Abstract
Priority effects, in which the order of species arrival dictates community assembly, can have a major influence on species diversity, but the genetic basis of priority effects remains unknown. Here, we suggest that nitrogen scavenging genes previously considered responsible for starvation avoidance may drive priority effects by causing rapid resource depletion. Using single-molecule sequencing, we de novo assembled the genome of the nectar-colonizing yeast, Metschnikowia reukaufii, across eight scaffolds and complete mitochondrion, with gap-free coverage over gene spaces. We found a high rate of tandem gene duplication in this genome, enriched for nitrogen metabolism and transport. Both high-capacity amino acid importers, GAP1 and PUT4, present as tandem gene arrays, were highly expressed in synthetic nectar and regulated by the availability and quality of amino acids. In experiments with competitive nectar yeast, Candida rancensis, amino acid addition alleviated suppression of C. rancensis by early arrival of M. reukaufii, corroborating that amino acid scavenging may contribute to priority effects. Because niche pre-emption via rapid resource depletion may underlie priority effects in a broad range of microbial, plant and animal communities, nutrient scavenging genes like the ones we considered here may be broadly relevant to understanding priority effects.
Keywords: species interaction, competition, resource pre-emption, community assembly, tandem gene duplication
1. Introduction
Many processes affect species diversity in ecological communities, but one process that is receiving renewed interest is priority effects, where the order of species arrival determines community assembly [1–3]. In communities of plants (e.g. [4]), animals (e.g. [5]), fungi (e.g. [6]), and microbes (e.g. [7]), species that arrive at newly disturbed or formed habitats often inhibit colonization by late-arriving species via resource pre-emption and/or modification [8]. Growing evidence indicates that these inhibitory priority effects limit species diversity of the local community via competitive exclusion [8]. At the same time, species diversity at the regional scale is enhanced by priority effects causing local communities to diverge in species composition as a result of variable arrival history [9]. To our knowledge, however, no study has explicitly identified genes affecting species traits responsible for resource pre-emption or modification, leaving the genetic mechanisms behind priority effects unknown.
One system where priority effects have been systematically studied is the microbial communities that develop in floral nectar [10–12]. Nectar is a ubiquitous resource not just for pollinating animals, but also for the microbes that are introduced via pollinators [13]. A small number of yeast and bacterial species, which comprise the nectar microbiome, can tolerate the osmotic pressure caused by high sugar concentrations and the nutritional scarcity caused by low amino acid concentrations [13,14]. Recent studies show that these species engage in resource competition, resulting in strong and pervasive priority effects [10,11]. Even complete exclusion by early arriving species is frequently observed, with direct consequences for microbial species diversity within and across flowers and their mediation of plant–pollinator mutualism [10,11,15]. Because population growth in nectar appears limited by the availability of amino acids [10,12], which the microbes deplete rapidly [8], pre-emption of amino acids by early colonizers is a plausible explanation for priority effects in this system. Moreover, the strength of priority effects can be predicted by the sensitivity of each species' growth rate to amino acid concentration as well as the amount of overlap in amino acid utilization between species [11], further implicating amino acids as the limiting resource that drives priority effects in this system. However, little is known about the genes involved in resource pre-emption or modification, leaving the molecular mechanisms behind priority effects elusive.
In this study, we focus on Metschnikowia reukaufii (Saccharomycetales: Metschnikowiaceae), a ubiquitous nectar yeast [16–18] that exhibits particularly strong priority effects in competition with other yeasts and bacteria [11,12]. We first present a high-quality draft genome of M. reukaufii and compare its characteristics to other yeast genomes. Minimal genetic data are currently available for nectar yeasts (but see [19]), but other yeasts, including Saccharomyces cerevisiae, Clavispora lusitaniae, and Debaryomyces hansenii, have long been used as model organisms for molecular genetics. Although not closely related to nectar yeasts, these well-studied genomes provide a basis that can inform the analysis of nectar yeasts. Additionally, we compare the M. reukaufii genome to those of several other species that are not as well characterized but are more closely related to M. reukaufii (see also the electronic supplementary material). These comparative analyses facilitated identification of candidate genes that may determine how M. reukaufii interacts with other inhabitants of the nectar microbiome. To provide supportive evidence for these genes, we also report the results of gene expression assays and interspecific competition experiments. Together, our results suggest that extensive gene duplication that enables efficient amino acid scavenging may underlie the strong priority effects exhibited by M. reukaufii.
2. Results and discussion
(a). Sequencing and assembly
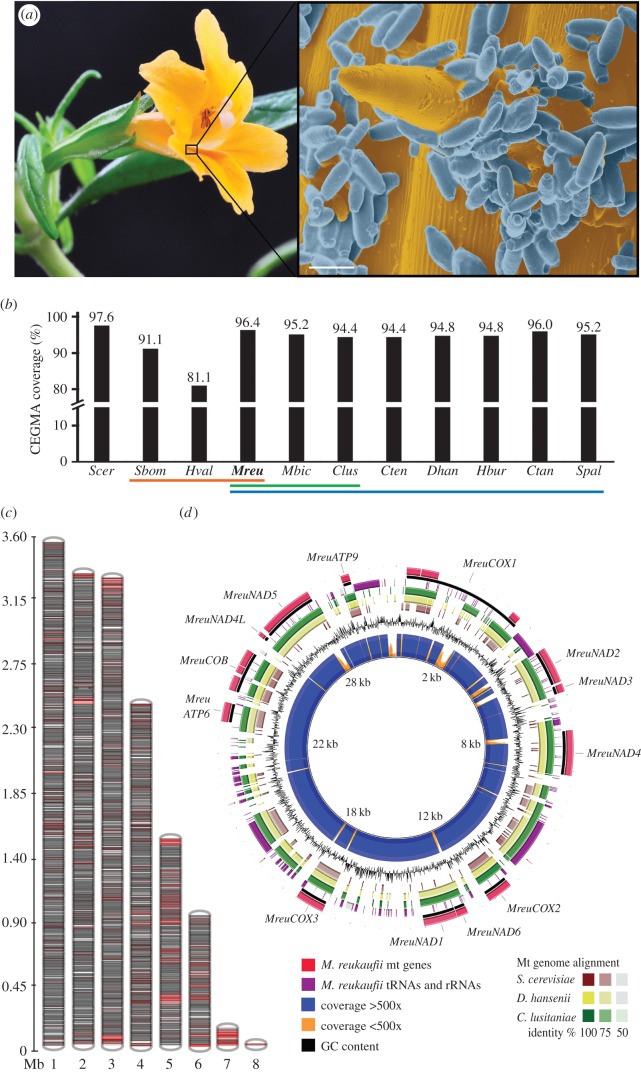
Genomic DNA from M. reukaufii strain MR1 was sequenced using high coverage (82×) single-molecule PacBio sequencing, supplemented by Illumina short-read sequencing (35×; electronic supplementary material, figure S1). PacBio sequencing yielded a total of 1.6 billion filtered bases and an average read length of more than 12 kb (electronic supplementary material, figure S2). The initial diploid draft assembly had a total size of 19.2 Mb (kmer size estimation = 18.4 Mb), an N50 of 1.2 Mb, and high-consensus accuracy of 99.994% (electronic supplementary material, figure S2), but was fragmented into 128 contigs. PacBio long read sequencing alleviates the assembly of complex and repetitive regions, but high levels of heterozygosity commonly found in wild yeasts [20] pose a challenge [21]. Indeed, not only was 80% of the M. reukaufii genome contained in the 17 largest contigs, but smaller 120 contigs also self-aligned within the remaining eight. To demarcate the haplotypes within this diploid genome, rectify base-level errors, and correct putative misjoins, we used a custom pipeline optimized for highly polymorphic genomes and included organelle assembly (electronic supplementary material, figure S1). Integration of a haplo-aware assembly improved the overall quality of the M. reukaufii reference genome. Resolved into eight scaffolds, the haploid reference genome had a total length of 15 522 805 bp and a tripled N50 of 3.4 Mb (figure 1c; electronic supplementary material, figure S3). At 15.5 Mb, similar to M. bicuspidata, we found evidence of genome expansion in this species, when compared with C. lusitaniae (see the electronic supplementary material, figure S4a). Telomeres are complex repeats and difficult to assemble, but in our case, the majority of the scaffolds contained a telomeric core motif (5′-AAGATAAATCAGTACATCCCT-3′) at one or both ends, illustrating high contiguity.
