Abstract
Hepatitis C virus (HCV) is a leading cause of chronic viral hepatitis worldwide. The study of antibody-mediated virus neutralization has been hampered by the lack of an efficient and high-throughput cell culture system for the study of virus neutralization. The HCV structural proteins have been shown to assemble into noninfectious HCV-like particles (HCV-LPs). Similar to serum-derived virions, HCV-LPs bind and enter human hepatocytes and hepatoma cell lines. In this study, we developed an HCV-LP-based model system for a systematic functional analysis of antiviral antibodies from patients with acute or chronic hepatitis C. We demonstrate that cellular HCV-LP binding was specifically inhibited by antiviral antibodies from patients with acute or chronic hepatitis C in a dose-dependent manner. Using a library of homologous overlapping envelope peptides covering the entire HCV envelope, we identified an epitope in the N-terminal E2 region (SQKIQLVNTNGSWHI; amino acid positions 408 to 422) as one target of human antiviral antibodies inhibiting cellular particle binding. Using a large panel of serum samples from patients with acute and chronic hepatitis C, we demonstrated that the presence of antibodies with inhibition of binding activity was not associated with viral clearance. In conclusion, antibody-mediated inhibition of cellular HCV-LP binding represents a convenient system for the functional characterization of human anti-HCV antibodies, allowing the mapping of envelope neutralization epitopes targeted by naturally occurring antiviral antibodies.
Hepatitis C virus (HCV), a member of the Flaviviridae, is a major cause of chronic viral hepatitis in the world (28, 31). Progression to chronic disease occurs in the majority of HCV-infected persons, and end-stage liver disease due to chronic HCV infection has become the main indication for liver transplantation (28, 31). Resolution of chronic infection is extremely rare; more often, chronic infection results in chronic hepatitis, liver cirrhosis, or hepatocellular carcinoma (23). Worldwide, an estimated 170 million people are infected with HCV. Treatment options for chronic HCV infection are limited, and a vaccine to prevent HCV infection is not available (15, 23).
Although the humoral and cellular immune responses induced by HCV have been analyzed in great detail, the mechanisms of viral clearance and persistence are still poorly understood (46). HCV can establish persistent infection despite a humoral and cellular immune response that is generally targeted against all viral proteins (30, 46). A strong cellular immune response appears to be important for virus clearance (28, 46). Chronic HCV infection results in the induction of a strong humoral immune response (30), and anti-HCV antibodies can be detected easily by using synthetic peptides or recombinant proteins in serologic assays (41). Using the chimpanzee model, antibodies with neutralizing properties have been described (18). These antibodies were directed against epitopes in the hypervariable region of the envelope glycoprotein 2 (E2) and appeared to be isolate specific. Antibody-mediated neutralization is also suggested by a study of patients undergoing liver transplantation for HCV- and hepatitis B virus-related liver cirrhosis. Infusion of anti-HBs hyperimmune globulin containing anti-HCV appeared to reduce HCV infection in the transplanted liver (19). Due to the pending development of convenient and efficient model systems for the analysis of large serum panels for virus neutralization, a systematic analysis of antibodies with neutralizing properties obtained from patients with acute or chronic HCV infection is lacking.
Several groups have recently described the synthesis of HCV-like particles (HCV-LPs) in insect cells using a recombinant baculovirus containing the cDNA of the HCV structural proteins core, E1, and E2 (6, 7, 13, 44, 45, 50). Recently, the formation of HCV-LPs in mammalian cells has also been described (10, 17). HCV-LPs exhibit morphological, biophysical, and antigenic properties similar to those of putative virions isolated from HCV-infected patients. In contrast to recombinant C-terminally truncated HCV E2 protein, the envelope proteins of HCV-LPs are presumably presented in a native, virion-like conformation. They may, therefore, interact with anti-HCV antibodies directed against nonlinear or conformational epitopes of HCV envelope proteins that may represent neutralizing epitopes (8, 13, 45). Indeed, a previous study demonstrated that antiviral antibodies in acute and chronic HCV infections interact with HCV-LPs when used as the capture antigen in an enzyme-linked immunosorbent assay (ELISA). Furthermore, HCV-LPs induce potent humoral and cellular immune responses in vivo and therefore offer a promising approach for vaccine development (7, 29, 33, 37).
