Abstract
Epstein-Barr virus nuclear antigen 3C (EBNA 3C) is essential for B-cell immortalization and functions as a regulator of viral and cellular transcription. EBNA 3C contains glutamine-rich and proline-rich domains and a region in the N terminus consisting of a stretch of basic residues followed by a run of leucine residues spaced seven amino acids apart. This N-terminal domain is widely believed to represent a leucine zipper dimerization motif (bZIP). We have performed the first structural and functional analysis of this motif and demonstrated that this domain is not capable of forming stable homodimers. Peptides encompassing the EBNA 3C zipper domain are approximately 54 to 67% α-helical in solution but cannot form dimers at physiologically relevant concentrations. Moreover, the EBNA 3C leucine zipper cannot functionally substitute for another homodimerizing zipper domain in domain-swapping experiments. Our data indicate, however, that the EBNA 3C zipper domain behaves as an atypical bZIP domain and is capable of self-associating to form higher-order α-helical oligomers. Using directed mutagenesis, we also identified a new role for the bZIP domain in maintaining the interaction between EBNA 3C and RBP-Jκ in vivo. Disruption of the helical nature of the zipper domain by the introduction of proline residues reduces the ability of EBNA 3C to inhibit EBNA 2 activation and interact with RBP-Jκ in vivo by 50%, and perturbation of the charge on the basic region completely abolishes this function of EBNA 3C.
Epstein-Barr virus (EBV) is a human gammaherpesvirus implicated in the development of a large number of malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, certain B- and T-cell lymphomas, and possibly breast cancer. EBV has the capacity to infect and immortalize B cells in vitro and establish a latent infection that is accompanied by the expression of a limited number of viral gene products. These include the EBV nuclear antigens (EBNAs) 1, 2, 3A, 3B, 3C, and LP, the membrane antigens LMPs 1, 2A, and 2B, and a class of small untranslated RNA molecules, the EBERs. All of the nuclear antigens have been shown to function as transcriptional regulators, and the regulation of viral and cellular gene expression by these proteins appears to be crucial to the immortalization process since viruses defective for EBNA 1, 2, 3A, or 3C expression are transformation deficient (4, 15, 34, 36). EBNA 1 also performs an essential role in maintaining the viral genome in its episomal form (22, 48).
Although EBNA 1 binds to specific DNA sequences in the oriP region of the EBV genome (29), the remaining EBNAs do not appear to bind to DNA in a sequence-specific manner but mediate their transcriptional effects through DNA-binding cellular cofactors. EBNA 2 is targeted to its response elements in the viral C and LMP promoters and the cellular CD21 and CD23 promoters through an interaction with the DNA-binding protein RBP-Jκ (6, 8, 40). Additional interactions between EBNA 2 and other cellular DNA-binding proteins from the Spi-1/Spi-B (PU.1) family have also been shown to play a role in the activation of the LMP 1 promoter by EBNA 2 (11, 33). The EBNA 3 family of proteins (EBNAs 3A, 3B, and 3C) also bind to RBP-Jκ, but this interaction results in the repression of transcription from EBNA 2-activated promoters (16, 30). It appears that, when bound to one of the EBNA 3 proteins, RBP-Jκ is no longer able to interact with EBNA 2 or bind to DNA (30, 41).
The first evidence implicating a member of the EBNA 3 family in the regulation of transcription, however, came from gene transfection experiments with EBNA 3C, which pointed to a role for this protein as a transcriptional activator. The expression of EBNA 3C in an EBV-negative Burkitt's lymphoma cell line resulted in increased expression of the EBV receptor CD21 (CR2) on the cell surface (44). Further evidence for the role of EBNA 3C as a transcriptional activator came from experiments in which EBNA 3C was shown to maintain the levels of LMP 1 in cells that had reached saturation density (1) and from Gal4 fusion experiments that identified a putative activation domain in the C terminus of the protein (amino acids 724 to 826) (18) (Fig. 1A). EBNA 3C has also been shown to augment the activation of the LMP 1 promoter by EBNA 2 in a manner that is dependent on the presence of the Spi-1 and Spi-B (PU.1) sites (51). Since EBNA 3C has been shown to associate with p300 and prothymosin α, it is possible that the transcriptional activation function of this protein is mediated by the recruitment of coactivators that promote the acetylation of histone tails and disruption of chromatin structure at the promoter (35).
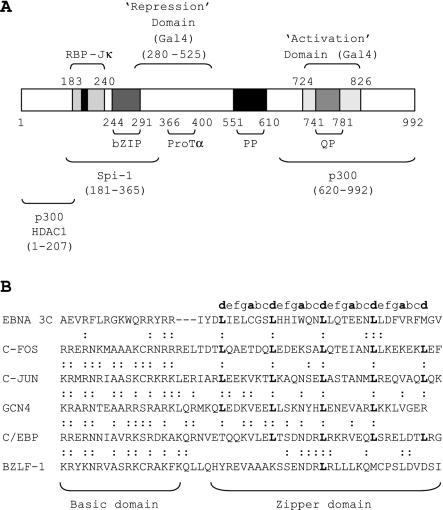
FIG. 1.
(A) Diagram of the EBNA 3C protein, indicating the location of domains with homology to other transcription factors and domains mapped in functional or biochemical assays. The black box in the center of the RBP-Jκ binding domain represents the core TFGC sequence required for RBP-Jκ binding at amino acids 209 to 212. (B) Sequence alignment of the putative EBNA 3C bZIP domain with the bZIP domains of other transcription factors. Dots indicate conserved residues (R-to-K and K-to-R substitutions in the basic region are classed as conserved). Letters indicate the assignment of heptad positions in the sequences; the hydrophobic a and d positions are shown in bold. Note that three spaces have been introduced in the EBNA 3C sequence in order to obtain the best alignment of the basic residues with other basic domains.
EBNA 3C is also able to function as a repressor of transcription from the viral C promoter in the absence of EBNA 2, an effect that is likely to be mediated through the recruitment of multiple corepressors, including histone deacetylases 1 and 2, mSin3A, NCoR, and CtBP to the promoter (12, 28, 37). Tethering experiments identified two domains of EBNA 3C, amino acids 280 to 525 and amino acids 580 to 992, that are able to function as cell type-independent and promoter-independent repressors of transcription when expressed as Gal4 fusion proteins, although the C-terminal domain has only modest activity (2) (Fig. 1A). The ability of unfused EBNA 3C to repress the viral C promoter, however, requires the presence of amino acids 207 to 368, residues that are dispensable when the protein is targeted to DNA as a fusion protein (27).
Sequence analysis of EBNA 3C has identified a number of domains commonly found in both viral and cellular transcription factors (Fig. 1A). These include proline-rich and glutamine-proline-rich domains and a region with some homology to the bZIP (basic zipper) domains found in the AP-1 family of transcription factors, e.g., c-Fos and c-Jun (Fig. 1B). The glutamine-proline-rich domain forms part of the region of EBNA 3C that has been shown to mediate transcriptional activation in Gal4 fusion assays (18). A region encompassing the bZIP domain has been shown to bind to the Spi-1 and Spi-B proteins in vitro, and it has been suggested that this interaction may mediate the coactivation function of EBNA 3C at the LMP1 promoter (51). bZIP domains normally consist of a region rich in basic amino acids required for binding to DNA, followed by an amphipathic α-helical dimerization motif (the zipper). bZIP proteins recognize bipartite DNA sequences in their target promoters and bind to DNA as parallel homo- or heterodimers. Dimerization between two zipper domains results in the formation of a coiled-coil structure in which hydrophobic residues from the two amphipathic helices interdigitate to form a tightly packed interface (23). Such dimeric coiled-coil motifs are usually characterized by the presence of a heptad repeat sequence containing a run of four or more leucine residues spaced seven residues apart (Fig. 1B) and are referred to as leucine zippers. The presence of a bZIP domain in EBNA 3C is perplexing because, unlike other members of the bZIP family, EBNA 3C does not appear to possess sequence-specific DNA-binding activity.
In this study, we used a combination of biophysical techniques and in vitro assays to test the hypothesis that the putative bZIP domain of EBNA 3C represents a functional leucine zipper dimerization motif. Using a rational mutagenesis approach, we also examined the role played by this domain in the transcriptional function of EBNA 3C in vivo.
MATERIALS AND METHODS
Plasmids.
