Abstract
Ecological research often assumes that species are adapted to their current climatic environments. However, climate fluctuations over geologic timescales have influenced species dispersal and extinction, which in turn may affect community structure. Modern community structure is likely to be the product of both palaeoclimate and modern climate, with the relative degrees of influence of past and present climates unknown. Here, we assessed the influence of climate at different time periods on the phylogenetic and functional trait structure of 203 African mammal communities. We found that the climate of the mid-Holocene (approx. 6000 years ago) and Last Glacial Maximum (approx. 22 000 years ago) were frequently better predictors of community structure than modern climate for mammals overall, carnivorans and ungulates. Primate communities were more strongly influenced by modern climate than palaeoclimate. Overall, community structure of African mammals appears to be related to the ecological flexibility of the groups considered here and the regions of continental Africa that they occupy. Our results indicate that the future redistribution, expansion and contraction of particular biomes due to human activity, such as climate and land-use change, will differentially affect mammal groups that vary in their sensitivity to environmental change.
Keywords: community assembly, palaeoclimate, functional traits, phylogenetic structure
1. Introduction
Understanding the factors that shape biological communities, and the relative strength of those factors, is a central goal of ecology. Among potential factors, it is widely accepted that climate plays a major role in shaping communities, and thus the interaction between climate and community structure patterns is a principal focus of ecological research [1–5]. Much of this research operates under the assumptions that (i) species are fundamentally in equilibrium with their current environment [6] and (ii) species are adapted to current climatic conditions [7]. These two related assumptions, however, are not always met, and can affect both applied and theoretical conclusions. For example, most species distribution models project future species distributions based on the climatic conditions where a species is currently found [6,8], implicitly or explicitly assuming that species are in equilibrium with current climatic conditions [7] and accordingly will track climate change in real time. It is possible, however, that species distributions are more strongly shaped by factors other than today's climate, such as palaeoclimate, and that models built only on modern climate will have limited predictive power. Likewise, an organism or community's functional traits may be adapted to past climatic conditions and have been retained despite climate change. When an anticipated relationship between functional traits and modern climate is not found, many studies invoke ‘phylogenetic inertia’ or some other non-adaptive mechanism to explain their findings [9–11], even when palaeoclimatic influences remain unexplored and may offer insight into observed patterns. Few studies have assessed the palaeoclimatic influence on functional diversity, as recently noted by Svenning et al. [12].
Analyses of the phylogenetic and functional trait structure of communities have been used as a proxy to understand the relative roles of assembly processes that structure communities, primarily competition and habitat filtering [1,3,13,14]. These metrics are sensitive to the ‘clustering’ or ‘overdispersion’ of related taxa or traits within a community and are based on ecological theory [2,5,13]. For example, the outcome of competition within a community is often niche partitioning or niche differentiation between competitors, resulting in an ‘even spacing’ pattern of close relatives or traits [1,13,15,16]. Conversely, habitat filtering is based on the concept that environmental gradients across space may serve as a filter because only species with specialized physiological or ecological traits can successfully inhabit particular, often stressful, environments [17]. The resulting pattern is one where closely related taxa or similar traits are ‘clustered’. Thus, it can be expected that competition is a strong factor in community assembly in environments where numerous taxa can successfully persist, while habitat filtering is more likely to be strong in environments where only a few taxa with specialized adaptations can successfully persist.
Here we analyse the relative influence of modern climate and palaeoclimate on the phylogenetic and functional trait structure of terrestrial African mammal communities. Mammals are an excellent faunal group for such a study, as their modern distributions are relatively well known and data on species traits are more readily available than for other animal clades. Mammals vary considerably in their functional traits, such as substrate use, dispersal ability, body size and diet, which may in turn affect underlying community assembly processes [18]. Africa is also home to a great diversity of species in 17 of the world's 20 orders of terrestrial mammals [19–21], and is virtually the only continent relatively unscathed by the late Quaternary extinction events [22,23]. Nevertheless, late Quaternary palaeoclimate change has been noted as a particularly important time period in the evolution and biogeography of African mammals [24–35], especially recurrent expansions and contractions of major vegetation biomes as climates changed [29,36–40].
We quantified the phylogenetic and functional trait structure of 203 African mammal communities and predicted these community structure metrics using both modern and palaeoclimatic datasets of the Last Glacial Maximum (LGM; approx. 22 000 years ago) and mid-Holocene (approx. 6000 years ago) with linear models and multimodel inference. Using these analyses, we address the question ‘what are the relative influences of modern and palaeoclimate on the phylogenetic and trait structure of African mammal communities?’ We predict that if mammal taxa strongly track climatic and environmental change, then modern climate should be the best predictor of community structure. Alternatively, if taxa are more ecologically flexible and/or if palaeoenvironmental history is important, then community structure may be better predicted by palaeoclimate.
2. Material and methods
(a). Community and trait data
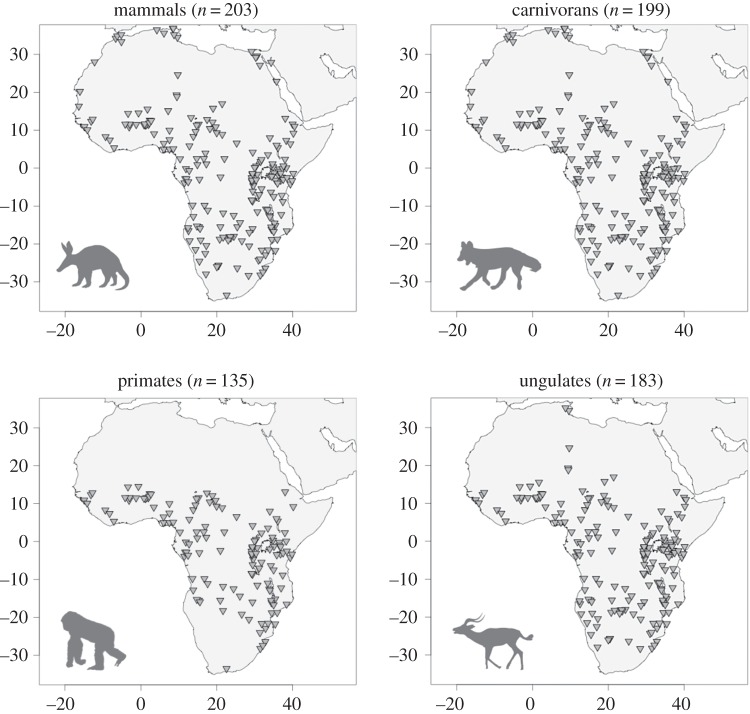
We compiled species lists of terrestrial mammal communities for 203 communities from national parks, game reserves and protected areas, spanning the entire African continent (figure 1; electronic supplementary material, table S1). Data came from published field surveys and park lists, existing databases and primary literature; we avoided using range maps because they often overestimate species occurrences [41,42]. Only terrestrial mammals weighing more than 500 g were included, as data for micromammals (e.g. trapping of rodent species) and bats are generally less available and less reliable. The overall mammal dataset included representatives from nine mammalian orders (electronic supplementary material, table S2). We then subdivided three groups from the overall mammal data: carnivorans (order Carnivora), primates (order Primates) and ungulates (orders Artiodactyla and Perissodactyla). We use the gradistic term ‘ungulate’ to include all African species of Artiodactlya and Perissodactyla, as the species in these orders broadly overlap in body size and dietary ecology [43] and thus probably influence one another more so than other groups during community assembly processes. All communities included a minimum of four species. Our final database consisted of 203 mammal, 199 carnivoran, 135 primate and 183 ungulate communities.
