Abstract
DNA vaccines have been successful in eliciting potent immune responses in mice. Their efficiency, however, is restricted in larger animals. One reason for the limited performance of the DNA vaccines is the lack of molecular strategies to enhance immune responses. Additionally, genes directly cloned from pathogenic organisms may not be efficiently translated in a heterologous host expression system as a consequence of codon bias. To evaluate the influence of codon optimization on the immune response, we elected to use the Tat antigens of human immunodeficiency virus type 1 (HIV-1) (subtype C) and HIV-2, as these viral antigens are poorly immunogenic in natural infection and in experimental immunization and they are functionally important in viral infectivity and pathogenesis. Substituting codons that are optimally used in the mammalian system, we synthetically assembled Tat genes and compared them with the wild-type counterparts in two different mouse strains. Codon-optimized Tat genes induced qualitatively and quantitatively superior immune responses as measured in a T-cell proliferation assay, enzyme-linked immunospot assay, and chromium release assay. Importantly, while the wild-type genes promoted a mixed Th1-Th2-type cytokine profile, the codon-optimized genes induced a predominantly Th1 profile. Using a pepscan strategy, we mapped an immunodominant T-helper epitope to the core and basic domains of HIV-1 Tat. We also identified cross-clade immune responses between HIV-1 subtype B and C Tat proteins mapped to this T-helper epitope. Developing molecular strategies to optimize the immunogenicity of DNA vaccines is critical for inducing strong immune responses, especially to antigens like Tat. Our identification of a highly conserved T-helper epitope in the first exon of HIV-1 Tat of subtype C and the demonstration of a cross-clade immune response between subtypes B and C are important for a more rational design of an HIV vaccine.
DNA vaccine technology has emerged as a novel mode of vaccination where a naked DNA construct, encoding one or more foreign proteins or epitopes, is used for immunization. When injected into a host, the DNA vector elicits a cellular or humoral immune response or both against the encoded antigen. Nucleic acid immunization offers several technical advantages over other formats of vaccination at the level of immunological outcome (25, 40). When administered intramuscularly, DNA vaccines elicit a predominantly T-helper cell Th1-type immune response, which is believed to be critical for conferring protection against several pathogens, especially viruses. Application of DNA vaccines, however, is limited, as they are usually unsuccessful in inducing strong immune responses in larger animals (60, 97). Various molecular approaches have been explored to elicit potent immune responses through genetic immunization. These approaches include coadministration of cytokines, such as interleukin-2 (IL-2), IL-15, gamma interferon (IFN-γ), RANTES, and IL-18 (8, 49, 103, 104); coexpression of costimulatory molecules such as CD40L, CD86, and CTLA-4 (44, 48, 93); engineering CpG motifs into the plasmid vectors (51, 52); expression of antigens as fusion proteins with molecular adjuvants, such as ubiquitin (34, 79), heat shock proteins (19), l-selectine (29), Flt3 ligand (84), and C3d (39, 80); adaptation of the prime-boost immunization strategies involving other vaccine formats in combination with DNA (41, 57); and many others (21, 85). Codon optimization of the antigen-encoding gene is a powerful strategy to maximize protein expression in a heterologous expression system that consequently leads to enhanced immune response (20, 94, 107).
Selective use of specific codons for protein translation is a characteristic feature of several species, a phenomenon called codon bias (87). Direct cloning of pathogen-derived genes into expression cassettes often leads to suboptimal expression of the wild-type genes in a heterologous system and may fail to stimulate strong immune responses. In a natural infection, codon bias of the wild-type genes may help reduce the magnitude of the immune surveillance due to suboptimal antigen expression in a host system, thus circumventing the induction of strong immune responses against the pathogenic organism. Immunization strategies using genetic vaccines, therefore, must replace these suboptimal codons with those more frequently used in the host system to elicit strong immune responses (20, 23, 91, 107).
Immunization with codon-optimized env (6) and gag (27, 107) genes of human immunodeficiency virus type 1 (HIV-1) led to enhanced expression of the genes and improved immune responses against the antigens. Similar studies conducted with a variety of other pathogenic organisms, such as Listeria (65), bacteria producing tetanus toxin (91), Plasmodium (65), human papillomavirus (20, 59), and others (40), ascertained the potential of codon optimization to enhance the efficiency of the DNA vaccines. The foreign genes or epitopes used in several of these studies were inherently immunodominant, thus possibly underestimating the outcome of codon optimization on the immune responses generated.
In an attempt to evaluate the influence of codon optimization on the immune response, we sought to use an inherently nonimmunodominant antigen in our studies. We opted for the transactivator protein (Tat) of HIV, as this viral antigen offers several technical advantages. Most important, the Tat proteins of HIV-1 and HIV-2 are small molecules consisting of 101 and 130 amino acids, respectively. The first exon of HIV-1 Tat (Tat-1), consisting of 72 amino acids, is functionally competent for viral transactivation and a wide variety of other functions ascribed to this protein (98). The small size of the Tat protein offers a practical advantage of uncomplicated chemical synthesis of the expression cassette essential for codon optimization. Tat is expressed early in the viral life cycle and is functionally important for its infectivity and pathogenicity (47). In addition to regulating viral gene expression, Tat modulates expression of various genes of the host. Additionally, Tat is secreted extracellularly, and the extracellular Tat governs viral latency and contributes to disease progression (66). Despite the functional importance of Tat for the virus, this antigen is not immunodominant either in natural infection (53, 56, 58, 78, 101) or in experimental immunization, especially when delivered as a genetic vaccine (8, 15). Inducing cellular and humoral immune responses against Tat is critical because of its early expression in the viral life cycle and its extracellular secretion (37, 82). At the genetic level, the Tat gene is conserved to a greater extent than that of the env gene, thus permitting induction of cross-clade immune responses (54). Last, as a consequence of its pleiotropic biological functions, a variety of functional assays are available for Tat to study the inhibitory effect of immune components on its biological functions.
The overall genetic content of HIV is AT-rich, suggesting suboptimal codon usage of its genes in the human host. The AT content of Tat-1 is approximately 54.3%, while that of HIV-2 Tat (Tat-2) is 51.8%. This is in contrast to the relatively low AT content (30 to 40%) in the coding regions of the host human genome (86). To enhance the translational efficacy and immunogenicity of HIV-1 and HIV-2 Tat proteins, we synthetically assembled the gene expression cassettes. We employed an amplification-based approach to generate codon-optimized Tat constructs. While designing the expression cassettes, codons frequently used in the mammalian system were substituted for the original AT-rich codons. The synthetic Tat constructs were evaluated in the mouse model for the generation of immune responses.
MATERIALS AND METHODS
Assembly of codon-optimized Tat-1 and Tat-2 genes.
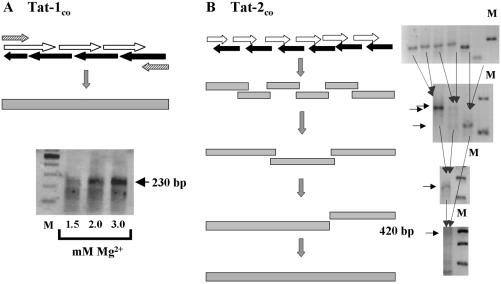
The synthetic and codon-optimized HIV-1 Tat (Tat-1co) gene, corresponding to the first exon of the consensus HIV-1 subtype C sequence, was assembled in a single-step PCR using overlapping oligonucleotides (Fig. 1A). The oligonucleotides (Table 1) were designed to replace the AT-rich codons of the wild-type gene with the synonymous GC-rich codons that corresponded to the most frequently used mammalian (human) codons. A total of 29 of the 72 codons were replaced in the Tat-1 gene. The oligonucleotides spanning the length of exon 1 of the Tat-1 gene were mostly 45 nucleotides long with a 25-bp overlap with the corresponding reverse oligonucleotide (Fig. 1A, schematic). The amplification reaction mixture of 100 μl contained 25 nM concentrations of each oligonucleotide and 250 nM concentrations of each primer (forward N298 [5′-CGG TAA GGT ACC GCC GCC GCC ATG GAG CCA GTA GAT CCT AAC CT] and reverse N180 [5′-TTT TCT AGA CTA TTG CTT TGA TAT AAG ATT TTG ATG]). The reaction mixture also contained 100 μM (each) deoxynucleoside triphosphates (dNTPs), 1.25 U of Taq DNA polymerase, and 3.0 mM Mg2+. Amplification was performed using the following cycling conditions: (i) 2 min at 94°C; (ii) 2 cycles, with 1 cycle consisting of 30 s at 42°C and 30 s at 72°C, followed by 2 min at 94°C; (iii) 2 cycles, with 1 cycle consisting of 30 s at 50°C and 30 s at 72°C, followed by 30 s at 94°C; (iv) 35 cycles, with 1 cycle consisting of 30 s at 56°C and 30 s at 72°C. The amplified product was purified using a column commercial kit (Qiagen), cloned directionally between KpnI and XbaI into pDV2, a mammalian expression vector containing a cytomegalovirus (CMV) promoter.
FIG. 1.
Assembly of synthetic Tat-1 and Tat-2 genes. (A) Assembly of Tat-1co. Exon 1 of HIV-1 C-Tat was assembled in a single-step PCR using different concentrations of Mg2+ as shown. The amplicon of 3 mM Mg2+ reaction was cloned into pDV2 vector under the control of a CMV promoter. The white and solid black horizontal arrows represent the sense and antisense oligonucleotides that constitute exon 1, respectively. The hatched arrows represent primers for amplification. A stop codon was introduced in the reverse primer. (B) Assembly of Tat-2co. HIV-2 Tat was synthesized using a sequential assembly approach. The oligonucleotides used in each PCR overlap with each other by 12 bp. Horizontal arrows mark the amplicons of desired length, and vertical arrows indicate the template combinations for subsequent PCRs. Lanes M, 100-bp DNA molecular size standards (MBI Fermentas).
TABLE 1.
Sequences of oligonucleotides used to assemble exon 1 of C-Tat-1
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| Sense | |
| N223 | CATGGAGCCAGTAGATCCTAACCTGGAGCCCTGGAACCACCCTGGC |
| N224 | AGCCAGCCCAAGACCGCCTGCAACAACTGCTACTGCAAGCACTGC |
| N225 | AGCTACCACTGCCTGGTGTGCTTCCAGACCAAGGGCCTGGGCATC |
| N226 | AGCTACGGCCGGAAGAAGCGGCGCCAGCGCCGGAGCGCTCCTCCA |
| N227 | AGCAGCGAGGACCACCAAAATCTTATATCAAAGCAGTAAT |
| Antisensea | |
| N228 | CTAGATTACTGCTTTGATATAAGAT |
| N229 | TTTGGTGGTCCTCGCTGCTTGGAGGAGCGCTCCGGCGCTGGC |
| N230 | GCCGCTTCTTCCGGCCGTAGCTGATGCCCAGGCCCTTGGTCTGGA |
| N231 | AGCACACCAGGCAGTGGTAGCTGCAGTGCTTGCAGTAGCAGTTGTT |
| N232 | GCAGGCGGTCTTGGGCTGGCTGCCAGGGTGGTTCCAGGGCT |
| N233 | CCAGGTTAGGATCTACTGGCTCCATGGTAC |
The antisense oligonucleotides are presented as the sequences complementary to the sense oligonucleotides.
Synthetic and codon-optimized HIV-2 Tat (Tat-2co) corresponding to the full-length sequence of an Indian HIV-2 isolate was assembled using six pairs of codon-optimized oligonucleotides (Table 2). To assemble the Tat-2 gene, we used a sequential amplification strategy. The complementary oligonucleotides of each pair overlapped with each other by 12 bp. Six individual amplification reactions were performed to amplify as many regions of Tat-2 (Fig. 1B). The adjoining amplification products also overlapped with each other by 12 bp. The six first-round PCR products served as templates for three second-round amplifications. Each of the second-round amplification mixtures contained two adjacent first-round amplicons and the two terminal oligonucleotides for primers. One-microliter portions from each of the first-round reaction mixtures were used as templates for the second-round reactions. Further rounds of amplification were performed until the full-length Tat-2 product was obtained (Fig. 1B). A total of 81 of the 130 codons of the wild-type Tat-2 gene (Tat-2wt) were substituted in the Tat-2co gene. A proofreading polymerase, Elongase (Gibco), was used to ensure the fidelity of amplification. The reaction conditions were as follows: preamplification denaturation (30 s at 94°C), followed by 40 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 20°C, and 30 s at 68°C. A 50-μl reaction mixture contained 1 μl of Elongase enzyme mixture (Gibco), 25 nM concentration of each oligonucleotide, 100 μM dNTPs, and 1.8 mM Mg2+. The resulting amplicons from each step served as templates for the subsequent reactions. The final 420-bp amplicon was cloned directionally between KpnI and XbaI sites on pDV2. The authenticity of the assembled expression cassettes was confirmed by restriction digestion analysis and sequencing.