Plasmids for use in domain-swapping experiments were constructed by cloning PCR-amplified fragments from pSG5 EBNA3C-FL or pSP64 GCN4 (gift from T. Kouzarides) between the PstI and HincII sites of pRSETA BZLF-1 (His tagged) to replace the BZLF-1 zipper domain. The entire BZLF-1 coding region of the hybrid pRSETA BZLF-1 constructs was then subcloned into pSP64 as a BamHI/EcoRI fragment for the production of untagged hybrid proteins.
pSG5 EBNA 3C-FL was generated from the previously described pSG5 EBNA 3C (27) that was found to have a deletion in a repeat region of EBNA 3C (amino acids 571 to 610) (20). The 1.48-kb BglII fragment of pSG5 EBNA3C was replaced with the corresponding 1.6-kb BglII fragment from EBNA 3C-pZip-neoSV (26) to repair the deletion and generate pSG5 EBNA3C-FL. pRSETA BZLF-1 was generated by PCR amplifying the coding region of BZLF-1 from pSP64 EB1 (31) with primers designed to introduce a BamHI site immediately adjacent to the start codon (5′-GGGGATCCATGATGGACCCAAACTCGAC-3′) and an EcoRI site immediately adjacent to the stop codon (5′-GGGAATTCTTAGAAATTTAAGAGATCCTCG-3′). This fragment was then inserted into the BamHI and EcoRI sites of pRSETA (Invitrogen).
pRSETA BZLF-1/3C‘d’ was generated with forward (5′-AAAACTGCAGGATTTGATAGAAC-3′) and reverse (5′-GAAACGCACGAAATCTAAAAGG-3′) primers to amplify the zipper region of EBNA 3C and introduce a PstI site at the 5′ end (bold) but generate a 3′ blunt end. Following digestion with PstI and treatment with polynucleotide kinase to phosphorylate the 3′ end, this PCR fragment was cloned between the PstI and HincII sites of pRSETA BZLF-1. The frame of the substitution was designed to place the leucine residues of EBNA 3C in the d position of the BZLF-1 heptad.
pRSETA BZLF-1 3C‘a’ was generated in a similar way, with the forward and reverse primers 5′-AAAACTGCAGCGAAGAATCTATG-3′ and 5′-ATCTAAAAGGTTCTCCTCGGTCTGG-3′, respectively, and designed to place the leucine residues of EBNA 3C in the a position of the BZLF-1 heptad.
pRSETA BZLF-1/GCN4 was generated with the forward and reverse primers 5′-AAAACTGCAGCAACTTGAAGACAAG-3′ and 5′-TTCGCCAACTAATTTCTTTAATCTGG-3′, respectively, to amplify the GCN4 zipper region from pSP64 GCN4. The leucine residues of the GCN4 zipper were placed in the d positions of the BZLF-1 heptad.
To generate pRSETA BZLF-1/3C-INI, sequential site-directed mutagenesis was first carried out on pSG5 EBNA 3C-FL to introduce the specific mutations W274N, E281I, and C267I. In the first step, complementary forward 5′-CTGCACCACATCAATCAAAACTTGCTCC-3′ and reverse 5′-GGAGCAAGTTTTGATTGATGTGGTGCAG-3′ primers were used to introduce the W274N substitution (italic). The resulting plasmid containing this substitution was then used as the template in a second round of mutagenesis to introduce the E281I substitution with the complementary forward (5′-CTTGCTCCAGACCATCGAGAACCTTTTAGATTTC-3′) and reverse (5′-GAAATCTAAAAGGTTCTCGATGGTCTGGAGCAAG-3′) primers. A further round of mutagenesis was performed with the plasmid containing these two substitutions with a further set of primers (forward, 5′-GATTTGATAGAACTGATTGGCTCTCTGCACCAC-3′, and reverse, 5′-GTGGTGCAGAGAGCCAATCAGTTCTATCAAATC-3′) to introduce the final C267I substitution. The mutated leucine zipper sequence was then amplified with the same primers designed to generate the pRSETA BZLF-1/3C‘d’ construct and cloned into pRSETA BZLF-1.
pSP64 BZLF-1/3C-MG was generated by using one forward (5′-GATTTCGTGCGTTTCATGGGCATTATCCCCCGGACACCAG-3′) and one reverse (5′-CTGGTGTCCGGGGGATAATGCCCATGAAACGCACGAAATC-3′) primerin site-directed mutagenesis on pSP64 BZLF-1/3‘d’ to introduce the D228M and S229G substitutions at the 3′ end of the BZLF-1 zipper.
Site-directed mutagenesis was carried out with a PCR-based method (Stratagene). Reactions contained 50 ng of template plasmid, 125 ng of each primer, 1× Pfx buffer (Invitrogen), 1 mM MgSO4, 3× Enhancer (Invitrogen), and 2 U of Pfx polymerase (Invitrogen) in a total volume of 50 μl. Following an initial incubation at 95°C for 3 min to inactivate antibodies present in the Pfx enzyme mix, reactions were carried out at 95°C for 30 s, 36 to 45°C (template dependent) for 1 min, and 45 to 60°C for 15 min (template dependent) for a total of 18 cycles. Template plasmid was digested with the methylation-sensitive enzyme DpnI, and the mutated plasmid was transformed into Escherichia coli DH5α.
Leucine-to-proline mutations were introduced into pSG5 EBNA3C-FL by site-directed mutagenesis as described above with the following sets of primers: pSG5 EBNA3C-P1+2 (L263P,L270P), forward 5′-GAATCTATGATCCGATAGAACTGTGTGGCTCTCCGCACCACATC-3′ and reverse 5′-GATGTGGTGCGGAGAGCCACACAGTTCTATCGGATCATAGATTC-3′, and pSG5 EBNA3C-P3+4 (L277P,L284P), forward 5′-TGGCAAAACCCGCTCCAGACCGAGGAGAACCCTTTAGATTTCG-3′ and reverse 5′-CGAAATCTAAAGGGTTCTCCTCGGTCTGGAGCGGGTTTTGCCA-3′. pSG5 EBNA3C-P1-4 (L263P,L270P,L277P,L284P) was generated by mutating pSG5 EBNA3C-P1+2 with the primers for pSG5 EBNA3C-P3+4.
Basic region mutations were introduced into pSG5 EBNA3C-FL by site-directed mutagenesis with the following sets of primers: pSG5 EBNA 3C-bR-A (R254A, R255A, R257A, R258A), forward 5′-CGTGGTAAATGGCAGGCGGCGTACGCAGCAATCTATGATTTG-3′ and reverse 5′-CAAATCATAGATTGCTGCGTACGCCGCCTGCCATTTACCACG-3′, and pSG5 EBNA 3C-bR-E (R254E, R255E, R257E, R258E), forward 5′-CCTTCGTGGTAAATGGCAGGAGGAGTACGAAGAAATCTATGATTTG-3′ and reverse 5′-CAAATCATAGATTTCTTCGTACTCCTCCTGCCATTTACCACGAAGG-3′. pSG5 EBNA 3C-bA (R247A, R250A, K251A, R254A, R255A, R257A, R258A) was generated by mutating pSG5 EBNA 3C-bR-A with the primers forward 5′-CGGGAAGCCGAGGTAGCCTTCCTTGCTGGTGCATGGCAGGCGGCGTAC-3′ and reverse 5′-GTACGCCGCCTGCCATGCACCAGCAAGGAAGGCTACCTCGGCTTCCCG-3′.
pSG5 EBNA 3C-bE (R247E, R250E, K252E, R255E, R256E, R258E, R259E) was generated by mutating pSG5 EBNA 3C-bR-E with the primers forward 5′-CGGGAAGCCGAGGTAGAGTTCCTTGAGGGTGAATGGCAGGAGGAGTAC-3′ and reverse 5′-GTACTCCTCCTGCCATTCACCCTCAAGGAACTCTACCTCGGCTTCCCG-3′. pSG5 EBNA3C-mJκ (T209A, F210A, G211A, C212A) was generated with the primers forward 5′-CATCATGTTAACTGCCGCAGCTGCAGCCCAAAATGCGGCAC-3′ and reverse 5′-GTGCCGCATTTTGGGCTGCAGCTGCGGCAGTTAACATGATG-3′.
For all zipper mutations, after sequencing to verify the mutation, the HpaI/SpeI fragment of the new plasmid was cloned back into pSG5 EBNA 3C-FL to avoid other PCR-induced mutations in the vector.
Cell culture.
The EBV-negative Burkitt's lymphoma cell line DG75 (gift from M. Rowe) was maintained in suspension culture in RPMI medium supplemented with 10% fetal bovine serum, 292 μg of glutamine per ml, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Invitrogen). Cells were passaged twice weekly and kept at 37°C in a humidified incubator containing 5% CO2.
Transient transfections.
DG75 cells were transfected by electroporation essentially as described previously (45). Briefly, cells were diluted 1:3 24 h prior to transfection, and 107 cells in serum-free medium were mixed with DNA and electroporated at 260 V and 950 μF (Bio-Rad Genepulser III). Transfections contained 0 or 10 μg of pSG52A (38), 0, 2, or 5 μg of pSG5 EBNA 3C plasmid, 2 μg of Cp-1425-GL2 (contains the SauIIIA fragment of the C promoter [EBV nucleotides 9911 to 11340] cloned into the BglII site of pGL2-basic), and 2 μg of pRL-CMV (Promega). DNA levels were kept constant with pSG5 vector where necessary.
Luciferase assays.