Figure 1.
Genome of Metschnikowia reukaufii. (a) Flower of host plant sticky monkeyflower (M. aurantiacus), zoomed inset, scanning electron micrograph of M. reukaufii cells (blue) attached to cells and trichome of sticky monkeyflower floral tube that contains nectar (gold), Scale bar, 5 µm, false-coloured (b) CEGMA completion of M. reukaufii (Mreu, this study), compared to published yeast genomes, from nectar specialists (yellow: S. bombicola (Sbom), H. valbyensis (Hval), Metschnikowiaceace species (green: M. bicuspidata (Mbic), Cl. lusitaniae (Clus)) and closely related CUG-Ser clade members (blue: C. tenuis (Cten), D. hansenii (Dhan), H. burtonii (Hbur), C. tanzawaensis (Ctan), Sp. passalidarum (Spal) and S. cerevisiae S288c reference genome (Scer). (c) Scaffolds representing nuclear genome: predicted genes (black lines) and predicted repeats (red lines). (d) Circular mitochondrial genome: predicted genes, tRNAs and rRNAs, sequencing coverage, GC content and sequence similarity to C. lusitaniae, D. hansenii and S. cerevisiae mitochondrial genomes.
To estimate the completeness of our assembly, we used the core set of highly conserved eukaryotic genes (CEG) [21]. With 96.4% of the CEG (239/248) present, the M. reukaufii genome ranks similar to other reference yeast genomes, with only some well-curated genomes (e.g. S. cerevisiae, 97.6%) showing slightly higher coverage (figure 1b).
(b). Annotation and alternative codon usage
Maker-predicted 6106 gene models of which 83% were supported by homology evidence from ESTs and/or proteins [22]. The average gene length was 1771 bp. Compared with S. cerevisiae S288c (12.3 Mb, 5404 genes) and C. lusitaniae (12.1 Mb, 5926 genes), M. reukaufii genome has a typical number of predicted protein-coding genes. Gene ontology mapping obtained hits for 5266 genes (86%), with 4689 genes (77%) assigned to a functional category and the remaining 577 genes uncharacterized.
Yeasts that belong to the CUG-Ser clade encode a unique seryl-tRNACAG, through which the CUG codon is encoded as serine (Ser) and rarely as the standard leucine (Leu) [23]. Because sister Metschnikowiaceae species, C. lusitaniae and M. bicuspidata, belong to this clade [23], we investigated whether M. reukaufii may encode CUG as Ser instead of Leu. Purine 33 (G33) in the C. albicans Ser-tRNACAG anticodon loop, which replaces a conserved pyrimidine (U33) found in all other tRNAs, is the main element that lowers the rate of leucylation [24]. Structural analysis of M. reukaufii tRNAs revealed that the predicted CUG-tRNAs carry G instead of U at position 33 as well as the Ser discriminator base G73, as observed in C. albicans (electronic supplementary material, figure S4b). However, a third Ser-identity element, G3C3 run in the variable loop, is missing similar to M. bicuspidata. This supports the recently proposed stepwise accumulation of tRNACAGSer features in the evolution of alternative CUG translation [23]. Alignment of conserved CUG-sites further supports the alternative codon usage in M. reukaufii. Using the Bagheera pipeline, we predicted CUG-usage for 113 sites across 30 conserved proteins [25]. Seventy-two per cent of these sites suggested alternative codon usage (electronic supplementary material, table S3). However, without direct measures such as sequencing of tryptic peptides, the actual rate of leucylation in M. reukaufii remains to be determined.
The initial genome assembly contained multiple incomplete mitochondrial (mt) genomes mis-assembled as forward and reverse repeats, probably due to the large differences in sequence coverage (40× of nuclear DNA) and GC content. Subsequent integration of a bait-mapping approach optimized for organelles assembled the 29 534 bp mitochondrion (figure 1d). Of the 14 core mitochondrial-encoded genes found in most fungi [26], the M. reukaufii mitochondrion harbours 13, including COX1, NAD2, NAD3, NAD4, COX2, NAD6, NAD1,COX3, ATP6, COB, NAD4 L, NAD5, ATP9 (figure 1d; electronic supplementary material, figure S5), with ATP8 absent. These genes, in addition to 23 tRNAs and four rRNAs (missing tRNAy and four rRNAs), comprise the mitochondrion (GC content = 33%).
