Abstract
Recombinase enzymes promote DNA repair by homologous recombination. The genes that encode them are ancestral to life, occurring in all known dominions: viruses, Eubacteria, Archaea and Eukaryota. Bacterial recombinases are also present in viruses and eukaryotic groups (supergroups), presumably via ancestral events of lateral gene transfer. The eukaryotic recA genes have two distinct origins (mitochondrial and plastidial), whose acquisition by eukaryotes was possible via primary (bacteria–eukaryote) and/or secondary (eukaryote–eukaryote) endosymbiotic gene transfers (EGTs). Here we present a comprehensive phylogenetic analysis of the recA genealogy, with substantially increased taxonomic sampling in the bacteria, viruses, eukaryotes and a special focus on the key eukaryotic supergroup Amoebozoa, earlier represented only by Dictyostelium. We demonstrate that several major eukaryotic lineages have lost the bacterial recombinases (including Opisthokonta and Excavata), whereas others have retained them (Amoebozoa, Archaeplastida and the SAR-supergroups). When absent, the bacterial recA homologues may have been lost entirely (secondary loss of canonical mitochondria) or replaced by other eukaryotic recombinases. RecA proteins have a transit peptide for organellar import, where they act. The reconstruction of the RecA phylogeny with its EGT events presented here retells the intertwined evolutionary history of eukaryotes and bacteria, while further illuminating the events of endosymbiosis in eukaryotes by expanding the collection of widespread genes that provide insight to this deep history.
Keywords: Amoebozoa, DNA repair, endosymbiotic gene transfer, mitochondria, recombinase, recA
1. Introduction
Recombinases are a family of enzymes responsible for DNA repair via homologous recombination [1]. These proteins are widely common in genomes of diverse organisms, including bacteria, Archaea, eukaryotes and even viruses [2]. The most relevant homologous groups are referred to as RecA in bacteria, UvsX in viruses, RADA and RADB in Archaea and RAD51X in eukaryotes, collectively addressed as recA superfamily [3]. Eukaryotes in general present a wide range of recombinases (RAD51A, DMC1, RAD51B, RAD51C, etc.), which arose by means of several duplication events, most of them probably occurring before the last eukaryotic common ancestor [4]. Owing to its near universality, the recA superfamily has received significant attention and has been implicated in recent attempts to discover new domains of life [3], as a protein model to research metagenomic data from oceans [5], and as a model for evolution by gene duplication and endosymbiotic gene transfer (EGT) [2,4].
The bacterial form of the recA gene is present in eukaryotic genomes because they were acquired via EGT in conjunction with the uptake of the mitochondrion and plastid [4]. Mitochondria are descendants of bacterial endosymbionts probably acquired before the last eukaryotic common ancestors, plastids being acquired later in evolution [6].
During the processes of both primary endosymbioses, extensive lateral gene transfer (EGT) took place: from the bacterial genomes to the nuclear genome [7,8]. The resulting organelles have extremely reduced genomes, coding only a few proteins, rRNAs and tRNAs, probably because these entities cannot be easily imported by the organelle if synthesized outside the organellar space [8]. As a result of EGT, eukaryotic RecA proteins are encoded in the nuclear genome, yet active inside organelles. These proteins are imported through the organellar membrane, after recognition by an N-terminus signalling transit peptide, which is cleaved in the organelle yielding the active protein [9–11].
Bacterial recA is widespread in eukaryotic genomes, but some lineages have secondarily lost the gene. An example is the Opisthokonta, because neither metazoa nor fungi have the genes [4]. Homologous recombination in the mitochondrial genome is carried out in humans by RAD51-group proteins [12], which probably replaced the eubacterial homologue RecA.
Here we present a comprehensive phylogenetic reconstruction of the recA genealogy, including 225 taxa among bacteria, eukaryotes and viruses. We show that, in the Amoebozoa, a sister-group to Opisthokonta, bacterial (mitochondrial recA) recAmt is ancestrally present in the nuclear genomes, in the same way as in Thecamonas trahens, greens plants and several SAR lineages, such as Oomycetes, Blastocystis, Cafeteria and other groups. The most parsimonious interpretation of these data indicates that recA is ancestral in eukaryotes, being lost in a few lineages.
2. Material and methods
(a). Amoebozoan sequences
Echinosteliopsis oligospora was isolated from dead leaf litter collected from Sam D. Hamilton Noxubee National Wildlife Refuge. Schizoplasmodiopsis vulgaris was isolated from dead leaf litter collected from North Vietnam. Other cultures were obtained from the Culture Collection of Algae and Protozoa (CCAP, Scotland, UK) or American type culture (ATCC; Manassas, VA).