Cells were harvested 48 h posttransfection, washed in phosphate-buffered saline (PBS), and then lysed in 100 μl of 1× PLB (Promega); 10-μl aliquots of cleared lysate were then assayed in duplicate in a 96-well plate with 50 μl of LarII followed by 50 μl of Stop and Glo solutions (Promega luciferase dual assay kit) with the sequential injection system on a Lucy 2 luminometer (LabTech). The firefly luciferase signal (pCp1425-GL2) was adjusted for transfection efficiency with the Renilla luciferase signal from the control plasmid pRL-CMV.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), plasmids pSP64 EB1 (wild-type BZLF-1) (31), pSP64 BZLF-1/3Cd, pSP64 BZLF-1/3Ca, pSP64 BZLF-1/3C-INI, pSP64 BZLF-1/3C-MG, and pSP64 BZLF-1/GCN4 were linearized with EcoRI and used as templates to generate RNA in in vitro transcription reactions (SP6 Ribomax kit; Promega). RNA (4 μg) was in vitro translated (wheat germ extract; Promega) in the presence of [35S]methionine to generate labeled proteins. Samples of in vitro-translated protein were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantitated with PhosphorImager software (Imagequant; Molecular Dynamics). After adjusting the signal to take into account the number of methionine residues in each protein, equivalent amounts of each protein were used in EMSAs. Volumes were kept constant with a no-RNA in vitro-translated control sample.
An oligonucleotide consisting of a consensus AP-1 site (AP-1a, 5′-GATCCATGACTCAGAGGAAAACATACG-3′) was labeled at the 5′ end with [γ-32P]ATP and then annealed to an unlabeled complementary oligonucleotide (AP-1b, 5′-CGTATGTTTTCCTCTGAGTCATGGATC-3′) by heating and slow cooling to generate a labeled double-stranded AP-1 probe for use in EMSAs.
Binding reactions contained up to 2 μl of in vitro-translated protein, 1× gel shift binding buffer (Promega), supplementary dithiothreitol (DTT) to obtain a final concentration of 5.5 mM, and 60 pg of double-stranded AP-1 probe in a total volume of 10 μl. Reactions were incubated for 30 min at 26°C prior to the addition of 2 μl of 4× TBE loading buffer (50% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol and 4× Tris-borate-EDTA). Protein-DNA complexes were analyzed by electrophoresis on a 6% TBE retardation gel (Novex; Invitrogen) with 0.5× TBE as the buffer. The gel was then fixed in 40% methanol and 10% acetic acid and dried under vacuum, and bands were visualized by autoradiography.
Native gel electrophoresis.
In vitro-translated protein samples were incubated at 26°C for 30 min in 1× gel shift binding buffer plus supplementary DTT in a total volume of 10 μl as for EMSA. SDS was included in the buffer at a final concentration of 0.5% where required. Native gel buffer was then added to samples to a final concentration of 1× (40 mM Tris-HCl [pH 7.5], 4% glycerol, 0.01% bromophenol blue) and samples were loaded on a prerun nondenaturing 7% Tris-acetate gel (Novex; Invitrogen). Electrophoresis was carried out with Tris-glycine native gel running buffer (Novex; Invitrogen), and gels were fixed, dried, and analyzed with a phosphorimager.
Immunoprecipitation and immunoblotting.
Cells were harvested 48 h posttransfection, washed in PBS, and then resuspended in 0.5 ml of EBC lysis buffer (120 mM NaCl, 50 mM Tris [pH 8.0], 5 mM DTT, 0.5% NP-40, 0.1% SDS, 2 mM phenylmethylsulfonyl fluoride, and EDTA-free protease inhibitor cocktail [Roche]). Cells were lysed for 30 min on ice, and the lysates were sonicated. Following centrifugation at 13,000 rpm to remove insoluble debris, the lysate was precleared with 10 μl of normal rabbit serum (Sigma). Preclearing antibodies and associated proteins were removed by incubating with bovine serum albumin-coated protein A-Sepharose beads (Sigma) for 30 min at 4°C with rotation, followed by brief centrifugation (10 s at 13,000 rpm). The precleared lysate was then incubated with 2 μl of rabbit polyclonal RBP-Jκ antibody STL84 (gift from E. Kieff) for 30 min on ice. Antibody complexes were isolated by incubation with bovine serum albumin-coated protein A-Sepharose beads for 2 h at 4°C with rotation. The beads were washed four times with 0.5 ml of EBC wash buffer (120 mM NaCl, 50 mM Tris [pH 8.0], 5 mM DTT, 0.5% NP-40, 0.5% SDS) and then heated to 95°C in gel sample buffer (46) to release associated proteins.
Samples were separated by SDS-PAGE with a 3 to 8% Tris-acetate gel and Tris-acetate running buffer (Novex). Proteins were transferred onto Protran nitrocellulose membranes (Schleicher and Schuell). EBNA 3C was detected by probing overnight at 4°C with the EBNA 3C monoclonal antibody E3cA10 (19) (gift from M. Rowe) at a concentration of 7 μg/ml in 5% milk. Following incubation with horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin secondary antibodies (Dako), membranes were washed three times with PBS-Tween, incubated with chemiluminescence reagents, and exposed to Hyperfilm (Amersham Pharmacia Biotech) to detect proteins.
Immunofluorescence.
Transfected DG75 cells were resuspended at 107 cells/ml in PBS, and the cell suspension was pipetted onto microdot slides (ICN) and then removed to leave a thin film of cells on the slide. Slides were fixed in methanol-acetone (1:1, vol/vol) at −20°C for 10 min, air dried, and then stored at −20°C until required. Slides were rehydrated and blocked with 20% normal goat serum (Sigma) in PBS and then stained with human serum monospecific for EBNA 3C (diluted to a final concentration of 1:40 in 20% normal goat serum in PBS) (M. S. serum; gift from M. Rowe). Slides were washed in three changes of PBS and EBNA 3C and then detected with fluorescein isothiocyanate-conjugated rabbit anti-human immunoglobulin G (Dako) diluted 1:50 in 20% normal goat serum in PBS. After further washing, nuclei were stained with 12.5 ng of 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI) (Sigma) per ml for 5 min. After a further two washes, coverslips were mounted on microscope slides with Mowiol mounting solution [0.2 M Tris (pH 8.5), 33% (wt/vol) glycerol, 13% (wt/vol) Mowiol, 2.5% (wt/vol) 1,4-diazobicyol (2,2,2)-octane (Dabco)] and sealed with clear nail polish. Slides were analyzed with a Zeiss Axioscope 2 microscope equipped with a 63× oil immersion objective and fitted with the appropriate filter sets. Images were captured with a Photometrics Quantix digital camera. Images were processed with Metamorph imaging software (Universal Imaging Corp.).
CD spectroscopy.
Two peptides encompassing the EBNA 3C zipper region, EBNA 3C-p1 (RYRRIYDLIELCGSLHHIWQNLLQTEENLLDFVRF) and EBNA 3C-p2 (acetyl-RYRRIYDLIELCGSLHHIWQNLLQTEENLLDFVRFMG-amide), were synthesized, purified by high-pressure liquid chromatography, and verified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Alta Biosciences). Because these peptides were insoluble in water, they were dissolved in 10% acetic acid to generate millimolar stock solutions whose concentrations were determined by the absorbance at 280 nm, assuming a molar extinction coefficient of 8,480 M−1 cm−1 for both peptides. Circular dichroism (CD) spectroscopy was carried out with a Jasco J-715 spectropolarimeter fitted with a Peltier temperature controller. In initial experiments, EBNA 3C-p1 was diluted from the stock solution into 10 mM HEPES (pH 7.9)-2 mM DTT. The pH was then gradually adjusted to pH 7.4 with 2 M KOH. For subsequent experiments, EBNA 3C-p1 and EBNA 3C-p2 solutions were prepared in 10 mM formate (pH 3.7)-2 mM DTT, and the pH was readjusted for the higher peptide concentrations with KOH. All spectra were measured at 20°C. The concentration of formate buffer in the 10 μM, 5 μM, and 1 μM peptide samples was reduced to 5 mM to reduce noise in the CD spectra.
For thermal unfolding experiments, the signal at 222 nm was measured as the temperature was increased in a stepwise manner at a rate of 1°C/min over the range from 5 to 85 or 90°C. The midpoint temperatures of unfolding were taken as the maximum of the first derivatives of the melting curves obtained.
Analytical ultracentrifugation.
Sedimentation equilibrium studies were conducted at 20°C in a Beckman Optima XL-I analytical ultracentrifuge fitted with an An-60 Ti rotor. For EBNA 3C-p1, a 100-μl sample of peptide (at a starting concentration of 110 μM peptide [pH 3.7], 10 mM formate, 2 mM DTT) was used with a 110-μl sample of matched buffer as the reference. The sample was equilibrated for 48 to 72 h at rotor speeds of 30,000, 40,000, 50,000, and 60,000 rpm. Sedimentation curves were recorded at 280 nm. The resulting data sets were fitted simultaneously with routines from the Beckman-Origin analysis software (version 6.0). Two fitting methods were used: the first assumed a single ideal species, and the second assumed a monomer-dimer equilibrium. The molecular weight (4,405 Da), molar extinction coefficient (8,480 M−1 cm−1), and partial specific volume (0.7398 ml mg−1) were calculated from the amino acid composition of EBNA 3C-p1. The density of the solvent was calculated as 0.9987 mg ml−1. Similar experiments were performed for EBNA 3C-p2. These data were only fitted to single-species models because they showed clear evidence of a large aggregated state on the order of 80 kDa in size; further detailed analyses of these data was not deemed useful.
