Abstract
Psittacine beak and feather disease (PBFD), caused by Beak and feather disease virus (BFDV), is the most significant infectious disease in psittacines. PBFD is thought to have originated in Australia but is now found worldwide; in Africa, it threatens the survival of the indigenous endangered Cape parrot and the vulnerable black-cheeked lovebird. We investigated the genetic diversity of putative BFDVs from southern Africa. Feathers and heparinized blood samples were collected from 27 birds representing 9 psittacine species, all showing clinical signs of PBFD. DNA extracted from these samples was used for PCR amplification of the putative BFDV coat protein (CP) gene. The nucleotide sequences of the CP genes of 19 unique BFDV isolates were determined and compared with the 24 previously described sequences of BFDV isolates from Australasia and America. Phylogenetic analysis revealed eight BFDV lineages, with the southern African isolates representing at least three distinctly unique genotypes; 10 complete genome sequences were determined, representing at least one of every distinct lineage. The nucleotide diversity of the southern African isolates was calculated to be 6.4% and is comparable to that found in Australia and New Zealand. BFDVs in southern Africa have, however, diverged substantially from viruses found in other parts of the world, as the average distance between the southern African isolates and BFDV isolates from Australia ranged from 8.3 to 10.8%. In addition to point mutations, recombination was found to contribute substantially to the level of genetic variation among BFDVs, with evidence of recombination in all but one of the genomes analyzed.
Psittacine beak and feather disease (PBFD) was first described in various species of Australian cockatoos in 1975 by Ross Perry and has since been recognized as the most significant infectious disease in psittacine birds (20). The onset of disease is characterized by the development of lethargy, depression, and severe anemia in affected birds. Clinical symptoms generally progress to weight loss, feather dystrophy and loss and, in severe cases, deformities of the beak and claws (16). PBFD is commonly associated with immunodeficiency-related diseases caused by depletion of lymphoid tissue (21), with damage to the lymphoreticular tissue being most pronounced in the bursa of Fabricius and the thymus (9). Affected birds frequently succumb to secondary infections, such as peritonitis, chlamydiosis, and mycotic ventriculitis (28).
The causative agent of PBFD, Beak and feather disease virus (BFDV), is a member of the genus Circovirus in the family Circoviridae. As with all other described circoviruses, BFDV has a small, single-stranded circular DNA genome of approximately 2 kb, the smallest genome of any known pathogenic virus, encapsidated in a spherical capsid with a diameter of 14 to 21 nm (21). The genome of BFDV contains two major open reading frames (ORFs), encoding the replication-associated protein (Rep) and the capsid protein (CP) (12). A third ORF (V2), common to all BFDV isolates, has been described, but it is unclear what role the transcriptional product of this ORF may play in the life cycle of the virus (10). Circoviruses have been found in parrots, pigeons, Senegal doves, canaries, finches, geese, southern black-backed gulls, ostriches, pigs, and chickens (28).
PBFD is thought to have originated in Australia, and traditionally most of the research on BFDV has focused on isolates from Australasia. Australian isolates of BFDV are genetically diverse and group into four phylogenetic clusters (1). Phylogenetic analysis of isolates from New Zealand revealed a similar clustering pattern and an apparent genotypic association with specific psittacine species (22). The proposed host specificity of viral genotypes is based on the phylogenetic characteristics of a group of closely related viruses found only in lorikeets. These viruses differ significantly from other BFDVs, and the monophyly of the group is well supported. There is, however, no evidence of adaptive selection within BFDV populations, and the subdivisions are most likely the result of genetic drift of viral genotypes in different parrot populations. The similarity of isolates found in Australia and New Zealand suggests that the evolution of a genotypic association between viruses and their hosts predates their dissemination throughout the world (22).
Very little is known about BFDV isolates found outside Australasia. However, the disease is a major problem among bird breeders in South Africa, where approximately 10 to 20% of South African psittacine breeding stocks are lost due to the disease each year. The incidence of PBFD in parrot populations in southern Africa in which the disease is endemic remains largely unknown, but at least two threatened species, the endangered Cape parrot (Poicephalus robustus) and the vulnerable black-cheeked lovebird (Agapornis nigrigenis), are affected by the disease (29). In the present study, we report on the genetic diversity of viruses associated with PBFD in southern Africa and discuss the possible impact of the disease on conservation efforts directed toward protecting indigenous African parrots.
MATERIALS AND METHODS
Feathers and heparinized blood samples were collected from various psittacine species showing clinical signs of PBFD throughout southern Africa. Details of the samples are summarized in Table 1. DNA was extracted from these samples (25) and used for PCR amplification of the entire BFDV genome or parts of the genome.
TABLE 1.