Figure 1.
Map of mammal, carnivoran, primate and ungulate communities used in this study.
We used body mass and diet as functional traits, as these traits are important characteristics of a species’ niche, and therefore are likely strongly linked to processes determining community structure (e.g. habitat filtering, interspecific competition). For example, body mass is highly correlated with many physiological, life history and ecological traits in carnivorans, primates and ungulates [44–46]. Trait data for mammal species were collected from Kingdon et al. [47] for body mass and Kissling et al. [48] for dietary data. The Kissling et al. [48] dietary dataset consists of categorical dietary variables (e.g. ‘invertebrate’, ‘mammal’, ‘fruit’ or ‘seed’) that are ranked by importance for each species. These data were crosschecked and supplemented with sources from other databases and field guides [49–51] for consistency.
Body mass was recorded as a continuous variable in grams, based on the average female body mass per species. Diet was also measured as a continuous variable, obtained from the ordination of the Kissling et al. [48] dietary data matrix. These data were used in a correlation matrix-based principal components analysis (PCA) using the prcomp function in R (electronic supplementary material, figure S1). We used the first two principal components in all analyses (55.5% total variance explained), as these were the only components with eigenvalues greater than 1.0. The component loadings were used to determine the dietary signal (e.g. carnivory, frugivory, herbivory) that each axis captured. Therefore, each mammal's score from the ordination represented its diet relative to all of the other mammals in the analysis. In addition, we also calculated dietary axes using principal coordinates analysis (PCoA) with Gower distances. However, the dietary axes generated using PCoA were strongly related to those generated from the PCA (e.g. PCoA axis 1∼PCA axis 1 r2 = 0.98) and therefore, we only used the PCA dietary axes for analyses. Raw trait data are presented in electronic supplementary material, table S2.
(b). Climate data
A central geospatial coordinate (latitude, longitude) was collected for each community. Rasters of climate data for three periods (modern, mid-Holocene, LGM) were downloaded from ecoClimate [52] for seven different general circulation models (GCMs) from the Couple Model Intercomparison Project (CMIP5) and Paleoclimate Modeling Intercomparison Project (PMIP3) working groups. These GCMs are widely used in ecological studies involving palaeoclimate [53–57]. From each of these GCMs, six bioclim variables were extracted using the central point of each community: BIO1, annual mean temperature; BIO4, temperature seasonality; BIO6, minimum temperature of the coldest month; BIO12, mean annual precipitation; BIO14, precipitation of the driest month and BIO15, precipitation seasonality (electronic supplementary material, table S1). We used these variables because they were not highly correlated with one another and are important for structuring African mammal communities [5]. For the results presented in the main text of the paper, we averaged bioclim variables across all GCMs to create an ‘average’ model for each time period. However, we include results from the seven individual GCMs in the electronic supplementary material.
For each of the seven GCMs and the average model, the six climate variables were log transformed and used in a correlation matrix-based PCA using the prcomp function in R. Individual PCAs were used for each of the three climate datasets for each of the GCMs and the average model (electronic supplementary material, figures S2–S4). We used the first two principal components in all analyses, as these encompassed approximately 80% of the variation within the data and had eigenvalues greater than 1.0. The component loadings were used to determine the climatic signals along each axis.
(c). Community structure metrics
We used two metrics to characterize the phylogenetic structure of communities, the nearest taxon index (NTI) and the net relatedness index (NRI), following Webb et al. [13] (electronic supplementary material, table S3). NTI is calculated as the phylogenetic distance between the two most closely related co-occurring taxa in a community relative to the entire species pool (i.e. all species in all communities). NRI is calculated as the average phylogenetic distance among species in a community related to the species pool. These methods have been applied extensively to plant [1,58] and animal [2,3,59,60] communities. We used the PHYLOCOM software package [61] to calculate NTI and NRI using a null model of 4999 randomizations to standardize these metrics. Pausas & Verdu [62] and Miller et al. [63] provide overviews of the null model approach for analysing community phylogenetic structure. The mammalian phylogeny of Bininda-Emonds et al. [64] was used for all phylogenetic analyses, as this phylogeny contains all species in our dataset and has been widely used in previous research [2,5,65,66]. However, we also calculated NRI and NTI from the newly available Faurby & Svenning [67] species-level phylogeny of all extant mammals. The NRI and NTI values calculated from the Faurby & Svenning [67] phylogeny were in almost all cases highly correlated with those using the Bininda-Emonds et al. [64] phylogeny (mammals NRI, r2 = 0.97; carnivorans NRI, r2 = 0.88; primates NRI, r2 = 0.98; ungulates NRI, r2 = 0.97; mammals NTI, r2 = 0.81; carnivorans NTI, r2 = 0.68; primates NTI, r2 = 0.94; ungulates NTI, r2 = 0.84). The single exception was carnivoran NTI (r2 = 0.68) and therefore we re-ran our linear models only for carnivoran NTI using the new Faurby & Svenning [67] phylogeny.
We used four metrics outlined by Kraft et al. [16] and Kraft & Ackerly [1] to characterize the functional trait structure of communities: range, variance, the standard deviation of nearest neighbour distance divided by the overall trait range (SDNNr), and the standard deviation of neighbour distance divided by the overall trait range (SDNDr). Two of these metrics (range, variance) are sensitive to habitat filtering, while the other two metrics (SDNNr, SDNDr) detect even spacing—a common pattern resulting from interspecific competition and niche partitioning [1]. Range is calculated as the difference between maximum and minimum trait value for a community and variance measures how widely species' trait values deviate from the community mean. SDNNr measures how distant the two most similar pair of species are to other species in the community, while SDNDr measures how regularly spaced taxa in a community are across a given trait range. More details on these metrics are provided in Kraft et al. [16] and Kraft & Ackerly [1]. As with the phylogenetic structure metrics, we used a null model approach to generate random communities of equal richness by extracting taxa from the entire meta-community pool weighted by their frequency of occurrence in the pool. We used 4999 randomizations to standardize these metrics.