RESULTS
EBNA 3C leucine zipper peptides are α-helical in solution.
Biophysical analysis of synthetic peptides with techniques such as CD spectroscopy and analytical ultracentrifugation is used extensively to study the structural characteristics and oligomerization states of leucine zipper sequences (21, 24, 25). To test the hypothesis that EBNA 3C contains a genuine leucine zipper, we carried out biophysical analysis on two peptides encompassing the zipper region.
The first peptide analyzed (EBNA 3C-p1) was 35 amino acids in length and included all four leucine repeats. The folding and stability of EBNA 3C-p1 in solution were examined by CD spectroscopy. Since α-helical, β-sheet, and randomly coiled secondary structures have distinct CD profiles, the use of this technique enabled us to determine whether EBNA 3C-p1 adopted the α-helical structure characteristic of leucine zippers. At pH 7.4 in HEPES buffer, 60 μM EBNA 3C-p1 exhibited a CD spectrum indicating the presence of some α-helical structure; the expected minima at 208 and 222 nm were present, but the intensity of the signal at 222 nm suggested that this peptide was only approximately 10% helical (Fig. 2A, upper spectrum). Furthermore, under these conditions, EBNA 3C-p1 was only sparingly soluble (up to 60 μM), which hampered full characterization by CD spectroscopy. Therefore, we explored conditions under which the peptide was more soluble.
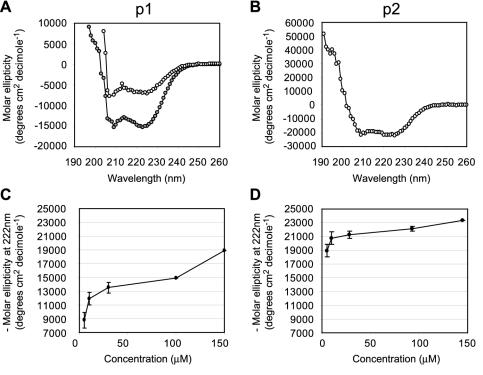
FIG. 2.
CD studies on the leucine zipper peptides EBNA 3C-p1 and EBNA 3C-p2. (A) CD spectra for 60 μM EBNA 3C-p1 in 10 mM HEPES-2 mM DTT, pH 7.4 (open circles), and 100 μM EBNA 3C-p1 in 10 mM formate-2 mM DTT, pH 3.7 (grey circles). (B) CD spectrum for 28 μM EBNA 3C-p2 in 10 mM formate-2 mM DTT, pH 3.7. (C) Graph showing the increase in CD signal at 222 nm with increasing EBNA 3C-p1 concentration. The results show the means and ranges of duplicate experiments. (D) Graph showing the increase in CD signal at 222 nm with increasing EBNA 3C-p2 concentration.
Consistent with other reports (21, 52), we found that at low pH (pH 3.7, 10 mM formate, and 2 mM DTT), the solubility and folding of the peptide improved. The insoluble nature of EBNA 3C-p1 contrasts sharply with the characteristics of typical leucine zipper peptides, which are usually fully soluble in water or standard buffers at neutral pH. Under low-pH conditions, we were able to obtain CD spectra at higher concentrations of EBNA 3C-p1. These spectra again displayed the expected minima at 208 and 222 nm, indicating the presence of α-helical structure (Fig. 2B, lower spectrum). We detected an increase in CD signal at 222 nm with increasing EBNA 3C-p1 concentrations up to a maximum of −18,900° cm−2 dmol−1 at 150 μM (Fig. 2C). Since a peptide containing 100% α-helix has a maximum molar ellipticity of approximately −35,000° cm−2 dmol−1 at 222 nm (24), we estimated that EBNA 3C-p1 is approximately 54% α-helical at 150 μM and pH 3.7.
Analysis of a second peptide (EBNA 3C-p2) was then carried out to determine whether the extension of the peptide sequence by two residues at the C terminus to include a methionine residue occupying a potential fifth position in the hydrophobic repeat improved its helical properties. This second peptide was also modified by acetylation and amidation at the N and C termini, respectively, to mimic the peptide bonds that exist in the intact natural protein. Since EBNA 3C-p2 was completely insoluble in water or HEPES buffer at pH 7, CD experiments were again carried out in formate buffer at pH 3.7. The CD spectra obtained under these conditions with EBNA 3C-p2 clearly indicated the presence of a large proportion of α-helical structure (Fig. 2B). EBNA 3C-p2 showed an increase in CD signal at 222 nm with increasing peptide concentrations, reaching a maximum of −23,300° cm−2 dmol−1 at 145 μM (Fig. 2D). We therefore estimated that EBNA 3C-p2 is 67% helical at this concentration. In contrast to EBNA 3C-p1, EBNA 3C-p2 was able to reach its maximum helical potential at lower peptide concentrations, suggesting that it is able to form more stably folded structures (Fig. 2D). The steady increase in CD signal that we observed with increasing EBNA 3C-p1 and EBNA 3C-p2 concentrations is consistent with the existence of an equilibrium between unfolded monomers and helical oligomers, suggesting that both of these peptides are able to self-associate.
EBNA 3C leucine zipper peptides form oligomers in vitro.
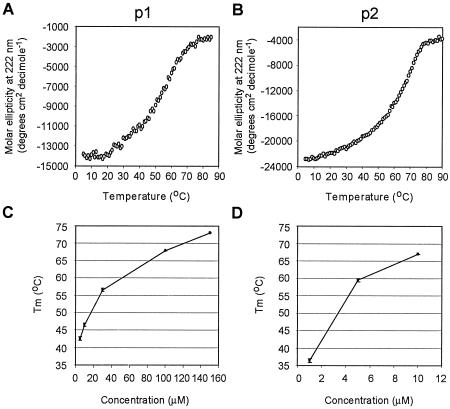
To probe the self-association of EBNA 3C-p1 and EBNA 3C-p2 further, we monitored the thermal denaturation of these peptides by CD spectroscopy. At a range of peptide concentrations, we observed sigmoidal melting curves typical of cooperatively folded protein structures such as coiled coils (Fig. 3A and B). In addition, we observed a concentration-dependent increase in the midpoint temperature of unfolding (Tm) of these structures indicating that these α-helices were forming multimeric structures (Fig. 3C and D); multimeric structures show increasing Tm with increasing concentration as a result of a shift in equilibria from unfolded monomers to folded multimers, whereas the Tm of nonoligomerizing systems does not change with concentration. Again it was apparent from these experiments that EBNA 3C-p2 was able to form more stably folded α-helical structures than EBNA 3C-p1 because the Tm values of EBNA 3C-p2 were higher than those obtained for EBNA 3C-p1 at similar concentrations (Fig. 3C and D). In fact, EBNA 3C-p2 was so stable that we were unable to obtain complete melting curves at peptide concentrations higher than 10 μM.
FIG. 3.
Thermal unfolding experiments on EBNA 3C-p1 and EBNA 3C-p2. (A) Melting curve of 30 μM EBNA 3C-p1. (B) Melting curve of 10 μM EBNA 3C-p2. (C) Graph showing the increase in the midpoint temperature of unfolding (Tm) of EBNA 3C-p1 with increasing peptide concentration. The results show the means and ranges of duplicate experiments. (D) Graph showing the increase in the midpoint temperature of unfolding (Tm) of EBNA 3C-p2 with increasing peptide concentration.
Sedimentation equilibrium experiments were then conducted in an attempt to determine the oligomerization state of EBNA 3C-p1 and EBNA 3C-p2 (Fig. 4). Data sets from experiments carried out with EBNA 3C-p1 were first analyzed by assuming a single species and gave a relative molecular size of 4,510 Da (with 95% confidence limits of 4,261 and 4,751). Within experimental error, this is consistent with the predominant species in solution being a monomer (molecular size of 4,405 Da). However, at high rotor speeds, we observed differences between the calculated fit assuming the existence of only monomers in solution (Fig. 4, lower panel, solid line) and the experimental sedimentation curves (Fig. 4, lower panel, crosses). These so-called residuals did not distribute evenly on either side of a straight line, as expected for a good fit, but showed a systematic error indicating deviation from the single-species model (Fig. 4, upper panel). Fitting the same data to a monomer-dimer association model gave a slightly improved fit (data not shown) and a dissociation constant of 2.18 mM (95% confidence limits, 2.65 and 1.33 mM), raising the possibility that a small proportion of EBNA 3C-p1 may be able to form very weak dimers.
FIG. 4.
Sedimentation equilibrium data for EBNA 3C-p1. The bottom trace shows the sedimentation curve recorded at a rotor speed of 60,000 rpm. Crosses mark the experimental data points, and the solid line shows the calculated fit generated assuming that a single ideal species is present in solution. The upper panel shows the residual signal (the difference between the calculated curve and the experimental data).