For E. oligospora, Clastostelium recurvatum, Cavostelium apophysatum, S. vulgaris, Crytodifflugia operculata, Vermamoeba vermiformis and Echinamoeba exudans, cells were grown on weak malt yeast agar (wMY; 0.002 g malt extract, 0.002 g yeast extract, 0.75 g K2HPO4, 15 g agar, 1.0 l deionized [DI] H2O) and Rhizamoeba saxonica was grown on a sterile artificial seawater wMY agar plate with various accompanying bacteria in culture. Arcella vulgaris was grown on sterile fresh water supplied with cereal grass medium and accompanying bacteria. Once amoeboid cells reached the dense culture stage, 2–3 ml of wMY liquid was poured over the agar plate. Subsequently, cells were scraped off and collected in a sterile 15 ml falcon tube. The cells were centrifuged at 4000g at 4°C for 5 min to pellet the cells. The pellet, which contained amoeboid cells, was subjected to cell lysis for RNA isolation. Total RNA was isolated, using TRIzol reagent (Sigma-Aldrich, St Louis, MO) according to the manufacturer's protocol (TRI reagent RNA isolation reagent). Quality of total RNA was assessed through electrophoresis in 1.8× Tris–borate–EDTA (TBE) agarose gel (Bioexpress, Kaysville, UT). The quantity of total RNA was diluted (1 : 200) and measured with fluorometry using the QubiT® (Life Technologies, Carlsbad, CA) high sensitivity RNA assays. The total RNA was further cleaned through ethanol precipitation. Total RNA with 0.25 M NaCl was spun down at 14 000g for 20 min at 4°C. The final pellet was washed with freshly made 75% ethyl alcohol. Double-stranded complementary DNA (dscDNA) synthesis was performed from 0.25 to 1.5 µg of total RNA using NEBNext® poly(A) mRNA magnetic isolation module followed by NEBNext® ultra RNA kit (New England Biolab (NEB), Ipswich, MA) according to the manufacturer's protocol.
Amoeba proteus was obtained from Carolina Biological Supply. Because Am. proteus grows in association with a eukaryotic flagellate as a food source, Chilomonas sp., a single cell was washed free of any associated eukaryotes by serial washes with spring water and starving the individual cell overnight in sterile spring water. Similarly, Difflugia USP was isolated from nature at the University of São Paulo campus, and single cells were serially washed with sterile water, and the individual cells were starved overnight. Subsequently, the cleaned cell was picked, using a micropipette into a 1.2 µl drop of sterile spring water. The reaction tube was then subjected to six freeze–thaw cycles in −80°C isopropanol and approximately 25°C DI H2O, respectively. Total RNA was isolated, and dscDNA was obtained using a modified version of Smart-seq2 [13]. The dscDNA was sheared using a Covaris S220 with the following settings: peak power 175 W, duty factor 10%, cycles per burst 200, mode frequency sweeping and duration of 30 s. The sequencing library was then created from the sheared dscDNA using NEBNext® Ultra DNA kit (New England Biolab (NEB)) according to the manufacturer's protocol.
Total RNA was extracted and converted to dscDNA from Ceratiomyxa fruticulosa, using a modified version of Smart-seq2 [13]. Approximately 200 spores were collected from a fresh fructification using a 0.008″ diameter platinum needle (Surepure Chemetals, Florham Park, NJ). Spores were then transferred into a PCR tube containing 1.2 µl liquid wMY (0.002 g yeast extract, 0.002 g malt extract, 0.75 g K2PO4 l−1 ddH2O) medium. After a 2.5 h incubation period at room temperature (approx. 21°C) cells were lysed by the addition of the Smart-seq2 cell lysis buffer and six rounds of a freeze–thaw cycle using −80°C isopropanol [13]. The resulting dscDNA was prepared for sequencing using a NexteraXT DNA library Prep kit (Illumina®, San Diego, CA).
Sequencing libraries was subjected to quality control (QC), using a combination of methods. The sequencing library concentrations were obtained with fluorometry using QubiT® high sensitivity dsDNA assays. First, the sequencing libraries were diluted (1 : 200) and then amplified using universal Illumina primers to estimate library sizes using electrophoresis in 1.8× TBE agarose gel. PCRs were composed of GoTaq® Green Master Mix (Promega, Madison, WI), IlluminaF (5′—AAT GAT ACG GCG ACC AC) at 10 µM and IlluminaR (3′—CAA GCA GAA GAC GGC AT) at 10 µM (oligonucleotide sequences © 2016 Illumina, Inc., all rights reserved), DNA template of adequate concentration and nuclease free water run under the following parameters: 5 min of initial denaturation at 94°C, followed by 20 cycles of 30 s of denaturation at 94°C, 25 s of annealing at 60°C and extension of 1 min at 72°C. Library molarities were calculated, using quantitative polymerase chain reaction (qPCR) of KAPA library Quant kit for Illumina (KAPA Biosystems, Boston, MA) according to the manufacturer's protocol. Additionally, the average molecular weight (MW) of each library is calculated by MW = (average library size in basepairs × 607.4 + 157.9). The nanomolarity of each library is calculated by nM = (MW/qubit concentration (in ng µl−1) × 1 000 000). Libraries molarities were subsequently diluted in 0.1× Tris–HCl EDTA pH 8.0 (TE) to the lowest molarity concentration in the set of libraries to be pooled together in equal volumes. All libraries were sequenced, using either the MiSeq or HiSeq 2000 platforms.
We passed the assembled transcriptome data through a series of QC steps to remove rRNA and bacterial contaminants [14]. The obtained reads were assembled, using Trinity RNA-Seq de novo assembly Trinity software [15]. TransDecoder (v. 2.0.1; https://transdecoder.github.io/) was used to predict coding peptide sequences from the baseline transcriptome contig sequences. Resulting amino acid sequences of 65 Amoebozoan representatives were concatenated to a single database for further analysis.