(d). Models
We used linear regression models to analyse the relationship between phylogenetic and functional trait metrics and modern and palaeoclimate variables. Each model contained up to six climate predictors, as represented by the first two principal components from the PCAs of the three climate datasets (modern, mid-Holocene and LGM). In addition, each model included latitude and longitude of communities as predictors. Including latitude and longitude as predictors allowed us to account for potential spatial autocorrelation in the models (e.g. [68–70]). Moran's I values generated from model residuals show very little spatial autocorrelation (electronic supplementary material, table S9).
We used the corrected Akaike information criterion (AICc) to determine the best models of community structure. In addition, we calculated the sum of AICc weights for each climate principal component (i.e. predictor) to determine which climate variables were the best predictor of community structure metrics. We produced these values from the dredge function in the package MuMIn [71]. We averaged the models within the top 95% of model weights following Burnham & Anderson [72]; the sum of AICc weights for variables was calculated from all possible models. In addition, as some of our climate PC axes were correlated (e.g. Mod1, Hol1, LGM1), we also examined whether models containing only palaeoclimate (e.g. trait ∼ LGM1 + LGM2) were stronger than those containing only modern climate (e.g. trait ∼ Mod1 + Mod2).
We ran a total of 56 models for each GCM and the average model (448 models total): for phylogenetic structure, a model was run for each group (mammals, carnivorans, primates and ungulates) and for each phylogenetic structure metric (NRI, NTI), resulting in eight total phylogenetic models per GCM and the average model; for functional traits, models were run for each group for four metrics (range, variance, SDNNr, SDNDr) of three traits (body mass, diet PC1, diet PC2), which resulted in a total of 48 functional trait models per GCM.
3. Results
(a). Principal components analysis loadings
The first principal component of the dietary PCA (diet PC1) represented animal matter versus plant matter in the diet. The second principal component of the dietary PCA (diet PC2) represented fruit consumption and distinguishes invertebrate from vertebrate animal matter in the diet (electronic supplementary material, figure S1).
For the climate PCAs, the first principal component of the modern climate variables (Mod1) represented temperature seasonality (BIO4) and mean annual precipitation (BIO12). The second principal component of the modern climate variables (Mod2) represented mean annual temperature (BIO1) and precipitation intensity (BIO14) (electronic supplementary material, figure S2). The first principal component of the Holocene climate variables (Hol1) represented variables related to temperature seasonality or variation (BIO4; BIO6), while the second principal component (Hol2) represented precipitation seasonality and variation (BIO14, BIO15) (electronic supplementary material, figure S3). The first principal component of the LGM climate variables (LGM1) represented temperature seasonality (BIO4) and mean annual precipitation (BIO12) and the second component (LGM2) represented mean annual temperature (BIO1) and precipitation variation (BIO14) (electronic supplementary material, figure S4).
(b). Phylogenetic structure metrics
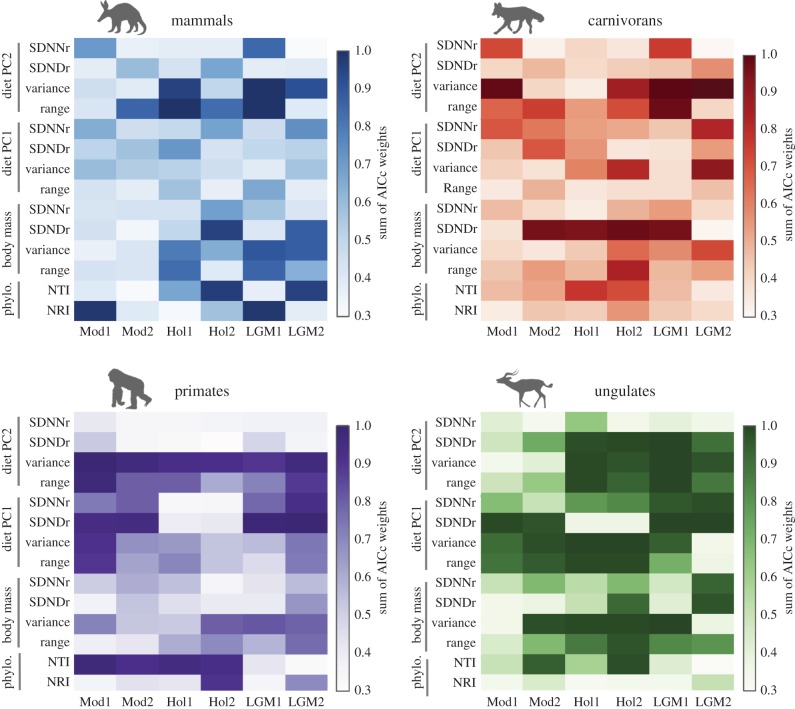
Mammal community phylogenetic structure was most strongly predicted by palaeoclimate, with LGM1 and Mod1 being the most important variables for NRI, while Hol2 and LGM2 strongly predicted NTI (figure 2; electronic supplementary material, figure S5 and table S5). Carnivoran phylogenetic structure was poorly predicted by climate variables overall, although mid-Holocene climate was the strongest predictor of both NRI and NTI (figure 2; electronic supplementary material, table S6). Primate phylogenetic community structure was influenced by both modern and palaeoclimate, as NRI was most strongly tied to Hol2, while NTI was strongly influenced by Mod1, although Mod2, Hol1 and Hol2 are also important variables (figure 2; electronic supplementary material, table S7). For ungulates, LGM2 was the most important variable for NRI, and Hol2 was the most important predictor of NTI (figure 2; electronic supplementary material, table S8).
Figure 2.
Heatmap of sum of Akaike information criterion (AICc) weights for each climate predictor across all phylogenetic and functional trait metrics. Climate data represent the first two principal component axes for an ordination of six climate variables per time period (see Material and methods). Phylogenetic structure metrics are net relatedness index (NRI) and nearest taxon index (NTI). Functional trait metrics include range, variance, the standard deviation of nearest neighbour distance divided by the overall trait range (SDNNr), and the standard deviation of neighbour distance divided by the overall trait range (SDNDr). (Online version in colour.)