During these experiments, however, we also observed the presence of a large species that appeared to sediment at low rotor speeds. Since our CD studies indicated that EBNA 3C-p2 was able to form more stable α-helical structures than EBNA 3C-p1, we performed further sedimentation equilibrium experiments with EBNA 3C-p2. In these experiments, we were unable to detect the sedimentation of any EBNA 3C-p2 dimers but detected the presence of a much larger species that sedimented with at approximately 80 kDa (data not shown). Together with the CD data, the area-under-the-curve data suggest that EBNA 3C-p2 is able to form α-helical oligomers that may represent the self-association of possibly 18 or more molecules of peptide. The formation of high-molecular-weight oligomers by the EBNA 3C zipper peptides is consistent with their high midpoint temperatures of unfolding (Fig. 3C and D).
The leucine zipper of EBNA 3C cannot substitute for the BZLF-1 coiled-coil domain.
Since our biophysical experiments raised the possibility that at least one of our zipper peptides (EBNA 3C-p1) was capable of forming very weak dimers in solution, we investigated whether the EBNA 3C zipper region was able to mediate homodimerization in domain-swapping experiments. The putative leucine zipper region of EBNA 3C was cloned in place of the dimerization domain of another EBV protein, BZLF-1 (Zta), which contains a well-characterized bZIP domain (9, 13). Hybrid proteins containing the EBNA 3C zipper region but retaining the DNA-binding domain of BZLF-1 were created (Fig. 5A). BZLF-1/3C‘d’ contained the leucines of the 3C zipper in the d positions of the BZLF-1 heptad. To explore the possibility that the leucine residues of the EBNA 3C zipper occupied the other hydrophobic position in the heptad (the a position), we also created BZLF-1/3C‘a’. The leucine zipper of the yeast transactivator GCN4 was cloned in place of the BZLF-1 zipper region (BZLF-1/GCN4) to create a control hybrid protein for use in these experiments (Fig. 5A).
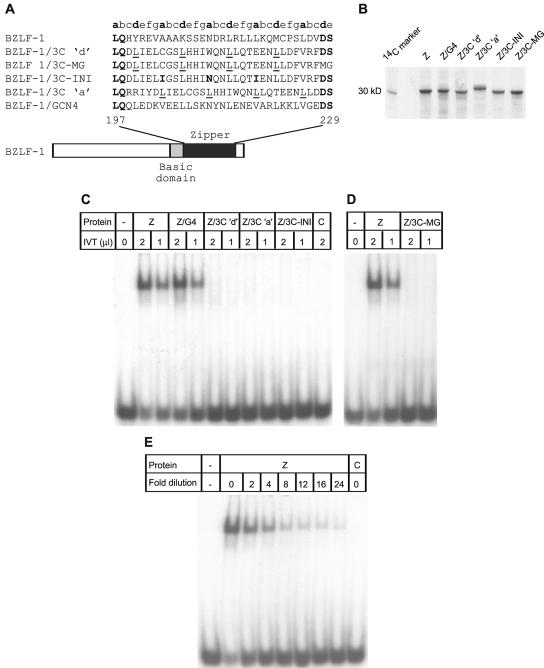
FIG. 5.
Domain-swapping experiments. (A) Diagram showing the hybrid proteins used. (B) SDS-PAGE of the [35S]methionine-labeled proteins used in EMSAs. Equal amounts of each protein were loaded on the gel. BZLF-1/3C ‘a’ consistently ran more slowly in SDS-PAGE, presumably as a result of differences in its amino acid composition. Intensity differences reflect the fact that wild-type BZLF-1 and BZLF-1/3C-MG contain three methionine residues and all other proteins contain two methionine residues. (C) EMSA with in vitro-translated (IVT) wild-type BZLF-1 (Z) and hybrid proteins. Control lanes (C) contained no RNA in vitro-translated controls. (D) EMSA with in vitro-translated BZLF-1/3C-MG. (E) EMSA with a series of dilutions of BZLF-1 protein. Lanes 1 and 9 contain 2 μl of undiluted BZLF-1 and 2 μl of the no-RNA in vitro-translated control, respectively.
Since BZLF-1 only binds DNA as a dimer, we were able to assess the dimerization potential of the EBNA 3C zipper domain by measuring the ability of hybrid proteins to bind to a BZLF-1 consensus binding site (AP-1) in EMSAs. Hybrid proteins were in vitro translated in the presence of [35S]methionine, and equal amounts of each protein were used for EMSAs (Fig. 5B). The results obtained from EMSAs demonstrated that neither the BZLF-1/3C‘d’ nor the BZLF-1/3C‘a’ hybrid protein was able to bind to a labeled AP-1 site DNA probe (Fig. 5B). In contrast, the BZLF-1/GCN4 hybrid protein bound to DNA as efficiently as wild-type BZLF-1 (Fig. 5B). On examination of the EBNA 3C leucine zipper sequence, we noted that three of the four residues occupying the a positions in the EBNA 3C heptad repeat were not normally found in leucine zipper sequences (47) (Fig. 1B) (see Discussion).
To investigate the possibility that these atypical residues were preventing the EBNA 3C zipper domain from functioning as an efficient dimerization interface, cysteine 267, tryptophan 274, and glutamic acid 281 were replaced with isoleucine (I), asparagine (N), and isoleucine (I), respectively, to create BZLF-1/3C-INI. This combination of residues at the a positions has been found to be optimal for dimer formation and is similar to that found in GCN4 (5, 7, 17, 47). This hybrid protein also failed to bind to DNA in EMSAs (Fig. 5C). We also extended the zipper fragment of EBNA 3C in the hybrid protein to include an additional methionine residue (occupying a potential fifth position in the repeat) and an additional glycine residue in the EBNA 3C-p2 peptide (BZLF-1/3C-MG). This hybrid protein was again unable to bind to DNA (Fig. 5D). To confirm that our assays were sensitive enough to detect even low levels of DNA binding activity by our hybrid proteins, we performed EMSAs with a series of dilutions of the wild-type BZLF-1 protein (Fig. 5E). We were able to detect binding of BZLF-1 to DNA clearly even when the protein was diluted 24-fold (Fig. 5E).
The results from these domain-swapping experiments demonstrate that the EBNA 3C leucine zipper is unable to function as a classical leucine zipper motif and cannot functionally replace the zipper domain of BZLF-1 to direct the homodimerization required for DNA binding. These results are consistent with our biophysical data, which indicated that the EBNA 3C zipper domain was incapable of efficient homodimerization.
EBNA 3C leucine zipper directs the formation of high-molecular-weight complexes.
Since our biophysical experiments indicated that peptides encompassing the zipper region of EBNA 3C were capable of self-associating to form high-molecular-weight complexes, we examined the association state of our BZLF-1/EBNA 3C leucine zipper hybrid proteins. Although our EMSA results showed that the EBNA 3C zipper domain was not capable of mediating homodimerization (Fig. 5), it was possible that this domain was able to direct the formation of higher-order protein complexes that would be incapable of binding to the bipartite BZLF-1 DNA recognition sequence. We therefore carried out native gel electrophoresis on in vitro-translated BZLF-1 and BZLF-1/3C zipper hybrid proteins (Fig. 6). We found that hybrid BZLF-1 proteins containing the BZLF-1 leucine zipper region migrated as high-molecular-weight species that ran very close to the top of the gel (Fig. 6 and data not shown). In contrast, the wild-type BZLF-1 protein ran much further into the gel, consistent with the existence of smaller dimeric BZLF-1 complexes. When samples were incubated with 0.5% SDS prior to loading on the native gel, BZLF-1 resolved as a lower-molecular-weight form consistent with its dissociation into a monomeric form (Fig. 6). Treatment of the BZLF-1/3C zipper hybrid protein with 0.5% SDS caused some dissociation of the high-molecular-weight complexes but did not completely dissociate these complexes into monomers, suggesting that the complexes formed are very stable (Fig. 6). The BZLF-1/GCN4 control hybrid protein behaved in the same manner as BZLF-1 (data not shown). These results are consistent with the data that we obtained from our biophysical studies and provide further evidence that the EBNA 3C zipper region alone is capable of directing the formation of high-molecular-weight complexes.
FIG. 6.
Native gel electrophoresis. [35S]methionine-labeled in vitro-translated proteins were incubated in gel shift binding buffer in the absence or presence of 0.5% SDS prior to being loaded on a native polyacrylamide gel. The positions of the likely dimeric (D) and monomeric (M) forms of BZLF-1 (Z) are indicated. The high-molecular-weight complexes formed by BZLF-1/3C‘d’ (H) are also indicated on the gel.
Leucine-to-proline substitutions in the zipper region reduce the ability of EBNA 3C to inhibit EBNA 2 activation.