(c). Tandem gene duplication
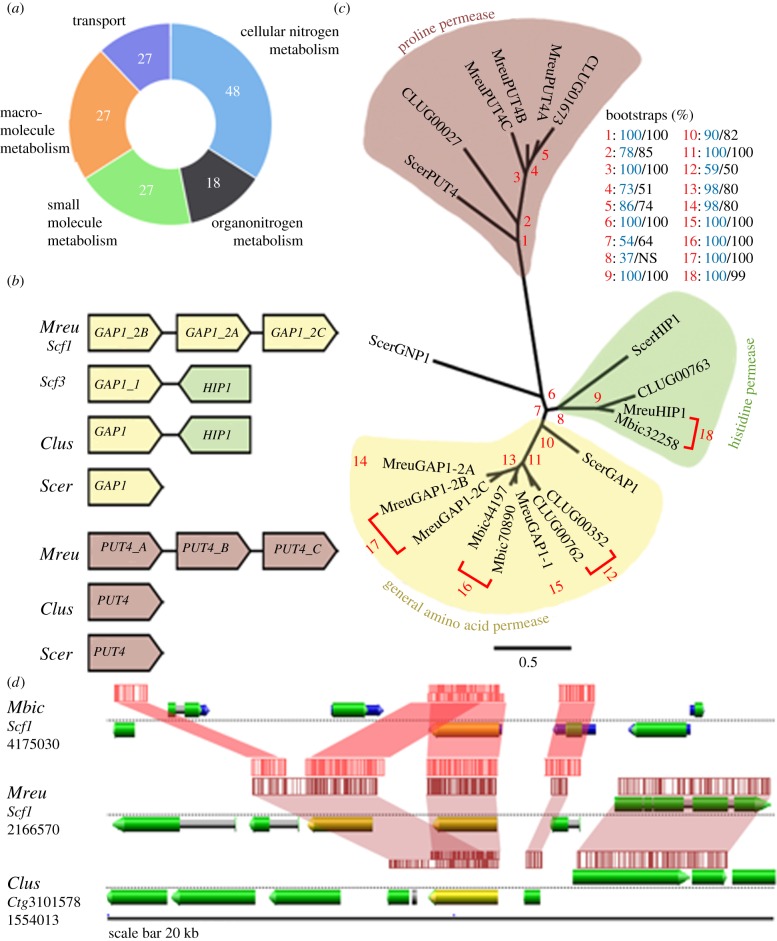
Genomic analyses of model organisms have shown that over a third of all protein-coding genes belong to multigene families that arise from duplications, e.g. whole-genome duplication, segmental duplication, and tandem gene duplication [27]. Most sequenced hemiascomycete yeast genomes harbour 1.5–2% of tandem gene arrays (TGAs), with some notable exceptions. Debaromyces hansenii, a marine yeast that tolerates extreme salt stress, has 4.4% of all genes arranged in TGAs [28]. Metschnikowia reukaufii showed a relatively high number of 363 TGAs, representing 5.9% of all annotated genes. Of these, 227 are novel TGAs compared to C. lusitaniae (electronic supplementary material, table S1). The largest fraction of these TGAs comprised genes involved in nitrogen metabolism, that of either cellular- (48 genes, 21%) or organo-nitrogen (18 genes, 8%, figure 2a). Another enriched fraction represented genes involved in cellular transport (27 genes, 12%, figure 2a). These transporters include homologues of ATP binding cassette and oligopeptide transporters, as well as amino acid permeases (electronic supplementary material, table S1).
Figure 2.
Tandem gene duplication for adaptation to low nitrogen environment. (a) Enriched gene ontology categories for the 227 unique tandem duplicated genes in M. reukaufii genome. (b) Schematic of GAP1 and PUT4 duplications compared to S. cerevisiae and C. lusitaniae homologues and (c) associated protein sequence phylogenetic tree showing expansion of amino acid permeases in M. reukaufii, with bootstrap supports from RaxML (blue) and PAUP* (black) associated with each numbered node (red). Panel (d) predicted amine oxidase (AOC) homologues (yellow) arranged in TGAs in M. reukaufii compared across syntenic regions on M. bicuspidata and C. lusitaniae. Shaded areas link conserved regions across the genomes of the three species.
(d). Duplication of nitrogen transport and metabolism genes
Two of the tandem duplicated genes, MreuGAP1-1 and MreuGAP1-2A, are of particular interest given that their closest homologue in S. cerevisiae is the general amino acid permease1 (GAP1). In fungi, amino acid uptake is mediated by yeast amino acid transporters (YATs, amino acid-polyamine-organocation superfamily), of which 18 have been functionally characterized in S. cerevisiae [29]. YAT members share a common topology with 12 trans-membrane domains and cytosolic N and C termini. Most YATs, such as the histidine permease1 (HIP1), have low capacity and are most active when amino acids are abundant in the growth medium [29]. They opportunistically transport specific amino acids for protein synthesis. By contrast, GAP1, a high-capacity transporter of all naturally occurring amino acids and analogues, is active when amino acids are limiting [29]. GAP1, therefore, functions as a scavenger of amino acids for nitrogen when supply is low. In fact, experimental evolution in S. cerevisiae has shown that GAP1 is a recurring locus for adaptation to nitrogen-limited environments [30,31] and additional GAP1 copies confer an average fitness advantage of 24–44% in nitrogen-limited media [31].
MreuGAP1-1 and 2A are predicted to encode 585 amino acids, located on the minus strand of scaffold 3 and 1, respectively (figure 2b). They share 64% and 62% protein identity with ScerGAP1, and 80% and 74% identity to C. lusitaniae's orthologue (CLUG_00762), respectively. In contrast to ScerGAP1's single-copy arrangement, M. reukaufii has two GAP1 TGAs. The first contains MreuGAP1-1 and 175 bp downstream a second transporter more closely related to ScerHIP1, with only 56% protein identity between them. Clavispora lusitaniae shares the same antiparallel TGA, suggesting that this duplication originated before the divergence of these Metschnikowiaceae species (figure 2b). However, the second GAP1 TGA with three additional GAP1 homologues (MreuGAP1-2A, 2B and 2C) is unique to M. reukaufii (figure 2b). Phylogenetic analysis supports the expansion of GAP1 in M. reukaufii (figure 2c). The fact that we identified numerous long PacBio reads contiguous over the GAP1 TGAs is evidence for the physical linkage of these genes, given the random nature of chimeric artefacts.
All four MreuGAP1 homologues share conserved LysP (COG0833) and AA permease domains (pfam00324) (electronic supplementary material, figure S9). The consensus amphipathic region (CAR domain) of ScerGAP1 forms the amino acid translocation channel with the critical residues N390, S391, S397 and R398 [31]. MreuGAP1-1, 2A to C, but not MreuHIP1, had all of these residues conserved (electronic supplementary material, figure S9). Similarly, the C-terminus, critical for transport and amino acid sensing in ScerGAP1, was highly conserved in MreuGAP1-1 and 2A to C (electronic supplementary material, figure S9). The conservation of all residues critical for amino acid transport in MreuGAP1-1 and 2A to C supports the preservation of function.