Viruses detected in PBFD-affected birds originating from diverse geographical and host origins in southern Africa
| Host species | Common name | Origin | Isolate | GenBank accession no. |
|---|---|---|---|---|
| Psittacus erithacus | African grey parrot | South Africa | AFG3-ZA | AY450443 |
| P. erithacus | African grey parrot | Gauteng, South Africa | AFG4-ZA | AY450434 |
| Cacatua alba | White cockatoo | Gauteng, South Africa | UC1-ZA | AY450436 |
| C. alba | White cockatoo | Western Cape, South Africa | UC2-ZA | AY450450 |
| Pionites leucogaster | White-bellied caique | Gauteng, South Africa | WBC1-ZA | AY450434 |
| Ara macao | Scarlet macaw | South Africa | SM1-ZA | AY450451 |
| Poicephalus rufiventris | African red-bellied parrot | KwaZulu-Natal, South Africa | ARB1-ZA | AY450452 |
| P. rufiventris | African red-bellied parrot | KwaZulu-Natal, South Africa | ARB2-ZA | AY450448 |
| P. rufiventris | African red-bellied parrot | KwaZulu-Natal, South Africa | ARB3-ZA | AY450449 |
| P. rufiventris | African red-bellied parrot | KwaZulu-Natal, South Africa | ARB4-ZA | AY450440 |
| Poicephalus gulielmi massaicus | Jardine parrot | KwaZulu-Natal, South Africa | GJP1-ZA | AY450441 |
| P. g. massaicus | Jardine parrot | KwaZulu-Natal, South Africa | GJP2-ZA | AY450447 |
| P. g. fantiensis | Jardine parrot | KwaZulu-Natal, South Africa | LJP1-ZA | AY450446 |
| P. g. gulielmi | Jardine parrot | KwaZulu-Natal, South Africa | BWJ1-ZA | AY450445 |
| P. g. fantiensis | Jardine parrot | KwaZulu-Natal, South Africa | LJP2-ZA | AY450444 |
| Poicephalus rueppellii | Rüppell's parrot | KwaZulu-Natal, South Africa | RP1-ZA | AY450439 |
| Poicephalus robustus | Cape parrot | KwaZulu-Natal, South Africa | CPA8-ZA | AY450437 |
| P. robustus | Cape parrot | KwaZulu-Natal, South Africa | CPA7-ZA | AY450438 |
| Agapornis nigrigenis | Black-cheeked lovebird | Zambia | BCL1-ZAM | AY450442 |
The full-length BFDV genome was amplified from individual blood samples by inverse PCR with primers 5′-GGATCCA(G/T)CCGGTTCTGGC(G/A)-3′ and 5′-GGATCCCACTACAAAGGAGGACCC-3′ (complementary sequences corresponding to the BamHI endonuclease restriction site are underlined). These partly complementary opposing sense primers were designed to straddle the BamHI endonuclease recognition site situated within the CP gene. Amplification of the circular BFDV genome would result in a linear representation of the entire genome. PCR products were purified by using High Pure PCR purification columns (Roche) and cloned into plasmid pGEM-T-Easy (Promega). Recombinant clones were end sequenced with commercially available M13 primers (Perkin-Elmer) by using a DYEnamic ET dye terminator cycle sequencing kit and analyzed with a MegaBACE 500 automated sequencer (Amersham Bioscience). Based on previously published sequence data (1), three additional primers (5′-GTATCGCCTGATGTGACGTCTG-3′, 5′-CTGGACATTGTGGCGAGAGAC-3′, and 5′-GACCGTTACCACCATAAAGTG-3′) were designed for sequencing of the remainder of the genome. At least two clones for each isolate were independently sequenced to ensure the validity of the data.
A 765-nucleotide (nt) fragment of the genome comprising the CP gene (nt 1228 to 1977 of the genome [GenBank accession number AF080560]) was amplified by PCR with primers 5′-GCGGCCGCATGCTGTGGGGCACCTCTAACTGC-3′ and 5′-CTCGAGTCTTTATTAAGTACTGGGATTG-3′ (sequences engineered to create endonuclease restriction sites are underlined). PCR products were cloned into plasmid pGEM-T-Easy and sequenced in both directions with commercially available M13 primers.
All nucleotide sequences determined in this study were aligned with published BFDV sequences (Table 2) by using ClustalX (27). The multiple sequence alignments of the complete BFDV genome sequences, as well as the major ORFs of the virus, were used to determine the phylogenetic relationships of the viruses. Phylogenetic reconstruction was performed by using the maximum-likelihood method implemented in PHYLIP 3.6 (2a). Bootstrap values were calculated by using the neighbor-joining method based on the two-parameter corrected distance matrix in MEGA 2.1 (7). The phylogenetic trees were rooted with sequences from nonpsittacine avian circoviruses.
TABLE 2.