(c). Functional trait structure metrics
Functional trait structure of mammal communities was strongly influenced by palaeoclimate variables, especially LGM climate (figure 2; electronic supplementary material, figure S5 and tables S4–S8). LGM climate most strongly influenced mammal body mass and the second principal component of the dietary PCA (diet PC2), while the first dietary component (diet PC1) was not strongly influenced by any climate variable, although mid-Holocene and LGM climate variables were the most important. Carnivoran functional trait structure was also best predicted by palaeoclimate overall, with mid-Holocene and LGM variables being the most important for body mass and diet PC2, while diet PC1 was influenced by both modern and LGM climate. The functional trait structure of primate communities, unlike the other groups, was similarly influenced by both modern and palaeoclimate, with Mod1 and LGM2 being especially strong. Conversely, ungulate communities showed the strongest influence of palaeoclimate as mid-Holocene and LGM climate were the top variables for all metrics and traits, but also had many more influential climate variables overall.
4. Discussion
We quantified several aspects of the phylogenetic and functional trait structure of modern African mammal communities and found that mid-Holocene and LGM climate were often equivalent if not better predictors of community structure than modern climate (table 1). The finding that African mammal community structure overall has been strongly influenced by palaeoclimate implies that many species are ecologically flexible, and/or that dispersal limitation has been strong enough to have prevented climate tracking over the last several thousand years.
Table 1.
Mean of sum of Akaike information criterion (AICc) weights for climate predictors across all phylogenetic and functional trait metrics.
| Mod1 | Mod2 | Hol1 | Hol2 | LGM1 | LGM2 | |
|---|---|---|---|---|---|---|
| mammals | 0.52 | 0.46 | 0.62 | 0.61 | 0.67 | 0.60 |
| carnivorans | 0.53 | 0.54 | 0.53 | 0.63 | 0.60 | 0.57 |
| primates | 0.68 | 0.63 | 0.55 | 0.56 | 0.57 | 0.69 |
| ungulates | 0.54 | 0.68 | 0.75 | 0.81 | 0.75 | 0.69 |
The relative strength of modern climate and palaeoclimate varied between mammalian groups (table 1). Most notably, primate community structure was strongly influenced by modern climate, whereas mammals, carnivorans and ungulates were more strongly influenced by palaeoclimate. A potential explanation for this pattern may lie in differences in ecological flexibility between mammal groups and the biomes of Africa they occupy. Primates are largely biome specific [73] with most of the African primate radiation being dependent on forest blocks in equatorial West and Central Africa [74–76]. LGM and mid-Holocene climate change are known to have significantly altered the distribution of forests across Africa, with forests contracting into small refugia during the arid LGM and subsequently expanding during the mid-Holocene as wetter conditions prevailed [37,77–79]. Molecular evidence suggests that shifts in forest vegetation during glacial and interglacial climate change strongly influenced African primates, including vicariance and dispersal in guenons [38,70] and gorillas [36], and demographic patterns in chimpanzees [80] and mandrills [81]. In addition, bioclimatic envelope models of African mammals and birds provide evidence for three major forest refugia in Central and West Africa [40], which would have represented the only viable habitats for most of Africa's primates during glacial periods. The combination of molecular and biogeographic evidence suggests that the highly forest-dependent primate radiation is a sensitive group to climatic and environmental change. Primate communities experienced local extinction and range contraction during glacial periods, whereas warm and wet interglacial periods provided opportunities for dispersal via the expanding forest biome. The ‘tracking’ of forests explains why biome-dependent primate communities are most closely tied to modern climate, whereas the comparatively ecologically resilient ungulate communities are more strongly predicted by palaeoclimate.
Ungulate communities were the most strongly influenced by palaeoclimate among all mammalian groups in our study (table 1). The connection between modern ungulate communities and palaeoclimate may be related to the ecological flexibility of ungulate species and the relative stability of savannah habitats compared with forests during the late Quaternary [37,77–79]. Ungulates are medium to large-sized mammals and ecological flexibility increases with body size in African mammals in general [73] and African ungulates specifically [46]. In addition, ungulates rely on leaves and/or grasses as a primary food source and these types of foods are widely distributed across several biomes ranging from semi-desert to forest ecotones [82]. Conversely, fruits play an important role in most primate diets and therefore primates are more closely tied to a single biome (forests) where fruit production is more plentiful [82]. Thus, ungulate communities, when compared with primates, are more ‘ecologically resilient’ and could have persisted in a greater diversity of biomes both across space and through time during Quaternary climate change. This ecological resilience of mammal communities has also been shown by Rodriguez [83,84] using fossil assemblages. Additionally, in contrast with forests, the savannahs of southern and eastern Africa were relatively stable throughout the Quaternary, experiencing only moderate encroachment from deserts along its perimeters during glacials [79,82] and expansion of forests during interglacials [77]. The stability of savannah environments extends deeper in time than the Quaternary, as fossil mammals and palaeoenvironmental proxies from palaeontological sites dating from the late Miocene onwards document a persistence of savannah mosaics over at least the last six to seven million years in southern and eastern Africa [85–90]. Today, these savannahs are home to the vast majority of the continent's ungulate diversity [75,91], and thus ungulate communities here experienced less habitat change than other areas of the continent. Furthermore, equatorial forests today and in the past are characterized by less seasonal temperature and rainfall regimes than savannahs, suggesting that ungulates may be more tolerant of climate and habitat shifts over longer timescales than primate species that are adapted to relatively stable forests environments.
That carnivoran communities are also best predicted by palaeoclimate is likely related to the patterning of mammal predator and prey species richness at macroscales. As with ungulates, carnivoran species richness is concentrated in the savannahs of eastern and southern Africa [75]. Mammal predator–prey species richness in Africa is only tightly linked in open habitats (e.g. savannahs); forests are skewed towards higher predator–prey ratios [92]. This discrepancy probably arises as the result of different environmental histories of savannahs and forests in Africa [92], but also from the fact that many predators of mammal species in forests are non-mammalian, such as snakes and raptors, which are major predators of primates [93–95]. Furthermore, carnivoran species are largely secondary and tertiary consumers and are not as dependent on local vegetation for subsistence as primates and ungulates. It is also clear that carnivoran communities have been greatly influenced by human activity today [96] and, at least, since the early Pleistocene during which the genus Homo evolved derived dietary strategies that placed hominins in direct conflict with Africa's diverse carnivore guild [97,98]. Thus, although ungulate and primate communities probably have also been influenced by humans, the fossil record demonstrates that human influence on carnivoran communities has been occurring for millions of years and therefore the anthropogenic impact on these communities may be greater than for other taxa.