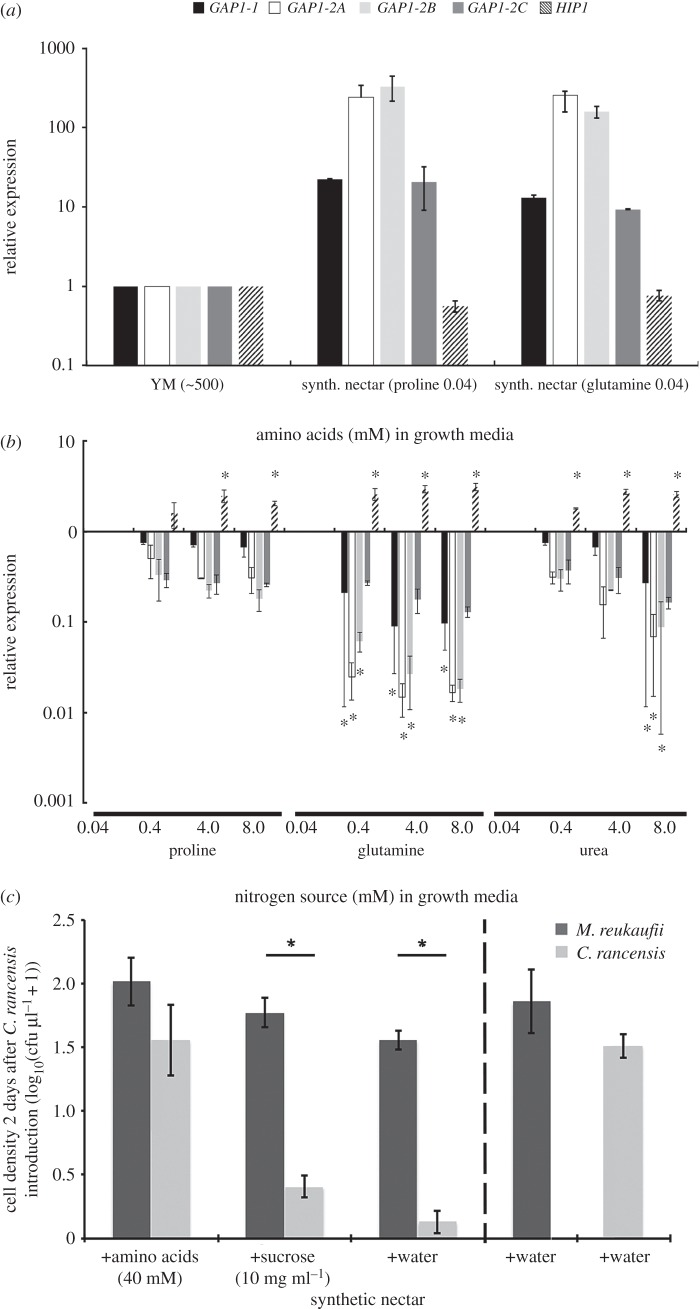
In order to ascertain the function of the MreuGAP1 genes, we quantified their expression under conditions that mimic floral nectar. Nitrogen resources are commonly low in sticky monkeyflower nectar, from which the M. reukaufii strain was isolated, with amino acids such as proline and glutamine ranging between 30 and 50 µM [10]. Quantitative RT-PCR showed that all MreuGAP1 homologues were expressed under synthetic nectar environments with low amino acid availability (figure 3a,b).
Figure 3.
Amino acid scavenging genes for priority effects caused by nitrogen pre-emption. (a) Expression of four GAP1 homologues of M. reukaufii cells growing for 4 h in either synthetic nectar (20% sucrose) with 0.04 mM proline or glutamine as nitrogen source or YM (approx. 400 mM mixed amino acids) compared to HIP1. All GAP1 homologues are expressed in nectar, but repressed in YM media. (b) NCR of GAP1 genes in M. reukaufii cells growing for 4 h in 20% sucrose supplemented with 0.04, 0.4, 4 or 8 mM of the poor (proline), good (glutamine) and non-amino acid (urea) nitrogen source. Expression of GAP1 homologues, but not HIP1, is strongly repressed by glutamine, and to a much lower extent by proline and urea. Data show mean ± s.d. relative to 0.04 mM concentrations; n = 3 biological replica each with two technical replica. (c) (left panel) Growth of the 2-day late-arriving C. rancensis was significantly repressed in the presence of early arriving M. reukaufii when either no nutrients were restocked (Padj = 5 × 10−7) or only sucrose was restocked (Padj = 15 × 10−6) (10 mg ml−1 per day after C. rancensis arrival). This effect was reversed when 40 mM amino acids were supplied daily post C. rancensis arrival (Padj = 0.646). No observed significant difference between their population densities when grown alone (Padj = 0.221) (right panel). Data show mean ± s.e.
Excessive uptake of amino acids can be detrimental to yeast growth [32]. To prevent toxic intracellular accumulation, ScerGAP1 is under nitrogen catabolite repression (NCR), which controls its abundance transcriptionally and post-translationally [32], a mechanism not shared by low-capacity permeases such as HIP1. Indeed, the promoter regions of MreuGAP1 homologues harboured UASNTR GLN3/GAT1 [33] activator binding site (GATAAG) upstream of their transcriptional start. Consistent with this regulation, the expression of all four MreuGAP1 homologues was strongly induced (20- to 350-fold) under synthetic nectar, compared with conditions with abundant nitrogen sources (YM) (figure 3a). Because the overall nutrient composition of YM differs from synthetic nectar, we evaluated the regulation of MreuGAP1 homologues in nectar with varying levels of high- and low-quality nitrogen sources, with MreuHIP1 used as a reference. As predicted, a good nitrogen source, glutamine, suppressed the expression of all MreuGAP1 homologues, with GAP1-2A inhibited by 97.5% at 10-fold (400 µM) the typical standing concentration found in sticky monkeyflower nectar (approx. 40 µM) [10]. By contrast, the mRNA abundance of MreuHIP1 increased by 259% under the same conditions (figure 3b). At high concentrations, both proline (poor nitrogen source) and urea (non-amino acid nitrogen source) repressed MreuGAP1-1 and 2A-C transcription, but the relative repression of each homologue was lower compared with glutamine. With proline as sole nitrogen source, even at 200 times (8 mM) the steady-state nectar concentration, GAP1-2A expression was reduced by only 30.5% (figure 3b). MreuHIP1 expression, on the other hand, increased twofold (figure 3b).