Circovirus reference sequences used in this study
| Host species | Common name | Origin | Isolate | GenBank accession no. |
|---|---|---|---|---|
| Psephotus haematogaster | Bluebonnet | Australia | BB-WA | AF311295 |
| Trichoglossu haematodus | Rainbow lorikeet | Australia | LK-VIC | AF311299 |
| Cacatua leabeateri | Major Mitchell's cockatoo | Australia | MMC-WA | AF311300 |
| Cacatua galerita | Sulfur-crested cockatoo | Australia | SCC1-WA | AF311302 |
| Cacatua galerita | Sulfur-crested cockatoo | Australia | SCC-NT | AF311301 |
| Agapornis roseicollis | Rosy-faced lovebird | Australia | LB-WA | AF311296 |
| Eolophus roseicapillus | Galah | Australia | Galah-WA | AF311298 |
| Cacatua tenuirostris | Eastern long-billed corella | Australia | ELBC-SA | AF311297 |
| Cacatua galerita | Sulfur-crested cockatoo | Australia | BFDV-AUS | AF080560 |
| T. haematodus | Rainbow lorikeet | New Zealand | RL2-NZ | AY148294 |
| T. haematodus | Rainbow lorikeet | New Zealand | RL3-NZ | AY148295 |
| T. haematodus | Rainbow lorikeet | New Zealand | RL5-NZ | AY148293 |
| T. haematodus | Rainbow lorikeet | New Zealand | RL6-NZ | AY148300 |
| Trichoglossu haematodus rubritorquis | Red-collared lorikeet | New Zealand | RCL-NZ | AY148291 |
| Lorius chlorocercus | Yellow-bib lorikeet | New Zealand | YBL1-NZ | AY148292 |
| L. chlorocercus | Yellow-bib lorikeet | New Zealand | YBL2-NZ | AY148299 |
| Psitteuteles goldiei | Goldie's lorikeet | New Zealand | GL-NZ | AY148298 |
| Eos reticulata | Blue-streak lorikeet | New Zealand | BSL1-NZ | AY148296 |
| E. reticulata | Blue-streak lorikeet | New Zealand | BSL2-NZ | AY148297 |
| C. tenirostitus | Longbill corella | New Zealand | LC1-NZ | AY148289 |
| C. galerita | Sulfur-crested cockatoo | New Zealand | SCC1-NZ | AY148285 |
| C. galerita | Sulfur-crested cockatoo | New Zealand | SCC2-NZ | AY148286 |
| C. galerita | Sulfur-crested cockatoo | New Zealand | SCC3-NZ | AY148287 |
| Unknown | Pooled virus | United States | BFDV-USA | AF071878 |
Detection of potential recombinant sequences, identification of likely parent sequences, and localization of possible recombination breakpoints were done by using RDP (10), Geneconv (15), MaxChi (11, 17), and bootscanning (24).
Base frequencies, nucleotide diversity, and tranversion/transition (ti/tv) ratios were calculated by using either PAUP* 4.1 (26) or MEGA 2.1. The average numbers of synonymous changes per synonymous site and nonsynonymous changes per nonsynonymous site along all ORFs were calculated by using DnaSP 3.1 (23). The distribution of synonymous and nonsynonymous substitutions along the viral genes was determined by using the average number of substitutions per codon position (site). The significance of clustering in the distribution was tested by using a permutation test with a sliding window ranging from 5 to 20 sites per window. Sites under positive selection were identified by using the likelihood-based models described by Yang et al. (31), which assign each site to one of a number of estimated values of nonsynonymous and synonymous substitution ratios. The nomenclature used here to describe each of the models is consistent with that proposed by Yang et al. (31). The results from nested models were compared by using a likelihood ratio test. Twice the likelihood ratio difference was compared with a χ2 distribution having a degree of freedom equal to the difference in the number of parameters in the respective models.
Nucleotide sequence accession numbers.
All nucleotide sequences determined in this study have been submitted to GenBank under the accession numbers listed in Table 1.
RESULTS
Phylogenetic relationships and recombination.
The nucleotide sequences of 10 putative BFDV isolates obtained from diverse psittacine species at different geographical locations in southern Africa were determined and aligned withpreviously published sequences (http://www.mcb.uct.ac.za/BFDV%20Genomes.txt). All southern African isolates displayed the same basic genome structure as that previously described for BFDV (12), including the positions of the ORFs and the stem-loop structure located between the Rep and CP genes. The genome sizes ranged from 1,988 to 2,002 nt (AFG4-ZA, UC1-ZA, and WBC1-ZA, 1,988 nt; AFRG3-ZA, 1,993 nt; BCL1-ZAM, 1,996 nt; GJP1-ZA, RP1-ZA, ARB4-ZA, CPA7-ZA, and CPA8-ZA, 2,002 nt). The nucleotide diversity (mean and standard deviation) of the southern African isolates was calculated to be 0.064 ± 0.004 (n = 10), and the average distance between the southern African isolates and BFDV-AUS ranged from 8.3 to 10.8%.