TABLE 2.
Sequences of oligonucleotides used to assemble Tat-2
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| Sense | |
| N267 | AGATCTGGTACCATGGAGACCCCCCTGAAGGCCCCCGAGAGCAGCCTGGA |
| N269 | CCAGCGAGCAGGACGTCGCCACCCAGGGCAGCGCTAGCCAGGGCGAGGAG |
| N270 | GAGGCCTGCGCCTGCGCCAGCACCTGCTACTGCAAGCTGTGCTGCTACCA |
| N271 | GCCTGGGCATCTGCTACGAGCGGAAGGGCCGGCGGCGGCGGACCCCCAAG |
| N272 | GTCCGACAAGAGCATCAGCACCCGGACCTGGAACAGCCAGCCCGAGAAGG |
| N273 | GGAGGCCACCGTGGAGACCGACACCGGCCCCGGCCG |
| Antisensea | |
| N275 | CCTGCTCGCTGGTGTGGCTGAAGGGCTCGTTGCAGCTCTCCAGGCTGCTC |
| N276 | GGCGCAGGCCTCCAGGGGCCGGTACAGCTGGCTCAGGATCTCCTCGCCCT |
| N277 | AGATGCCCAGGCCCTTCTGCAGGAAGCAGAGCTGGCAGTGGTAGCAGCAC |
| N278 | CTCTTGTCGGACGTCAACGTTGGGTGGGTCTTGGTCTTCTTGGGGGTCCG |
| N279 | ACGGTGGCCTCCACGGGCTTCTTCTGCTCCTTCTCGGGCT |
| N280 | GGATCCTCTAGATCACCGGCCGGGGCCG |
The antisense oligonucleotides are presented as the sequences complementary to the sense oligonucleotides.
Histidine (His)-tagged subtype C Tat-1 (C-Tat-1) was amplified from a primary Indian HIV isolate and cloned into the bacterial expression vector pET28A. Glutathione S-transferase (GST)-tagged Tat-2 (GST-Tat-2) expression vector was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The wild-type HIV-2 gene was a gift from Robin Mukhopadhyaya.
SEAP assay.
293 cells were cotransfected with HIV secreted alkaline phosphatase (SEAP) (a gift from Bryan Cullen) and HIV-1 Tat expression vectors by the CaCl2 method (18). The culture supernatant (200 μl) was sampled at regular intervals and stored at −20°C until use. The reporter SEAP activity was measured using 4-nitrophenyl phosphate as the substrate (24). In all transfections, we used CMV-β-galactosidase reporter vector as an internal standard for normalization. β-Galactosidase activity in cell extracts was measured by a colorimetric assay (90).
Western blot analysis.
293 cells were transiently transfected with Tat expression vectors, and the cells were harvested 48 h after transfection. The cells were lysed in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer, cell extract was resolved on an SDS-12% polyacrylamide gel, and the protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Immun-Blot PVDF; Bio-Rad) using a semidry transfer apparatus (Trans-Blot SD; Bio-Rad) at 25 V for 90 min. The cell extract was independently probed for β-actin (Oncogene), following the manufacturer's instructions, to serve as a loading control. HIV-1 Tat was detected using a monoclonal antibody (2D1.1; catalog no. 4138, National Institutes of Health AIDS Research and Reference Reagent Program) raised against the N-terminal 15 amino acid residues of the antigen (28). The membranes were incubated overnight at 4°C in 5 ml of monoclonal antibody diluted 10,000 times in 0.5% skim milk powder solution in phosphate-buffered saline (PBS). HIV-2 Tat was detected using a mouse antiserum raised against the full-length protein in our laboratory. After the membranes were washed three times with Tris-buffered saline (pH 8.0), they were incubated with 5 ml of anti-mouse horseradish peroxidase conjugate (BD Pharmingen) (diluted 1:1,000) for 1 h at room temperature. The blots were developed by chemiluminescence reaction using a commercial kit (ECL+plus; Amersham).
HLM1 transfection and p24 ELISA.
HLM1 cells, which contain a Tat-defective provirus, were transfected with HIV-1 Tat constructs using a cationic lipid (Geneporter; Gene Therapy Systems). The culture supernatant was sampled at regular intervals and stored at −20°C until use. Empigen was added to each sample to a final concentration of 1%, and the samples were incubated at 56°C for 30 min to inactivate the virus and to release the p24 antigen from the viral particles. The quantity of p24 in the samples was measured using an in-house capture enzyme-linked immunosorbent assay (ELISA). Briefly, 0.5 μg of capture antibody (ADP 410; National Institute for Biological Standards and Control) in 100 μl of carbonate buffer was added to each well of a 96-well plate (Greiner), and the plates were incubated overnight at room temperature. The plates were blocked with 5% skim milk powder in PBS for 1 h at room temperature. Solutions containing the antigen were added to each well (100 μl/well), and the plates were incubated for 4 h at 37°C. A biotinylated monoclonal antibody (ARP 454; National Institute for Biological Standards and Control) specific for p24 was diluted to a final concentration of 1 μg/ml using a buffer containing Tris (10 mM), Empigen (0.1%), Tween 20 (0.05%), and sheep serum (5%). Wells containing 100 μl of the detection antibody were incubated at 37°C for 1 h. A streptavidin peroxidase conjugate (Sigma) was added to each well at a dilution of 1:1,000 and incubated for 1 h. One hundred microliters of the substrate solution containing o-phenylenediamine was added to each well, plates were incubated for 15 min at room temperature, and 50 μl of 1 N HCl was added to each well to stop the enzyme reaction. The optical density was measured at 490 nm using an ELISA reader (Molecular Dynamics).
Semiquantitative RT-PCR.
Reverse transcriptase PCR (RT-PCR) was performed using random hexamers for reverse transcription and specific primers for DNA PCR. Total RNA was isolated using TRIzol reagent (Gibco) from 293 cells transiently transfected with the Tat expression vectors. For amplification, total RNA was used as a template at three different concentrations, 1.0, 0.33, and 0.11 μg per reaction mixture. Reverse transcription was performed using 1 μM concentrations of random hexamers, 1 mM dNTPs, 3 mM Mg2+, Moloney murine leukemia virus RT (Promega), and 100 U of RNase inhibitor (Promega) in a volume of 20 μl. The RT reaction conditions were 35 min at 42°C and 5 min at 94°C. Five-microliter portions of the RT reaction contents were transferred to the DNA PCR mixture of 50 μl. DNA PCR was performed with the primer pair N177 and N180 using 1.25 U of Taq DNA polymerase by hot start PCR. The PCR mixture contained 100 μM dNTPs, 250 nM concentrations of primers, and 1.8 mM Mg2+. The following reaction conditions were used for amplification: (i) 1 min at 94°C; (ii) 35 cycles, with 1 cycle consisting of 30 s at 56°C and 1 min at 72°C. β-Actin transcript amplification was used as a positive control employing the primers 139F (5′-GTG GGG CGC CCC AGG CAC CA) and 139R (5′-CTC CTT AAT GTC ACG CAC GAT TTC).
DNA immunization.
The plasmid DNA intended for immunization was prepared using Qiagen endotoxin-free Giga kits and resuspended in endotoxin-free PBS (Manukirti Biogems, Bangalore, India; <0.06 endotoxin unit). The endotoxin concentration was quantitated in a standard Limulus amebocyte lysate assay (QCL-1000; Biowhittaker) and found to be within recommended limits (<0.1 endotoxin unit/μg of DNA). One hundred micrograms of the DNA was injected into the tibialis anterior muscle of mice that were 8 to 12 weeks old. Each immunization consisted of four or five mice per group. The immunization schedule involved one primary immunization, followed by one or three booster immunizations spaced 2 weeks apart. Animals were housed and maintained in a facility adhering to the recommendations of the Committee for the Purpose of Control and Supervision of Experiments on Animals in India.
Lymphoproliferation assay.
Splenocytes (5 × 106) from immunized mice were incubated with 5 μg of recombinantly expressed antigen per ml for 5 days. Tat-1 was purified using the His tag by Ni-nitrilotriacetic acid (Qiagen) affinity chromatography, and Tat-2 was purified using the GST tag by glutathione-Sepharose (Amersham) affinity chromatography. The purity of the proteins was confirmed using SDS-polyacrylamide gel electrophoresis. Control proteins containing identical tags (His-p24 and GST-PC4) were also purified by similar means and used in the assays as controls for nonspecific proliferation. After incubation, the extent of cell proliferation was measured by adding [3H]thymidine (5 μCi/ml) (NET520A; Perkin Elmer Life Sciences) to the wells, and the cultures were incubated for 3 h at 37°C for incorporation of the label. The cells were harvested, washed, resuspended in 50 μl of PBS, and deposited onto filter paper disks (Whatman no. 3 filter paper). After thorough washing, the filters were dried, and radioactivity counts were determined using a β-scintillation counter (Wallac 1409). Concanavalin A (ConA) was used as a positive control for cell proliferation at a final concentration of 5 μg/ml.
Cytotoxicity assay.
Splenocytes harvested from immunized mice were incubated with irradiated syngeneic stimulators (P815 cells expressing Tat and EL-4 cells expressing Tat for BALB/c and C57BL/6 mice, respectively) for 5 days in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). After in vitro stimulation, the effector cells were incubated with 51Cr-labeled syngeneic target cells (104 cells/200 μl) at different effector-to-target cell ratios. Nontransfected parental cell lines (P815 or EL-4) served as controls. The effector and target cells were coincubated for 5 h at 37°C, and 100 μl of the supernatant was assayed using a gamma counter (LKB Rack gamma; Wallac). Counts from spontaneous lysis usually remained below 30% of the maximal lysis. Values for maximal lysis were obtained by incubating the labeled target cells in the presence of 2% Triton X-100. Percent specific lysis was calculated by the following formula: percent specific lysis = (experimental lysis − spontaneous lysis)/(maximal lysis − spontaneous lysis).
ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were performed for the Th1 cytokine IFN-γ (m IFNγ Eli-spot; DIACLONE Research) and the Th2 cytokine IL-4 (mouse IL-4 ELISPOT set; BD Pharmingen) before and after in vitro stimulation. Briefly, the cytokine-specific capture antibody (1 μg/100 μl of PBS) was adsorbed onto the PVDF-backed 96-well plates by incubating the antibody solution overnight at 4°C. The plates were blocked, and the primed splenocytes (0.5 × 106 cells) were added along with the stimulator cells (at a 1:3 ratio) in a final volume of 200 μl to the wells and incubated for 24 to 36 h. The cells were then lysed, cell debris was removed by extensive washing, and the wells were incubated with a biotinylated anticytokine antibody (0.5 μg/ml) for 2 h. The plates were washed extensively, and the wells were incubated with enzyme-conjugated avidin (HRP for IL-4 and alkaline phosphatase for IFN-γ) for 1 h. Spots were developed using the appropriate substrate (3-amino-9-ethylcarbazole and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate, respectively) and incubating the plates for 20 min at room temperature. ConA (5 μg/ml) was used as a positive control for IL-4 secretion, and phorbol myristate acetate (1 μg/ml) plus ionomycin (0.5 μg/ml) were used as a positive control for IFN-γ secretion. The spots were enumerated using a stereomicroscope (MZ6; Leica) at a magnification of ×16.
Tat-1 epitope mapping.
Six 20-mer peptides spanning exon 1 of HIV-1 Tat were synthesized (Peptron, Daejeon, Korea); these peptides overlapped each other by 10 amino acids. The peptides represented the consensus sequence of HIV-1 C-Tat. The peptides were used at a final concentration of 2 μg/ml. Pepscan analysis for IFN-γ ELISPOT assay was performed by incubating the peptides with 5 × 105 splenocytes from immunized BALB/c mice per well for 36 h at 37°C. The controls were normalized for dimethyl sulfoxide concentration. Eventually, additional peptides were designed for fine mapping of the identified epitope (see Fig. 7).
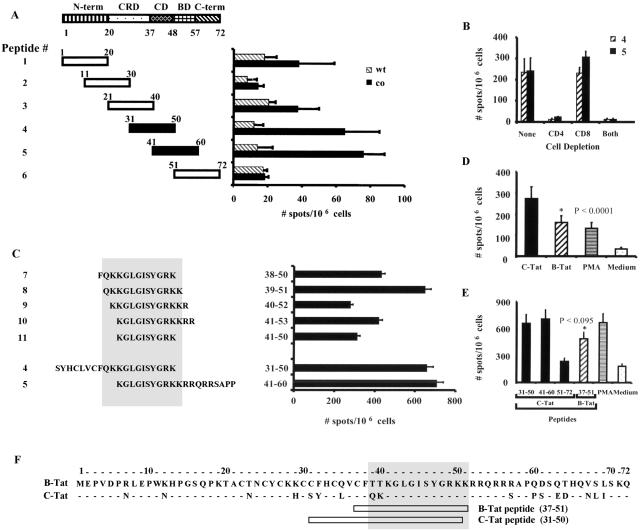
FIG. 7.