Despite the importance of mid-Holocene and LGM climate in our analyses, it is likely that we have underestimated the role of palaeoclimate. Some of the climate variables in our analyses were highly correlated (e.g. Mod1, Hol1, LGM1) and this could result in inflated type II error rates [99]. Yet, this issue should not impact our main findings because high levels of collinearity result in increased type II error rates for individual predictors, especially when correlated predictors contain different levels of unbiased error [99,100]. In our case, palaeoclimate variables certainly contain more error because they are reconstructed values, as opposed to modern climate data based on direct measurements or interpolation from weather stations [101]. Therefore, the palaeoclimate variables should be associated with increased type II error, yet were often found to be better predictors of community structure than modern climate variables. In addition, models containing only palaeoclimate variables (e.g. trait ∼ LGM1 or trait ∼ LGM1 + LGM2) generally had lower AICc values and higher model weights than those containing only modern climate (e.g. trait ∼ Mod1 or trait ∼ Mod1 + Mod2) (electronic supplementary material, table S9). Thus, even given potential multi-collinearity issues, palaeoclimate was often a stronger predictor of community structure metrics than modern climate.
In sum, we found a strong role of palaeoclimate on modern community patterns and found that modern climate alone was not sufficient to explain the total influence of climatic factors on community structure in African mammals. Our results have implications for predicting the future of tropical biodiversity, as the tropics are the heart of the World's mammalian diversity [19,20] but are threatened by human activity. Anthropogenic climate change represents a major threat to the World's remaining mammalian diversity as novel climates are projected to appear mainly in the tropics, with annual temperatures projected over the next century to be up to 7°C warmer than today [102,103]. Both temperature and precipitation changes will result in a redistribution of major vegetation biomes across Africa [104–106]. Climatically induced biome changes are being exacerbated by extensive land cultivation (e.g. agriculture, logging) [107] by an ever-expanding human population in Africa, which is expected to contribute to most of global population growth over the next century [108]. Our results indicate that the future redistribution, expansion and contraction of particular biomes due to human activity will differentially affect mammal communities that vary in their sensitivity to environmental change.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank S. Faurby, S. Varela and two anonymous reviewers whose detailed comments and suggestions greatly improved this manuscript. Thanks to editors D. Costa and R. Wilson. D. Feary provided helpful comments on this work, and J. Franklin, H. Glowacka, E. Locke, R. Merchant and I. Smail provided assistance, discussions, and comments on earlier versions of this manuscript. Thanks to J. DeBenny and J. Cohn for help collecting the community data.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
J.R., J.M.K., L.B. and K.E.R. designed the research; all authors performed the research and contributed to writing the paper.
Competing interests
We declare we have no competing interests.
Funding
J.R. was supported by a National Science Foundation Graduate Research Fellowship.
References
- 1.Kraft NJ, Ackerly DD. 2010. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. ( 10.1890/09-1672.1) [DOI] [Google Scholar]
- 2.Cardillo M. 2011. Phylogenetic structure of mammal assemblages at large geographical scales: linking phylogenetic community ecology with macroecology. Phil. Trans. R. Soc. B 366, 2545–2553. ( 10.1098/rstb.2011.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham CH, Parra JL, Tinoco BA, Stiles FG, McGuire JA. 2012. Untangling the influence of ecological and evolutionary factors on trait variation across hummingbird assemblages. Ecology 93, S99–S111. ( 10.1890/11-0493.1) [DOI] [Google Scholar]
- 4.Soudzilovskaia NA, Elumeeva TG, Onipchenko VG, Shidakov II, Salpagarova FS, Khubiev AB, Tekeev DK, Cornelissen JHC. 2013. Functional traits predict relationship between plant abundance dynamic and long-term climate warming. Proc. Natl Acad. Sci. USA 110, 18 180–18 184. ( 10.1073/pnas.1310700110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamilar JM, Beaudrot L, Reed KE. 2015. Climate and species richness predict the phylogenetic structure of African mammal communities. PLoS ONE 10, e0121808 ( 10.1371/journal.pone.0121808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin J. 2010. Mapping species distributions: spatial inference and prediction. Cambridge, UK: University Press. [Google Scholar]
- 7.Araújo MB, Peterson AT. 2012. Uses and misuses of bioclimatic envelope modeling. Ecology 93, 1527–1539. ( 10.1890/11-1930.1) [DOI] [PubMed] [Google Scholar]
- 8.Elith J, Leathwick JR. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Syst. 40, 677–697. ( 10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 9.Blomberg SP, Garland T. 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899–910. ( 10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 10.Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003. ( 10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 11.Kamilar JM, Cooper N. 2013. Phylogenetic signal in primate behaviour, ecology and life history. Phil. Trans. R. Soc. B. 368, 20120341 ( 10.1098/rstb.2012.0341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svenning JC, Eiserhardt WL, Normand S, Ordonez A, Sandel B. 2015. The influence of paleoclimate on present-day patterns in biodiversity and ecosystems. Annu. Rev. Ecol. Syst. 46, 551–572. ( 10.1146/annurev-ecolsys-112414-054314) [DOI] [Google Scholar]
- 13.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 14.Cavender-Bares J, Kozak KH, Fine PV, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. ( 10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 15.Stubbs WJ, Bastow Wilson J. 2004. Evidence for limiting similarity in a sand dune community. J. Ecol. 92, 557–567. ( 10.1111/j.0022-0477.2004.00898.x) [DOI] [Google Scholar]