The phylogenetic relationships of the 10 southern African isolates and 10 previously published BFDV isolates is shown in Fig. 1a. As previously reported, the BFDV isolates were grouped into clearly defined genetic clusters or lineages. For descriptive purposes, we have tentatively defined a genotype as any group of sequences that is at least 5% divergent from the next closest lineage. The southern African isolates were restricted to four out of the eight apparent BFDV genotypes. The sequences of three southern African isolates (AFG4-ZA, WBC1-ZA, and UC1-ZA) were found to be closely related to a genomic consensus sequence derived from pooled BFDV isolates from the United States (GenBank accession number AF071878). The remainder of the southern African isolates represented three distinctly unique genotypes.
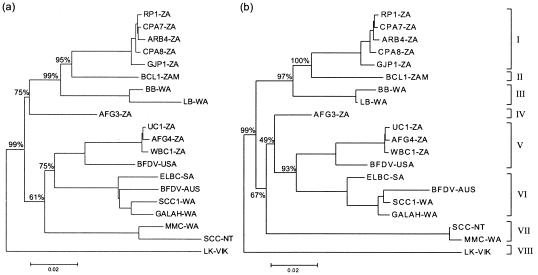
FIG. 1.
Maximum-likelihood trees depicting the phylogenetic relationships of 20 BFDV isolates ignoring recombination between isolates (a) and with major recombinant regions removed from individual sequences (b). The full-length tree was rooted by using three nonpsittacine avian circoviruses (canary circovirus [GenBank accession number AJ301633], columbid circovirus, [AJ298229], and goose circovirus [AJ304456]). Only the subtrees containing the BFDV isolates are shown. Bootstrap values were calculated by using the neighbor-joining method based on the two-parameter corrected distance matrix. The major virus genotypes are bracketed and identified by roman numerals I to VIII.
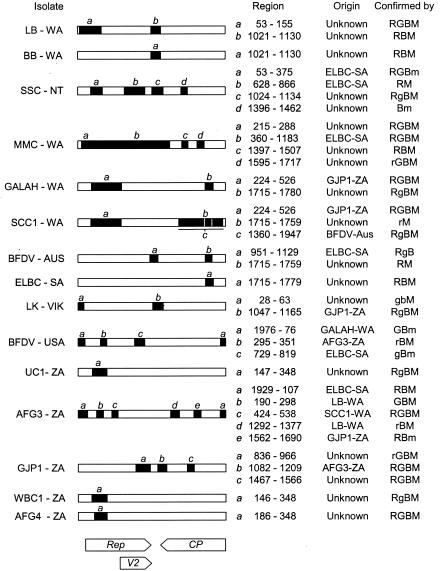
The relative positions of some viruses within the phylogenetic tree were found to vary when different parts of the genome were used as the basis of the analyses. These data suggested that recombination may contribute significantly to the evolution of BFDV. A multiple alignment of the full-length BFDV genomes was used in conjunction with four independent recombination detection programs to detect potential recombination events. The results of the recombination analysis are shown in Fig. 2. Evidence of recombination was found in all but one of the sequences analyzed; no recombinant regions were identified within the genome of isolate BCL1-ZAM. Recombination events were distributed across the entire genome but were more frequent within the Rep gene. Interlineage and intralineage recombinations were detected. Most notably, two putative recombination events detected within the LK-VIC sequence each involved a second sequence outside the LK lineage (the LK lineage was previously described by Ritchie et al. [22]). Both events were statistically well supported and identified MMC-WA- and GJP1-ZA-like viruses as potential minor parents.
FIG. 2.
Recombinant regions detected within BFDV sequences. Region coordinates are nucleotide positions relative to the origin of virion strand replication. Rep, Rep gene; CP, CP gene; V2, second virion sense ORF. Wherever possible, the origin of the recombinant region is identified. Whenever the origin of a particular recombinant region is identified, the isolate named is a close relative of the parental virus of that region. Recombinant regions and parental viruses were identified by using the RDP (R), Geneconv (G), bootscanning (B), and MaxChi (M) methods. Uppercase letters indicate that a region was identified by a method with greater than 99% certainty (99% bootstrap support in the case of bootscanning). Lowercase letters indicate that a region was identified by a method with between 95 and 99% certainty (between 95 and 99% bootstrap support in the case of bootscanning).
It has long been recognized that recombination can drastically affect the accuracy of phylogenetic inference. For the BFDV isolates analyzed here, the branching pattern of the maximum-likelihood tree constructed from an alignment with the obvious recombinant regions removed (Fig. 1b) remained largely unchanged relative to that of a tree constructed from an alignment containing the recombinant regions, except for the AFG3-ZA, ELBC-SA, and BFDV-AUS viruses. All eight tentative genotypes remained clearly defined and well supported by significant bootstrap values. The branch lengths were, however, affected. In such situations, when the evolutionary history is affected by recombination, it would be more accurate to describe the relationships in terms of reticulate evolution; alternatively, the possible inaccuracy of phylogenetic estimates for samples with several underlying phylogenies can be addressed by defining data partitions (such as genes) and performing separate analyses of the partitions (30).
CP.