In S. cerevisiae, a second amino acid scavenger active under nitrogen limitation is the high-capacity proline utilization 4 permease (PUT4). PUT4 and GAP1 are the main importers of the imino acid proline [34,35]. Although less preferred than glutamine, proline is one of the most abundant nitrogen sources in many natural environments of yeasts, including sticky monkeyflower nectar [10,36]. The genome of M. reukaufii features three parallel duplicated PUT4 homologues on the minus strand of scaffold 6 (figure 2b). They are predicted to encode 567 (MreuPUT4A), 565 (MreuPUT4B) and 566 (MreuPUT4C) amino acid proteins, approximately 3800 bp and 1450 bp apart, respectively (figure 2b). Multiple PacBio long reads contiguous over the three PUT4 homologues support the physical linkage of these genes. The qRT-PCR showed that all three MreuPUT4 genes were expressed under synthetic nectar conditions (electronic supplementary material, figure S8). Well characterized in S. cerevisiae and Aspergillus nidulans, the amino acid residues critical for transport and substrate-specificity of PUT4 have been experimentally verified [37]. MreuPUT4A to C not only share all conserved residues essential for transport, but also those unique to PUT4 members of the YAT clade (electronic supplementary material, figure S9). Oligopeptide transporters were also tandem duplicated in the M. reukaufii genome, e.g. five copies of an OPT2 homologue (electronic supplementary material, table S1). Similar to GAP1 and PUT4, OPT2 has been shown critical for growth under nitrogen starvation [38].
More generally, we found that genes involved in nitrogen metabolism were highly enriched (21% and 8%, figure 2a) among TGAs identified in the M. reukaufii genome. One example is MreuAOC1A and B, which are homologous to the Cu2+ containing amine oxidase of Ogataea polymorpha (OpolAOC1) [39]. MreuAOC1A and B encode 609 and 620 amino acid proteins located 1729 bp apart on the minus strand of scaffold 1 (figure 2d) and share 78% and 77% protein identity to C. lusitaniae's homologue (CLUG_00771), respectively. OpolAOC1 has been studied thoroughly and its overexpression is linked to increased cell growth [39]. The complete list of M. reukaufii TGAs contains many more examples of genes with predicted functions in amino acid biosynthesis or catabolism (electronic supplementary material, table S1), indicating that these families may have undergone adaptive expansion.
(e). Nitrogen scavenging as a mechanism of priority effects
Based on our findings, we hypothesize that rapid depletion of amino acids promoted by gene duplications is a key mechanism of the priority effects observed in M. reukaufii. If M. reukaufii colonizes nectar prior to other yeasts, it should severely limit amino acid availability for late-arriving species.
To begin to test this hypothesis, we conducted competition experiments against one of the closest known relatives of M. reukaufii, Candida rancensis (Metschnikowiaceae) [40], commonly found in monkeyflower nectar [41]. Candida rancensis is one of the strongest competitors of M. reukaufii, likely due to resource-use overlap [10], but it is severely disadvantaged when M. reukaufii colonizes the nectar first [10]. Using established methods to quantify priority effects [10,11] and to simulate realistic nectar microbiome interactions, we inoculated M. reukaufii into synthetic nectar 2 days before C. rancensis. Consistent with previous studies [10–12], M. reukaufii suppressed the growth of C. rancensis when it was given a head start (figure 3c; Padj = 5 × 10−7). This effect remained even when 10 mg ml−1 of sucrose was replenished in the nectar medium every 24 h (figure 3c; Padj = 1.5 × 10−7). By contrast, when 40 mM of amino acids were supplied every 24 h, the negative effect of early M. reukaufii arrival was rescinded and the two species coexisted at similar population densities (figure 3c; Padj = 0.646). These results support the hypothesis of amino acid limitation as a cause of priority effects.
To test the hypothesis unequivocally, competition experiments using GAP1 loss of function mutants would be particularly powerful. In addition, genomic and competition analyses with other species of nectar-inhabiting yeasts should be informative because the strength of priority effects varies among these species [10–12]. Given that several nectar-inhabiting yeasts that exert priority effects with M. reukaufii are similar to M. reukaufii both phylogenetically [10] and ecologically [11], GAP1 duplication may well be a mechanism shared by many of the nectar yeast species in the Metschnikowiaceae family. One hypothesis that we believe is worth testing in future research is whether the GAP1 copy number varies among species and correlates with the strength of priority effects.
3. Conclusion
Our newly assembled M. reukaufii genome allowed us to identify candidate genes underlying the priority effects observed in nectar. In particular, we have highlighted the hypothesis that extensive genome expansion, especially in high-capacity amino acid transporter genes such as GAP1 and PUT4, allows M. reukaufii to exert strong priority effects against other nectar microbes. Further investigation will be required to unequivocally test this hypothesis. Nonetheless, the findings presented here lay a molecular foundation on which to build a better understanding of how species assemble into ecological communities. Niche pre-emption via rapid resource depletion may underlie priority effects in a broad range of microbial, plant and animal communities [8,42]. For this reason, nutrient scavenging genes like the ones considered in the yeast here may be broadly relevant to understanding priority effects.
4. Material and methods
(a). Strain
Metschnikowia reukaufii, strain MR1, was isolated from floral nectar of the sticky monkeyflower (Mimulus aurantiacus) at the Jasper Ridge Biological Preserve, Stanford, California, USA (37°24′29″ N and 122°13′39″ W) as previously described [10]. Overnight cultures (at 25°C) from single colonies grown in yeast media (YM) were used for DNA extraction process.
(b). DNA extraction, sequencing and genome assembly
PacBio: gDNA was extracted from 7 × 109 cells using the Genomic-tips 100/G platform (Qiagen), optimized for yeast cells. The gDNA was examined for bacterial contamination via PCR amplification of the 16S rRNA gene using universal primers (electronic supplementary material, table S2), but no amplicons were returned. SMRTbell libraries were obtained using the ‘Procedure and Checklist – 20 kb Template Preparation using BluePippin™ Size Selection’ protocol at the UC Davis Sequencing Core. The p5 sequencing polymerase and Magbeads (Pacific Biosciences, Menlo Park, CA, USA) were used to bind the SMRTbell templates annealed to sequencing primers. Two SMRT Cells were run on the PacBio RS II system using P6C4 chemistry, with an on-plate concentration of 150 pM and a 240-min data collection mode.
Illumina: gDNA was prepared from 106 cells using the DNeasy Blood & Tissue Kit (Qiagen). Similar to PacBio gDNA assessment, 16S rRNA PCR was used to verify the absence of bacterial contamination. Genome library was prepared using the Illumina Sequencing Library Preparation protocol as described previously [43]. The library was run on a single lane of an Illumina HiSeq 2500 sequencer with 101 bp paired-end mode.