- 16.Kraft NJ, Valencia R, Ackerly DD. 2008. Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322, 580–582. ( 10.1126/science.1160662) [DOI] [PubMed] [Google Scholar]
- 17.Cornwell WK, Schwilk DW, Ackerly DD. 2006. A trait-based test for habitat filtering: convex hull volume. Ecology 87, 1465–1471. ( 10.1890/0012-9658(2006)87%5B1465:ATTFHF%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 18.Qian H. 2009. Global comparisons of beta diversity among mammals, birds, reptiles, and amphibians across spatial scales and taxonomic ranks. J. Syst. Evol. 47, 509–514. ( 10.1111/j.1759-6831.2009.00043.x) [DOI] [Google Scholar]
- 19.Ceballos G, Ehrlich PR. 2006. Global mammal distributions, biodiversity hotspots, and conservation. Proc. Natl Acad. Sci. USA 103, 19 374–19 379. ( 10.1073/pnas.0609334103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley LB, et al. 2010. Phylogeny, niche conservatism and the latitudinal diversity gradient in mammals. Proc. R. Soc. B 277, 2131–2138. ( 10.1098/rspb.2010.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingdon J. 2013. Mammalian evolution in Africa. In Mammals of Africa volume I: introductory chapters and Afrotheria (eds Kingdon J, et al.), pp. 75–100. London, UK: Bloomsbury Publishing. [Google Scholar]
- 22.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. 2004. Assessing the causes of Late Pleistocene extinctions on the continents. Science 306, 70–75. ( 10.1126/science.1101476) [DOI] [PubMed] [Google Scholar]
- 23.Faurby S, Svenning JC. 2015. Historic and prehistoric human-driven extinctions have reshaped global mammal diversity patterns. Divers. Distrib. 21, 1155–1166. ( 10.1111/ddi.12369) [DOI] [Google Scholar]
- 24.Wallace AR. 1903. Man's place in the Universe. London, UK: Chapman and Hall. [Google Scholar]
- 25.Lydekker R. 1908. The game animals of Africa. London, UK: Roland Ward. [Google Scholar]
- 26.Lönnberg E. 1929. The development and distribution of the African fauna in connection with and depending upon climate changes. Ark. Zool. 21A, 1–33. [Google Scholar]
- 27.Wayland EJ. 1940. Desert versus forest in Eastern Africa. Geogr. J. 96, 329–340. ( 10.2307/1788805) [DOI] [Google Scholar]
- 28.Kingdon J. 1974. East African mammals: an atlas of evolution in Africa, volume I. Chicago, IL: University of Chicago Press. [Google Scholar]
- 29.Kingdon J. 1990. Island Africa: the evolution of Africa's rare animals and plants. London, UK: Collins. [Google Scholar]
- 30.Vrba ES. 1992. Mammals as a key to evolutionary theory. J. Mamm. 73, 1–28. ( 10.2307/1381862) [DOI] [Google Scholar]
- 31.Vrba ES. 1995. The fossil record of African antelopes (Mammalia, Bovidae) in relation to human evolution and paleoclimate. In Paleoclimate and evolution, with emphasis on human origins (eds Vrba ES, Denton GH, Partridge TC, Burckle LH), pp. 385–424. New Haven, CT: Yale University Press. [Google Scholar]
- 32.Faith JT, Potts R, Plummer TW, Bishop LC, Marean CW, Tryon CA. 2012. New perspectives on middle Pleistocene change in the large mammal faunas of East Africa: Damaliscus hypsodon sp. nov. (Mammalia, Artiodactyla) from Lainyamok, Kenya. Palaeogeogr. Palaeoclimatol. Palaeoecol. 361, 84–93. ( 10.1016/j.palaeo.2012.08.005) [DOI] [Google Scholar]
- 33.Faith JT, Tryon CA, Peppe DJ, Fox DL. 2013. The fossil history of Grévy's zebra (Equus grevyi) in equatorial East Africa. J. Biogeogr. 40, 359–369. ( 10.1111/j.1365-2699.2012.02796.x) [DOI] [Google Scholar]
- 34.Faith JT. 2014. Late Pleistocene and Holocene mammal extinctions on continental Africa. Earth Sci. Rev. 128, 105–121. ( 10.1016/j.earscirev.2013.10.009) [DOI] [Google Scholar]
- 35.Rowan J, Faith JT, Gebru Y, Fleagle JG. 2015. Taxonomy and paleoecology of fossil Bovidae (Mammalia, Artiodactyla) from the Kibish Formation, southern Ethiopia: Implications for dietary change, biogeography, and the structure of the living bovid faunas of East Africa. Palaeogeogr. Palaeoclimatol. Palaeoecol. 420, 210–222. ( 10.1016/j.palaeo.2014.12.017) [DOI] [Google Scholar]
- 36.Anthony NM, et al. 2007. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc. Natl Acad. Sci. USA 104, 20 432–20 436. ( 10.1073/pnas.0704816105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowling SA, Cox PM, Jones CD, Maslin MA, Peros M, Spall SA. 2008. Simulated glacial and interglacial vegetation across Africa: implications for species phylogenies and trans-African migration of plants and animals. Glob. Change Biol. 14, 827–840. ( 10.1111/j.1365-2486.2007.01524.x) [DOI] [Google Scholar]
- 38.Tosi AJ. 2008. Forest monkeys and Pleistocene refugia: a phylogeographic window onto the disjunct distribution of the Chlorocebus Ihoesti species group. Zool. J. Linn. Soc. 154, 408–418. ( 10.1111/j.1096-3642.2008.00419.x) [DOI] [Google Scholar]
- 39.Lorenzen ED, Heller R, Siegismund HR. 2012. Comparative phylogeography of African savannah ungulates. Mol. Ecol. 21, 3656–3670. ( 10.1111/j.1365-294X.2012.05650.x) [DOI] [PubMed] [Google Scholar]
- 40.Levinsky I, Araújo MB, Nogués-Bravo D, Haywood AM, Valdes PJ, Rahbek C. 2013. Climate envelope models suggest spatio-temporal co-occurrence of refugia of African birds and mammals. Glob. Ecol. Biogeogr. 22, 351–363. ( 10.1111/geb.12045) [DOI] [Google Scholar]
- 41.Hurlbert AH, White EP. 2005. Disparity between range map- and survey-based analyses of species richness: patterns, processes and implications. Ecol. Lett. 8, 319–327. ( 10.1111/j.1461-0248.2005.00726.x) [DOI] [Google Scholar]
- 42.Hurlbert AH, Jetz W. 2007. Species richness, hotspots, and the scale dependence of range maps in ecology and conservation. Proc. Natl Acad. Sci. USA 104, 13 384–13 389. ( 10.1073/pnas.0704469104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kingdon J. 2006. The Kingdon field guide to African mammals. London, UK: Bloomsbury Publishing. [Google Scholar]
- 44.Harvey PH, Clutton-Brock TH. 1985. Life history variation in primates. Evolution 39, 559–581. ( 10.2307/2408653) [DOI] [PubMed] [Google Scholar]
- 45.Gittleman JL. 1986. Carnivore brain size, behavioral ecology, and phylogeny. J. Mamm. 67, 23–36. ( 10.2307/1380998) [DOI] [Google Scholar]