Identification and characterization of a dominant T-helper epitope in Tat-1. (A) Identification of an epitope in the core and basic region of Tat-1. The domain structure of Tat and the amino acid coordinates of the domains are illustrated in the schematic diagram at the top. N-term, N terminus; CRD, cysteine-rich domain; CD, core domain; BD, basic domain; C-term, C terminus. Six synthetic peptides, spanning the entire length of exon 1 of Tat, were used in the ELISPOT assay for IFN-γ. The 20-mer peptides (except the last one, which is a 22-mer) overlap each other by 10 residues. The Tat gene used for immunization and the sequence of the peptides used in the analysis represent the consensus sequence of C-Tat. Spleen cells (5 × 106 cells/assay) collected from BALB/c mice immunized with the wild-type or codon-optimized Tat-1 (using the one-prime-one-boost immunization regimen and 100 μg of DNA/mouse) were pulsed with the peptides (2 μg/ml) and used in the assay. Spleens from three mice per group were pooled for the analysis. The ELISPOT assay was performed as described in Materials and Methods. Each bar represents the mean of three independent wells ± 1 standard deviation (error bar). The experiment was performed two times. The two immunodominant peptides, peptides 4 and 5, are shown as black bars for easy identification. co, codon-optimized Tat; wt, wild-type Tat. (B) The core or basic region epitope is a T-helper epitope. Primed splenocytes were initially incubated with monoclonal antibodies specific to mouse CD4 (GK1.5) or CD8 (3.155) or with both antibodies. Cells were subsequently incubated with fresh rabbit serum as a source of complement, and live cells were purified on a Histopaque gradient. Complement-treated or control splenocytes were stained with fluorescein isothiocyanate-conjugated anti-mouse CD4 or phycoerythrin-conjugated anti-mouse CD8-PE (BD Pharmingen) and analyzed by cytometry to confirm cell depletion. Splenocytes (0.5 × 106/assay) depleted of CD4+ or CD8+ cells or both subsets were incubated with peptides 4 and 5, and the ELISPOT assay was performed as described in Materials and Methods. Each bar represents the mean of three independent wells ± 1 standard deviation (error bar). The experiment was performed two times. (C) Identification of the core epitope. A new set of 13-mer peptides that span the core and basic domains was used for the IFN-γ ELISPOT assay. Peptide 4 (amino acid residues 31 to 50) and peptide 5 (amino acid residues 41 to 60) were included in the same assay for comparison. Pulsing of the target cells and ELISPOT assay were performed as described above for panel A. These mice, however, received two additional booster immunizations unlike the mice in panel A. The core epitope is shaded. (D) Cross-clade immune reactivity between C-Tat and B-Tat antigens. Mice (n = 4) were immunized with C-Tat DNA, and pooled splenocytes from these mice were stimulated in vitro with P815 cells expressing C-Tat or B-Tat antigen. The ELISPOT assay for IFN-γ was performed as described above for panel A. PMA, phorbol myristate acetate (5 μg/ml). (E) Cross-clade immune response against the core or basic region T-cell epitope. Pooled splenocytes from BALB/c mice (n = 3) immunized with C-Tat DNA were pulsed with 2 μg of the peptide per ml, and the cells were used in the ELISPOT assay for IFN-γ as described above in panel A. Each peptide represents the consensus amino acid sequence of the respective subtype. PMA, phorbol myristate acetate (5 μg/ml); Medium, culture medium only. (F) The core T-helper epitope is highly conserved. A pairwise alignment of exon 1 amino acid residues of subtypes B and C. Identical amino acids are indicated by the dashes. The core T-helper epitope exemplified by peptide 8 is shaded. Boxes below the sequence alignment depict the peptides used in the experiment. Except for the two amino acid residues at the N-terminal region, the epitope is highly conserved across all the major subtypes of HIV-1.
Complement-mediated cell depletion of T-cell subsets.
Splenocytes (40 × 106 cells) from three or more immunized BALB/c mice were pooled, and the pooled cells were incubated in 0.5 ml of culture supernatant of anti-mouse CD4 (GK1.5) or CD8 (3.155) hybridoma cell line for 1 h at 4°C. Serum samples collected from 4-week-old rabbits were added to the cells as a source of complement to a final 1:5 or 1:20 dilution and incubated for 30 min at 37°C. The cells were washed with RPMI 1640 medium containing 5% FBS. Lymphocytes were passed through a Ficoll gradient (Histopaque-1083; Sigma) to isolate viable cells. The cells were collected in 15-ml tubes (Corning), washed three times with 5 ml of RPMI 1640 medium supplemented with 5% FBS, and used for IFN-γ ELISPOT assay.
Statistical analysis.
Results of lymphoproliferative assays are presented as stimulation index values as the mean ± standard deviation of at least three independent experiments, each of which was performed in triplicate. Results of ELISPOT assays are expressed as the number of spots per 106 cells, means of triplicate wells. Comparisons between the Tat constructs were made using the Student's t test. P values less than 0.05 were considered statistically significant.
RESULTS
Codon optimization increases the translation efficiency of the Tat-1 and Tat-2 genes.
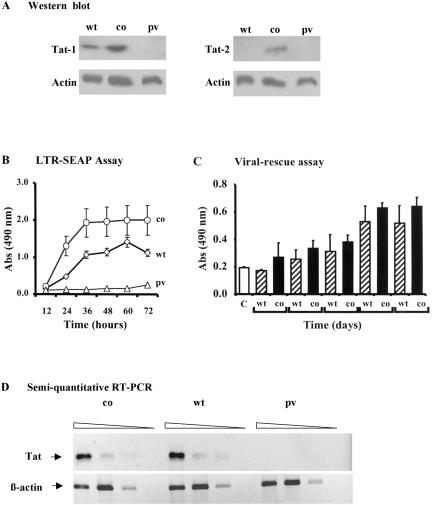
HIV-1 and HIV-2 Tat genes were synthetically assembled such that the suboptimal codons were substituted with codons frequently used in the mammalian system. As a result of codon optimization, the AT content of the Tat-1 and Tat-2 genes was reduced from 54.3 and 51.8% to 40.1 and 30.0%, respectively. The protein expression efficiencies of the synthetic genes were compared with those of the wild-type constructs in 293 cells. Western blot results demonstrated enhanced expression of synthetic genes, Tat-1co and Tat-2co, at significantly higher quantities than the wild-type genes (Tat-1wt and Tat-2wt genes, respectively) (Fig. 2A). Tat-1co expressed four times more antigen than Tat-1wt. Especially with Tat-2, the amount of antigen produced from the wild-type gene was so low that we failed to detect its presence in the Western blot. Expression of Tat-2 from this construct, however, was confirmed in a more sensitive reporter assay (results not shown).
FIG. 2.
Functional analyses of Tat expression vectors. (A) Western blot analysis for Tat-1 and Tat-2. 293 cells were transfected with 10 μg of plasmid DNA by the CaCl2 method. The cells were harvested 48 h after transfection and lysed, and the lysate was resolved on an SDS-12% polyacrylamide gel. Tat-1 was detected using a monoclonal antibody (2D1.1; AIDS Research and Reference Reagent Program). β-Actin served as a loading control and probed using a commercial kit (Oncogene). Tat-2 was detected using a mouse antiserum raised in the laboratory against GST-Tat-2 protein. co, codon-optimized Tat; wt, wild-type Tat; pv, parental vector. (B) Expression of SEAP from 293 cells transiently transfected with 1 μg of LTR-SEAP and 10 μg of Tat-1 expression vectors by the CaCl2 method. Culture supernatants were collected after different periods of time, and a colorimetric assay was used to quantitate SEAP in culture supernatants (24). CMV-β-galactosidase vector was used in all transfections to control for differences in the transfection efficiency. β-Galactosidase levels in the cell extracts were quantitated by a colorimetric assay (90). Abs, absorbance. (C) Rescue of a Tat-defective provirus. HLM-1 cells, containing a single copy of a Tat-defective provirus, were transfected with 1 μg of Tat expression vectors using a commercial lipid formulation. Spent medium was collected at different times, and p24 concentration in the samples was measured using an in-house antigen capture assay. Results are presented as means ± 1 standard deviation (error bars) of three samples, and the data are representative of two independent experiments. CMV-SEAP (0.3 μg) was cotransfected to serve as an internal control for normalization of the data. C, empty vector. (D) Semiquantitative RT-PCR analysis of Tat-1 constructs. 293 cells in 100-mm-diameter dishes were transfected with plasmid DNA (10 μg) by the CaCl2 method. Cells were harvested 48 h after transfection, and total RNA was isolated using TRIzol reagent (Gibco). RNA was diluted in serial threefold dilutions starting at 1 μg, and each dilution served as a template for Tat gene and β-actin RT-PCRs using specific primers as described in Materials and Methods. The amplicons were resolved on a 1% agarose gel.
Transient transfection of 293 cells with Tat expression vectors and HIV-1 long terminal repeat (LTR)-green fluorescent protein (GFP) reporter plasmid demonstrated that the synthetic Tat constructs were transcriptionally functional (data not shown). Transfection of HOS-CD4-LTR-GFP cells with Tat-2 expression vectors by electroporation confirmed the functional integrity of the Tat-2 constructs (data not shown). For a quantitative evaluation of Tat-1 expression, 293 cells were cotransfected with different Tat vectors and a reporter vector expressing SEAP, HIV-1 LTR-SEAP. Culture supernatants were collected at different periods, and the SEAP concentration was determined in a colorimetric assay (24). SEAP was detected in culture medium starting from 12 h, and its concentration gradually increased up to 72 h (Fig. 2B). At every point of sampling, the quantity of SEAP released into the medium was significantly higher from Tat-1co than from the wild-type vector. Reporter expression from Tat-1co also reached saturation earlier, suggesting higher levels of protein synthesis. These results are in agreement with the Western blot analysis (Fig. 2A) and illustrated the functional integrity of the synthetic Tat constructs.
Codon-optimized Tat-1 is functional in the virological context.
Reporter gene analysis is useful in evaluating Tat-mediated transactivation of the viral LTR. This analysis, however, may not directly correlate with the function of Tat in the context of the viral life cycle. We sought to examine whether codon-optimized Tat-1 could also lead to enhanced virion production. HLM-1 cells, which contain a single copy of a Tat-defective provirus (83), were used in the transfection experiments. Tat-1wt and Tat-1co were delivered to the cells using a commercial lipid formulation (Geneporter; Gene Therapy Systems), and the concentration of p24 in the medium was quantitated using an in-house sandwich ELISA. Both Tat-1wt and Tat-1co successfully complemented the defective provirus and released p24 into the medium. Although synthetic Tat-1 consistently released more p24 at every time point and earlier than the wild-type gene, the difference between the two constructs was not statistically significant (Fig. 2C). This observation indicated that an intracellular concentration of the transactivator protein above a threshold level is sufficient to drive virion production and that expression levels of Tat above this threshold need not translate to increased virion production.
Enhanced protein synthesis is not due to increased transcription.
To examine whether the increased level of protein synthesis observed from synthetic Tat genes was the outcome of gene expression regulation at the level of transcription or translation or both, we performed a semiquantitative RT-PCR. Total RNA was extracted from 293 cells transfected with the Tat-1wt or Tat-1co construct. RT-PCR was performed on serially diluted RNA samples as described in Materials and Methods. As expected, the degrees of amplification of these two Tat constructs were not significantly different (Fig. 2D). This observation, therefore, supported our premise that the increased protein synthesis from the synthetic Tat vector was most probably a result of enhanced protein translation, not more efficient transcription.
Immunological evaluation.
We evaluated the immunogenic potential of the wild-type and codon-optimized Tat genes in mice. Plasmid DNA, extracted using Qiagen columns, was used in all the immunization protocols. Mice (C57BL/6 and BALB/c mice, four or five animals per group) were immunized intramuscularly using a one-prime-one-boost or one-prime-three-boost regimen. DNA was administered at a low (10 μg/mouse) or high (100 μg/mouse) dose to study the influence of the dose on the immune response. After immunization, the animals were sacrificed, and the splenocytes were used in various immunological analyses.
Enhanced cell proliferation in mice immunized with codon-optimized Tat.
Splenocytes harvested from mice immunized with the wild-type or codon-optimized Tat expression vectors or the parental plasmid were incubated in vitro with recombinantly expressed antigen, and the extent of antigen-specific cell proliferation was measured using the [3H]thymidine incorporation assay (Fig. 3). We observed antigen-specific and dose-dependent cell proliferation in response to both Tat-1 and Tat-2 antigens in C57BL/6 (Fig. 3) and BALB/c (data not shown) mice. Cell proliferation was significantly higher in mice immunized with 100 μg of DNA than in mice immunized with 10 μg of DNA with both the wild-type and codon-optimized Tat constructs. Although cell proliferation in mice immunized with 100 μg of Tat-1co was higher than that of Tat-1wt, this difference was not statistically significant. Importantly, the effect of codon optimization of Tat was striking at the lower DNA concentration. At this dose, while splenocytes of Tat-1co immunization proliferated, splenocytes of Tat-1wt immunization failed to respond to Tat protein, indicating a lack of optimal antigen expression to prime an efficient immune response (Fig. 3A). Cell proliferation was not observed in lymphocytes harvested from control mice injected with the parental vector or when nonspecific antigens were employed in the experiment, demonstrating the specificity of the assay.