To determine whether the helical nature of the leucine zipper domain of EBNA 3C was important for the transcriptional function of EBNA 3C, we performed directed mutagenesis to replace two or more of the leucine residues in the zipper with proline residues (Fig. 7A). We found that substitution of the first two (P1+2), the last two (P3+4), or all four (P1-4) of the leucines in the EBNA 3C zipper sequence reduced the ability of EBNA 3C to inhibit the activation of a reporter construct containing the viral C promoter by EBNA 2 (Fig. 7B). EBNA 2 was able to activate the C promoter by 7.3-fold in the absence of EBNA 3C, but in the presence of 2 μg of wild-type EBNA 3C-expressing plasmid, this transactivation was reduced to 1.8-fold (Fig. 7B). In contrast, in the presence of 2 μg of the P1+2-, P3+4-, or P1-4-expressing plasmid, EBNA 2 was still able to activate the C promoter by 4.9-, 3.6-, and 4.2-fold, respectively (Fig. 7B). This represents a reduction in repression activity of 56, 32, and 43%, respectively. All zipper mutants were expressed at levels equivalent to that of the wild-type EBNA 3C protein (Fig. 7B). These results therefore show that disruption of the helical nature of the zipper region reduces the ability of EBNA 3C to repress EBNA 2 activation of the C promoter, possibly by reducing the efficiency of its interaction with RBP-Jκ. It is therefore possible that the self-association of EBNA 3C via the zipper domain is important in enabling it to bind and sequester RBP-Jκ efficiently. It is also possible that the helical nature of the EBNA 3C zipper domain is important in maintaining the N-terminal structure of EBNA 3C so that the RBP-Jκ binding site is accessible.
FIG. 7.
Analysis of the effects of zipper and basic region mutations on the ability of EBNA 3C to repress EBNA 2 activation of the C promoter in vivo. (A) Amino acid sequence of the EBNA 3C bZIP region, showing the locations of the mutations introduced. (B) Effect of leucine-to-proline substitutions in the zipper region on the repression activity of EBNA 3C. DG75 cells were transiently transfected with the indicated amounts of plasmids pSG52A and pSG5EBNA3C in addition to 2 μg of C promoter reporter (pCp1425-GL2) and 2 μg of the control plasmid pRL-CMV. The Renilla luciferase activity (pRL-CMV) was used to correct for transfection efficiency. Results are expressed as transactivation by EBNA 2 relative to the level of transcription obtained in the absence of EBNA 2 and EBNA 3C. The bar charts show the mean ± standard deviation of three independent experiments. Samples of cell lysates (adjusted for transfection efficiency) were separated by SDS-PAGE and Western blotted, and EBNA 3C expression was detected with the EBNA 3C monoclonal antibody E3CA10. (C) Effect of basic domain mutations on the repression activity of EBNA 3C. (D) Effect of mutating the RBP-Jκ binding site (TFGC to AAAA at positions 209 to 212) on the repression activity of EBNA 3C.
The charge on the basic region of EBNA 3C is required for the inhibition of EBNA 2 activation.
Since the basic residues found N-terminal to the zipper domain of EBNA 3C are well conserved between viral isolates (see Discussion) even though the DNA-binding function of this region appears to be redundant, we next investigated whether the charge on the basic region of EBNA 3C was required for the function of EBNA 3C as an inhibitor of EBNA 2 activation. Site-directed mutagenesis was used to replace a cluster of arginine residues located next to the zipper domain (arginines 254, 255, 257, and 258) with either uncharged alanine residues (bR-A) or negatively charged glutamic acid residues (bR-E) (Fig. 7A). We found that substitution of these arginine residues dramatically reduced the ability of EBNA 3C to inhibit the activation of the viral C promoter (Cp) by EBNA 2 in transient-transfection assays (Fig. 7C). Although 2 μg of wild-type EBNA 3C-expressing plasmid was able to reduce C promoter transactivation by EBNA 2 from 8.2-fold to 1.6-fold, in the presence of the same amount of bR-A or bR-E EBNA 3C-expressing plasmids, EBNA 2 was still able to activate the C promoter efficiently (7- and 6.6-fold, respectively) (Fig. 7C). We found that substitution of further positively charged residues within the basic domain (arginines 247 and 250 and lysine 252) with alanine (bA) or glutamic acid (bE) impaired the repression function of EBNA 3C even further (Fig. 7C). In the presence of 2 μg of plasmid expressing the bA or bE EBNA 3C mutant, EBNA 2 was still able to activate the C promoter to maximum levels (8.1- and 9.2-fold, respectively) (Fig. 7C). Western blotting analysis of transfected-cell lysates demonstrated that all mutants were expressed at levels equivalent to that of the wild-type EBNA 3C protein (Fig. 7C).
The ability of EBNA 3C to inhibit the activation of transcription by EBNA 2 is dependent on the interaction of EBNA 3C with EBNA 2's cellular DNA-targeting partner, RBP-Jκ. The interaction of EBNA 3C with RBP-Jκ seems to prevent RBP-Jκ from interacting with EBNA 2 or with DNA (16, 30, 41). It is striking that the mutation of seven basic residues in the basic domain of EBNA 3C (bA and bE) abolished the ability of EBNA 3C to inhibit the activation of transcription by EBNA 2 to the same extent as mutations in the previously mapped RBP-Jκ binding motif at amino acids 209 to 212 (50) (Fig. 7D). Our results therefore show that residues in the basic domain of EBNA 3C are required for the interaction of EBNA 3C with RBP-Jκ and identify a new role for this region of EBNA 3C.
We can therefore conclude that residues outside of the region previously shown to be required for the interaction with RBP-Jκ (50) are important for the function of EBNA 3C as a repressor of EBNA 2 activation.
Mutations in the EBNA 3C bZIP domain do not affect the nuclear localization of EBNA 3C.
In order to rule out the possibility that the mutations that we had introduced in the bZIP region of EBNA 3C were affecting the transcriptional activity of the protein by preventing its correct nuclear localization, we examined the localization of our EBNA 3C mutants within cells (Fig. 8). DG75 cells were transiently transfected with EBNA 3C-expressing plasmids (Fig. 7A), and slides prepared from these cells were then stained with EBNA 3C-specific human serum followed by fluorescein isothiocyanate-conjugated secondary antibodies (Fig. 8, left panel). Cells were also stained with DAPI to visualize cell nuclei (Fig. 8, right panel). All of the EBNA 3C proteins used in these studies were able to localize to the nucleus and were not present in the cytoplasm (Fig. 8 and data not shown). Not all cells stained positive for EBNA 3C, since the efficiency of transfection was between 1.5 and 8%. These observations are consistent with a recent report demonstrating that the nuclear localization signals for EBNA 3C lie between residues 72 and 80, 412 and 418, and 939 and 945 and not within the bZIP region (14).
FIG. 8.
Immunofluorescence staining for EBNA 3C. Cell smears of DG75 cells transiently transfected with constructs expressing wild-type (WT) and mutant EBNA 3C proteins (see Fig. 7A) were fixed and then stained with human serum monospecific for EBNA 3C followed by fluorescein isothiocyanate-conjugated rabbit anti-human immunoglobulin G (left panel). The same cells were also stained with DAPI to visualize cell nuclei (right panel).
Mutation of the basic domain of EBNA 3C interferes with RBP-Jκ binding.
To confirm that the reduction in EBNA 3C repression activity that we observed in our EBNA 2 activation assays resulted from the decreased association of our mutated EBNA 3C proteins with RBP-Jκ, we performed coimmunoprecipitation experiments. Transient transfections were carried out as above, and the amount of EBNA 3C that associated with RBP-Jκ was determined (Fig. 9). We found that substitution of all arginine and lysine residues in the basic domain with glutamic acid residues (bE) reduced the ability of EBNA 3C to associate with RBP-Jκ to an even greater extent than mutation of the core RBP-Jκ binding motif (mJκ) (Fig. 9B). Mutation of all four leucine residues in the zipper domain to prolines (P1-4) also reduced the amount of EBNA 3C found associated with RBP-Jκ to approximately 50% of that observed for the wild-type protein (Fig. 9B). These results are completely consistent with the results that we obtained in the C promoter transactivation assays (Fig. 7) and confirm that the reduced ability of these mutant proteins to inhibit the activation of the C promoter by EBNA 2 stems from a decrease in their association with RBP-Jκ.
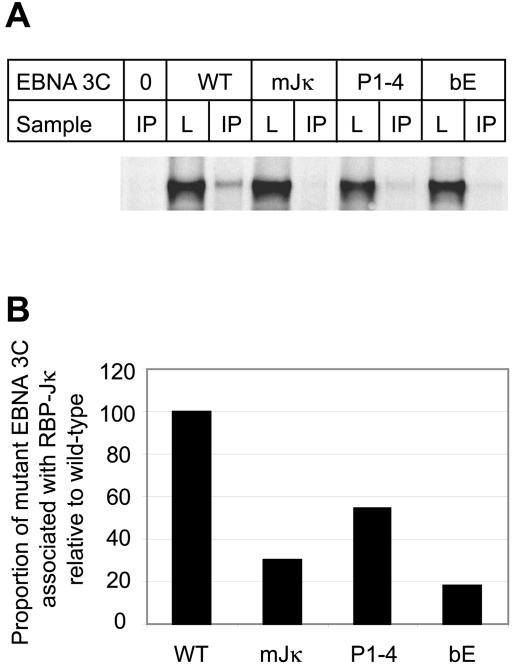
FIG. 9.
