathic effect: development of a new method for screening of anti-FIV substances in vitro. Antivir. Res. 19:161-172. [DOI] [PubMed] [Google Scholar]
- 41.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 42.Verschoor, E. J., L. A. Boven, H. Blaak, A. L. van Vliet, M. C. Horzinek, and A. de Ronde. 1995. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J. Virol. 69:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willett, B. J., K. Adema, N. Heveker, A. Brelot, L. Picard, M. Alizon, J. D. Turner, J. A. Hoxie, S. Peiper, J. C. Neil, and M. J. Hosie. 1998. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J. Virol. 72:6475-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willett, B. J., and M. J. Hosie. 1999. The role of the chemokine receptor CXCR4 in infection with feline immunodeficiency virus. Mol. Membr. Biol. 16:67-72. [DOI] [PubMed] [Google Scholar]
- 45.Willett, B. J., L. Picard, M. J. Hosie, J. D. Turner, K. Adema, and P. R. Clapham. 1997. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J. Virol. 71:6407-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]