(i). Genome assembly
The combined raw reads of the two SMRT cells were filtered and trimmed to remove low-quality sequences. We ranked different PacBio de novo assemblies generated by SMRTAnalysis (v. 1.2, patch 5), Celera (v. 8.3 RC2) and DipSPAdes (v. 3.6.2), based on their N50 value, number of contigs, average contig size and mapping quality scores against approximately 35× coverage MR1 Illumina reads. The draft assembly generated by SMRTAnalysis outperformed all other assemblies and was therefore chosen for the assembly pipeline (electronic supplementary material, figure S2). The SMRTAnalysis whitelisting protocol was used to exclude possible bacteria-contaminated reads from the filtered sub-reads pool. The initial MR1_a1 draft consisted of 128 contigs, totalling 19.56 Mb with a consensus concordance of 99.994% (electronic supplementary material, figure S2). We used Pilon (v. 1.16) with five consecutive runs to error correct the MR1_a1 diploid draft. SSPACE basic (Illumina 101 bp-paired end, v. 3.0) and SSPACE-long (PacBio error-corrected CLR, v. 1-1) were used for scaffolding. This final version of the diploid genome (MR1_a10) was then used to determine the haploid genome. To resolve the putative haplotypes, we employed a modified Haplomerger2 (v. 20151124) pipeline (electronic supplementary material, figure S1). The haploid genome was manually curated to resolve assembly errors and identify the mitochondrial sequence.
(ii). Mitochondrion assembly
The haploid assembly contained a partial mitochondrial sequence but showed assembly errors with direct and inverted repeats assembled into three separate contigs of 29, 48 and 128 kb length. To assemble the MR1 mitochondrion, we made use of a custom pipeline (electronic supplementary material, figure S1). The partial mitochondrial sequence from the haploid assembly contained the conserved COX1 sequence, which was used for a bait-mapping approach with the Illumina reads. The generated sequence was used with MITObim (v. 1.8) and MIRA4 for further mapping for 31 iterations to construct the mitochondrial sequence. Additionally, we used the mitochondrial sequence of C. lusitaniae as reference for bait-mapping with MIRA4 to construct the draft MR1 mitochondrion sequence, followed by 21 iterations of MITObim/MIRA4 to improve the final MR1 mitochondrion sequence. The consensus sequence was further error-corrected with Pilon.
The integrative pipeline presented here makes use of three successive tiers, diploid draft, haplotype reconstruction and independent organelle assembly. It allows de novo assembly efforts, especially for organisms with high rates of heterozygosity, to near-complete genomes with minimal need for manual curation, ideal for downstream applications such as automated gene annotation/ontology, phylogenetics and resequencing/variant discovery.
(c). Genome annotation, gene ontology and genetic features
We used the ab initio gene prediction pipeline MAKER (v. 2.31.8) to annotate the genome [44]. EST evidence from M. fructicola and M. bicuspidata was used to train MAKER [23,45]. Protein evidence from M. bicuspidata, D. hansenii, Candida tenuis and C. lusitaniae were also used [23,44,46]. Within the MAKER pipeline, RepeatMasker (v. 4.0.6) [22] using the latest RepBase repeat libraries was employed for soft masking. To refine initial gene models, CEGMA (v. 2.5) was used in conjunction with geneID (v. 1.4), genewise (v. 2.2.3-rc7) and HMMER (v. 3.1b2) to identify highly conserved eukaryotic ‘core’ genes. The identified core genes were used to subsequently train SNAPhmm (release 2013-11-29). In addition, AUGUSTUS (v. 3.0.2) and GeneMarkHMM (v. 4.21), were invoked by MAKER for gene prediction. Only genes with predicted complete open reading frames were retained. Gene calls were generated using SNAPhmm, AUGUSTUS and Exonerate (v. 2.2.0), using the evidence sets detailed above. Six consecutive MAKER runs were used to further train prediction tools and improve gene model quality. The consensus mitochondrial sequence was annotated using the MITOS web portal. We selected a range of species in a closely related sister clade (CUG-Ser) for which published genomes were available, other ascomycetous nectar yeasts, and the manually created reference genome of S. cerevisiae in order to evaluate the assembly completeness of M. reukaufii genome, using CEGMA genes as reference.
Blast2GO (v. 2.7.2) [47] was used with BLASTP searches against the NCBI fungi dataset, filtered using Blast2GO annotation algorithm, and GO, and enzyme code were annotated using the GO database. Interproscan results were imported into Blast2GO and merged with GO annotations. Annotation statistics were produced using Eval (v. 2.2.8) and Geneious v. 8.0.2. Repeat identification and annotation was generated using the REPET pipeline [48].
Whole genomes of M. bicuspidata NRRL YB-4993 [23] and C. lusitaniae ATCC 42720 [44] were compared against the genome of M. reukaufii MR1_a14. Pairwise alignments were performed for coding sequences of predicted gene models using adaptive seeds [49]. The SynFind synteny search pipeline identified syntenic blocks by chaining the hits from large-scale alignment tool (LAST) with a distance cut-off of 20 genes apart, and with at least four gene pairs per syntenic block [50]. The syntenic blocks were screened further using QUOTA-ALIGN to retain one-to-one blocks and remove weak blocks [50]. The outputs were visually inspected to confirm the structural similarity of the M. reukaufii genome to other genomes. Resulting syntenic gene blocks were used to identify orthologues between the genome pairs. Duplicated genes (TGAs) were mapped with Blast2GO to classify their biological function.
Manually selected tandem duplicated genes such as GAP1 and PUT4 were analysed further. Pairwise alignments of amino acid sequences from previously discussed species with those of M. reukaufii genes of interest were performed in Geneious using the Blosum62 cost matrix with free end gaps, followed by manual curation. Maximum parsimony and likelihood trees were calculated using PAUP* (heuristic search with TBR branch swapping and 100 bootstraps) [51] and RaxML 7.2.8 (GAMMA BLOSUM62 protein model, 100 bootstraps) [52].
(d). RNA isolation and qRT-PCR
Total RNA was isolated from M. reukaufii as described [53], with the following modifications: post supernatant removal, cell pellet was resuspended in 100 µl water, immediately frozen in liquid N and stored at −80°C. Frozen cells were re-suspended in 1.5 ml 50 : 50 (v/v) RNA buffer and phenol/chlorophorm/isoamyl alcohol (PCI, 25 : 24 : 1 (v/v/v)), mixed and incubated at 65°C for 1 h, mixing every 15 min. After centrifugation, supernatant was washed twice with PCI and incubated with one volume 8 M lithium chloride for 2.5 h (−20°C) for precipitation, followed by two washes in cold 80% ethanol. Final pellets were re-suspended in 100 µl TE buffer. For qRT-PCR, RNA was pre-treated with TURBO DNA-free DNase I (ThermoFisher), and 2.5 µg RNA/sample was synthesized to cDNA by reverse transcriptase (Maxima First Strand Kit, ThermoFisher). The TAF10 gene (ScerTAF10, Mreu_scf5_1.979) was used as internal control. qRT-PCR was performed with SensiMix SYBR & Fluorescein Kit (Bioline) in a LightCycler 480 Instrument II (Roche). Developed qRT-PCRs were tested for efficiency using a dilution series of target cDNA and their amplification efficiencies ranged between 90–105% (R2 > 0.95) and single melting peak observed for each amplicon (electronic supplementary material, figure S10 and table S2). All qRT-PCR reactions included three biological replicates and two technical replicates each.