The capsid protein genes of nine additional southern African BFDV isolates were amplified by PCR, sequenced, and aligned with the corresponding regions of the 20 full-length genome sequences described above. Of the 753 nt constituting the gene, 484 (64.3%) were conserved across all taxa. At the amino acid level, this means that 148 of a total of 251 amino acids (58%) were completely conserved. The nucleotide diversity was calculated to be 0.101 ± 0.007 (n = 29). The sequences displayed unequal base frequencies for each of the three codon positions, and the rate of substitution was significantly biased toward transitions (ti/tv ratio, 1.94).
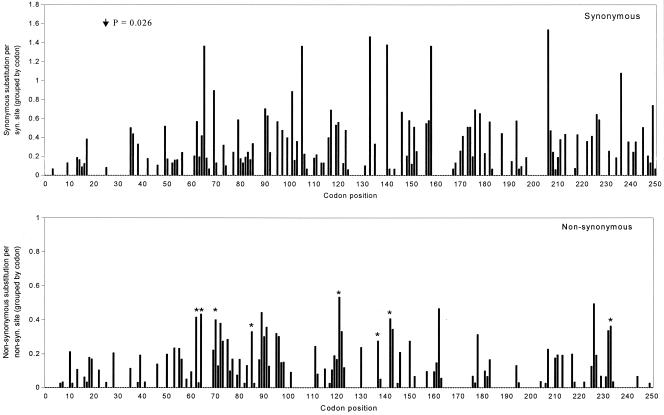
The distribution of nucleotide substitutions along the CP gene is shown in Fig. 3. The distribution of nonsynonymous sites showed no evidence of clustering irrespective of the window size, while the synonymous profile contained a single significant mutational “cold spot” (i.e., gap) centered at position 25 (window size, 15) (P = 0.026). Interestingly, the position of the cold spot corresponds to one of two putative bipartite nuclear localization signals situated between positions 15 and 32 and positions 23 to 40 (data not shown). The presumably vital role of this region of CP in nuclear transport and assembly may severely limit the number of possible mutations.
FIG. 3.
Distributions of nucleotide substitutions along the BFDV CP gene. The y axis indicates the number of substitutions per site averaged over all possible pairwise sequences. The synonymous mutational cold spot centered at position 25 is indicated by an arrow. Nonsynonymous substitutions identified as being under positive selection are marked by asterisks.
The ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS ratio) across the entire CP gene was estimated to be 0.35, suggesting that the gene was not under positive selective pressure. Calculating dN/dS ratios across an entire gene may, however, be too insensitive for detecting positive selection acting on only a few sites, resulting in the underestimation of the overall selective pressure exerted on the protein (13). This problem can be partially overcome by codon-based methods implemented within a maximum-likelihood framework which allows for heterogeneous ratios among sites (14). Analyses of the CP gene data set with models M3 (discrete) and M8 (β and ω), implemented in the codeml component of the PAML program package, provided consistent evidence of positive selection in the gene. Approximately 8% of sites were estimated to be under positive selection (M3: p2 = 0.083, ω2 = 2.91) (Table 3).
TABLE 3.
Likelihood values and parameter estimates for BFDV CP
| Model code | na | Likelihoodb | dN/dS ratio | Estimates of parametersc |
|---|---|---|---|---|
| Ignoring recombination | ||||
| M3 (3 categories) | 29 | −3,379.21 | 0.534 | p2 = 0.083, ω2 = 2.91 |
| M7 | 29 | −3,399.08 | 0.365 | p = 0.102, q = 0.177 |
| M8 | 29 | −3,381.51 | 0.549 | p0 = 0.84579, p = 0.23812, q = 0.84027, p1 = 0.154, ω = 2.356 |
| Recombinant regions removed | ||||
| M3 (3 categories) | 28 | −3,383.78 | 0.537 | p2 = 0.057, ω2 = 3.44 |
| M7 | 28 | −3,398.29 | 0.392 | p = 0.09449, q = 0.14654 |
| M8 | 28 | −3,384.28 | 0.558 | p0 = 0.90511, p = 0.15727, q = 0.33863, p1 = 0.09489, ω = 2.85629 |
Number of sequences in the data set.
Log likelihood of the fitted model.
For each model, the proportion of sites in the ith category (i = 0, 1, or 2) is reported (pi), together with the values of dN/dS assigned to each category (ωi). p and q are parameters that determine the shape of the beta distribution of ω values in models M7 and M8.
Individual sites under positive selection were identified by a Bayesian approach. Sixty-six sites for which p was >0.5 (ω > 1) were identified by model M3. Only 37 sites were identified by model M8; all were included in the set identified by the discrete model. When more stringent criteria (p > 0.95, ω > 1) were used, only nine sites (sites 62, 64, 69, 70, 85, 121, 137, 142, and 233) common to both models were identified.