Recent studies have demonstrated that HCV-LPs interact with defined human cell lines and hepatocytes similar to viral particles isolated from human serum. The interaction of HCV-LPs with target cell lines therefore represents a novel model system for the study of viral binding and entry (4, 44, 48, 50). In this study, we demonstrate that the HCV-LP-based system can be conveniently used for the functional characterization of anti-HCV antibodies in acute and chronic hepatitis C. We show that cellular binding of HCV-LPs is inhibited by human anti-HCV antibodies, and we define the envelope epitopes targeted by these antibodies. Furthermore, we use this system as a surrogate model to study the roles of neutralizing antibodies in viral clearance and the clinical outcome of HCV infection.
MATERIALS AND METHODS
Patients.
Serial anti-HCV-positive serum samples (n = 93; sampled at time points from 0 to 18 months following the diagnosis of HCV infection) were obtained from 21 patients with acute symptomatic hepatitis C (8 patients with acute self-limited hepatitis and viral clearance and 13 patients with acute hepatitis that progressed to chronic infection who were prospectively followed at the Department of Medicine II, Klinikum Grosshadern, University of Munich, Munich, Germany, between 1995 and 1999 [22]). The diagnosis of acute hepatitis C was based on the following criteria (22): (i) elevated alanine aminotransferase levels at least 20 times the upper limit of normal; (ii) seroconversion to anti-HCV-positive status by second- or third-generation ELISA or recombinant immunoblot assay II (EIA II [Abbott Laboratories, Chicago, Ill.] and RIBA II [Ortho Diagnostics, Raritan, N.J.]), respectively; (iii) positive PCR for HCV RNA (Amplicor; Roche Diagnostics, Branchburg, N.J.); and (iv) a history of sudden onset of liver disease in previously healthy individuals. Potential causes of acute hepatitis, such as other forms of viral hepatitis, autoimmune hepatitis, alcoholic liver disease, toxins, or metabolic etiologies, were ruled out. In addition, sera (n = 10; sampled >6 months following the diagnosis of HCV infection) from 10 patients with chronic HCV infection were obtained from the Department of Medicine II, University of Freiburg, Freiburg, Germany. All patients were serologically negative for hepatitis B virus and human immunodeficiency virus infection. HCV genotypes were determined by INNO-LiPAHCV II (Innogenetics, Ghent, Belgium) or VERSANT(r) HCV Genotype Assay (Bayer HealthCare LLC, Morristown, N.J.). Sera from patients with acute symptomatic hepatitis C were analyzed for anti-core, -NS3, -NS4, and -NS5 antibodies using the INNOTESTHCV Ab III assay (Innogenetics). The sera were also analyzed for anti-envelope antibodies using recombinant C-terminally truncated envelope glycoproteins E1 and E2 or a panel of envelope-specific peptides as described previously (45).
Synthesis and purification of HCV-LPs.
Procedures for the expression and purification of HCV-LPs have been described previously (6, 48). HCV-LPs of genotype 1b were derived from the cDNA of the HCV-J strain (26), and HCV-LPs of genotype 1a were obtained from the infectious clone H77C (51). Control preparations were derived from insect cells infected with a recombinant baculovirus containing the cDNA for β-glucuronidase (GUS). Insect cell control preparations served as negative controls in all binding experiments. The HCV-LP E2 concentration was determined by an E2-specific ELISA, as described recently (48).
Cell lines.
HuH-7 and HepG2 cells were maintained in Dulbecco's modified Eagle's medium (Gibco Life Technologies, Gaithersburg, Md.) containing 10% fetal calf serum (PAA Laboratories, Linz, Austria). The maintenance of Spodoptera frugiperda Sf9 insect cells has been described in detail (6).
Analysis of cellular HCV-LP binding by flow cytometry.