Dictyostelium discoideum RecAmt peptide sequence (GeneBank FAA00018) was used as the query in searches with tBlastn algorithm [16] and an arbitrary expected value threshold of e-40 maximum was established. Sequences were deposited in GenBank (electronic supplementary material, S3).
(b). Sequences for diverse eukaryotes
The D. discoideum RecAmt protein was used as a query for searches in GenBank for similar proteins from other groups of organisms by tBlastn and Blastp algorithms [16] with arbitrary e-value threshold of maximum e-40. The bacterial RecA representatives were chosen with a phylogenetic strategy. Big bacterial lineages were targeted in the construction of the datasets. We adopted the phylogenetic proposal of bacterial relationships as in Battistuzzi & Hedges [17]. Another set of genes was obtained from the marine microbial eukaryotic transcriptome sequencing project (MMETSP project) [18]. The translated databases were screened using the hmmsearch tool of HMMER package (https://hmmer.org). Best hits were captured from databases by the FAST program [19]. All sequences resulting from all different sources were gathered in a single matrix for further phylogenetic reconstruction.
(c). Experimental design and phylogenetic reconstruction
The goal of this survey was to determine the pattern of presence/absence of recA in major eukaryotic lineages, as well as clarify events of lateral gene transfer. While a number of methods have been proposed for efficient experimental design in phylogenetic reconstructions, there are no canonically accepted methods to reconstruct the deep history of a single gene family. Some of the proposed approaches are restricted to nucleotide sequences [20,21] and would not be directly applicable for deep reconstructions where amino acid sequences are used. Others might be employed when analysing protein sequences, but more adequate for comparative analysis between two or more different candidate proteins [22,23]. In order to better resolve the splits on the tree, we tried to sample the most diverse dataset as possible to avoid long branches and to add taxa that would connect near internal nodes, following previously recommended practices [24,25].
Several rounds of alignments for RecA were constructed in Seaview [26,27] with alignment algorithm MAFFT, using the L-INS-I setting [28]. The resulting matrix had their least probable homologous sites and unpaired site removed by the Gblocks algorithm [29] and fine adjusted manually. This strategy was followed by PhyML [30] analysis, using maximum-likelihood (ML) as the optimality criterion in order to assess the quality of the sequences and visual inspections were done in order to reveal contaminants. For the final tree, a MAFFT alignment was used to construct a HMM-profile with the hmmbuild algorithm of HMMER and the whole set of homologues sequences was aligned with the hmmalign algorithm of HMMER package (https://hmmer.org). The resulting matrix had the least probable homologous sites and unpaired sites removed by the Gblocks algorithm [29] and fine adjusted manually (only sites with a probability of homology p ≥ 0.8 were included). The resulting matrix of aligned and trimmed sequences was used as input for RAxML software [31,32], which performed an ML phylogenetic analysis with 120 independent initial searches using the PROTGAMMALGI molecular evolution model. Independently, to establish support, 1200 non-parametric bootstrap pseudo-replicates were performed, using the PROTGAMMALGI model. The best-fit model (LG + G4 + I) was determined by online ProtTest software [33,34]. The final matrix is available in electronic supplementary material, S4.
A Bayesian analysis was performed with the same matrix subjected to PhyloBayes software [35]. For the analysis, five independent chains were run for 20 000 cycles using default priors, CAT model and LG substitution model. A burn-in of 2000 cycles (10%) was applied after determining that likelihood values had stabilized. A maxdiff parameter of less than 0.3 was attained as recommended by the PhyloBayes manual, which indicates that topologies on the five runs had converged acceptably to a single answer.
3. Results and discussion
(a). The eubacterial recA type gene has been transferred to eukaryotic genomes in at least two occasions
Recombinases are a highly conserved group of enzymes. The recA genes are characteristic of Eubacteria; Archaeal RADA groups with eukaryotic RAD51A and meiosis-specific DMC1, forming a well-defined group, RADα; finally, archaeal RADB groups with eukaryotic RAD51B, RAD51C and others, forming the RADβ group of genes [2,4]. Thus, the most parsimonious interpretation for the presence of eubacterial recA in eukaryotes is that this event was a lateral gene transfer.