- 46.du Toit JT, Owen-Smith N. 1989. Body size, population metabolism, and habitat specialization among large African herbivores. Am. Nat. 133, 736–740. ( 10.1086/284949) [DOI] [Google Scholar]
- 47.Kingdon J, et al. 2013. Mammals of Africa (6 volumes). London, UK: Bloomsbury Publishing. [Google Scholar]
- 48.Kissling WD, et al. 2014. Establishing macroecological trait datasets: digitalization, extrapolation, and validation of diet preferences in terrestrial mammals worldwide. Ecol. Evol. 4, 2913–2930. ( 10.1002/ece3.1136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Estes R. 1991. The behavior guide to African mammals. Berkeley, CA: University of California Press. [Google Scholar]
- 50.Skinner JD, Chimimba CT. 2005. The mammals of the Southern African sub-region. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 51.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: Ecological Archives E090-184. Ecology 90, 2648 ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 52.Lima-Ribeiro MS, Varela S, González-Hernández J, de Oliveira G, Diniz-Filho JAF, Terribile LC. 2015. EcoClimate: a database of climate data from multiple models for past, present, and future for macroecologists and biogeographers. Biodiv. Info. 10, 1–21. ( 10.17161/bi.v10i0.4955) [DOI] [Google Scholar]
- 53.Collevatti RG, Lima-Ribeiro MS, Diniz-Filho JAF, Oliveira G, Dobrovolski R, Terribile LC. 2013. Stability of Brazilian seasonally dry forests under climate change: inferences for long-term conservation. Am. J. Plant Sci. 4, 792–805. ( 10.4236/ajps.2013.44098) [DOI] [Google Scholar]
- 54.Gavin DG, et al. 2014. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytol. 204, 37–54. ( 10.1111/nph.12929) [DOI] [PubMed] [Google Scholar]
- 55.Feng G, Mao L, Sandel B, Swenson NG, Svenning JC. 2016. High plant endemism in China is partially linked to reduced glacial–interglacial climate change. J. Biogeogr. 43, 145–154. ( 10.1111/jbi.12613) [DOI] [Google Scholar]
- 56.Ordonez A, Svenning JC. 2016. Strong paleoclimatic legacies in current plant functional diversity patterns across Europe. Ecol. Evol. 6, 3405–3416. ( 10.1002/ece3.2131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ordonez A, Svenning JC. 2016. Functional diversity of North American broad-leaved trees is codetermined by past and current environmental factors. Ecosphere 7, 1–14. ( 10.1002/ecs2.1237) [DOI] [Google Scholar]
- 58.Kraft NJ, Cornwell WK, Webb CO, Ackerly DD. 2007. Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 170, 271–283. ( 10.1086/519400) [DOI] [PubMed] [Google Scholar]
- 59.Kamilar JM, Guidi LM. 2010. The phylogenetic structure of primate communities: variation within and across continents. J. Biogeogr. 37, 801–813. ( 10.1111/j.1365-2699.2009.02267.x) [DOI] [Google Scholar]
- 60.Cantalapiedra JL, Fernández MH, Morales J. 2014. The biogeographic history of ruminant faunas determines the phylogenetic structure of their assemblages at different scales. Ecography 37, 1–9. ( 10.1111/j.1600-0587.2013.00236.x) [DOI] [Google Scholar]
- 61.Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. ( 10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- 62.Pausas JG, Verdú M. 2010. The jungle of methods for evaluating phenotypic and phylogenetic structure of communities. BioScience 60, 614–625. ( 10.1525/bio.2010.60.8.7) [DOI] [Google Scholar]
- 63.Miller ET, Farine DR, Trisos CH. 2015. Phylogenetic community structure metrics and null models: a review with new methods and software. Ecography 39, 1–17. [Google Scholar]
- 64.Bininda-Emonds OR, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 65.Cooper N, Freckleton RP, Jetz W. 2011. Phylogenetic conservatism of environmental niches in mammals. Proc. R. Soc. B 278, 2384–2391. ( 10.1098/rspb.2010.2207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pointer MA, Kamilar JM, Warmuth V, Chester SGB, Delsuc F, Mundy NI, Asher RJ, Bradley BJ. 2012. Runx2 tandem repeats and the evolution of facial length in placental mammals. BMC Evol. Biol. 12, 103 ( 10.1186/1471-2148-12-103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faurby S, Svenning JC. 2015. A species-level phylogeny of all extant and late Quaternary extinct mammals using a novel heuristic-hierarchical Bayesian approach. Mol. Phylogenet. Evol. 84, 14–26. ( 10.1016/j.ympev.2014.11.001) [DOI] [PubMed] [Google Scholar]
- 68.Borcard D, Legendre P, Drapeau P. 1992. Partialling out the spatial component of ecological variation. Ecology 73, 1045–1055. ( 10.2307/1940179) [DOI] [Google Scholar]
- 69.Legendre P, Borcard D, Peres-Neto PR. 2005. Analyzing beta diversity: partitioning the spatial variation of community composition data. Ecol. Monogr. 75, 435–450. ( 10.1890/05-0549) [DOI] [Google Scholar]
- 70.Kamilar JM, Martin SK, Tosi AJ. 2009. Combining biogeographic and phylogenetic data to examine primate speciation: an example using cercopithecin monkeys. Biotropica 41, 514–519. ( 10.1111/j.1744-7429.2009.00513.x) [DOI] [Google Scholar]
- 71.Bartoń K. 2014. MuMIn: multi-model inference. R package version, 1(5). [Google Scholar]
- 72.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. Berlin, Germany: Springer. [Google Scholar]
- 73.Hernandez-Fernández M, Vrba ES. 2005. Body size, biomic specialization and range size of African large mammals. J. Biogeogr. 32, 1243–1256. ( 10.1111/j.1365-2699.2005.01270.x) [DOI] [Google Scholar]
- 74.Eeley HA, Foley RA. 1999. Species richness, species range size and ecological specialization among African primates: geographical patterns and conservation implications. Biodiv. Conserv. 8, 1033–1056. ( 10.1023/A:1008831320469) [DOI] [Google Scholar]
- 75.Andrews P, O'Brien E. 2010. Mammal species richness in Africa. In Cenozoic mammals of Africa (eds Werdelin L, Sanders WJ), pp. 929–947. Berkeley, CA: University of California Press, Berkeley. [Google Scholar]
- 76.Gouveia SF, Villalobos F, Dobrovolski R, Beltrão-Mendes R, Ferrari SF. 2014. Forest structure drives global diversity of primates. J Anim. Ecol. 83, 1523–1530. ( 10.1111/1365-2656.12241) [DOI] [PubMed] [Google Scholar]
- 77.Jolly D, et al. 1998. Biome reconstruction from pollen and plant macrofossil data for Africa and the Arabian Peninsula at 0 and 6000 years. J. Biogeogr. 25, 1007–1027. ( 10.1046/j.1365-2699.1998.00238.x) [DOI] [Google Scholar]