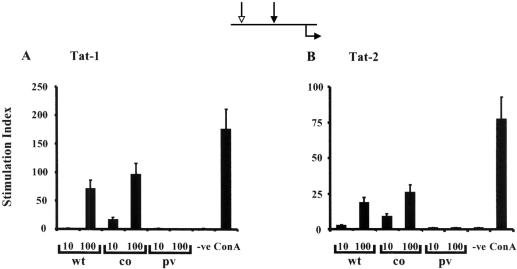
FIG. 3.
Tat-specific lymphoproliferative immune response. C57BL/6 mice (four mice per group) were genetically immunized with two different doses of Tat-1 (A) or Tat-2 (B) expression vector. The line diagram at the top of the figure depicts the immunization regimen; the white arrow represents the primary immunization, the black arrow represents the booster immunization, and the black arrow below the line represents the time of harvest. Splenocytes were harvested 1 week after the booster immunization. In a 24-well plate, 5 × 106 primed splenocytes were incubated with recombinantly expressed antigen, His-Tat-1, or GST-Tat-2 at a concentration of 5 μg/ml for 5 days. Following incubation, the cells were incubated with 5 μCi of [3H]thymidine per ml for 3 h at 37°C. A stimulation index above 3 was considered a positive response. Results are presented as the means of three samples ± 1 standard deviation (error bars), and the data are representative of two independent experiments. The mice had been immunized with 10 or 100 μg of wild-type Tat (wt), codon-optimized Tat (co), or parental vector (pv). The cells were grown in the absence (-ve) and presence of ConA as a control for cell proliferation.
Codon-optimized Tat constructs elicit stronger CTL responses.
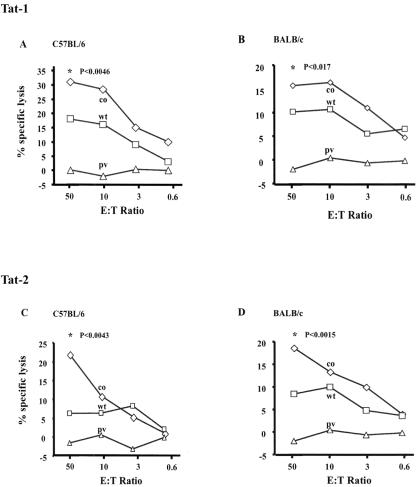
Considering the importance of Tat-specific cytotoxic T-lymphocyte (CTL) response to disease progression (1, 4, 30, 95), we sought to evaluate the magnitude of the CTL response generated in mice immunized with different Tat constructs. Conventional 51Cr release assay was used to compare the CTL responses elicited by the wild-type and codon-optimized constructs of Tat-1 and Tat-2 in C57BL/6 and BALB/c mice with or without in vitro stimulation. We observed antigen-specific and dose-dependent CTL responses in both mouse strains immunized with Tat-1 or Tat-2. Cell lysis was significantly higher at the larger dose, and only data from the 100-μg immunizations are presented (Fig. 4). DNA constructs encoding the synthetic Tat genes elicited significantly higher CTL responses than the wild-type counterparts. The difference in the magnitude of CTL responses between codon-optimized and wild-type vectors was statistically significant. No antigen-specific cell lysis was observed in mice immunized with the parental control vector.
FIG. 4.
CTL responses in C57BL/6 and BALB/c mice immunized with Tat-expressing DNA vectors. Mice (four or five mice per group) were immunized with two different quantities of DNA (10 and 100 μg/mouse); however, only data from the group immunized with 100 μg are presented. The mice were immunized with Tat-1 (A and B) or Tat-2 (C and D). The immunization schedule is depicted in the line diagram of Fig. 3. Splenocytes were stimulated in vitro with syngeneic cells stably expressing Tat-1 or Tat-2 antigen (EL-4 cells expressing Tat-1 or Tat-2 for C57BL/6 mice and P815 cells expressing Tat-1 or Tat-2 for BALB/c mice) at a 10:1 ratio for 5 days. Activated splenocytes were incubated with appropriate Tat-expressing EL-4 or P815 stable transfectants as target cells (104 labeled target cells per well) at different effector-to-target cell (E:T) ratios in a standard 5-h 51Cr release assay. Data are presented as the means of three samples, and the experiment was repeated two times. co, codon-optimized Tat; wt, wild-type Tat; pv, parental vector. An asterisk represents a statistically significant difference in the immune responses between the codon-optimized and corresponding wild-type Tat genes. The P values are shown.
Codon-optimized Tat vectors elicit optimal Th1 responses.
Immune responses skewed toward the T-helper Th1 cell type are considered to be immunoprotective in several infections, including HIV or AIDS (45, 50, 63). We sought to evaluate the nature of the cytokine profile generated, Th1 versus Th2, in mice immunized with two different doses of Tat-expressing DNA vectors. To this end, we used the ELISPOT technique to evaluate IFN-γ and IL-4 production, which are signature cytokines for Th1- and Th2-type responses, respectively. Primed splenocytes with or without in vitro stimulation were incubated with syngeneic target cells expressing Tat-1 or Tat-2 in a 96-well ELISPOT format.
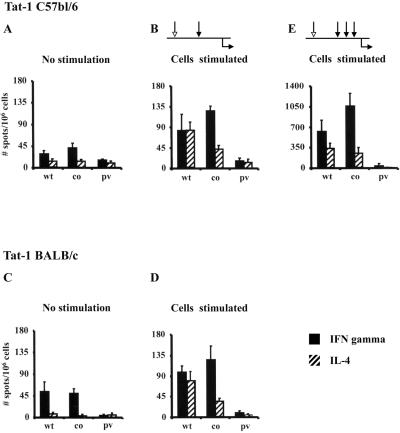
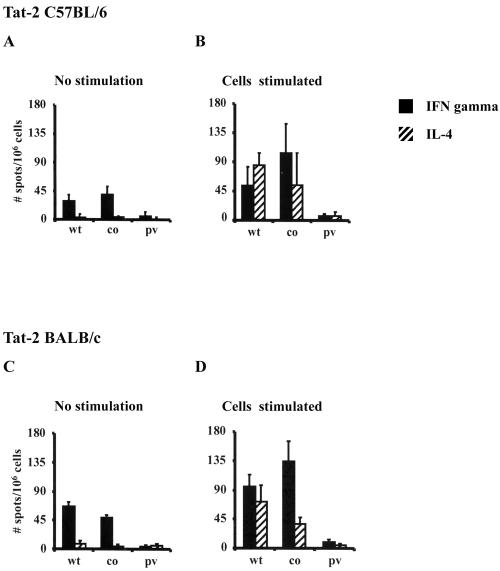
Wild-type and codon-optimized expression vectors of Tat-1 and Tat-2 induced antigen-specific immune responses in both mouse strains (C57BL/6 and BALB/c) (Fig. 5). The immune responses were proportional to the dose of DNA injected, and only the data from the 100-μg immunization are presented. Although the immune responses were detected without in vitro stimulation, antigen stimulation enhanced these responses to a greater magnitude. Of note, while IFN-γ-secreting splenocytes were identified prior to in vitro stimulation, IL-4-secreting cells were detected only after stimulation (Fig. 5B and D). Immunization of mice with wild-type Tat vectors (one-prime-one-boost regimen) demonstrated a mixed type of cytokine profile in which both IFN-γ- and IL-4-secreting cells were identified at comparable frequencies (Fig. 5B and D). Importantly, immunization with codon-optimized Tat vectors by the identical immunization scheme induced a more pronounced IFN-γ response. Although IL-4 producers were still present in these mice, there was a significant reduction in their frequency compared with that of wild-type Tat immunization (Fig. 5B and D). A switch in the cytokine profile from a mixed Th1-Th2 response to a more pronounced Th1-type was observed in both C57BL/6 (Fig. 5B) and BALB/c (Fig. 5D) mice. Identical results were observed with Tat-2 immunizations in both mouse strains (Fig. 6), suggesting functional similarity between the Tat-1 and Tat-2 antigens.
FIG. 5.
ELISPOT response to Tat-1 in C57BL/6 (A, B, and E) and BALB/c (C and D) mice. Mice (four or five mice per group) were injected with two different quantities of DNA (10 and 100 μg/mouse); however, only data from the 100-μg immunization group are presented. The immunization schedules are shown in the line diagrams above the bar graphs; the white arrow represents the primary immunization, the black arrows represent the booster immunizations, and the black arrow below the line represents the time of harvest. The assay was performed using cells without stimulation or after in vitro stimulation with syngeneic cells stably expressing the Tat-1 antigen (EL-4 cells expressing Tat-1 for C57BL/6 mice and P815 cells expressing Tat-1 for BALB/c mice). For stimulation, splenocytes were incubated with the syngeneic antigen-presenting cells at a 10:1 ratio for 5 days. A cell pool consisting of 0.5 × 106 primed splenocytes and 0.16 × 106 syngeneic stable transfectant cells was added to the ELISPOT wells, and the assay was performed as described in Materials and Methods. Each bar represents the mean for three individual wells ± 1 standard deviation (error bar), and the experiment was repeated two times. co, codon-optimized Tat; wt, wild-type Tat; pv, parental vector. (E) A switch in the Th profile due to additional booster immunizations (see the line diagram) with Tat-1wt. C57BL/6 mice (four mice per group) were administered three booster immunizations after priming. The successive injections were spaced 2 weeks apart, and all the injections contained 100 μg/dose. Note that the y axis is different from that of the other graphs due to elevated immune response induced as a result of additional booster immunizations. The experiment was performed twice.
FIG. 6.
ELISPOT response to Tat-2 in C57BL/6 (A and B) and BALB/c (C and D) mice. Mice (four or five mice per group) were injected with two different quantities of DNA (10 and 100 μg/mouse); however, only data from the 100-μg DNA immunization group are presented. The immunization schedule used was the one-prime-one-boost immunization regimen depicted in Fig. 5. The assay was performed on cells without stimulation or after in vitro stimulation with syngeneic cells (EL-4 cells expressing Tat-2 for C57BL/6 mice and P815 cells expressing Tat-2 for BALB/c mice) stably expressing Tat-2. The experimental details are provided in the legend to Fig. 5. Each bar represents the mean of three individual wells ± standard deviation (error bar), and the experiment was performed two times. co, codon-optimized Tat; wt, wild-type Tat; pv, parental vector.
Triggering a switch in the cytokine profile by the codon-optimized Tat vectors alluded to the possibility of larger quantities of protein synthesized from these vectors that could have caused the differential cytokine profile. Western blot results identified enhanced levels of protein synthesis from the codon-optimized vectors (Fig. 2). To test the hypothesis that increasing the number of booster immunizations with wild-type Tat vectors, thereby exposing the immune system to more antigen, could also bring about a similar switch in the cytokine profile, we immunized C57BL/6 mice with wild-type or codon-optimized Tat-1 expression vectors. Groups of mice received either one or three booster immunizations after the primary immunization. All the immunizations (100 μg per mouse) were spaced 2 weeks apart, and the splenocytes were harvested 1 week after the last booster immunization. The splenocytes were stimulated in vitro with syngeneic cells expressing Tat-1 (EL-4/Tat-1) and tested for antigen-specific cytokine secretion.
Several important features emerged from this analysis. The overall frequency of antigen-specific cells increased significantly with increasing number of booster immunizations (compare Fig. 5B and E). The Tat-1co construct elicited a higher immune response by both immunization schemes. A Th1-type immune response was observed with the synthetic Tat DNA by both immunization schemes. In contrast, the wild-type Tat gene elicited a mixed Th1-Th2 immune response when a single booster dose was administered. Importantly, the immune response was skewed toward a more desirable Th1-type immune response when more booster doses were administered with the wild-type Tat-1 DNA (Fig. 5E). Additional booster doses, however, could have exposed the mice not only to larger quantities of synthesized Tat antigen but also to higher amounts of plasmid DNA. When delivered intramuscularly, plasmid DNA could promote a Th1-type cytokine profile. Although our data would not permit us to assess the relative contribution of these two factors, we believe that it is the larger quantities of the Tat antigen, not DNA, that tilted the cytokine profile from a mixed Th1-Th2 immune response to a predominantly Th1-type immune response. We suggest this on the basis of the observation that a single booster immunization with the codon-optimized Tat-1 vectors induced the optimal cytokine response.