Binding of HCV-LPs to cell lines was performed as described recently (38). Cells (1.5 × 105 per assay) were incubated with HCV-LPs or a control insect cell preparation (GUS) in phosphate-buffered saline (PBS)-2% bovine serum albumin (BSA; pH 5.2) (A 3912; Sigma-Aldrich, St. Louis, Mo.) (final volume, 100 μl) at various concentrations for 1 h at 4°C. After the removal of nonbound HCV-LPs by three washes with PBS-2% BSA, the cells were incubated for 40 min at 4°C with the mouse monoclonal anti-E2 antibodies 16A6 (48) or AP33 (44) or human polyclonal anti-HCV antibody diluted in 50 μl of PBS-2% BSA (1:100). Human polyclonal anti-HCV antibody was obtained from the serum of a 50-year-old female patient with chronic hepatitis C (HCV genotype 1a) followed at the Department of Medicine of the University of Freiburg and is available on request. Titration by an E2-specific ELISA (48) revealed an anti-E2 antibody titer of 1:25,600. The anti-HCV-LP titer was 1:512,000, as determined by an HCV-LP-specific ELISA (8). The cells were washed three times in PBS-2% BSA at 4°C. The cells were incubated for 30 min at 4°C with phycoerythrin (PE)-conjugated anti-mouse immunoglobulin G (IgG) antibody (Jackson ImmunoResearch, West Grove, Pa.) or fluorescein isothiocyanate (FITC)-conjugated anti-human IgG antibody (ICN Biomedicals Inc., Aurora, Ohio) diluted in 50 μl of PBS-2% BSA (1:100). The cells were washed three times in PBS at 4°C. Cell-bound fluorescence was analyzed with a FACScalibur flow cytometer (Becton Dickinson) using Cellquest version 3.11 and CellquestPro software. These programs produce histograms of each sample and calculate the mean fluorescence intensity (MFI) of the cell population, which relates directly to the surface density of PE-labeled E2 bound to hepatocytes (38).
Analysis of HCV-LP binding and internalization using immunofluorescence and laser scanning confocal microscopy.
For the study of cellular HCV-LP binding, HuH-7 cells (0.5 × 105 per assay) were incubated with HCV-LPs (genotype 1a) at 4°C for 40 min. For the study of temperature-dependent HCV-LP entry (44), cells were incubated for an additional 60 min at 37°C. After removal of nonbound HCV-LPs by washing the cells with ice-cold PBS, the cells were added to poly-l-lysine-coated cover slides, fixed with PBS containing 3.5% paraformaldehyde, and permeabilized using PBS containing 0.1% Triton X-100. The cells were stained for HCV-LP binding and entry using the mouse anti-E2 antibody 16A6 (dilution, 1:100 in PBS) and Cy3-conjugated anti-mouse IgG (dilution, 1:250 in PBS; Jackson ImmunoResearch). For costaining of the cytoskeleton, cells were coincubated with a polyclonal rabbit anti-actin antibody (dilution, 1:200 in PBS; Sigma-Aldrich) and FITC-conjugated anti-rabbit IgG (dilution, 1:500 in PBS; ICN Biomedicals). Between antibody incubations (1 h at room temperature [RT]), the cells were washed three times in PBS. For staining of the nucleus, cells were incubated with DRAQ-5 (Biostatus, Shepshed, Leicestershire, United Kingdom), a highly permeable DNA-interactive agent, according to the manufacturer's protocol. Prior to analysis by confocal laser scanning microscopy, the cover slides were mounted in antifade reagent (Fluoroguard; Bio-Rad Laboratories, Hercules, Calif.) to minimize photobleaching. The stained cells were analyzed in cross section using an LSM 410 laser scanning confocal microscope (Carl Zeiss Corp., Jena, Germany) with argon (488-nm wavelength) and helium-neon (543- and 633-nm wavelength) lasers. Digitalized images were analyzed using Zeiss-LSM Image Browser version 2.8.
Antibody-mediated inhibition of cellular HCV-LP binding.
To study whether HCV-LP binding can be inhibited by serum-derived antibodies, HCV-LPs (0.1 μg of HCV-LP E2/ml) were mixed with human serum in various dilutions and incubated for 1 h at 37°C. HuH-7 cells (1.5 × 105) were added to the mixture and incubated for 1 h at 4°C to allow HCV-LP binding. After the cells were washed, the amount of HCV-LP bound to the cells was assessed as described above. All sera were screened for inhibition of HCV-LP binding using both mouse monoclonal anti-E2 and human polyclonal anti-HCV antibodies. Only sera demonstrating a decrease in cellular binding of >50% in both assays were considered inhibitory. Since the supply of human polyclonal anti-HCV antibody was limited, mouse monoclonal anti-E2 antibody was used for further characterization of inhibition of HCV-LP binding following the assessment of samples in screening assays. MFI values from HCV-LPs incubated with control serum (positive control), with anti-HCV-positive serum (experimental values), and control preparations (GUS) with control serum (negative control) were measured, and the specific neutralization was determined as described previously (38), according to the following equation: specific neutralization = [(positive control MFI − experimental MFI)/(positive control MFI − negative control MFI)] × 100. The inhibition of the binding titer was defined as the serum dilution that showed <50% inhibition of cellular HCV-LP binding.