(e). Competition experiments
Sterile synthetic nectar was prepared as previously described [11], with 4 mM amino acids. On day 0, 200 M. reukaufii cells were introduced into sterile microcosms containing 9 µl of nectar. To induce priority effects, 200 cells of Candida rancensis were introduced to half of the microcosms after 48 h. Thereafter, nutrient supplements of either 40 mM amino acids, 10 mg ml−1 sucrose or water were provided every 24 h. Monocultures of M. reukaufii and C. rancensis in conditions described above were maintained as positive controls, as well as negative controls (no yeast). The experiment was independently replicated twice, consisting of six biological replicates/treatment. At the end of day 4, 0.5 µl nectar from each treatment microcosm was plated onto YM agar and cfu µl−1 calculated (cfu = colony forming units). ANOVA followed by Tukey's HSD test for post hoc comparisons was performed on log-transformed data in R v. 3.3.2.
Supplementary Material
Acknowledgements
We thank Michael Banf, Giltae Song, Chris Hittinger and Garret Huntress for computational assistance, Lily Cheung and Sandeep Venkataram for discussion, Xuhuai Ji and Lutz Froenicke for sequencing assistance, Lydia-Marie Joubert for SEM assistance, and two anonymous reviewers, Melissa Dsouza, Andrew Letten and members of the community ecology group at Stanford for comments.
Data accessibility
Genome assembly, maker-predicted annotation and raw reads are available on GenBank (project accession PRJNA336445) and GAP1 and PUT4 alignment has been submitted to the Dryad Digital Repository [54]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MDYR00000000-MDYS00000000. The version described in this paper is version MDYR00000000-MDYS00000000.
Authors' contributions
M.K.D. and T.F. conceived the study; M.K.D., T.H. and T.F. designed the experiments; M.K.D. and T.H. performed the experiments and M.K.D. and T.H. analysed the data. M.K.D. wrote the initial manuscript, and all authors contributed to revising the manuscript.
Competing interests
We declare we have no competing interests.
Funding
The National Science Foundation (award no.: DEB 1149600), the Department of Biology and the Terman Fellowship of Stanford University and the Carnegie Endowment fund (T.H.) supported this research.
References
- 1.Drake JA. 1991. Community-assembly mechanics and the structure of an experimental species ensemble. Am. Nat. 137, 1–26. ( 10.1086/285143) [DOI] [Google Scholar]
- 2.Sutherland JP. 1974. Multiple stable points in natural communities. Am. Nat. 108, 859–873. ( 10.1086/282961) [DOI] [Google Scholar]
- 3.Chase JM. 2003. Community assembly: when should history matter? Oecologia 136, 489–498. ( 10.1007/s00442-003-1311-7) [DOI] [PubMed] [Google Scholar]
- 4.Ejrnæs R, Bruun HH, Graae BJ. 2006. Community assembly in experimental grasslands: suitable environment or timely arrival? Ecology 87, 1225–1233. ( 10.1890/0012-9658(2006)87%5B1225:CAIEGS%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 5.Wilbur HM, Alford RA. 1985. Priority effects in experimental pond communities: responses of Hyla to Bufo and Rana. Ecology 66, 1106–1114. ( 10.2307/1939162) [DOI] [Google Scholar]
- 6.Hiscox J, Savoury M, Müller CT, Lindahl BD, Rogers HJ, Boddy L. 2015. Priority effects during fungal community establishment in beech wood. ISME J. 9, 2246–2260. ( 10.1038/ismej.2015.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devevey G, Dang T, Graves CJ, Murray S, Brisson D. 2015. First arrived takes all: inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains. BMC Microbiol. 15, 61 ( 10.1186/s12866-015-0381-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukami T. 2015. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23. ( 10.1146/annurev-ecolsys-110411-160340) [DOI] [Google Scholar]
- 9.Martin LM, Wilsey BJ. 2012. Assembly history alters alpha and beta diversity, exotic–native proportions and functioning of restored prairie plant communities. J. Appl. Ecol. 49, 1436–1445. ( 10.1111/j.1365-2664.2012.02202.x) [DOI] [Google Scholar]
- 10.Peay KG, Belisle M, Fukami T. 2012. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc. R. Soc. B 279, 749–758. ( 10.1098/rspb.2011.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vannette RL, Fukami T. 2014. Historical contingency in species interactions: towards niche-based predictions. Ecol. Lett. 17, 115–124. ( 10.1111/ele.12204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker CM, Fukami T. 2014. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. Proc. R. Soc. B 281, 20132637 ( 10.1098/rspb.2013.2637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera CM, Canto A, Pozo MI, Bazaga P. 2010. Inhospitable sweetness: nectar filtering of pollinator-borne inocula leads to impoverished, phylogenetically clustered yeast communities. Proc. R. Soc. B 277, 747–754. ( 10.1098/rspb.2009.1485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pozo MI, Herrera CM, Lachance M-A, Verstrepen K, Lievens B, Jacquemyn H. 2015. Species coexistence in simple microbial communities: unravelling the phenotypic landscape of co-occurring Metschnikowia species in floral nectar. Environ. Microbiol. 18, 1850–1862. ( 10.1111/1462-2920.13037) [DOI] [PubMed] [Google Scholar]
- 15.Vannette RL, Gauthier M-PL, Fukami T. 2013. Nectar bacteria, but not yeast, weaken a plant–pollinator mutualism. Proc. R. Soc. B 280, 20122601 ( 10.1098/rspb.2012.2601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaeffer R, Irwin RE. 2014. Yeasts in nectar enhance male fitness in a montane perennial herb. Ecology 95, 1792–1798. ( 10.1890/13-1740.1) [DOI] [PubMed] [Google Scholar]
- 17.Brysch-Herzberg M. 2004. Ecology of yeasts in plant–bumblebee mutualism in Central Europe. FEMS Microbiol. Ecol. 50, 87–100. ( 10.1016/j.femsec.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 18.Lachance M-A, Starmer WT, Rosa CA, Bowles JM, Barker JSF, Janzen DH. 2001. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1, 1–8. ( 10.1111/j.1567-1364.2001.tb00007.x) [DOI] [PubMed] [Google Scholar]
- 19.Herrera CM, Pozo MI, Bazaga P. 2012. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Mol. Ecol. 21, 2602–2616. ( 10.1111/j.1365-294X.2011.05402.x) [DOI] [PubMed] [Google Scholar]