Codon-optimized Tat vectors fail to elicit humoral immune responses.
Eliciting a humoral immune response against Tat may be important to attenuate various pathogenic properties ascribed to the extracellular form of this viral factor (35, 37). DNA vaccines are intrinsically deficient in eliciting humoral immune responses to the encoded antigen, despite stimulating strong cellular immune responses (15, 43). Since Tat DNA vectors elicited potent cell-mediated immune responses in our experiments, we sought to evaluate whether the immunizations also stimulated antibodies to Tat. Sera were collected from mice (C57BL/6 and BALB/c) immunized with Tat-1 DNA (100 μg/immunization/mouse) using the one-prime-three-boost regimen. Anti-Tat antibodies were quantified in an indirect ELISA. Both the Tat constructs elicited a low-level humoral immune response (data not shown). The response elicited by the synthetic construct was marginally higher than that of the wild-type DNA. This difference, however, was not statistically significant. Unlike others who reported antibody response to a Tat DNA vaccine in mice (17), we did not use bupivacain in our immunizations.
Immune responses are focused on the core region of Tat-1.
Our studies with Tat-expressing DNA vectors generated potent cellular immune responses in BALB/c and C57BL/6 mice. Most of the previous studies of Tat protein immunization identified the N-terminal domain of Tat as the immunodominant region (67). In contrast, DNA immunizations elicited a broader immune response targeting several domains of Tat (42). We sought to identify and characterize the domains that are immunodominant in C-Tat. This analysis was necessary considering the number of natural variations identified in C-Tat (73b). We chemically synthesized six 20-mer peptides spanning the entire length of exon 1 of Tat-1. The peptides overlapped with each other by 10 amino acid residues. A consensus amino acid sequence of C-Tat protein, the same sequence as that of the DNA vaccine, was used to design the peptides. Splenocytes from BALB/c mice that were primed with wild-type or codon-optimized Tat-1 vectors using a one-prime-one-boost immunization regimen were analyzed for cytokine production against individual peptides in an IFN-γ ELISPOT assay.
Results from these experiments demonstrated that the magnitude of the immune response in mice immunized with the codon-optimized vectors was higher than the response in mice immunized with wild-type Tat-1. Immunizations with the wild-type vector appeared to have recognized several domains of Tat uniformly, although at a lower magnitude. Immunization with the codon-optimized Tat, on the other hand, predominantly recognized peptides 4 (residues 31 to 50) and 5 (residues 41 to 60). The overlapping 10-amino-acid stretch between these two peptides, encompassing the immunodominant epitope (Fig. 7A), corresponds to the core region of Tat-1. This result was significant, as the core region represents a motif conserved across the viral subtypes. In addition to the core region, immune responses of significant magnitude were also directed against two other peptides, peptides 1 and 3, representing the amino-terminal and cysteine-rich domains of Tat, respectively. This pattern of immune response, broader spreading of the epitopes, is a characteristic feature of genetic immunization (17, 42).
CD4+ T cells mediate immune responses against the core region of Tat.
Since both CD4+ and CD8+ T cells could secrete IFN-γ following antigenic stimulation, we wanted to know which of the two subsets was the primary source of IFN-γ secretion. To answer this question, we used the strategy of complement-mediated cell depletion to remove the CD4+ or CD8+ subset or both subsets from spleen cells and performed the cytokine analysis. Immunodominant peptides 4 (residues 31 to 50) and 5 (residues 41 to 60) were used in this experiment. Monoclonal antibodies specific to mouse CD4 or CD8, in combination with rabbit complement, were used for cell depletion, and cell depletion was confirmed by flow cytometry analysis (data not presented). CD4+ cell depletion of the splenocytes completely eliminated IFN-γ-secreting cells in response to both the immunodominant peptides (Fig. 7B). In contrast, CD8 cell depletion of the splenocytes did not influence the frequency of the cytokine-secreting cells. These results indicated that CD4+ T cells mediated the peptide-specific responses and that the epitope was likely to be a T-helper epitope.
To further define the epitope encompassed by peptides 4 and 5, we designed a new set of 13-mer peptides (peptides 7 through 11) centered on the core region of Tat. Pepscan analysis of these peptides using the IFN-γ ELISPOT assay identified peptide 8 (residues 39 to 51) as the one against which maximal responses were observed, indicating that the optimal epitope was characterized by this peptide (Fig. 7C). A 10-mer peptide (residues 41 to 50) encompassing the amino acids shared by peptides 4 and 5 elicited a partial immune response, indicating that the flanking residues are important for the optimal epitope identified.
C-Tat-1 elicits cross-clade reactive immune responses.
An ideal HIV vaccine must elicit cross-clade reactive immune response to confer protection against a wide range of viral subtypes and recombinants. Since C-Tat of HIV-1 shares significant homology (76% identity in exon 1) with subtype B Tat (B-Tat), we sought to examine the extent of cross-reactivity between these two antigens in an IFN-γ ELISPOT assay. BALB/c mice were genetically immunized with C-Tat-1co using the one-prime-three-boost immunization regimen. Primed splenocytes were assayed for IFN-γ secretion in response to activation by P815 cells expressing C-Tat-1 (homologous activation) or P815 cells stably expressing B-Tat (heterologous activation). The full-length Tat (86 amino acids) was cloned from HXB2 molecular clone. Under these experimental conditions, we identified significant levels of IFN-γ-secreting cells when C-Tat-primed splenocytes were activated with B-Tat, although to a lesser extent than that of the C-Tat-specific response (Fig. 7D). This observation demonstrated the considerable magnitude of cross-clade reactivity between B- and C-Tat antigens.
Given that the overall immune response to Tat in a genetic immunization is broadly distributed across multiple epitopes and that the difference in the magnitude of immunoreactivity between B-Tat-1 and C-Tat-1 antigens is statistically significant, we sought to identify the level of cross-reactivity between these two antigens in the context of the immunodominant core or basic region epitope. For this comparison, we used B- and C-Tat peptides encompassing the core sequence of the T-cell epitope (residues 37 to 51 for B-Tat and 31 to 50 for C-Tat). Each peptide represented a consensus amino acid sequence of the respective subtype. P815 cells pulsed with the C- or B-Tat peptide were used in the assay as targets for the splenocytes from mice immunized with C-Tat DNA. In this analysis, although B-Tat peptide induced fewer IFN-γ-secreting cells than the C-Tat peptide did (Fig. 7E), the difference was not statistically significant, suggesting a higher level of conservation of the epitope. A greater level of cross-reactivity between B- and C-Tat peptides, in contrast to that of antiprotein immune response, may be a reflection of the highly conserved nature of the T-helper epitope. The core epitope is conserved not only between HIV subtype B and C clades but also among other subtypes. While 17 of the 72 amino acid residues varied between the consensus C-Tat and B-Tat sequences used in this study and 5 of the 21 residues varied between these two peptides, only 2 of the 13 residues within the core epitope varied between the two peptides (Fig. 7F). These data indicate that the amino acid residues contributing to major histocompatibility complex (MHC) recognition and anchorage of the peptides are possibly conserved among the subtypes and that this functionally important region of Tat contained a highly conserved and immunologically important epitope.
DISCUSSION
Successful DNA immunization requires high-level expression of pathogen-derived genes in mammalian cells. One potential obstacle for efficient expression of a heterologous gene in the host is the interspecies difference in codon usage (87). Pathogens, especially viruses, are subjected to selection pressures to conserve such codons that are suboptimally utilized in the host. Direct cloning of viral genes into the mammalian expression vectors for the purpose of genetic immunization could limit protein synthesis in vivo and fail to stimulate strong immune responses against the encoded antigens as a result of codon bias. We attempted to address the problem of suboptimal codon usage in the context of a DNA vaccine by using the transactivator proteins of HIV-1 and HIV-2.
Codon usage of Tat is strikingly different from that of the human genes, making Tat protein suitable for evaluating the influence of codon optimization on genetic immunization. Importantly, Tat antigens usually do not induce strong immune responses in a natural infection (53, 56, 58, 78, 92, 101). Additionally, when delivered through different formats, Tat elicited a limited magnitude of immune responses in humans (15) and experimental animals (8, 73). Other factors in favor of selecting Tat for this study are the functional importance of this viral antigen to the infectivity of the virus (35, 47, 81, 82) and the existence of an inverse correlation between immune responses to Tat and disease progression (4, 75, 76, 78, 95, 106).
To increase the immunopotency of Tat, we synthetically assembled the Tat genes and optimized the codons for mammalian expression. The AT content of the Tat genes of HIV-1 and HIV-2 was significantly reduced owing to codon optimization. This also led to enhanced translational efficiency in mammalian cells without modulating the transcription rate (Fig. 2). Having characterized the Tat expression vectors at the functional and biochemical levels, we evaluated the antigenic potential of the DNA vectors in a mouse model. Two different mouse strains, two different DNA dosages, and two different immunization regimens were evaluated in these studies.
Overall, the codon-optimized Tat-1 and Tat-2 genes elicited significantly higher levels of cellular immune responses. Importantly, the codon-optimized vectors promoted a predominantly Th1-type immune response in all the immunizations. In contrast, wild-type Tat vectors preferentially promoted a mixed type of cytokine profile, in a one-prime-one-boost immunization regimen. This was unexpected, as the mode of immunization, intramuscular administration of the DNA, has been shown to skew the immune response predominantly toward a Th1-type response (33, 55, 71, 99). Additionally, Tat has been documented to function as a T-cell adjuvant with a propensity to tilt the immune response in favor of a Th1-type profile (31, 32). In contrast, experimental evidence also suggested that Tat could promote antigen-specific immunosuppression by directly modulating T-cell function (22, 96, 105) or indirectly modulating T-cell function by stimulating the expression of cytokines, such as IL-10 (9), transforming growth factor β1 (77), and tumor necrosis factor (12), or by suppressing the expression of cytokines, such as IL-12 (46).
These reports broadly suggested that Tat could select a Th2-type cytokine profile in natural infection and in vaccine immunizations. Promotion of a Th2-type cytokine profile in intramuscular inoculation of DNA has not been documented extensively. We ascribe the induction of a mixed Th1-Th2-type cytokine profile in our experiments by wild-type Tat to low-level protein expression from the wild-type expression vectors, as demonstrated by the Western blot analysis (Fig. 2), rather than to the quantity of DNA delivered. The observation that administration of additional booster immunizations (one-prime-three-boost immunization regimen) with the wild-type vectors tilts the cytokine profile in favor of the Th1-type immune response lends support to this hypothesis. On the basis of our data, it is tempting to propose that Tat preferentially promotes a Th1-type cytokine profile only above a certain threshold concentration (immunizations with codon-optimized Tat vectors) and not below this threshold (immunizations with wild-type Tat). Of note, promotion of a mixed cytokine profile by wild-type genes and reversal of the profile to a Th1 type by codon-optimized genes was consistent with two independent antigens, Tat-1 and Tat-2, validating our observation. Our results with the wild-type Tat constructs may partly explain why a mixed type of cytokine profile is predominant in the natural course of HIV-1 infection (7).
It is difficult to speculate what type of cytokine profile Tat would promote in natural infections as a viral protein secreted into the body fluids. A reliable measure of the concentrations of Tat in the body fluids of HIV-seropositive subjects is not available. Although in a minority of the subjects Tat was identified at a concentration as high as 1 to 40 ng/ml (100, 102), it is not known if a correlation exists between the Th profile observed in disease progression and Tat concentration in the body fluids.
In the present work, we employed only exon 1 of Tat-1, as this part of the viral antigen is adequate for supporting a plethora of the biological functions. Exon 2 of Tat-1 is not only considered less significant for immune response but also is genetically more heterogeneous. However, a recent study demonstrated, for the first time, that exon 2 of Tat-1 could play an important role in immune surveillance and viral escape (89). In light of this finding, it is important to evaluate the immunogenicity of the full-length Tat antigen. In our study, however, we also used expression vectors encoding the full-length Tat-2 antigen consisting of both the exons. Importantly, both Tat-1 and the full-length Tat-2 expression vectors essentially induced identical patterns of immune response.
Identification of immunodominant domains within Tat is of interest for vaccine development and for the study of host immune response. In Tat, several T-cell, B-cell, and CTL epitopes have been identified in natural infections (10, 26, 62, 67, 68, 74) and in experimental immunizations of mice (11, 17, 42, 64) and rabbits (38). While studies of the immune response to protein immunization with Tat primarily focused on the N-terminal region of Tat (17, 67), Tat genetic immunizations broadly recognized several epitopes within the amino-terminal, cysteine-rich, and core domains of the viral antigen (17, 42). Importantly, the core domain has been demonstrated to be immunodominant in several studies, and overlapping B-cell, T-cell, and CTL epitopes have been mapped to this domain in humans and experimental animals (1, 10, 42, 68, 74). Considering the small size of Tat, it is not surprising that several of the epitopes overlap with one another. Of the various domains, the core and basic domains of Tat are the regions that are highly conserved among viral clades. Most of the previous studies, however, characterized the epitope profile of Tat antigen of HIV subtype B origin. Although Tat is considered to be conserved to a greater extent, a previous analysis from our laboratory identified a minimum of six natural variations within the first exon of C-Tat (73b). One of the variations at position 39 is located in the core domain of Tat.