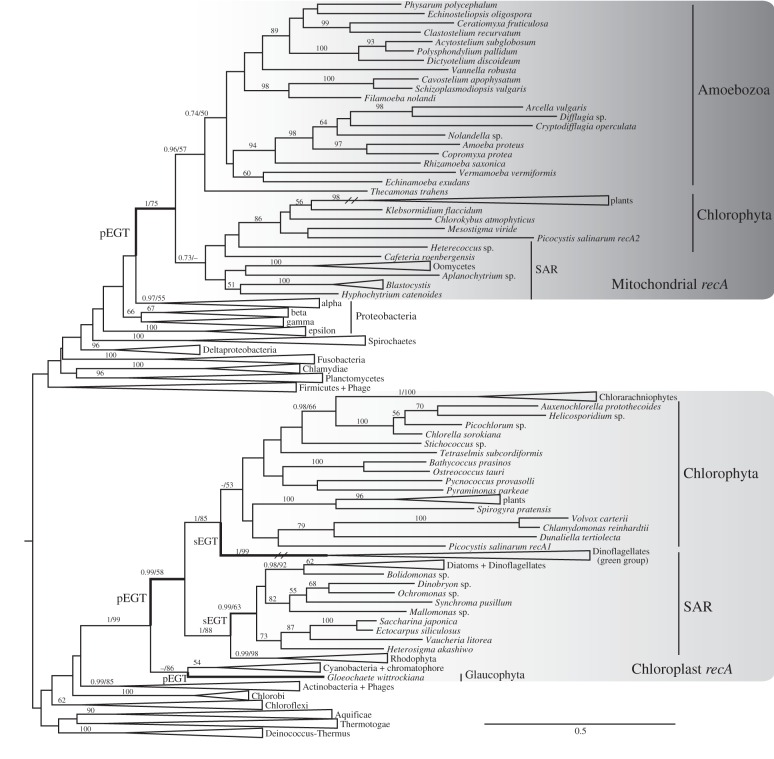
We performed a screening of both novel and available transcriptomes of microbial eukaryotes searching for previously unidentified recA in a wide range of deep level lineages. We have combined these into a broad bacterial taxonomic sampling and reconstructed a comprehensive gene genealogy of recA, upon which the general history of EGT can be investigated (figure 1 and electronic supplementary material, figures S1 and S2). The phylogenetic reconstruction reveals two independent primary endosymbiotic gene transfers (pEGT) from Eubacteria into the eukaryotic nucleus, one related to mitochondrial origin and the other to plastidial origin. The same topology still reveals the occurrence of secondary endosymbiotic gene transfers (sEGT), plastid-type bacterial genes being transferred from the red algal secondary endosymbiont to a lineage of Stramenopiles and from green algae to a group of dinoflagellates (figure 1).
Figure 1.
Phylogenetic tree of the bacterial homologue recA. Major group branches are collapsed. Eukaryotes received recA genes by two independent events of primary endosymbiotic gene transfers, with subsequent events of secondary endosymbiotic gene transfer. Full ML tree is available as electronic supplementary material, figure S1 and tree S1; full Bayesian tree is available as electronic supplementary material, figure S2 and tree S2. Mitochondrial plant clade and dinoflagellate branches are represented as half-length.
The tree obtained recovers several well-established deep relationships within bacteria, plants and Amoebozoa. The possibility of recovering such deep relationships, the universality of the recombinases among organisms and the abundance of available sequences suggest that the recA superfamily might be employed in helping resolving deep branching relationships, also along other genetic markers.
Our Bayesian and ML analyses converged on most of the topologies obtained, with small differences observed: dinoflagellates are associated with Chlorophyta in ML and nest within Chlorophyta in Bayesian analysis; the glaucophyte Gloeochaete wittrockiana is a sister-group to cyanobacteria in the ML analysis and nested within Cyanobacteria in the Bayesian (figure 1 and electronic supplementary material, figure S2).
(b). The mitochondrial recA type (recAmt) was present in the genome of the last eukaryotic common ancestor and has been lost in several lineages
Although only one protein was used, canonical relationships were recovered, even if with low support in some cases. The Alphaproteobacteria was recovered in all reconstructions as sister-group to a monophyletic mitochondrial clade, which is the currently accepted relationship [36]. Amoebozoa, Chlorophyta and non-photosynthetic SAR-supergroup members share a mitochondrial recA gene (bootstrap support (BS) and Bayesian posterior probabilities (PP) of 75/1, respectively, figure 1).
Through our deep sampling of genomic-level data of Amoebozoa, we find that recAmt is pervasive in the lineage (figure 1). The presence of the gene was already assessed and documented in the model organism D. discoideum [10,37]. However, here we demonstrate that D. discoideum is not an isolated Amoebozoan in the recA tree as previously considered [2,4]. On the contrary, it is only one instance within the entire Amoebozoa supergroup (figure 1). The class of genes is robustly present in Amoebozoa, even though absent in a few lineages. For instance, Entamoeba probably lost recAmt owing to the atrophy of mitochondrial organelles into anaerobic mitosomes [38] and in Acanthamoeba, we infer that the gene was replaced by an alternative eukaryotic RAD51 homologue, as in Opisthokonta. Taken as a whole, our sampling demonstrates that recAmt is present in Tubulinea, Arcellinida, Flabellinida, Dictyosteliida, Myxogastria and other groups, which make up the majority of the Amoebozoa clade [39]. Thus, the most parsimonious interpretation is that recAmt was present in the last common ancestor of the Amoebozoa.
Chintapalli et al. [2] suggested a hypothetical transfer of the recA to Amoebozoa from cyanobacteria. Our results show otherwise, the Amoebozoan recA are derived from Alphaproteobacteria, i.e. from mitochondria (figure 1). Evidence supporting our hypothesis includes: (i) proteins are targeted to mitochondria, where they are active, and (ii) Amoebozoan RecA proteins group with Oomycetes + plant RecA in a well-supported, mitochondrially derived clade. Another proposition by Chintapalli et al. is an EGT from brown, red algae and green plants recA to ‘plants’. In fact, the EGT flux is different: a gene influx from red algae to stramenopiles, brown algae and relatives (BS 88/PP 1; figure 1). The phylogenetic reconstruction provided by them lacks resolution, being unable to differentiate between recAmt and recAp (discussed below), which were available in their dataset. The misrepresentation of relationships is a result of poor taxon-sampling as well as reconstruction of historical relationships using an optimality criterion that is widely known to be prone to topology errors (i.e. the neighbour joining methodology, see Farris et al. [40] for a discussion).