Significantly high recombination rates can lead to an overestimation of substitutions, since the likelihood models for the detection of positively selected sites rely on the phylogenetic relationships among sequences (4). To test whether recombination affected the identification of sites under positive selection, the analyses were repeated with a modified data set in which the major recombinant regions within individual sequences were removed. Estimates of the parameters with the revised tree topology in model M3 did not differ significantly from those presented above (Table 3). The identification of sites under positive selection was also not influenced by recombination within the data sets (results not shown).
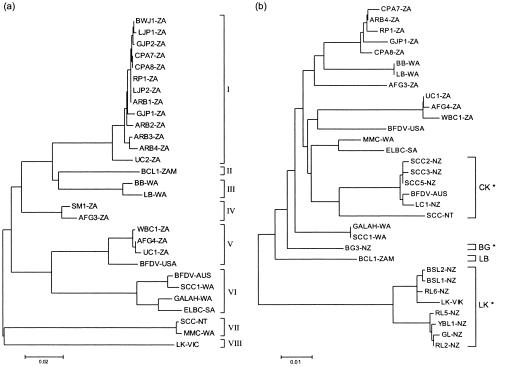
A maximum-likelihood tree depicting the phylogenetic relationships of the CP sequences is shown in Fig. 4a. The topology of the CP phylogeny closely resembles that reconstructed from the full genome sequences, with eight clearly defined genetic clusters. The southern African isolates were restricted to lineages I, II, IV, and V. We found no evidence to support any relationship between the genetic variation and the regional distribution of the southern African isolates. Southern African lineages also could not be associated with specific psittacine species. At least three southern African genotypes consisted of closely related viruses isolated from species representative of both families, Psittacidae (parrots) and Cacatuidea (cockatoos), within the order Psittaciformes.
FIG. 4.
Maximum-likelihood trees depicting the phylogenetic relationships of CP (a) and Rep (b) of BFDV. Lineages supported by significant bootstrap values are bracketed. The CK, LB, and LK lineages within the Rep phylogeny (marked with an asterisk) were previously described by Ritchie et al. (22) and refer to the group of host species associated with the genotype: CK, cockatoos; BG, budgerigars; and LK, lorikeets. The LB lineage described here similarly refers to the host species associated with the genotype: LB, lovebird.
Rep.
Portions of the Rep genes from 32 isolates, corresponding to nt 198 to 759 of the BFDV genome represented by GenBank accession number AF080560, were aligned. As with the CP gene, unequal base frequencies were observed within the Rep gene for all three codon positions, and transitions were favored above transversions (ti/tv ratio, 1.94). The genetic variation within the Rep gene was, however, found to be considerably less than the variation within the CP gene (0.068 ± 0.007; n = 32). The dN/dS ratio across the Rep gene was estimated to be 0.17. The discrete model (M3) fitted the data well; fewer than 1% of sites had a ω of >1. The beta mixture models (M8 to M11) did not fit the data any better than the simple beta model (M7), the parameter estimates of which suggested a highly skewed L shape for the ω distribution. These results suggest that there are no obvious sites within the Rep gene that are under positive selection. Removing recombinant regions within individual sequences did not significantly affect the outcome of the analyses (data not shown).
The phylogenetic relationships of the Rep sequences are shown in Fig. 4b. The topology of the maximum-likelihood tree based on the Rep sequences differed slightly from that of the tree based on the CP phylogeny. In addition to the three lineages described by Ritchie et al. (22), only one additional lineage was supported by significant bootstrap values. This fourth lineage (LB) was identified from a single sequence isolated from a black-cheeked lovebird from which samples were obtained in the mid-Machile River area in Zambia. The Rep sequence of this isolate (BCL1-ZAM) differed by at least 5% from all of the other sequences.
The differences in the topologies of the trees are partly due to recombination within Rep. Posada and Crandall (18) demonstrated that the effect of recombination on phylogenies is dependent on the relatedness of the sequences involved and the sizes of the recombinant regions. When recombination occurs between closely related taxa or when recombination is ancient, the phylogeny under which the majority of the sites have evolved will generally be recovered. However, when the recombinational breakpoint divides the region into two parts of similar lengths, an inaccurate phylogeny underlying the data will most likely be inferred (19). This scenario is most evident in our data when the relative positions of isolates MMC-WA and SCC-NT within each of the respective trees are considered. These isolates represent a unique lineage within the CP phylogeny (lineage VII) which is not represented in the tree based on the Rep sequences. As shown in Fig. 2, the genomes of both isolates are recombinant for large portions of Rep. The frequent occurrence of recombination within the Rep gene, with recombinant tracts often exceeding 50% of the length of the gene, will most likely seriously compromise the reliability of BFDV phylogenies and phylogenetic inferences based on this gene alone.
DISCUSSION
It is evident that indigenous psittacine species in southern Africa are at risk from PBFD and that to ignore the impact of BFDV on both captive and wild populations of African parrots might severely hamper efforts to conserve this natural heritage. As part of an integrated approach to the management of the threat posed by BFDV, we have investigated the genetic diversity of putative BFDV isolates in southern Africa.