Given the importance of Tat for viral pathogenesis and C-Tat containing considerable number of variations compared to the well-studied B-Tat, we sought to characterize the immunodominant epitope(s) in C-Tat using the pepscan strategy for IFN-γ secretion in an ELISPOT assay. In agreement with previous reports (17, 42), we observed that in BALB/c mice, multiple regions of Tat were targeted in genetic immunization, including the amino-terminal, cysteine-rich, and the core domains. Among these domains, the core region of the Tat antigen elicited the strongest immune response and contained a T-cell epitope. Complement-mediated depletion of the CD4+ cells, but not the CD8+ cells, from splenocytes completely eliminated the IFN-γ-secreting cells, indicating that the putative T-cell epitope has a T-helper nature.
Further characterization of the epitope, using 13-mer peptides within the core and basic region identified peptide 8, spanning residues 39 to 51, as the one containing the core epitope. A comparison of immune responses to peptides 7 and 8 on one hand and peptides 8 and 9 on the other hand revealed that the optimal epitope is preferably flanked by a glutamine at positions 39 and a lysine at 51 in C-Tat. Surprisingly, although peptide 4 lacks lysine 51 and peptide 5 lacks glutamine 39 and lysine 40, they still induced immune responses at levels nearly comparable to that of the optimal peptide 8 (Fig. 7C). On the basis of this observation, we speculate that the optimal size of the epitope is longer than 13 amino acids and the flanking amino acid residues play a significant role in immune recognition.
Peptides binding MHC class II molecules are usually longer than 13 amino acids, and they are held in the groove of the MHC molecule by a series of bonds distributed along the length of the peptide backbone. While the peptide is immobilized in this position through the interactions of the anchoring residues within the core epitope, the two ends of the peptide are not bound but protrude out of the groove and are of variable length. Peptide 8 (residues 39 to 51) probably defines the core of the T-helper epitope of the Tat core domain. Of the 13 amino acids of this T-helper epitope, 10 residues at the N-terminal region are mapped to the core domain, while the remaining 3 residues map to the basic domain of Tat.
Most of the amino acid residues in the T-helper epitope that we identified here are highly conserved across the subtypes of HIV-1 with the exception of the two N-terminal residues at positions 39 and 40. Although several amino acids are found at position 39 in Tat of non-subtype C origin, nearly 89% of the strains designated as subtype C in the HIV-1 sequence database contained only a glutamine at this position. However, less than 1% of the non-subtype C strains contain a glutamine at this location, and only the subtype A strains have a glutamine here. Considering the terminal location of these two residues and their highly polymorphic nature, it is possible that these amino acids may constitute the T-cell receptor binding region and may also determine antigen specificity.
To evaluate this possibility, we compared the cross-reactive immune response between B-Tat and C-Tat antigens and the corresponding peptides. We observed considerable numbers of IFN-γ-secreting cells in response to heterologous antigen activation (C-Tat priming and B-Tat activation), either in the context of the Tat protein (Fig. 7D) or the peptides (Fig. 7E). The difference between homologous and heterologous antigen activation, however, was statistically significant only in the context of the protein, not the epitope-containing peptide. In either case, we identified considerable magnitude of cross-clade immune reactivity between B- and C-Tat antigens. It would be interesting to see whether the cross-reactive immune responses would be sufficient to confer cross-clade immune protection.
Tat-1 and Tat-2 have a considerable degree of homology (∼36% in exon 1), and it is possible that the basic domain of Tat-2 might also harbor a T-cell epitope. Since we did not have Tat-2-expressing target cells, we tested a possible cross-reactivity between Tat-1 and Tat-2 using a different experimental format. Splenocytes isolated from mice immunized with Tat-2 were used in a CTL experiment with Tat-1-expressing target cells. Under these experimental conditions, we did not observe significant levels of cell lysis, suggesting that the immunodominant regions of Tat-1 and Tat-2 may be different (73a). However, performing experiments with Tat-2 immunizations and Tat-1- or Tat-2-expressing target cells is necessary to map immunodominant regions of Tat-2 and to evaluate the extent of cross-reactivity between these antigens.
Although the application of Tat as a potential candidate vaccine for HIV has been explored extensively, the efficacy of Tat vaccine in viral challenge studies is controversial. While some studies reported complete (14, 30, 69) or partial (2, 3, 13, 70) protection, others failed to observe such protection (3, 61, 73, 88). Especially when delivered as a DNA, most of the studies used the Tat gene directly cloned from the virus, which may have limited the efficacy of the vaccines. Although some of these studies reported strong immune responses, the cytokine profile generated was not analyzed. While several DNA vaccine studies encoding other viral proteins, such as Env, Gag, Pol, and Nef, used human versions of these genes (6, 23, 27, 36, 107), only one such study reported the use of codon-optimized B-Tat (2). Despite the limited success reported by DNA vaccination approaches in general and Tat DNA vaccines in particular, the most promising approach thus far appears to be priming with DNA, followed by booster immunization with modified vaccinia virus Ankara (5). The application of the prime-boost approach, however, may be limited because of the safety concerns of the viral vectors on one hand and the preexisting immunity to viruses in the populations on the other hand. Considering the functional importance of Tat for viral pathogenicity (82, 98), the existence of an inverse correlation between disease progression and anti-Tat humoral immunity (53, 76, 78, 106) and the identification of mutations in Tat CTL epitopes in viral escape mutants (4, 16), the potential of Tat as a candidate vaccine cannot be undermined. Developing molecular strategies, such as the one reported here and other approaches (8, 21, 72), is essential for DNA vaccine design to immunopotentiate nonimmunodominant antigens like Tat. Importantly, the demonstration that a single booster immunization with the codon-optimized Tat genes could elicit a rapid Th1-type cytokine response is important for vaccine design.
In summary, we have demonstrated that codon optimization of an otherwise weak viral antigen could make it immunodominant. While the wild-type Tat-1 and Tat-2 antigens induced a mixed Th1-Th2-type immune profile, codon-optimized genes elicited a more desirable Th1 response. We also identified an immunodominant epitope in the core and basic region of Tat-1 that is highly conserved among the viral subtypes. Although subtype C strains of HIV-1 cause more than 50% of the total infections globally and more than 95% infections in India, few studies have attempted to characterize the HLA haplotype of the seropositive individuals and the cognate viral epitopes recognized by them. In the absence of such information, identification of immunodominant epitopes highly conserved among viral subtypes is useful for HIV vaccine design with broader application. Molecular approaches that immunopotentiate DNA vaccines are essential for optimal performance of this technology.
Acknowledgments
U.R. acknowledges financial support from the Department of Biotechnology of the Government of India (grant BT/MED/HIV/05/99). L.R. and K.K.A. are recipients of the C.S.I.R. fellowship from the Government of India.
We thank Karna Venkata Ramana for critically reading the manuscript and Nagendran Ramalingam for technical assistance. A number of reagents used in this study were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health of the United States and the Centralised Facility for AIDS Reagents, National Institute for Biological Standards and Control, United Nations Programme on HIV/AIDS.
REFERENCES
- 1.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, and B. D. Walker. 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agwale, S. M., M. T. Shata, M. S. Reitz, Jr., V. S. Kalyanaraman, R. C. Gallo, M. Popovic, and D. M. Hone. 2002. A Tat subunit vaccine confers protective immunity against the immune-modulating activity of the human immunodeficiency virus type-1 Tat protein in mice. Proc. Natl. Acad. Sci. USA 99:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., L. Mortara, B. R. Mothe, M. Liebl, P. Jing, B. Calore, M. Piekarczyk, R. Ruddersdorf, D. H. O'Connor, X. Wang, C. Wang, D. B. Allison, J. D. Altman, A. Sette, R. C. Desrosiers, G. Sutter, and D. I. Watkins. 2002. Tat-vaccinated macaques do not control simian immunodeficiency virus SIVmac239 replication. J. Virol. 76:4108-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 5.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 6.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barcellini, W., G. P. Rizzardi, M. O. Borghi, C. Fain, A. Lazzarin, and P. L. Meroni. 1994. TH1 and TH2 cytokine production by peripheral blood mononuclear cells from HIV-infected patients. AIDS 8:757-762. [DOI] [PubMed] [Google Scholar]
- 8.Billaut-Mulot, O., T. Idziorek, M. Loyens, A. Capron, and G. M. Bahr. 2001. Modulation of cellular and humoral immune responses to a multiepitopic HIV-1 DNA vaccine by interleukin-18 DNA immunization/viral protein boost. Vaccine 19:2803-2811. [DOI] [PubMed] [Google Scholar]
- 9.Blazevic, V., M. Heino, A. Lagerstedt, A. Ranki, and K. J. Krohn. 1996. Interleukin-10 gene expression induced by HIV-1 Tat and Rev in the cells of HIV-1 infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:208-214. [DOI] [PubMed] [Google Scholar]
- 10.Blazevic, V., A. Ranki, S. Mattinen, S. L. Valle, S. Koskimies, G. Jung, and K. J. Krohn. 1993. Helper T-cell recognition of HIV-1 Tat synthetic peptides. J. Acquir. Immune Defic. Syndr. 6:881-890. [PubMed] [Google Scholar]
- 11.Brake, D. A., J. Goudsmit, W. J. Krone, P. Schammel, N. Appleby, R. H. Meloen, and C. Debouck. 1990. Characterization of murine monoclonal antibodies to the Tat protein from human immunodeficiency virus type 1. J. Virol. 64:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buonaguro, L., G. Barillari, H. K. Chang, C. A. Bohan, V. Kao, R. Morgan, R. C. Gallo, and B. Ensoli. 1992. Effects of the human immunodeficiency virus type 1 Tat protein on the expression of inflammatory cytokines. J. Virol. 66:7159-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cafaro, A., A. Caputo, C. Fracasso, M. T. Maggiorella, D. Goletti, S. Baroncelli, M. Pace, L. Sernicola, M. L. Koanga-Mogtomo, M. Betti, A. Borsetti, R. Belli, L. Akerblom, F. Corrias, S. Butto, J. Heeney, P. Verani, F. Titti, and B. Ensoli. 1999. Control of SHIV-89.6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nat. Med. 5:643-650. [DOI] [PubMed] [Google Scholar]
- 14.Cafaro, A., F. Titti, C. Fracasso, M. T. Maggiorella, S. Baroncelli, A. Caputo, D. Goletti, A. Borsetti, M. Pace, E. Fanales-Belasio, B. Ridolfi, D. R. Negri, L. Sernicola, R. Belli, F. Corrias, I. Macchia, P. Leone, Z. Michelini, P. ten Haaft, S. Butto, P. Verani, and B. Ensoli. 2001. Vaccination with DNA containing tat coding sequences and unmethylated CpG motifs protects cynomolgus monkeys upon infection with simian/human immunodeficiency virus (SHIV89.6P). Vaccine 19:2862-2877. [DOI] [PubMed] [Google Scholar]