Effect of mutations in the bZIP domain on the ability of EBNA 3C to bind to RBP-Jκ. Cells were transiently transfected as in Fig. 7 with 0 or 10 μg of pSG52A and 0 or 2 μg of each of the pSG5 EBNA3C-expressing plasmids. Cells were then lysed, and RBP-Jκ was immunoprecipitated with an RBP-Jκ rabbit polyclonal antibody. Following pulldown of the immunoprecipitated complexes with protein A-Sepharose beads, the released proteins were analyzed by SDS-PAGE followed by Western blotting with the EBNA 3C monoclonal antibody E3cA10 (19). A sample of each lysate (L) representing approximately 3% of the total was run alongside each immunoprecipitation (IP). (B) Quantitation of the gel shown in panel A. The proportion of EBNA 3C associating with RBP-Jκ was expressed as a percentage of the amount of EBNA 3C present in the lysate, and the percent association of mutant proteins was then expressed relative to the percent association detected for wild-type EBNA 3C.
DISCUSSION
Although it is has become widely accepted in the field that the N terminus of the essential EBV protein EBNA 3C contains a bZIP (basic zipper) domain, no study to date has investigated whether this domain actually displays the structural and functional properties of a genuine α-helical leucine zipper dimerization motif. The presence of a bZIP domain in EBNA 3C is somewhat perplexing because no sequence-specific DNA-binding activity has ever been attributed to EBNA 3C, although this protein is capable of binding to DNA-cellulose columns in a nonspecific manner (32). It is noteworthy that the EBNA 3C basic region shows very little homology to the basic domains found in other bZIP proteins (Fig. 1), and it is possible that any DNA-binding function of this domain of EBNA 3C has been lost during the course of evolution. Nonetheless, the zipper domain of EBNA 3C does contain leucine residues at every seventh position, in common with other leucine zipper motifs (Fig. 1). Using biophysical and domain-swapping techniques, we set out to determine whether the EBNA 3C zipper domain functions as a bona fide α-helical dimerization motif.
The efficient folding of a zipper region into an α-helix is a prerequisite for the formation of the coiled-coil dimer represented by leucine zippers, and circular dichroism studies have shown that most leucine zipper peptides are 70 to 100% helical in the 30 to 50 μM range at pH 7 (24, 25). Biophysical analysis of the EBNA 3C leucine zipper domain proved troublesome because peptides encompassing this region were either sparingly soluble or insoluble at neutral pH. These are unusual characteristics for leucine zipper peptides; zipper peptides are normally fully soluble in water or standard buffers at pH 7. Nonetheless, by solubilizing our peptides at low pH, we were able to obtain consistent CD data to show that peptides encompassing the EBNA 3C zipper domain (EBNA 3C-p1 and EBNA 3C-p2) were approximately 50 to 70% α-helical in solution at pH 3.7.
EBNA 3C-p2, which was two amino acids longer than EBNA 3C-p1 and was “capped” to mimic the peptide bonds that exist in the intact protein, exhibited the most helical content at low concentrations and was approximately 60% helical at 28 μM. These data raised the possibility that the EBNA 3C leucine zipper could represent an, albeit atypical, α-helical dimerization domain. However, although both EBNA 3C-p1 and EBNA 3C-p2 exhibited a concentration-dependent increase in their midpoint temperatures of unfolding, indicative of self-association, sedimentation equilibrium experiments were only able to detect the formation of some very weak dimers by EBNA 3C-p1. Since the estimated dissociation constant of these dimers was approximately 2 mM, it is unlikely that they would be formed in vivo.
Consistent with our biophysical data, we found that the leucine zipper domain of EBNA 3C was unable to functionally replace the zipper domain of BZLF-1 to direct efficient homodimerization and DNA binding in domain-swapping experiments. However, in sedimentation equilibrium experiments carried out on both zipper peptides, we observed sedimentation of a high-molecular-weight species. For EBNA 3C-p2, we estimated that this high-molecular-weight complex could correspond to the self-association of approximately 18 or more molecules of peptide. Moreover, using native gel electrophoresis, we observed that BZLF-1 proteins containing the EBNA 3C leucine zipper no longer migrated as dimers but were visible as high-molecular-weight complexes on the gel. From our biophysical and domain-swapping data, we can therefore conclude that the EBNA 3C leucine zipper domain is not capable of forming stable homodimers but may instead direct the formation of large multimers.
Although we have clearly shown that the EBNA 3C leucine zipper is not capable of efficient homodimerization, our experiments cannot rule out the possibility that this domain is able to heterodimerize with other leucine zipper-containing proteins. However, our data do suggest that this is unlikely to be the case. The archetypal heterodimeric leucine zipper from c-Fos preferentially forms dimers with c-Jun but is still able to form homodimers with an estimated dissociation constant of 6 μM at concentrations of 40 μM and above (25). Since the only dimers that we could detect in our biophysical experiments had an estimated dissociation constant of around 2 mM, it is very likely that the EBNA 3C leucine zipper is unable to form either homo- or heterodimers efficiently.
The lack of dimerizing activity exhibited by the EBNA 3C zipper domain can be explained by a further examination of the EBNA 3C zipper sequence (Fig. 1B). We note that although this region of the protein contains a run of four leucine residues spaced seven amino acids apart, followed by a methionine residue in a possible fifth position, it does not contain the correct complement of residues that normally make up the full heptad repeat sequence (abcdefg) found in other leucine zippers (Fig. 1B). In fact, the leucine zipper domain of EBNA 3C is not predicted to form a coiled coil at all by a computer prediction program (maximum coils score of 0.001). Three of the four residues occupying the a positions in the EBNA 3C zipper (cysteine, tryptophan, and glutamic acid) are not usually found in this position in dimeric structures because they do not have small hydrophobic side chains (Fig. 1B) (47).
In addition, in stable leucine zippers, the e and g positions in the heptad are frequently occupied by charged amino acids. Electrostatic interactions between oppositely charged amino acids in the g position of the heptad in one helix and the e position of the following heptad in the partnering helix act to stabilize dimerization. In most human bZIP proteins (76%), the g and e positions are occupied by either glutamic acid, arginine, lysine, or glutamine (39). By contrast, in EBNA 3C, these heptad positions are frequently occupied by uncharged hydrophobic amino acids (leucine and isoleucine). The presence of unfavorable amino acids in three of the four a positions in the EBNA 3C zipper sequence and the lack of stabilizing residues in the e and g positions is likely to explain why the EBNA 3C leucine zipper does not form stable dimers. It is interesting that even the substitution of the atypical a position residues with amino acids that have been shown to be optimal for dimer formation (isoleucine, asparagine, and isoleucine) (5, 7, 17, 47) did not improve the dimerization potential of the EBNA 3C zipper region in domain-swapping experiments (Fig. 5). It is therefore likely that further changes in the EBNA 3C sequence, possibly the introduction of the correctly charged amino acids at the e and g positions, would be required to make this domain capable of homodimerization. It appears that the atypical residues in the EBNA 3C zipper sequence are tolerated in the higher-order EBNA 3C zipper multimers that we detected in our studies.
Using in vivo assays to measure the transcriptional repression activity of EBNA 3C, we also identified an important new role for the bZIP domain of EBNA 3C in maintaining the interaction between EBNA 3C and RBP-Jκ. Disruption of the helical nature of the zipper region by the introduction of proline residues at the d positions reduced the transcriptional repression function of EBNA 3C by 50%, and substitution of a series of positively charged amino acids in the putative basic domain virtually abolished this function of the protein. The interaction of EBNA 3C with RBP-Jκ is required to inhibit the activation of the C promoter and the LMP 1, 2A, and 2B promoters by EBNA 2 (30, 41). The regulation of EBNA 2 transcriptional activity by EBNA 3C in this way is likely to be important in preventing the toxic overexpression of LMP 1, (42-44) and in maintaining the correct level of EBNA transcription from the C promoter.
The core consensus sequence required for RBP-Jκ binding has been mapped to amino acids 209 to 212 of the EBNA 3C protein, a region that lies outside the bZIP motif (50). It is clear that although mutation of these four amino acids in EBNA 3C blocks RBP-Jκ binding and prevents the inhibition of EBNA 2 activation (50), our new data show that the bZIP domain also plays a role in maintaining the efficiency of the EBNA 3C-RBP-Jκ interaction. It is possible that the self-association of EBNA 3C via the zipper domain is important in enabling EBNA 3C to bind and sequester RBP-Jκ efficiently. Alternatively, the helical nature of the zipper domain could be important in maintaining the N-terminal structure of EBNA 3C so that the RBP-Jκ binding site is accessible. Our results also indicate that the basic residues adjacent to the zipper, although apparently nonfunctional as a site-specific DNA-binding domain, are required to maintain the transcriptional repression activity of EBNA 3C.