IgG purification from human serum.
In order to demonstrate that inhibition of binding was mediated by anti-HCV antibodies, IgG was purified from HCV-positive and HCV-negative control sera using a MabTrap kit (Amersham Biosciences, Freiburg, Germany) (9). Briefly, a HiTrap protein G column was equilibrated with 3 ml of binding buffer. Serum samples were diluted 1:1 in binding buffer and applied to the column (total volume, 1 ml). Following washing of the column with 7 ml of binding buffer, the purified IgG was eluted using 5 ml of elution buffer and neutralized using 75 μl of neutralizing buffer according to the manufacturer's protocol. Fractions were analyzed for the presence of anti-HCV antibodies using an HCV-LP-based ELISA (8). Subsequently, IgG-containing fractions were pooled and desalted using a HiTrap desalting column (Amersham Biosciences). The purified IgG was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining. The concentration of purified IgG was determined by Bradford assay using a Bio-Rad protein assay. The serum IgG concentration was determined using a turbidimetric immunoassay (Roche Diagnostics, Mannheim, Germany).
Epitope mapping of antibodies inhibiting cellular binding of HCV-LPs.
To identify HCV-LP envelope epitopes targeted by human antibodies inhibiting cellular HCV-LP binding, an anti-HCV-positive serum with strong inhibition of binding activity was incubated with overlapping 15-mer peptides of the HCV envelope glycoproteins (comprising amino acid positions 201 to 758) derived from the HCV-J strain (26) or PBS (as a control) for 1 h at RT (peptide concentration, 100 μg/ml; serum dilution, 1:50). The peptides were provided through the European Research Network HCVacc (QLK2-1999-00356) and were synthesized by Clonestar Corp. (Brno, Czech Republic). HCV-LPs (derived from the homologous isolate as the peptides) were then added to the peptide-antibody complexes and incubated for 1 h at 37°C. Finally the HCV-LPs, antibodies, and peptides were added to HuH-7 cells and incubated for 1 h at 4°C. Binding of HCV-LPs was detected by flow cytometry using the monoclonal anti-E2 antibody AP33 as described above. For the assessment of concentration-dependent reversion of HCV-LP inhibition of binding activity, an anti-HCV-positive serum (dilution, 1:50) was incubated in the presence of various concentrations of E2 peptide 408 (amino acid sequence, SQKIQLVNTNGSWHI, corresponding to amino acid positions 408 to 422). Reversion of HCV-LP inhibition of binding activity was calculated by the following equation: ΔMFI = experimental MFI (anti-HCV-positive serum + HCV envelope peptide) − positive control MFI (anti-HCV positive serum + PBS).
Statistical analysis.
Comparison between subgroups was performed by the chi-square test using the SAS Analyst statistical software package, version 6.12 (SAS Institute, Cary, N.C.).
RESULTS
HCV-LP-based model system for viral envelope binding and entry.
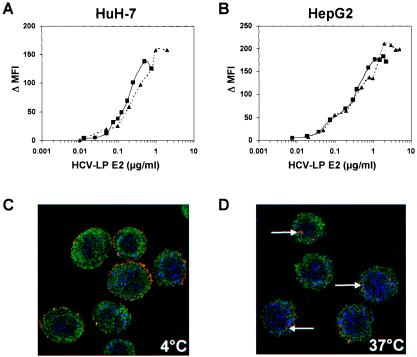
To establish a convenient model system for the study of antibody-mediated inhibition of viral envelope binding to target cells, we first characterized the interaction of HCV-LPs with the hepatoma cell lines HuH-7 and HepG2. These liver-derived cell lines have been used as model systems to study the interaction of viral envelope glycoproteins with the host cell membrane (4, 20, 40, 44, 48). Furthermore, defined hepatoma cells have been shown to allow the entry of virus, virus-like particles, and retroviral HCV pseudoparticles (2, 5, 24, 44), and they may support low-level HCV infection (42). HCV-LPs of genotypes 1a and 1b were synthesized using recombinant baculoviruses containing the cDNA for the HCV structural proteins of strains HCV-J and HCV-H77. Using flow cytometry and anti-envelope antibodies for the detection of cellular HCV-LP binding, HCV-LPs demonstrated concentration-dependent and saturable binding to HuH-7 and HepG2 cells, respectively (Fig. 1A and B). This side-by-side analysis of HCV-LPs of genotypes 1a and 1b extends a previous study describing the binding of HCV-LPs of the infectious clone H77C to target cells (48).