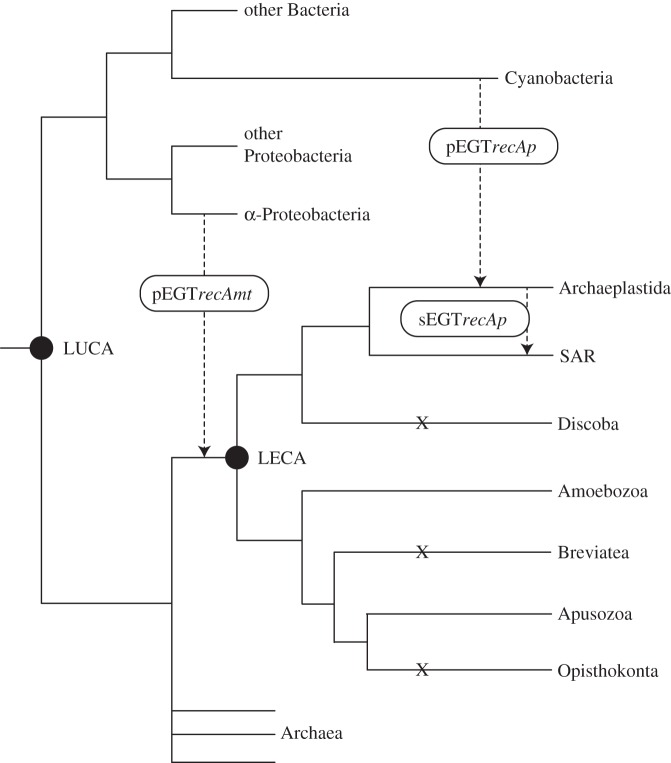
Several other major eukaryotic groups seem to have secondarily lost their bacterial recAmt homologues, as can be seen in Opisthokonta, Discoba and Alveolata (figure 2). Opisthokonta is part of a larger group, Obazoa, that includes anaerobic amoeboflagellates (Breviatea) and aerobic flagellates (Apusozoa) [43]. In our analyses, the genome of Thecamonas trahens (Apusozoa) has a recAmt that groups with Amoebozoa with moderate support (BS 57/PP 0.96; figure 1). However, we were not able to recover recAmt in the transcriptome of Pygsuia biforma (Breviatea), which is probably owing to its loss in the evolution of anaerobiosis within the breviates [43]. Opisthokonta, along with other obazoans are the sister-group to Amoebozoa and lack bacterial recombinases entirely. The loss probably occurred in the ancestral opisthokont, as neither Nucletmycea (Fungi + protistan relatives) nor Holozoa (Metazoa + protistan relatives) present any recA genes. Presumably, eukaryotic recombinases replaced the bacterial ones. For instance, RAD51C protein is imported by mitochondria and participates in mitochondrial DNA repair in Homo sapiens [12]. We performed extensive searches for recA among animals in GenBank returning only a handful of hits scattered through Metazoa. When analysed in our phylogenetic framework, these appear to be contaminants in non-curated databases.
Figure 2.
Three-domain depiction of the Tree of Life, with proposed acquisition and secondary loss events of bacterial recA homologues by eukaryotic groups. Major branches are based in Williams et al. [41] and eukaryotic relationships are based in Derelle et al. [42].
Plants present a large group with retained recAmt genes. The green plants not only kept the mitochondrial recombinases, but also went through several rounds of gene duplication after EGT and diversification of eukaryotes (especially in the angiosperms; electronic supplementary material, figure S1). The evolutionary history of land plants is marked by events of polyploidization by whole-genome duplication. One event of polyploidization has probably occurred in the ancestor of the angiosperms, prior to divergence of monocots and eudicots [44,45] and other events followed after the split of these lineages [45]. These facts would explain the pattern observed here, which is congruent with genome duplication events in plants. Presumably, this substantial expansion correlates with the gains of new functions or maintenance of the original function with differential expression by tissue or life cycle specificity [46]. Duplication of recAmt in angiosperms may be an effect of genome-wide duplications in this lineage. The sampled species (Zea mays, Oryza sativa, Arabidopsis thaliana, Populus trichocarpa and Ricinus comunis) present two to four duplications of recAmt homologues in their genomes, at least one happening before the monocots/eudicots split, followed by subsequent lineage specific duplication events (electronic supplementary material, figure S1).
Heterotrophic stramenopiles show robust evidence for the presence of nuclear encoded recAmt. Their bacterial recombinases are clearly mitochondrial derived (figure 1). Oomycetes and several other Stramenopiles lineages, such as Blastocystis and Hyphochytrium, the flagellated bicoecid Cafeteria roenbergensis, the labyrinthulid Aplanochytrium, all of them SAR members, present mitochondrial originated recA genes. The photosynthesizing SAR lineages seem to have lost the recAmt, which was probably replaced by the plastid form (recAp).