- 15.Calarota, S. A., A. C. Leandersson, G. Bratt, J. Hinkula, D. M. Klinman, K. J. Weinhold, E. Sandstrom, and B. Wahren. 1999. Immune responses in asymptomatic HIV-1-infected patients after HIV-DNA immunization followed by highly active antiretroviral treatment. J. Immunol. 163:2330-2338. [PubMed] [Google Scholar]
- 16.Cao, J., J. McNevin, U. Malhotra, and M. J. McElrath. 2003. Evolution of CD8+ T cell immunity and viral escape following acute HIV-1 infection. J. Immunol. 171:3837-3846. [DOI] [PubMed] [Google Scholar]
- 17.Caselli, E., M. Betti, M. P. Grossi, P. G. Balboni, C. Rossi, C. Boarini, A. Cafaro, G. Barbanti-Brodano, B. Ensoli, and A. Caputo. 1999. DNA immunization with HIV-1 tat mutated in the trans activation domain induces humoral and cellular immune responses against wild-type Tat. J. Immunol. 162:5631-5638. [PubMed] [Google Scholar]
- 18.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, C. H., T. L. Wang, C. F. Hung, Y. Yang, R. A. Young, D. M. Pardoll, and T. C. Wu. 2000. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 60:1035-1042. [PubMed] [Google Scholar]
- 20.Cid-Arregui, A., V. Juarez, and H. zur Hausen. 2003. A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J. Virol. 77:4928-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen, A. D., J. D. Boyer, and D. B. Weiner. 1998. Modulating the immune response to genetic immunization. FASEB J. 12:1611-1626. [PubMed] [Google Scholar]
- 22.Cohen, S. S., C. Li, L. Ding, Y. Cao, A. B. Pardee, E. M. Shevach, and D. I. Cohen. 1999. Pronounced acute immunosuppression in vivo mediated by HIV tat challenge. Proc. Natl. Acad. Sci. USA 96:10842-10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbet, S., L. Vinner, D. M. Hougaard, K. Bryder, H. V. Nielsen, C. Nielsen, and A. Fomsgaard. 2000. Construction, biological activity, and immunogenicity of synthetic envelope DNA vaccines based on a primary, CCR5-tropic, early HIV type 1 isolate (BX08) with human codons. AIDS Res. Hum. Retrovir. 16:1997-2008. [DOI] [PubMed] [Google Scholar]
- 24.Cullen, B. R., and M. H. Malim. 1992. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 216:362-368. [DOI] [PubMed] [Google Scholar]
- 25.Davis, H. L. 1997. Plasmid DNA expression systems for the purpose of immunization. Curr. Opin. Biotechnol. 8:635-646. [DOI] [PubMed] [Google Scholar]
- 26.Demirhan, I., A. Chandra, O. Hasselmayer, P. Biberfeld, and P. Chandra. 1999. Detection of distinct patterns of anti-tat antibodies in HIV-infected individuals with or without Kaposi's sarcoma. J. Acquir. Immune Defic. Syndr. 22:364-368. [DOI] [PubMed] [Google Scholar]
- 27.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drew, D. R., J. S. Boyle, A. M. Lew, M. W. Lightowlers, P. J. Chaplin, and R. A. Strugnell. 2001. The comparative efficacy of CTLA-4 and L-selectin targeted DNA vaccines in mice and sheep. Vaccine 19:4417-4428. [DOI] [PubMed] [Google Scholar]
- 30.Ensoli, B., and A. Cafaro. 2000. Control of viral replication and disease onset in cynomolgus monkeys by HIV-1 TAT vaccine. J. Biol. Regul. Homeost. Agents 14:22-26. [PubMed] [Google Scholar]
- 31.Fanales-Belasio, E., A. Cafaro, A. Cara, D. R. Negri, V. Fiorelli, S. Butto, S. Moretti, M. T. Maggiorella, S. Baroncelli, Z. Michelini, A. Tripiciano, L. Sernicola, A. Scoglio, A. Borsetti, B. Ridolfi, R. Bona, P. ten Haaft, I. Macchia, P. Leone, M. R. Pavone-Cossut, F. Nappi, E. Vardas, M. Magnani, E. Laguardia, A. Caputo, F. Titti, and B. Ensoli. 2002. HIV-1 Tat-based vaccines: from basic science to clinical trials. DNA Cell Biol. 21:599-610. [DOI] [PubMed] [Google Scholar]
- 32.Fanales-Belasio, E., S. Moretti, F. Nappi, G. Barillari, F. Micheletti, A. Cafaro, and B. Ensoli. 2002. Native HIV-1 Tat protein targets monocyte-derived dendritic cells and enhances their maturation, function, and antigen-specific T cell responses. J. Immunol. 168:197-206. [DOI] [PubMed] [Google Scholar]
- 33.Feltquate, D. M., S. Heaney, R. G. Webster, and H. L. Robinson. 1997. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J. Immunol. 158:2278-2284. [PubMed] [Google Scholar]
- 34.Fu, T. M., L. Guan, A. Friedman, J. B. Ulmer, M. A. Liu, and J. J. Donnelly. 1998. Induction of MHC class I-restricted CTL response by DNA immunization with ubiquitin-influenza virus nucleoprotein fusion antigens. Vaccine 16:1711-1717. [DOI] [PubMed] [Google Scholar]
- 35.Gallo, R. C. 1999. Tat as one key to HIV-induced immune pathogenesis and Tat toxoid as an important component of a vaccine. Proc. Natl. Acad. Sci. USA 96:8324-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao, F., Y. Li, J. M. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. R. Shaw, S. Allen, R. Musonda, G. M. Shaw, A. J. Zajac, N. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retrovir. 19:817-823. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein, G. 1996. HIV-1 Tat protein as a potential AIDS vaccine. Nat. Med. 2:960-964. [DOI] [PubMed] [Google Scholar]
- 38.Goldstein, G., G. Tribbick, and K. Manson. 2001. Two B cell epitopes of HIV-1 Tat protein have limited antigenic polymorphism in geographically diverse HIV-1 strains. Vaccine 19:1738-1746. [DOI] [PubMed] [Google Scholar]
- 39.Green, T. D., B. R. Newton, P. A. Rota, Y. Xu, H. L. Robinson, and T. M. Ross. 2001. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine 20:242-248. [DOI] [PubMed] [Google Scholar]
- 40.Gurunathan, S., D. M. Klinman, and R. A. Seder. 2000. DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol. 18:927-974. [DOI] [PubMed] [Google Scholar]
- 41.Hanke, T., T. J. Blanchard, J. Schneider, C. M. Hannan, M. Becker, S. C. Gilbert, A. V. Hill, G. L. Smith, and A. McMichael. 1998. Enhancement of MHC class I-restricted peptide-specific T cell induction by a DNA prime/MVA boost vaccination regime. Vaccine 16:439-445. [DOI] [PubMed] [Google Scholar]
- 42.Hinkula, J., C. Svanholm, S. Schwartz, P. Lundholm, M. Brytting, G. Engstrom, R. Benthin, H. Glaser, G. Sutter, B. Kohleisen, V. Erfle, K. Okuda, H. Wigzell, and B. Wahren. 1997. Recognition of prominent viral epitopes induced by immunization with human immunodeficiency virus type 1 regulatory genes. J. Virol. 71:5528-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu, G. J., R. Y. Wang, D. S. Han, H. J. Alter, and J. W. Shih. 1999. Characterization of the humoral and cellular immune responses against hepatitis C virus core induced by DNA-based immunization. Vaccine 17:3160-3170. [DOI] [PubMed] [Google Scholar]
- 44.Ihata, A., S. Watabe, S. Sasaki, A. Shirai, J. Fukushima, K. Hamajima, J. Inoue, and K. Okuda. 1999. Immunomodulatory effect of a plasmid expressing CD40 ligand on DNA vaccination against human immunodeficiency virus type-1. Immunology 98:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imami, N., A. Pires, G. Hardy, J. Wilson, B. Gazzard, and F. Gotch. 2002. A balanced type 1/type 2 response is associated with long-term nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 76:9011-9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito, M., T. Ishida, L. He, F. Tanabe, Y. Rongge, Y. Miyakawa, and H. Terunuma. 1998. HIV type 1 Tat protein inhibits interleukin 12 production by human peripheral blood mononuclear cells. AIDS Res. Hum. Retrovir. 14:845-849. [DOI] [PubMed] [Google Scholar]
- 47.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 48.Kim, J. J., L. K. Nottingham, D. M. Wilson, M. L. Bagarazzi, A. Tsai, L. D. Morrison, A. Javadian, A. A. Chalian, M. G. Agadjanyan, and D. B. Weiner. 1998. Engineering DNA vaccines via co-delivery of co-stimulatory molecule genes. Vaccine 16:1828-1835. [DOI] [PubMed] [Google Scholar]
- 49.Kim, J. J., J. Yang, K. H. Manson, and D. B. Weiner. 2001. Modulation of antigen-specific cellular immune responses to DNA vaccination in rhesus macaques through the use of IL-2, IFN-γ, or IL-4 gene adjuvants. Vaccine 19:2496-2505. [DOI] [PubMed] [Google Scholar]
- 50.Klein, S. A., J. M. Dobmeyer, T. S. Dobmeyer, M. Pape, O. G. Ottmann, E. B. Helm, D. Hoelzer, and R. Rossol. 1997. Demonstration of the Th1 to Th2 cytokine shift during the course of HIV-1 infection using cytoplasmic cytokine detection on single cell level by flow cytometry. AIDS 11:1111-1118. [DOI] [PubMed] [Google Scholar]
- 51.Kojima, Y., K. Q. Xin, T. Ooki, K. Hamajima, T. Oikawa, K. Shinoda, T. Ozaki, Y. Hoshino, N. Jounai, M. Nakazawa, D. Klinman, and K. Okuda. 2002. Adjuvant effect of multi-CpG motifs on an HIV-1 DNA vaccine. Vaccine 20:2857-2865. [DOI] [PubMed] [Google Scholar]
- 52.Krieg, A. M., A. K. Yi, J. Schorr, and H. L. Davis. 1998. The role of CpG dinucleotides in DNA vaccines. Trends Microbiol. 6:23-27. [DOI] [PubMed] [Google Scholar]
- 53.Krone, W. J., C. Debouck, L. G. Epstein, P. Heutink, R. Meloen, and J. Goudsmit. 1988. Natural antibodies to HIV-tat epitopes and expression of HIV-1 genes in vivo. J. Med. Virol. 26:261-270. [DOI] [PubMed] [Google Scholar]
- 54.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinsky. 2002. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 55.Kwissa, M., V. von Kampen, R. Zurbriggen, R. Gluck, J. Reimann, and R. Schirmbeck. 2000. Efficient vaccination by intradermal or intramuscular inoculation of plasmid DNA expressing hepatitis B surface antigen under desmin promoter/enhancer control. Vaccine 18:2337-2344. [DOI] [PubMed] [Google Scholar]
- 56.Lamhamedi-Cherradi, S., B. Culmann-Penciolelli, B. Guy, M. Kieny, F. Dreyfus, A. Saimot, D. Sereni, D. Sicard, J. Levy, and E. Gomard. 1992. Qualitative and quantitative analysis of human cytotoxic T-lymphocyte responses to HIV-1 proteins. AIDS 6:1249-1258. [DOI] [PubMed] [Google Scholar]
- 57.Letvin, N. L., D. C. Montefiori, Y. Yasutomi, H. C. Perry, M. Davies, C. Lekutis, M. Alroy, D. C. Freed, C. I. Lord, L. K. Handt, M. A. Liu, and J. W. Shiver. 1997. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc. Natl. Acad. Sci. USA 94:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieberman, J., J. A. Fabry, D. M. Fong, and G. R. Parkerson. 1997. Recognition of a small number of diverse epitopes dominates the cytotoxic T lymphocyte response to HIV type 1 in an infected individual. AIDS Res. Hum. Retrovir. 13:383-392. [DOI] [PubMed] [Google Scholar]
- 59.Liu, W., F. Gao, K. Zhao, W. Zhao, G. Fernando, R. Thomas, and I. Frazer. 2002. Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T-lymphocyte induction and anti-tumour activity. Virology 301:43-52. [DOI] [PubMed] [Google Scholar]
- 60.MacGregor, R. R., J. D. Boyer, K. E. Ugen, K. E. Lacy, S. J. Gluckman, M. L. Bagarazzi, M. A. Chattergoon, Y. Baine, T. J. Higgins, R. B. Ciccarelli, L. R. Coney, R. S. Ginsberg, and D. B. Weiner. 1998. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J. Infect. Dis. 178:92-100. [DOI] [PubMed] [Google Scholar]
- 61.Matano, T., M. Kano, A. Takeda, H. Nakamura, N. Nomura, Y. Furuta, T. Shioda, and Y. Nagai. 2003. No significant enhancement of protection by Tat-expressing Sendai viral vector-booster in a macaque AIDS model. AIDS 17:1392-1394. [DOI] [PubMed] [Google Scholar]
- 62.McPhee, D. A., B. E. Kemp, S. Cumming, D. Stapleton, I. D. Gust, and R. R. Doherty. 1988. Recognition of envelope and tat protein synthetic peptide analogs by HIV positive sera or plasma. FEBS Lett. 233:393-396. [DOI] [PubMed] [Google Scholar]
- 63.Meyaard, L., E. Hovenkamp, I. P. Keet, B. Hooibrink, I. H. de Jong, S. A. Otto, and F. Miedema. 1996. Single cell analysis of IL-4 and IFN-γ production by T cells from HIV-infected individuals: decreased IFN-γ in the presence of preserved IL-4 production. J. Immunol. 157:2712-2718. [PubMed] [Google Scholar]