The level of genetic diversity among southern African BFDV isolates is similar to that which has been described in Australia and New Zealand. The southern African isolates have apparently, however, diverged substantially from viruses found in other parts of the world and clustered into three unique genotypes. Assuming the disease is not endemic in Africa, the existence of these clearly defined subpopulations would suggest that BFDV was introduced into southern Africa on at least three separate occasions. In addition to these genotypes, a group of southern African isolates were found to be closely related to viruses isolated in North America. It is unclear whether this genotype was recently introduced into South Africa or whether it represents a fourth genotype that evolved in southern Africa and subsequently spread to other parts of the world.
The level of divergence between the genotypes in Africa and isolates found in other parts of the world does, however, suggest that the occurrence BFDV on the African continent is not due to recent introductions and that Australian and African BFDV populations have possibly diverged sufficiently to produce regionally distinct lineages. It furthermore draws in to question the theory that BFDV arose in Australia; it could be argued that the similar levels of genetic diversity of BFDV isolates in Africa and most of the Australian lineages and the clear separation of African from Australasian lineages indicate that these groups of viruses diverged at an very early stage in their evolutionary history. The existence of the BFDV lineage exclusively associated with lorikeets and thus far found only in Australasia adds an interesting dimension to this theory; this group of viruses could represent the “true Australian variant” of BFDV, with all other BFDV isolates evolving from a common ancestor that may have originated in Africa and subsequently spread to Australasia.
With the constant movement of birds across geographical borders through trade, there is an increasing risk of spreading the disease into new areas and populations. Coupled to this is the risk of generating unique viruses through recombination between established virus populations and newly introduced viruses. Recombination has been well documented as a key strategy for generating diversity in both RNA and DNA viruses (2, 8, 21, 25) and appears to contribute substantially to the level of genetic variation among BFDVs; we found evidence of recombination in all but one of the viral genomes analyzed in this study. The failure to detect any recombinant regions within the genome of the Zambian isolate taken from a wild black-cheeked lovebird further suggests that separate and unique southern African genotypes may exist. Putative recombination events between viruses from Africa and Australia presumably occurred prior to the geographical isolation.
Mixed infections, a prerequisite for recombination, have never been shown to occur for BFDV, but the frequency of recombination detected among isolates suggests that it is a common feature of the disease. The putative recombination event between LK-VIC and viruses outside the LK lineage further implies that the speculative genotypic association of viral sequences and specific psittacine hosts does not exclude mixed infections involving diverse lineages. Several putative recombination events for which the possible parent sequences could not be identified were detected. This result suggests that not all circulating BFDV genotypes were represented in our data sets and that the full breadth of BFDV diversity is not accurately represented by the isolates described thus far.
The relative contributions of mutation, reassortment, and recombination to genetic diversity within viral genomes vary depending on the characteristics of the particular virus and gene products involved (5, 6). Interestingly, we found no evidence of sites under diversifying selective pressure within the Rep gene. The functional role of Rep may limit the number of nonsynonymous substitutions that would not adversely affect its performance. This notion is reflected by the high proportion of sites within Rep with a ω value of <1 (M3: p0 = 0.647, ω0 = 0.00737), suggesting that the protein might be under purifying selective pressure. However, most of the detectable recombination events in our data set occurred within rep. These results suggest that recombination might contribute substantially more to the level of variation within rep than genetic drift. This situation is similar to that found in plant-infecting geminiviruses (15). There is substantial phylogenetic evidence suggesting that BFDV-like circoviruses are descendants of a recombinant virus that inherited the 5′ portion of its rep and its origin of virion sense replication from a plant-infecting nanovirus or geminivirus (3, 12). Recombinational exchange of rep fragments between massively diverged or even unrelated virus lineages suggests that this gene and its product may have uniquely modular properties—a suggestion that is supported by the Rep recombinants detected in this study and a geminivirus recombination study (15).
CP of BFDV is a major constituent of infectious virus particles and is therefore a likely target of immune surveillance. This assumption is supported by the various ω ratios of individual sites across the gene. Comparison of relative fixation rates of synonymous and nonsynonymous mutations revealed statistically significant evidence that CP is under positive selection. The distribution of positively selected sites in structural and immunogenic proteins of various viruses, such as foot-and-mouth disease virus and human immunodeficiency virus, have been shown to correspond to known monoclonal antibody epitopes (4). The hypervariability of specific sites within CP of BFDV similarly could be the result of immune evasion. Different serotypes of BFDV have not yet been identified, but the possibility that antigenically distinct subgroups of BFDV could exist should not be ignored during the design of vaccines against the virus.