(c). A second paralogue, the plastid recA type (recAp), was obtained in the endosymbiotic plastid event
Further screening of the phylogeny reveals a second eukaryotic group of eukaryotic recA (figure 1). These are the plastid-related recAp. Again, a highly supported clade emerges with a rich diversity of photosynthesizing organisms, that is sister to the cyanobacterial recA (BS 99/PP 1; figure 1). The grouping of green plants, dinoflagellates, red algae, brown algae and diatoms indicates that these groups inherited RecA vertically from the single endosymbiotic origin of all known plastids, as earlier suggested [47,48]. However, the grouping of the glaucophyte Gloeochaete wittrockiana with cyanobacteria, either as a sister-group or even nested within them, may be interpreted either as lack of phylogenetic resolution in the current reconstruction, or as an independent acquisition of this particular gene in the glaucophytes. Another known exception is the chromatophore of the rhizarian Paulinella chromatophora (electronic supplementary material, figure S1), which represents clearly an independent primary endosymbiotic event [49–51], in which the recAp gene has not been transferred to the nucleus.
The close proximity between Rhodophyta and the photosynthesizing lineages of Stramenopiles (SAR; BS 88/PP 1; figure 1) reinforces the secondary endosymbiosis hypothesis and more, also demonstrates an sEGT (figure 2), a eukaryote–eukaryote transfer of a bacterially originated gene. As it seems, the photosynthesizing Stramenopiles (Bolidomonas, Diatoms, Phaeophyceae, Xanthophyceae and others) present functional forms of red algal-derived recAp, putatively from a secondary endosymbiotic event. Noteworthy is the absence of recAmt in the red algae and in the lineages that acquired the recAp from them. The plastidal form seems to have replaced the mitochondrial one, potentially playing a role in both organelles simultaneously. This is possible by means of a dual target system, i.e. the same protein may be addressed to both organelles [52,53].
Chlorophyta maintained their recAp, but differently from recAmt, without further replications (figure 1). This group, especially angiosperms, is the only one exhibiting both recAmt and recAp simultaneously, although either form may be lost in some lineages.
Dinoflagellates also present a recAp, but are divided into two groups: a diatom associated and a chlorophyte associated, with long branches in the latter. Presumably, these longer branches are owing to high evolutionary rates in dinoflagellates [54]. The highly supported association between dinoflagellates and chlorophytes (bootstrap and Bayesian support 85/1; figure 1) does not support the red algal origin for a big part of dinoflagellate plastids. A parallel can be traced with euglenids: both groups present three-layered chloroplasts, probably derived from secondary endosymbiotic events, involving chlorophytes in the case of euglenids [55]. There is also a rhizarian group nested among unicellular chlorophytes, the chlorarachniophytes (figure 1). These organisms clearly acquired their chloroplasts from the green group and even maintained a nucleomorph of the endosymbiont [55].
(d). Multiple gene transfer of recA have occurred in the history of life by endosymbiotic gene transfer, including multiple instances of bacteria to eukaryotic transfers and other instances of bacteria to virus transfers
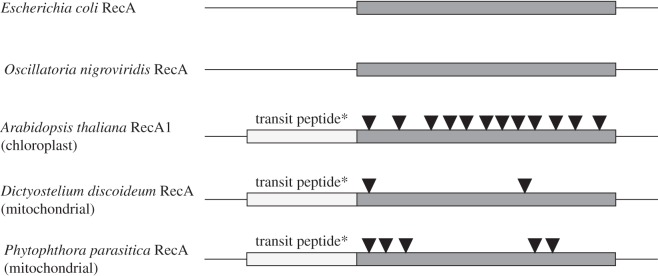
Amoebozoan, plants and Oomycetes RecA proteins are encoded with a signalling sequence before the active sites of the enzyme. This sequence is crucial for the import mechanism into organelles and is not found in bacterial homologues. Another striking difference is the presence of several introns in the recA found in eukaryotes, all of which must have been acquired after the EGT event as the bacterial forms are devoid of any introns (figure 3). Presumably, the organelle importing system must have been fully functional in the last eukaryotic common ancestor [36]. Most of transferred genes are vital to the organelle, and an importing system is a sine qua non conditio for successful EGT [56]. Once an importing system is fully functional, the organelle copy of the transferred gene may be lost by mutational decay.
Figure 3.
Comparison between sequence of original recA present in bacteria and their homologues transferred to eukaryotes. Eukaryotic forms have an approximately 50aa transit peptide in the N-terminal portion of the gene product, which is trimmed after import into the organelle; several introns were acquired after transfer to eukaryotes. Black triangles represent intron locations.
As a consequence, no organelle genome, from the approximately 7400 surveyed by us, keeps its original recA. This complete lack of recombinases in organelles suggests that EGT occurred only once in the ancestor of all eukaryotes for the recAmt and more than once for plastid homologues (at least a primary and a secondary EGT). Once established, the import mechanism paved a way for subsequent endosymbioses, most notably involving acquisition of photosynthesis by several groups. Additionally, it is possible also to verify the lateral gene transfer of recA from bacteria to some of their phage viruses, in this case Mycobacterium and Bacillus phages (electronic supplementary material, figure S1). As viruses are intracellular parasites, they interact very intimately with their hosts and some genes are prone to be transferred and may be fixed in the viral genomes.