- 78.Elenga H, et al. 2000. Pollen-based biome reconstruction for southern Europe and Africa 18 000 yr bp. J. Biogeogr. 27, 621–634. ( 10.1046/j.1365-2699.2000.00430.x) [DOI] [Google Scholar]
- 79.Anhuf D, et al. 2006. Paleo-environmental change in Amazonian and African rainforest during the LGM. Palaeogeogr. Palaeoclimatol. Palaeoecol. 239, 510–527. ( 10.1016/j.palaeo.2006.01.017) [DOI] [Google Scholar]
- 80.Hvilsom C, Carlsen F, Heller R, Jaffré N, Siegismund HR. 2014. Contrasting demographic histories of the neighboring bonobo and chimpanzee. Primates 55, 101–112. ( 10.1007/s10329-013-0373-3) [DOI] [PubMed] [Google Scholar]
- 81.Ting N, et al. 2012. Genetic signatures of a demographic collapse in a large-bodied forest dwelling primate (Mandrillus leucophaeus). Ecol. Evol. 2, 550–561. ( 10.1002/ece3.98) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White F. 1983. The vegetation of Africa: a descriptive memoir to accompany the UNESCO/AETFAT/UNSO vegetation map of Africa. Paris: UNESCO. [Google Scholar]
- 83.Rodríguez J. 2004. Stability in Pleistocene Mediterranean mammalian communities. Palaeogeogr. Palaeoclimatol. Palaeoecol. 207, 1–22. ( 10.1016/j.palaeo.2003.12.016) [DOI] [Google Scholar]
- 84.Rodríguez J. 2006. Structural continuity and multiple alternative stable states in Middle Pleistocene European mammalian communities. Palaeogeogr. Palaeoclimatol. Palaeoecol. 239, 355–373. ( 10.1016/j.palaeo.2006.02.001) [DOI] [Google Scholar]
- 85.Sikes NE. 1994. Early hominid habitat preferences in East Africa: paleosol carbon isotopic evidence. J. Hum. Evol. 27, 25–45. ( 10.1006/jhev.1994.1034) [DOI] [Google Scholar]
- 86.Reed KE. 1997. Early hominid evolution and ecological change through the African Plio-Pleistocene. J. Hum. Evol. 32, 289–322. ( 10.1006/jhev.1996.0106) [DOI] [PubMed] [Google Scholar]
- 87.Reed KE. 2008. Paleoecological patterns at the Hadar hominin site, Afar regional state, Ethiopia. J. Hum. Evol. 54, 743–768. ( 10.1016/j.jhevol.2007.08.013) [DOI] [PubMed] [Google Scholar]
- 88.de Ruiter DJ, Sponheimer M, Lee-Thorp JA. 2008. Indications of habitat association of Australopithecus robustus in the Bloubank Valley, South Africa. J. Hum. Evol. 55, 1015–1030. ( 10.1016/j.jhevol.2008.06.003) [DOI] [PubMed] [Google Scholar]
- 89.Cerling TE, et al. 2011. Woody cover and hominin environments in the past 6 million years. Nature 476, 51–56. ( 10.1038/nature10306) [DOI] [PubMed] [Google Scholar]
- 90.Rowan J, Reed KE. 2015. The paleoclimatic record and Plio-Pleistocene paleoenvironments. In Handbook of paleoanthropology (eds Henke W, Tattersall I), pp. 465–491. Berlin, Germany: Springer. [Google Scholar]
- 91.du Toit JT, Cumming DHM. 1999. Functional significance of ungulate diversity in African savannas and the ecological implications of the spread of pastoralism. Biodiv. Conserv. 8, 1643–1661. ( 10.1023/A:1008959721342) [DOI] [Google Scholar]
- 92.Sandom C, Dalby L, Fløjgaard C, Kissling WD, Lenoir J, Sandel B, Trøjelsgaard K, Ejrnæs R, Svenning J-C. 2013. Mammal predator and prey species richness are strongly linked at macroscales. Ecology 94, 1112–1122. ( 10.1890/12-1342.1) [DOI] [PubMed] [Google Scholar]
- 93.Isbell LA. 2006. Snakes as agents of evolutionary change in primate brains. J. Hum. Evol. 51, 1–35. ( 10.1016/j.jhevol.2005.12.012) [DOI] [PubMed] [Google Scholar]
- 94.Isbell LA. 2009. The fruit, the tree, and the serpent. Cambridge, MA: Harvard University Press. [Google Scholar]
- 95.Wheeler BC, Bradley BJ, Kamilar JM. 2011. Predictors of orbital convergence in primates: a test of the snake detection hypothesis of primate evolution. J. Hum. Evol. 61, 233–242. ( 10.1016/j.jhevol.2011.03.007) [DOI] [PubMed] [Google Scholar]
- 96.Ripple WJ, et al. 2014. Status and ecological effects of the world's largest carnivores. Science 343, 1241484 ( 10.1126/science.1241484) [DOI] [PubMed] [Google Scholar]
- 97.Lewis ME, Werdelin L. 2010. Carnivoran dispersal out of Africa during the early Pleistocene: relevance for hominins? In Out of Africa I (eds Fleagle JG, Shea JJ, Grine FE, Baden AL, Leakey RE), pp. 13–26. Berlin, Germany: Springer. [Google Scholar]
- 98.Werdelin L, Lewis ME. 2013. Temporal change in functional richness and evenness in the eastern African Plio-Pleistocene carnivoran guild. PLoS ONE 8, e57944 ( 10.1371/journal.pone.0057944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Freckleton RP. 2011. Dealing with collinearity in behavioural and ecological data: model averaging and the problems of measurement error. Behav. Ecol. Sociobiol. 65, 91–101. ( 10.1007/s00265-010-1045-6) [DOI] [Google Scholar]
- 100.Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 101.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. ( 10.1002/joc.1276) [DOI] [Google Scholar]
- 102.Williams JW, Jackson ST, Kutzbach JE. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738–5742. ( 10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Corlett RT. 2012. Climate change in the tropics: the end of the world as we know it? Biol. Conserv. 151, 22–25. ( 10.1016/j.biocon.2011.11.027) [DOI] [Google Scholar]
- 104.Scheiter S, Higgins SI. 2008. Impacts of climate change on the vegetation of Africa: an adaptive dynamic vegetation modelling approach. Glob. Change Biol. 15, 2224–2246. ( 10.1111/j.1365-2486.2008.01838.x) [DOI] [Google Scholar]
- 105.Midgley GF, Bond WJ. 2015. Future of African terrestrial biodiversity and ecosystems under anthropogenic climate change. Nat. Clim. 5, 823–829. ( 10.1038/nclimate2753) [DOI] [Google Scholar]
- 106.Moncrieff GR, Scheiter S, Bond WJ, Higgins SI. 2014. Increasing atmospheric CO2 overrides the historical legacy of multiple stable biome states in Africa. New Phytol. 201, 908–915. ( 10.1111/nph.12551) [DOI] [PubMed] [Google Scholar]
- 107.Schmitz C, et al. 2014. Land-use change trajectories up to 2050: insights from a global agro-economic model comparison. Agric. Econom. 45, 69–84. ( 10.1111/agec.12090) [DOI] [Google Scholar]
- 108.Gerland P, et al. 2014. World population stabilization unlikely this century. Science 346, 234–237. ( 10.1126/science.1257469) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.