- 20.Liti G, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458, 337–341. ( 10.1038/nature07743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, et al. 2014. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 5, 5896 ( 10.1038/ncomms6896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196. ( 10.1101/gr.6743907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley R, et al. 2016. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 113, 9882–9887. ( 10.1073/pnas.1603941113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos M, Perreau VM, Tuite MF. 1996. Transfer RNA structural change is a key element in the reassignment of the CUG codon in Candida albicans. EMBO J. 15, 5060–5068. [PMC free article] [PubMed] [Google Scholar]
- 25.Mülhausen S, Kollmar M. 2014. Predicting the fungal CUG codon translation with Bagheera. BMC Genomics 15, 411 ( 10.1186/1471-2164-15-411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang BF, Gray MW, Burger G. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397. ( 10.1146/annurev.genet.33.1.351) [DOI] [PubMed] [Google Scholar]
- 27.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. 2003. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA 100, 11 484–11 489. ( 10.1073/pnas.1932072100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Despons L, Baret PV, Frangeul L, Louis VL, Durrens P, Souciet J-L. 2010. Genome-wide computational prediction of tandem gene arrays: application in yeasts. BMC Genomics 11, 1–13. ( 10.1186/1471-2164-11-56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack DL, Paulsen IT, Saier MH. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146, 1797–1814. ( 10.1099/00221287-146-8-1797) [DOI] [PubMed] [Google Scholar]
- 30.Møller HD, Andersen KS, Regenberg B. 2013. A model for generating several adaptive phenotypes from a single genetic event: Saccharomyces cerevisiae GAP1 as a potential bet-hedging switch. Commun. Integr. Biol. 6, e23933 ( 10.4161/cib.23933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. 2010. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc. Natl. Acad. Sci. USA 107, 18 551–18 556. ( 10.1073/pnas.1014023107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofman-Bang J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12, 35–74. ( 10.1385/mb:12:1:35) [DOI] [PubMed] [Google Scholar]
- 33.Daugherty J, Rai R, el Berry HM, Cooper TG. 1993. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 175, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole KE, Walker M, Warren T, Gardner J, McBryde C, de Barros Lopes M, Jiranek V. 2009. Proline transport and stress tolerance of ammonia-insensitive mutants of the PUT4-encoded proline-specific permease in yeast. J. Gen. Appl. Microbiol. 55, 427–439. ( 10.2323/jgam.55.427) [DOI] [PubMed] [Google Scholar]
- 35.Donaton MCV, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelein JM. 2003. The GAP1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50, 911–929. ( 10.1046/j.1365-2958.2003.03732.x) [DOI] [PubMed] [Google Scholar]
- 36.Nepi M. 2014. Beyond nectar sweetness: the hidden ecological role of non-protein amino acids in nectar. J. Ecol. 102, 108–115. ( 10.1111/1365-2745.12170) [DOI] [Google Scholar]
- 37.Gournas C, Evangelidis T, Athanasopoulos A, Mikros E, Sophianopoulou V. 2015. The Aspergillus nidulans proline permease as a model for understanding the factors determining substrate binding and specificity of fungal amino acid transporters. J. Biol. Chem. 290, 6141–6155. ( 10.1074/jbc.M114.612069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunkel N, Hertlein T, Franz R, Reuß O, Sasse C, Schäfer T, Ohlsen K, Morschhäuser J. 2013. Roles of different peptide transporters in nutrient acquisition in Candida albicans. Eukaryot. Cell 12, 520–528. ( 10.1128/EC.00008-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber KN, Haima P, Gietl C, Harder W, Ab G, Veenhuis M. 1994. The methylotrophic yeast Hansenula polymorpha contains an inducible import pathway for peroxisomal matrix proteins with an N-terminal targeting signal (PTS2 proteins). Proc. Natl. Acad. Sci. USA 91, 12 985–12 989. ( 10.1073/pnas.91.26.12985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzman B, Lachance MA, Herrera CM. 2013. Phylogenetic analysis of the angiosperm–floricolous insect–yeast association: have yeast and angiosperm lineages co-diversified? Mol. Phylogenet. Evol. 68, 161–175. ( 10.1016/j.ympev.2013.04.003) [DOI] [PubMed] [Google Scholar]
- 41.Belisle M, Peay KG, Fukami T. 2012. Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microb. Ecol. 63, 711–718. ( 10.1007/s00248-011-9975-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Meester L, Vanoverbeke J, Kilsdonk LJ, Urban MC. 2016. Evolving perspectives on monopolization and priority effects. Trends Ecol. Evol. 31, 136–146. ( 10.1016/j.tree.2015.12.009) [DOI] [PubMed] [Google Scholar]
- 43.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 ( 10.1371/journal.pone.0128036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler G, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662. ( 10.1038/nature08064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hershkovitz V, et al. 2013. De-novo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC Genomics 14, 1–13. ( 10.1186/1471-2164-14-168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dujon B, et al. 2004. Genome evolution in yeasts. Nature 430, 35–44. ( 10.1038/nature02579) [DOI] [PubMed] [Google Scholar]
- 47.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 48.Flutre T, Duprat E, Feuillet C, Quesneville H. 2011. Considering transposable element diversification in de novo annotation approaches. PLoS ONE 6, e16526 ( 10.1371/journal.pone.0016526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493. ( 10.1101/gr.113985.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H, Bomhoff MD, Briones E, Zhang L, Schnable JC, Lyons E. 2015. SynFind: compiling syntenic regions across any set of genomes on demand. Genome Biol. Evol. 7, 3286–3298. ( 10.1093/gbe/evv219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 52.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collart MA, Oliviero S. 2001. Preparation of yeast RNA. In Current protocols in molecular biology, vol. 23, series vol. IV, pp. 13.12:13.12.1–13.12.5. New York, NY: John Wiley and Sons, Inc ( 10.1002/0471142727.mb1312s23) [DOI] [PubMed]
- 54.Dhami MK, Hartwig T, Fukami T. 2016. Data from: Genetic basis of priority effects: insights from nectar yeast. Dryad Digital Repository ( 10.5061/dryad.5h17h) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assembly, maker-predicted annotation and raw reads are available on GenBank (project accession PRJNA336445) and GAP1 and PUT4 alignment has been submitted to the Dryad Digital Repository [54]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MDYR00000000-MDYS00000000. The version described in this paper is version MDYR00000000-MDYS00000000.