The genetic diversity of the global virus population, compounded by the difficulties in effectively curbing illegal trade in wild-caught birds, has serious implications for the control of PBFD in southern Africa. Continued monitoring of both wild and captive populations will be a central feature of efforts to understand BFDV epidemiology and effectively address the threat posed by the virus to endangered parrot populations. Since all eight putative genotypes of BFDV are clearly defined by the relationships between their CP sequences, molecular characterization of the CP gene can potentially be used to trace the origin of PBFD outbreaks at a global level. However, the effects of recombination on direct phylogenetic reconstruction or phylogenetically based analyses should not be ignored if accurate inferences about the evolutionary history of the viruses are to be made.
Acknowledgments
We thank Daniel Haydon (University of Guelph, Guelph, Ontario, Canada) for assistance and insight concerning the aspects of selection. Some of the blood samples were kindly supplied by Lesley Kenmuir and Ben Minnaar. The careful proofreading of the manuscript prior to submission by Antoinette van Schalkwyk is also greatly appreciated.
This project was partly funded by the Loro Parque Foundation.
REFERENCES
- 1.Bassami, M. R., I. Ypelaar, D. Berryman, G. E. Wilcox, and S. R. Raidal. 2001. Genetic diversity of beak and feather disease virus detected in psittacine species in Australia. Virology 279:392-400. [DOI] [PubMed] [Google Scholar]
- 2.Chenault, K. D., and U. Melcher. 1994. Phylogenetic relationships reveal recombination among isolates of cauliflower mosaic virus. J. Mol. Evol. 39:496-505. [DOI] [PubMed] [Google Scholar]
- 2a.Felsenstein, J. 2004. PHYLIP: phylogenetic inference package, version 3.6. Department of Genome Sciences, University of Washington, Seattle. (Distributed by the author.)
- 3.Gibbs, M. J., and G. F. Weiller. 1999. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc. Natl. Acad. Sci. USA 96:8022-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haydon, D. T., A. D. Bastos, N. Knowles, and A. R. Samuel. 2001. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holland, J. D. E. 1998. Origin and evolution of viruses. Virus Genes 16:13-21. [DOI] [PubMed] [Google Scholar]
- 6.Keese, P., and A. Gibbs. 1993. Plant viruses: master explorers of evolutionary space. Curr. Opin. Genet. Dev. 3:873-877. [DOI] [PubMed] [Google Scholar]
- 7.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA 2: Molecular Evolution Genetics Analysis software. Bioinformatics 17:1242-1243. [DOI] [PubMed] [Google Scholar]
- 8.Lai, M. M. C. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latimer, K. S., P. M. Rakich, F. D. Niagro, B. E. Ritchie, W. L. Steffens, R. P. Campagnoli, D. A. Pesti, and P. D. Lukert. 1991. An updated review of psittacine beak and feather disease. J. Assoc. Avian Vet. 5:211-220. [Google Scholar]
- 10.Martin, D. P., and E. P. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 11.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 12.Niagro, F. D., A. N. Forsthoefel, R. P. Lawther, L. Kamalanathan, B. W. Ritchie, K. S. Latimer, and P. D. Lukert. 1998. Beak and feather disease virus and porcine circovirus genomes: intermediates between geminiviruses and plant circoviruses. Arch. Virol. 143:1723-1744. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen, R. 1997. The ration of replacement to silent divergence and tests of neutrality. J. Evol. Biol. 10:217-231. [Google Scholar]
- 14.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and pplications to HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 16.Pass, D. A., and R. A. Perry. 1984. The pathology of psittacine beak and feather disease. Aust. Vet. J. 61:69-74. [DOI] [PubMed] [Google Scholar]
- 17.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posada, D., and K. A. Crandall. 2002. The effect of recombination in phylogeny reconstruction. J. Mol. Evol. 54:396-402. [DOI] [PubMed] [Google Scholar]
- 19.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 20.Raidal, S. R., C. L. McElnea, and G. M. Cross. 1993. Seroprevalence of psittacine beak and feather disease virus in wild psittacine birds in New South Wales. Aust. Vet. J. 70:137-139. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie, B. W., F. D. Niagro, P. D. Lukert, W. L. Steffens, and K. S. Latimer. 1989. Characterization of a new virus derived from cockatoos with psittacine beak and feather disease. Virology 171:83-88. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie, P. A., I. L. Anderson, and D. M. Lambert. 2003. Evidence of specificity of psittacine beak and feather disease viruses among avian hosts. Virology 306:109-115. [DOI] [PubMed] [Google Scholar]
- 23.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 24.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Swofford, D. L. 2001. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b8. Sinauer Associates, Sunderland, Mass.
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd, D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29:373-394. [DOI] [PubMed] [Google Scholar]
- 29.Warburton, L. S., and M. R. Perrin. 2002. PBFDV bei frei lebenden Ruβköpfchen in Sambia. Papageien 5:166-169. [Google Scholar]
- 30.Wiens, J. J. 1998. Combining data sets with different phylogenetic histories. Syst. Biol. 47:568-581. [DOI] [PubMed] [Google Scholar]
- 31.Yang, Z., R. Nielsen, N. Goldman, and A.-M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]