Lastly, recA is present in the genome of the chromatophore, the photosynthetic organelle, of Paulinella chromatophora. This endosymbiosis between a cyanobacterium and an amoeboid rhizarian occurred independently from other primary endosymbioses [49]. The same trend of EGT is observable in this case, as only about 26% of its genes remain in the organelle [50], but the recA gene has not been transferred to the nucleus yet.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the Core Facility for Scientific Research—University of São Paulo (CEFAP-USP/GENIAL facility) for Illumina sequencing. We are grateful to two anonymous referees for their helpful comments on an earlier version of the manuscript.
Data accessibility
Alignment matrix, phylogenetic trees and accession numbers for new sequences are available as the electronic supplementary material.
Authors' contributions
P.G.H. and D.J.G.L. designed the experiments; A.K.T., D.J.G.L., M.W.B. and S.K. obtained and assembled Amoebozoan transcriptomes; P.G.H. performed all computational analysis; D.J.G.L., M.W.B. and P.G.H. interpreted results of analysis; A.K.T., D.J.G.L., M.W.B., P.G.H. and S.K. wrote the manuscript. All authors read and approved the final version of this manuscript.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by a FAPESP Doctorate Fellowship to P.G.H. (no. 2015/06306-0), a FAPESP Young Investigator Award to D.J.G.L. (no. 013/04585) and an NSF Award to M.W.B. (DEB 1456054).
References
- 1.Hiom K. 2012. Homologous recombination: how RecA finds the perfect partner. Curr. Biol. 22, R275–R278. ( 10.1016/j.cub.2012.03.002) [DOI] [PubMed] [Google Scholar]
- 2.Chintapalli SV, et al. 2013. Reevaluation of the evolutionary events within recA/RAD51 phylogeny. BMC Genomics 14, 240 ( 10.1186/1471-2164-14-240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M, Venter JC, Eisen JA. 2011. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE 6, e18011 ( 10.1371/journal.pone.0018011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Z, Kong H, Nei M, Ma H. 2006. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl Acad. Sci. USA 103, 10 328–10 333. ( 10.1073/pnas.0604232103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. ( 10.1126/science.1093857) [DOI] [PubMed] [Google Scholar]
- 6.Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theories for eukaryote origin. Phil. Trans. R. Soc. B 370, 20140330 ( 10.1098/rstb.2014.0330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin W. 2003. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc. Natl Acad. Sci. USA 100, 8612–8614. ( 10.1073/pnas.1633606100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams KL, Palmer JD. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395. ( 10.1016/S1055-7903(03)00194-5) [DOI] [PubMed] [Google Scholar]
- 9.Khazi FR, Edmondson AC, Nielsen BL. 2003. An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol. Genet. Genomics 269, 454–463. ( 10.1007/s00438-003-0859-6) [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa Y, Wakabayashi M, Nakamura S, Kodaira K, Shinohara H, Yasukawa H. 2004. A homolog of Escherichia coli RecA in mitochondria of the cellular slime mold Dictyostelium discoideum. DNA Repair 3, 515–525. ( 10.1016/j.dnarep.2004.01.014) [DOI] [PubMed] [Google Scholar]
- 11.Rowan BA, Oldenburg DJ, Bendich AJ. 2010. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J. Exp. Bot. 61, 2575–2588. ( 10.1093/jxb/erq088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sage JM, Knight KL. 2013. Human Rad51 promotes mitochondrial DNA synthesis under conditions of increased replication stress. Mitochondrion 13, 350–356. ( 10.1016/j.mito.2013.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. ( 10.1038/nprot.2014.006) [DOI] [PubMed] [Google Scholar]
- 14.Grant JR, Lahr DJG, Rey FE, Burleigh JG, Gordon JI, Knight R, Molestina RE, Katz LA. 2012. Gene discovery from a pilot study of the transcriptomes from three diverse microbial eukaryotes: Corallomyxa tenera, Chilodonella uncinata, and Subulatomonas tetraspora. Protist Genomics 1, 3–18. ( 10.2478/prge-2012-0002) [DOI] [Google Scholar]
- 15.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 17.Battistuzzi FU, Hedges SB. 2009. A major clade of prokaryotes with ancient adaptations to life on land. Mol. Biol. Evol. 26, 335–343. ( 10.1093/molbev/msn247) [DOI] [PubMed] [Google Scholar]
- 18.Keeling PJ, et al. 2014. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 ( 10.1371/journal.pbio.1001889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence TJ, Kauffman KT, Amrine KCH, Carper DL, Lee RS, Becich PJ, Canales CJ, Ardell DH. 2015. FAST: FAST analysis of sequences toolbox. Front. Genet. 6, 172 ( 10.3389/fgene.2015.00172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman N. 1998. Phylogenetic information and experimental design in molecular systematics. Proc. R. Soc. Lond. B 265, 1779–1786. ( 10.1098/rspb.1998.0502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z. 1998. On the best evolutionary rate for phylogenetic analysis. Syst. Biol. 47, 125–133. ( 10.1080/106351598261067) [DOI] [PubMed] [Google Scholar]
- 22.Townsend JP. 2007. Profiling phylogenetic informativeness. Syst. Biol. 56, 222–231. ( 10.1080/10635150701311362) [DOI] [PubMed] [Google Scholar]