FIG. 1.
Concentration-dependent and saturable binding of HCV-LPs to and entry into human hepatoma cells. (A and B) Binding of HCV-LPs to target cells is concentration-dependent and saturable. HuH-7 (A) and HepG2 (B) cells were incubated with increasing concentrations of HCV-LPs of genotype 1a (triangles), as well as genotype 1b (squares), and particle binding was analyzed by flow cytometry as described in Materials and Methods. On the y axis, net MFI values for each HCV-LP E2 concentration were calculated by subtracting the MFI of the negative control with anti-E2 and PE-conjugated anti-mouse IgG antibodies from that obtained with the respective HCV-LP concentration (x axis). (C and D) Analysis of HCV-LP entry into target cells using anti-E2-specific immunofluorescence and cross section laser scanning confocal microscopy. (C) Cellular binding of HCV-LP was assessed by incubation of HuH-7 cells with HCV-LPs (genotype 1a) at 4°C for 40 min. (D) For temperature-dependent HCV-LP entry, cells were incubated for an additional 60 min at 37°C. After removal of nonbound HCV-LPs by washing the cells with ice-cold PBS, the cells were added to poly-l-lysine-coated cover slides, fixed, permeabilized, and stained for HCV-LPs using anti-E2 and Cy3-conjugated anti-mouse IgG antibodies (arrows). For the costaining of the cytoskeleton, cells were coincubated with a rabbit anti-actin and FITC-conjugated anti-rabbit IgG antibody. For costaining of the nucleus, the cells were incubated with DRAQ-5, a highly permeable DNA-interactive agent.
The two hepatoma cell lines exhibited similar HCV-LP binding profiles, although saturation of HCV-LP binding to HuH-7 cells was reached at a slightly lower HCV-LP E2 concentration than for HepG2 cells. Saturation of HCV-LP binding was reached at an E2 concentration of ∼0.5 to 1 μg/ml for HuH-7 cells (Fig. 1A) and 1 to 2 μg/ml for HepG2 cells (Fig. 1B). These data may indicate a higher number of HCV-LP E2 binding sites on HepG2 than on HuH-7 cells. HCV-LPs of genotypes 1a and 1b exhibited similar binding profiles (saturation of binding at 0.5 and 1 μg of E2/ml for HCV-LPs of genotype 1a or 1b and HuH-7 cells, respectively). Half-maximal saturation of HCV-LP binding was present at E2 concentrations between 0.2 and 0.4 μg/ml.
The next step after particle binding is uptake, or entry of particles into the cell. To investigate whether HCV-LPs were internalized into HuH-7 cells following binding to the cell surface, we performed anti-E2-specific immunofluorescence and confocal laser scanning microscopy of HuH-7 cells incubated with HCV-LPs. As shown in Fig. 1C and D, incubation of cells with HCV-LPs (genotype 1a) at 4°C resulted in the exclusive detection of HCV-LP E2 at the cell surface, consistent with HCV-LP binding to the cell membrane. By contrast, incubation of HuH-7 cells with HCV-LPs at 37°C resulted in the translocation of E2 immunoreactivity inside the cell, consistent with temperature-dependent cellular HCV-LP entry. These data indicate that HCV-LPs are taken up by HuH-7 in a temperature-dependent manner following cell surface binding. These findings corroborate and extend previous experiments using dye-labeled HCV-LPs (44).
Inhibition of cellular HCV-LP binding by serum-derived anti-HCV antibodies.