- 64.Morris, C. B., A. Thanawastien, D. E. Sullivan, and J. D. Clements. 2001. Identification of a peptide capable of inducing an HIV-1 Tat-specific CTL response. Vaccine 20:12-15. [DOI] [PubMed] [Google Scholar]
- 65.Nagata, T., M. Uchijima, A. Yoshida, M. Kawashima, and Y. Koide. 1999. Codon optimization effect on translational efficiency of DNA vaccine in mammalian cells: analysis of plasmid DNA encoding a CTL epitope derived from microorganisms. Biochem. Biophys. Res. Commun. 261:445-451. [DOI] [PubMed] [Google Scholar]
- 66.Noonan, D., and A. Albini. 2000. From the outside in: extracellular activities of HIV Tat. Adv. Pharmacol. 48:229-250. [DOI] [PubMed] [Google Scholar]
- 67.Noonan, D. M., A. Gringeri, R. Meazza, O. Rosso, S. Mazza, M. Muca-Perja, H. Le Buanec, R. S. Accolla, A. Albini, and S. Ferrini. 2003. Identification of immunodominant epitopes in inactivated Tat-vaccinated healthy and HIV-1-infected volunteers. J. Acquir. Immune Defic. Syndr. 33:47-55. [DOI] [PubMed] [Google Scholar]
- 68.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific Elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osterhaus, A. D., C. A. van Baalen, R. A. Gruters, M. Schutten, C. H. Siebelink, E. G. Hulskotte, E. J. Tijhaar, R. E. Randall, G. van Amerongen, A. Fleuchaus, V. Erfle, and G. Sutter. 1999. Vaccination with Rev and Tat against AIDS. Vaccine 17:2713-2714. [DOI] [PubMed] [Google Scholar]
- 70.Pauza, C. D., P. Trivedi, M. Wallace, T. J. Ruckwardt, H. Le Buanec, W. Lu, B. Bizzini, A. Burny, D. Zagury, and R. C. Gallo. 2000. Vaccination with tat toxoid attenuates disease in simian/HIV-challenged macaques. Proc. Natl. Acad. Sci. USA 97:3515-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pertmer, T. M., T. R. Roberts, and J. R. Haynes. 1996. Influenza virus nucleoprotein-specific immunoglobulin G subclass and cytokine responses elicited by DNA vaccination are dependent on the route of vector DNA delivery. J. Virol. 70:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters, B. S. 2001. The basis for HIV immunotherapeutic vaccines. Vaccine 20:688-705. [DOI] [PubMed] [Google Scholar]
- 73.Putkonen, P., M. Quesada-Rolander, A. Leandersson, S. Schwartz, R. Thorstensson, K. Okuda, B. Wahren, and J. Hinkula. 1998. Immune responses but no protection against SHIV by gene-gun delivery of HIV-1 DNA followed by recombinant subunit protein boosts. Virology 250:293-301. [DOI] [PubMed] [Google Scholar]
- 73a.Ramakrishna, L., K. K. Anand, M. Mahalingam, K. M. Mohankumar, R. Shilpa, N. B. Siddappa, and U. Ranga. 2004. Codon optimization and ubiquitin conjugation of human immunodeficiency virus-1 Tat lead to enhanced cell mediated immune responses. Vaccine 22:2586-2598. [DOI] [PubMed] [Google Scholar]
- 73b.Ranga, U., R. Shankarappa, N. B. Siddappa, L. Ramakrishna, R. Nagendran, M. Mahalingam, A. Mahadevan, J. Narayana, P. Satishchandra, S. K. Shankar, and V. R. Prasad. 2004. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J. Virol. 78:2586-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ranki, A., J. Suni, V. Blazevic, P. Holmstrom, S. Mattinen, K. Krohn, and S. L. Valle. 1997. T-cell recognition of HIV antigens in HIV-seroreverted persons. AIDS 11:132-133. [PubMed] [Google Scholar]
- 75.Re, M. C., G. Furlini, M. Vignoli, E. Ramazzotti, G. Roderigo, V. De Rosa, G. Zauli, S. Lolli, S. Capitani, and M. La Placa. 1995. Effect of antibody to HIV-1 Tat protein on viral replication in vitro and progression of HIV-1 disease in vivo. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10:408-416. [DOI] [PubMed] [Google Scholar]
- 76.Re, M. C., M. Vignoli, G. Furlini, D. Gibellini, V. Colangeli, F. Vitone, and M. La Placa. 2001. Antibodies against full-length Tat protein and some low-molecular-weight Tat-peptides correlate with low or undetectable viral load in HIV-1 seropositive patients. J. Clin. Virol. 21:81-89. [DOI] [PubMed] [Google Scholar]
- 77.Reinhold, D., S. Wrenger, T. Kahne, and S. Ansorge. 1999. HIV-1 Tat: immunosuppression via TGF-β1 induction. Immunol. Today 20:384-385. [DOI] [PubMed] [Google Scholar]
- 78.Reiss, P., J. M. Lange, A. De Ronde, F. de Wolf, J. Dekker, C. Debouck, and J. Goudsmit. 1990. Speed of progression to AIDS and degree of antibody response to accessory gene products of HIV-1. J. Med. Virol. 30:163-168. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez, F., J. Zhang, and J. L. Whitton. 1997. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J. Virol. 71:8497-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ross, T. M., Y. Xu, T. D. Green, D. C. Montefiori, and H. L. Robinson. 2001. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with gp120-C3d fusion proteins. AIDS Res. Hum. Retrovir. 17:829-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubartelli, A., A. Poggi, R. Sitia, and M. R. Zocchi. 1998. HIV-1 Tat: a polypeptide for all seasons. Immunol. Today 19:543-545. [DOI] [PubMed] [Google Scholar]
- 82.Rusnati, M., and M. Presta. 2002. HIV-1 Tat protein: a target for the development of anti-AIDS therapies. Drugs Fut. 27:481-493. [Google Scholar]
- 83.Sadaie, M. R., and G. L. Hager. 1994. Induction of developmentally programmed cell death and activation of HIV by sodium butyrate. Virology 202:513-518. [DOI] [PubMed] [Google Scholar]
- 84.Sailaja, G., S. Husain, B. P. Nayak, and A. M. Jabbar. 2003. Long-term maintenance of gp120-specific immune responses by genetic vaccination with the HIV-1 envelope genes linked to the gene encoding Flt-3 ligand. J. Immunol. 170:2496-2507. [DOI] [PubMed] [Google Scholar]
- 85.Scheerlinck, J. Y. 2001. Genetic adjuvants for DNA vaccines. Vaccine 19:2647-2656. [DOI] [PubMed] [Google Scholar]
- 86.Sharp, P. M., M. Averof, A. T. Lloyd, G. Matassi, and J. F. Peden. 1995. DNA sequence evolution: the sounds of silence. Philos. Trans. R. Soc. Lond. B 349:241-247. [DOI] [PubMed] [Google Scholar]
- 87.Sharp, P. M., E. Cowe, D. G. Higgins, D. C. Shields, K. H. Wolfe, and F. Wright. 1988. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens: a review of the considerable within-species diversity. Nucleic Acids Res. 16:8207-8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Silvera, P., M. W. Richardson, J. Greenhouse, J. Yalley-Ogunro, N. Shaw, J. Mirchandani, K. Khalili, J. F. Zagury, M. G. Lewis, and J. Rappaport. 2002. Outcome of simian-human immunodeficiency virus strain 89.6p challenge following vaccination of rhesus macaques with human immunodeficiency virus Tat protein. J. Virol. 76:3800-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith, S. M., S. Pentlicky, Z. Klase, M. Singh, C. Neuveut, C. Y. Lu, M. S. Reitz, Jr., R. Yarchoan, P. A. Marx, and K. T. Jeang. 2003. An in vivo replication-important function in the second coding exon of Tat is constrained against mutation despite cytotoxic T lymphocyte selection. J. Biol. Chem. 278:44816-44825. [DOI] [PubMed] [Google Scholar]
- 90.Spear, B. T., T. Longley, S. Moulder, S. L. Wang, and M. L. Peterson. 1995. A sensitive lacZ-based expression vector for analyzing transcriptional control elements in eukaryotic cells. DNA Cell Biol. 14:635-642. [DOI] [PubMed] [Google Scholar]
- 91.Stratford, R., G. Douce, L. Zhang-Barber, N. Fairweather, J. Eskola, and G. Dougan. 2000. Influence of codon usage on the immunogenicity of a DNA vaccine against tetanus. Vaccine 19:810-815. [DOI] [PubMed] [Google Scholar]
- 92.Tahtinen, M., A. Ranki, S. L. Valle, V. Ovod, and K. Krohn. 1997. B-cell epitopes in HIV-1 Tat and Rev proteins colocalize with T-cell epitopes and with functional domains. Biomed. Pharmacother. 51:480-487. [DOI] [PubMed] [Google Scholar]
- 93.Tsuji, T., K. Hamajima, N. Ishii, I. Aoki, J. Fukushima, K. Q. Xin, S. Kawamoto, S. Sasaki, K. Matsunaga, Y. Ishigatsubo, K. Tani, T. Okubo, and K. Okuda. 1997. Immunomodulatory effects of a plasmid expressing B7-2 on human immunodeficiency virus-1-specific cell-mediated immunity induced by a plasmid encoding the viral antigen. Eur. J. Immunol. 27:782-787. [DOI] [PubMed] [Google Scholar]
- 94.Uchijima, M., A. Yoshida, T. Nagata, and Y. Koide. 1998. Optimization of codon usage of plasmid DNA vaccine is required for the effective MHC class I-restricted T cell responses against an intracellular bacterium. J. Immunol. 161:5594-5599. [PubMed] [Google Scholar]
- 95.van Baalen, C. A., O. Pontesilli, R. C. Huisman, A. M. Geretti, M. R. Klein, F. de Wolf, F. Miedema, R. A. Gruters, and A. D. Osterhaus. 1997. Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J. Gen. Virol. 78:1913-1918. [DOI] [PubMed] [Google Scholar]
- 96.Viscidi, R. P., K. Mayur, H. M. Lederman, and A. D. Frankel. 1989. Inhibition of antigen-induced lymphocyte proliferation by Tat protein from HIV-1. Science 246:1606-1608. [DOI] [PubMed] [Google Scholar]
- 97.Wang, S., X. Liu, K. Fisher, J. G. Smith, F. Chen, T. W. Tobery, J. B. Ulmer, R. K. Evans, and M. J. Caulfield. 2000. Enhanced type I immune response to a hepatitis B DNA vaccine by formulation with calcium- or aluminum phosphate. Vaccine 18:1227-1235. [DOI] [PubMed] [Google Scholar]
- 98.Watson, K., and R. J. Edwards. 1999. HIV-1-trans-activating (Tat) protein: both a target and a tool in therapeutic approaches. Biochem. Pharmacol. 58:1521-1528. [DOI] [PubMed] [Google Scholar]
- 99.Weiss, R., S. Scheiblhofer, J. Freund, F. Ferreira, I. Livey, and J. Thalhamer. 2002. Gene gun bombardment with gold particles displays a particular Th2-promoting signal that overrules the Th1-inducing effect of immunostimulatory CpG motifs in DNA vaccines. Vaccine 20:3148-3154. [DOI] [PubMed] [Google Scholar]
- 100.Westendorp, M. O., R. Frank, C. Ochsenbauer, K. Stricker, J. Dhein, H. Walczak, K. M. Debatin, and P. H. Krammer. 1995. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375:497-500. [DOI] [PubMed] [Google Scholar]
- 101.Wieland, U., J. E. Kuhn, C. Jassoy, H. Rubsamen-Waigmann, V. Wolber, and R. W. Braun. 1990. Antibodies to recombinant HIV-1 vif, tat, and nef proteins in human sera. Med. Microbiol. Immunol. 179:1-11. [DOI] [PubMed] [Google Scholar]
- 102.Xiao, H., C. Neuveut, H. L. Tiffany, M. Benkirane, E. A. Rich, P. M. Murphy, and K. T. Jeang. 2000. Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc. Natl. Acad. Sci. USA 97:11466-11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xin, K. Q., K. Hamajima, S. Sasaki, T. Tsuji, S. Watabe, E. Okada, and K. Okuda. 1999. IL-15 expression plasmid enhances cell-mediated immunity induced by an HIV-1 DNA vaccine. Vaccine 17:858-866. [DOI] [PubMed] [Google Scholar]
- 104.Xin, K. Q., Y. Lu, K. Hamajima, J. Fukushima, J. Yang, K. Inamura, and K. Okuda. 1999. Immunization of RANTES expression plasmid with a DNA vaccine enhances HIV-1-specific immunity. Clin. Immunol. 92:90-96. [DOI] [PubMed] [Google Scholar]
- 105.Zagury, D., A. Lachgar, V. Chams, L. S. Fall, J. Bernard, J. F. Zagury, B. Bizzini, A. Gringeri, E. Santagostino, J. Rappaport, M. Feldman, A. Burny, and R. C. Gallo. 1998. Interferon alpha and Tat involvement in the immunosuppression of uninfected T cells and C-C chemokine decline in AIDS. Proc. Natl. Acad. Sci. USA 95:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zagury, J. F., A. Sill, W. Blattner, A. Lachgar, H. Le Buanec, M. Richardson, J. Rappaport, H. Hendel, B. Bizzini, A. Gringeri, M. Carcagno, M. Criscuolo, A. Burny, R. C. Gallo, and D. Zagury. 1998. Antibodies to the HIV-1 Tat protein correlated with nonprogression to AIDS: a rationale for the use of Tat toxoid as an HIV-1 vaccine. J. Hum. Virol. 1:282-292. [PubMed] [Google Scholar]
- 107.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]