The important roles played by the basic residues and the leucine residues in the function of EBNA 3C as a negative regulator of EBNA 2 transcription are supported by the conservation of these residues between viral isolates and to some extent across species. Sequence data from five representative type 1 strains (3; R. Tierney, personal communication) show that the entire zipper region, including all four leucine residues, is conserved in all strains (Fig. 10). In addition, although amino acid changes do occur in the basic region, these are conservative substitutions that maintain the basic charge in this region (arginine 259 to lysine in NW and LY, and arginines 255 and 259 to lysine in SC). The DH strain of EBV has additional substitutions in the basic region (tyrosines 257 and 261 are replaced by phenylalanine), although these again represent conservative changes. These changes are also found in the prototype type 2 EBV strain, AG876 (Fig. 10). Type 2 strains of EBV contain only three of the four leucines in the zipper region, the third leucine being replaced by another hydrophobic amino acid (methionine).
FIG. 10.
Comparison of the leucine zipper sequences from five representative type 1 EBV strains (RT, SC, NW, LY, and DH) and one type 2 strain (IM 12) with the prototype type 1 and type 2 strains B95-8 and AG876, respectively. The sequences of the rhesus lymphocryptovirus (RhLCV) and the baboon lymphocryptovirus (BaLCV) EBNA 3C homologues are also shown. Amino acid differences from the B95-8 sequence are shown in bold, and the conserved leucine residues are indicated by shaded boxes.
Recent sequencing data obtained from the rhesus and baboon lymphocryptoviruses also identified a partially conserved leucine zipper motif in the EBNA 3C homologues expressed by these viruses (10, 49). However, although the same three leucine residues conserved in type 2 viruses are also present in the rhesus and baboon lymphocryptovirus sequences, the third leucine is replaced by a nonconservative arginine residue (Fig. 9). The basic regions of the EBNA 3C homologues of these viruses are also conserved to some extent (Fig. 10). The EBNA 3C protein from rhesus lymphocryptovirus retains five of the seven basic residues and contains an additional arginine residue, and the basic region of the baboon lymphocryptovirus retains four of the seven basic amino acids. The homology in the bZIP region and the RBP-Jκ binding motif appears to be sufficient to allow both of these EBNA 3C homologues to interact efficiently with RBP-Jκ in vitro and to inhibit the activation of transcription by EBNA 2 (10, 49).
In summary, we have shown that the putative leucine zipper of EBNA 3C is not capable of forming stable homodimers and cannot substitute for a genuine homodimerizing zipper domain. In fact, our data indicate that the leucine zipper domain of EBNA 3C is able to assemble into higher-order oligomers that may represent the self-association of possibly 18 or more zipper domains. In addition, we have shown that disruption of the zipper helix interferes with the ability of EBNA 3C to inhibit EBNA 2 transactivation of the viral C promoter and that the charge on the basic domain of EBNA 3C is also required for the repression function of EBNA 3C. It is likely that the conservation of the basic charge on this region and the conservation of the leucine/hydrophobic repeat in different viral isolates reflects the importance of the bZIP region for efficient RBP-Jκ binding and the importance of regulation of the transcriptional activity of EBNA 2 by EBNA 3C in the viral life cycle.
Acknowledgments
This work was supported by the Wellcome Trust through a Research Career Development Fellowship (064014) awarded to M.J.W. D.N.W. acknowledges equipment grants from the BBSRC and the Wellcome Trust.
We thank Fiona Nitsche for generating the pGL2-Cp-1425 reporter construct used in these studies and Rosemary Tierney for providing us with additional EBNA 3C leucine zipper sequence data (CRUK Institute for Cancer Studies, Birmingham, United Kingdom). We also acknowledge Matt Hicks (A.J.S.'s laboratory) for generating the pRSETA BZLF-1 plasmid and Anna Kapferer (undergraduate project student in M.J.W.'s laboratory) for generating the pSP64 BZLF-1/GCN 4 and pSP64 BZLF-1/3C‘d’ constructs. Thank you to members of the D.N.W. laboratory for help with circular dichroism spectroscopy and to Mark Coldwell for help with fluorescence microscopy. We also thank Martin Rowe (Section of Infection and Immunity, University of Wales College of Medicine, Cardiff, United Kingdom) for the gift of the DG75 cell line, the EBNA 3C antibody (E3CA10) and the M.S. serum, Tony Kouzarides (Wellcome/CRC Institute, Cambridge, United Kingdom) for the gift of the pSP64GCN4 plasmid, and Elliott Kieff (Brigham and Women's Hospital and Harvard Medical School, Boston, Mass.) for providing the RBP-Jκ rabbit polyclonal antibody.
REFERENCES
- 1.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks, J. M., D. S. Croom-Carter, A. M. Leese, R. J. Tierney, G. Habeshaw, and A. B. Rickinson. 2000. Cytotoxic T-lymphocyte responses to a polymorphic Epstein-Barr virus epitope identify healthy carriers with coresident viral strains. J. Virol. 74:1801-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez, L., Jr., D. N. Woolfson, and T. Alber. 1996. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat. Struct. Biol. 3:1011-1018. [DOI] [PubMed] [Google Scholar]
- 6.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 8.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 9.Hicks, M. R., S. Balesaria, C. Medina-Palazon, M. J. Pandya, D. N. Woolfson, and A. J. Sinclair. 2001. Biophysical analysis of natural variants of the multimerization region of Epstein-Barr virus lytic-switch protein BZLF1. J. Virol. 75:5381-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, H., Y. G. Cho, and F. Wang. 2000. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J. Virol. 74:5921-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J.κ and PU. 1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight, J. S., K. Lan, C. Subramanian, and E. S. Robertson. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 14.Krauer, K., M. Buck, J. Flanagan, D. Belzer, and T. Sculley. 2004. Identification of the nuclear localization signals within the Epstein-Barr virus EBNA-6 protein. J. Gen. Virol. 85:165-172. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Roux, A., B. Kerdiles, D. Walls, J. F. Dedieu, and M. Perricaudet. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596-602. [DOI] [PubMed] [Google Scholar]
- 17.Lumb, K. J., and P. S. Kim. 1995. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry 34:8642-8648. [DOI] [PubMed] [Google Scholar]
- 18.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maunders, M. J., L. Petti, and M. Rowe. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J. Gen. Virol. 75:769-778. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, S. M., G. W. Wilkinson, E. Floettmann, N. Blake, and A. B. Rickinson. 1996. A recombinant adenovirus expressing an Epstein-Barr virus (EBV) target antigen can selectively reactivate rare components of EBV cytotoxic T-lymphocyte memory in vitro. J. Virol. 70:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhle-Goll, C., M. Nilges, and A. Pastore. 1995. The leucine zippers of the HLH-LZ proteins Max and c-Myc preferentially form heterodimers. Biochemistry 34:13554-13564. [DOI] [PubMed] [Google Scholar]
- 22.Nonoyama, M., and J. S. Pagano. 1972. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat. New Biol. 238:169-171. [DOI] [PubMed] [Google Scholar]
- 23.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 24.O'Shea, E. K., R. Rutkowski, and P. S. Kim. 1989. Evidence that the leucine zipper is a coiled coil. Science 243:538-542. [DOI] [PubMed] [Google Scholar]
- 25.O'Shea, E. K., R. Rutkowski, W. F. Stafford 3rd, and P. S. Kim. 1989. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 245:646-648. [DOI] [PubMed] [Google Scholar]
- 26.Petti, L., J. Sample, F. Wang, and E. Kieff. 1988. A fifth Epstein-Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J. Virol. 62:1330-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 30.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sample, C., and B. Parker. 1994. Biochemical characterization of Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology 205:534-539. [DOI] [PubMed] [Google Scholar]
- 33.Sjoblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76:2679-2692. [DOI] [PubMed] [Google Scholar]
- 34.Skare, J., J. Farley, J. L. Strominger, K. O. Fresen, M. S. Cho, and H. zur Hausen. 1985. Transformation by Epstein-Barr virus requires DNA sequences in the region of BamHI fragments Y and H. J. Virol. 55:286-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touitou, R., M. Hickabottom, G. Parker, T. Crook, and M. J. Allday. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75:7749-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsang, S. F., F. Wang, K. M. Izumi, and E. Kieff. 1991. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J. Virol. 65:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinson, C., M. Myakishev, A. Acharya, A. A. Mir, J. R. Moll, and M. Bonovich. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 22:6321-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human J. kappa recombination signal sequence binding protein (RBP-J. kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 43.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West, M. J., and J. Karn. 1999. Stimulation of Tat-associated kinase-independent transcriptional elongation from the human immunodeficiency virus type-1 long terminal repeat by a cellular enhancer. EMBO J. 18:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West, M. J., A. D. Lowe, and J. Karn. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 75:8524-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woolfson, D. N., and T. Alber. 1995. Predicting oligomerization states of coiled coils. Protein Sci. 4:1596-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, B., R. Dalbies-Tran, H. Jiang, I. K. Ruf, J. T. Sample, F. Wang, and C. E. Sample. 2003. Transcriptional regulatory properties of Epstein-Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J. Virol. 77:5639-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, N. E., C. M. Kay, and R. S. Hodges. 1994. The role of interhelical ionic interactions in controlling protein folding and stability. De novo designed synthetic two-stranded alpha-helical coiled-coils. J. Mol. Biol. 237:500-512. [DOI] [PubMed] [Google Scholar]