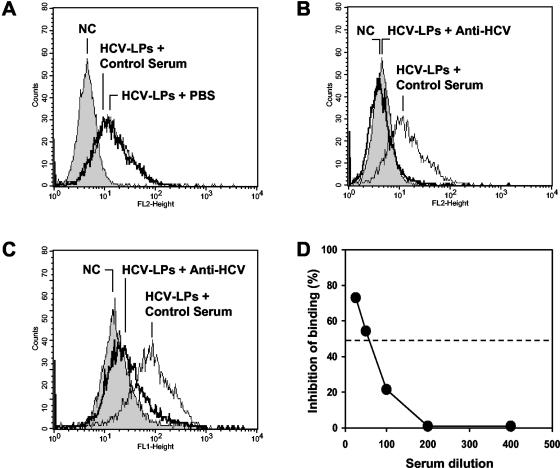
We next studied whether cellular HCV-LP binding is inhibited by sera from HCV-infected individuals. As shown in Fig. 2, anti-HCV-positive sera from patients with hepatitis C inhibited cellular HCV-LP binding (Fig. 2). To confirm that the observed inhibition of binding was due to anti-HCV antibodies present in these sera, cellular HCV-LP binding was studied in the presence of 10 anti-HCV-negative control sera. None of these control sera inhibited cellular HCV-LP binding (Fig. 2A and B). To exclude the possibility that the observed inhibition of HCV-LP binding was due to masking of HCV-LP epitopes by nonneutralizing anti-HCV antibodies present in the sera, inhibition of binding was studied using a human serum containing high-titer anti-E2 and anti-HCV-LP antibodies (human polyclonal anti-HCV) (Fig. 2C). Revealing the binding with antibodies from the same species as the neutralizing serum (human-human) is critical because nonneutralizing anti-envelope antibodies present in human serum could cover E2 after it is bound to target cells and could, therefore, interfere with the assessment of neutralization if the binding was revealed with an anti-E2 antibody from a different species (such as a mouse monoclonal antibody) (38). Only sera demonstrating a reproducible decrease in cellular binding of >50% were considered inhibitory.
FIG. 2.
Antibody-mediated inhibition of cellular HCV-LP binding. HCV-LPs were incubated with anti-HCV-positive serum or a pool of anti-HCV-negative control sera (dilution, 1:50). HCV-LP-antibody complexes were added to HuH-7 cells for 1 h at 4°C. After removal of nonbound HCV-LP-antibody complexes by washing the cells in PBS-2% BSA, the binding of HCV-LPs was detected by flow cytometry using the mouse monoclonal anti-E2 antibody AP33 and PE-conjugated anti-mouse IgG (A and B) or human polyclonal anti-HCV and FITC-conjugated anti-human IgG antibody (C). The fluorescence intensity and relative cell numbers (Counts) are shown on the x and y axes, respectively. NC, negative control, corresponding to HuH-7 cells incubated with control insect cell preparations (GUS) and control serum. (D) For the assessment of concentration-dependent antibody-mediated inhibition of binding, HCV-LPs (genotype 1b) were incubated in subsaturating concentrations with an anti-HCV-positive serum or a pool of anti-HCV-negative control sera (at various dilutions as indicated on the x axis) for 1 h at 37°C. HCV-LP-antibody complexes were added to HuH-7 cells for 1 h at 4°C. HCV-LP binding to the HuH-7 cells was detected by flow cytometry as described above. Inhibition of cellular HCV-LP binding (y axis) was calculated as described in Materials and Methods.
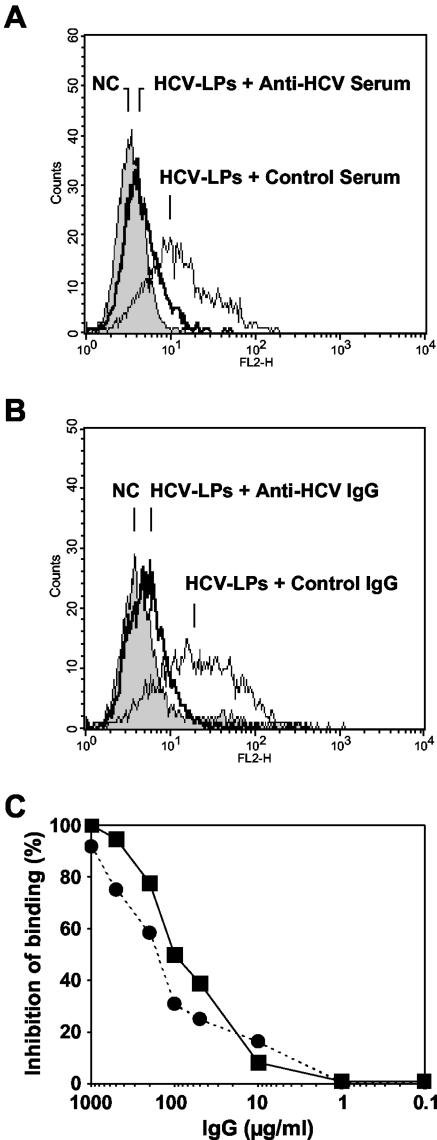
Inhibition of HCV-LP binding was dependent on the serum concentration (Fig. 2D), indicating a concentration-dependent effect of inhibitory activity by anti-HCV antibodies present in these sera. Titration of inhibition of binding activity by endpoint dilution revealed a maximal antibody titer of 1:200. In order to determine whether inhibition of binding was mediated by anti-HCV antibodies, IgG was purified from HCV-positive and HCV-negative control sera using protein G antibody affinity chromatography. Purified anti-HCV IgG inhibited cellular HCV-LP binding (Fig. 3B) similarly to anti-HCV serum (Fig. 3A). Inhibition of HCV-LP binding by purified anti-HCV IgG was concentration dependent, showing 50% inhibition of binding at concentrations of ∼100 μg/ml (Fig. 3C). Interestingly, concentrations of purified IgG and serum IgG exhibited similar levels of inhibition of binding profiles (Fig. 3C).