- 23.López-Giráldez F, Townsend JP. 2011. PhyDesign: an online application for profiling phylogenetic informativeness. BMC Evol. Biol. 11, 152 ( 10.1186/1471-2148-11-152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Susko E, Roger AJ. 2012. The probability of correctly resolving a split as an experimental design criterion in phylogenetics. Syst. Biol. 61, 811–821. ( 10.1093/sysbio/sys033) [DOI] [PubMed] [Google Scholar]
- 25.Geuten K, Massingham T, Darius P, Smets E, Goldman N. 2007. Experimental design criteria in phylogenetics: where to add taxa. Syst. Biol. 56, 609–622. ( 10.1080/10635150701499563) [DOI] [PubMed] [Google Scholar]
- 26.Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. CABIOS 12, 543–548. ( 10.1093/bioinformatics/12.6.543) [DOI] [PubMed] [Google Scholar]
- 27.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. ( 10.1093/molbev/msp259) [DOI] [PubMed] [Google Scholar]
- 28.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537, 39–64. ( 10.1007/978-1-59745-251-9_3) [DOI] [PubMed] [Google Scholar]
- 29.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. ( 10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 32.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 33.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. ( 10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 34.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. ( 10.1093/bioinformatics/btr088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288. ( 10.1093/bioinformatics/btp368) [DOI] [PubMed] [Google Scholar]
- 36.Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283, 1476–1481. ( 10.1126/science.283.5407.1476) [DOI] [PubMed] [Google Scholar]
- 37.Eichinger L, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57. ( 10.1038/nature03481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495. ( 10.1128/MMBR.05024-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahr DJG, Grant JR, Katz LA. 2013. Multigene phylogenetic reconstruction of the Tubulinea (Amoebozoa) corroborates four of the six major lineages, while additionally revealing that shell composition does not predict phylogeny in the Arcellinida. Protist 164, 323–339. ( 10.1016/j.protis.2013.02.003) [DOI] [PubMed] [Google Scholar]
- 40.Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. 1996. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124. ( 10.1111/j.1096-0031.1996.tb00196.x) [DOI] [PubMed] [Google Scholar]
- 41.Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236. ( 10.1038/nature12779). [DOI] [PubMed] [Google Scholar]
- 42.Derelle R, Torruella G, Klimeš V, Brinkmann H, Kim E, Vlček Č, Lang BF, Eliáš M. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl Acad. Sci. USA 112, E693–E699. ( 10.1073/pnas.1420657112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown MW, Sharpe SC, Silberman JD, Heiss AA, Lang BF, Simpson AGB, Roger AJ. 2013. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc. R. Soc. B 280, 20131755 ( 10.1098/rspb.2013.1755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Bodt S, Maere S, Van de Peer Y. 2005. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597. ( 10.1016/j.tree.2005.07.008) [DOI] [PubMed] [Google Scholar]
- 45.Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. ( 10.1038/nature09916) [DOI] [PubMed] [Google Scholar]
- 46.Miller-Messmer M, Kühn K, Bichara M, Le Ret M, Imbault P, Gualberto JM. 2012. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 159, 211–226. ( 10.1104/pp.112.194720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330. ( 10.1016/j.cub.2005.06.040) [DOI] [PubMed] [Google Scholar]
- 48.Reyes-Prieto A, Weber APM, Bhattacharya D. 2007. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168. ( 10.1146/annurev.genet.41.110306.130134) [DOI] [PubMed] [Google Scholar]
- 49.Marin B, Nowack ECM, Melkonian M. 2005. A plastid in the making: evidence for a second primary endosymbiosis. Protist 156, 425–432. ( 10.1016/j.protis.2005.09.001) [DOI] [PubMed] [Google Scholar]
- 50.Nowack ECM, Melkonian M, Glöckner G. 2008. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418. ( 10.1016/j.cub.2008.02.051) [DOI] [PubMed] [Google Scholar]
- 51.Reyes-Prieto A, Yoon HS, Moustafa A, Yang EC, Andersen RA, Boo SM, Nakayama T, Ishida K, Bhattacharya D. 2010. Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 27, 1530–1537. ( 10.1093/molbev/msq032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackenzie SA. 2005. Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol. 15, 548–554. ( 10.1016/j.tcb.2005.08.007) [DOI] [PubMed] [Google Scholar]
- 53.Millar AH, Whelan J, Small I. 2006. Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 9, 610–615. ( 10.1016/j.pbi.2006.09.002) [DOI] [PubMed] [Google Scholar]
- 54.Pochon X, Putnam HM, Gates RD. 2014. Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2, e394 ( 10.7717/peerj.394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Archibald JM. 2015. Genomic perspectives on the birth and spread of plastids. Proc. Natl Acad. Sci. USA 112, 10 147–10 153. ( 10.1073/pnas.1421374112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. ( 10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Alignment matrix, phylogenetic trees and accession numbers for new sequences are available as the electronic supplementary material.