FIG. 3.
Inhibition of HCV-LP binding by purified human anti-HCV IgG. (A) Inhibition of cellular HCV-LP binding by anti-HCV-positive or control serum (dilution, 1:25). (B) Inhibition of cellular HCV-LP binding by purified anti-HCV or control IgG (1 mg/ml) from the same sera shown in panel A. (C) Concentration-dependent inhibition of HCV-LP binding by purified anti-HCV IgG (squares) and serum containing anti-HCV antibodies (circles). Analysis of inhibition of HCV-LP binding was performed as described in the legend to Fig. 2. Serum IgG concentrations were determined as described in Materials and Methods.
Antibody-mediated inhibition of binding and viral clearance in acute and chronic hepatitis C.
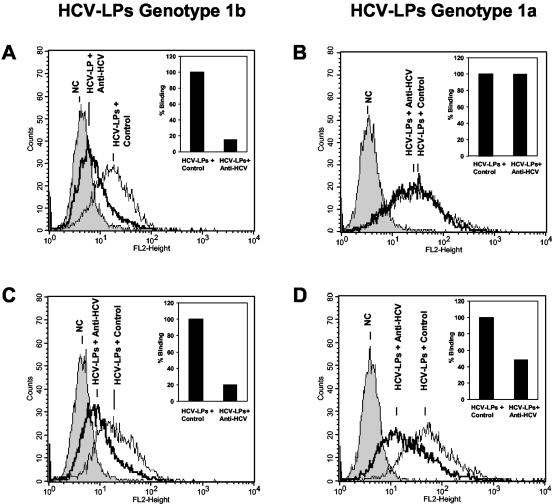
Among the 21 patients with acute symptomatic hepatitis C, 8 resolved acute infection; the other patients developed chronic infection. A total of 13 out of 21 patients with acute symptomatic hepatitis C demonstrated inhibition of cellular HCV-LP binding. Quantitative assessment of antibodies revealed inhibition of binding titers ranging between 1:50 and 1:200. To study whether antibody-mediated inhibition of HCV-LP was strain or isolate specific, we used HCV-LPs derived from genotypes 1a and 1b as ligands for cellular binding (Fig. 4). Two inhibitions of binding profiles were observed. The majority of patients (9 of 13) with acute symptomatic hepatitis C demonstrated strain-specific inhibition of HCV-LP binding (Fig. 4A and B). These patients were all infected with HCV genotype 1b and were characterized by antibody-mediated inhibition of HCV-LP binding restricted to HCV-LPs of genotype 1b. Four out of 13 patients with acute symptomatic hepatitis C (infected with HCV genotypes 1a, 1b, and 3) and inhibition of HCV-LP binding activity exhibited cross-strain inhibition of binding activity (Fig. 4C and D). Patients without inhibition of HCV-LP binding activity were infected with genotypes 1b (four of eight), 3 (three of eight), and 4 (one of eight).
FIG. 4.
Strain-dependent inhibition of cellular HCV-LP binding. For the assessment of antibody-mediated neutralization of binding, HCV-LPs derived from the HCV-J strain (genotype 1b) (A and C) or clone H77c (genotype 1a) (B and D) were incubated in subsaturating concentrations with anti-HCV-positive sera from two patients infected with HCV genotype 1a or 1b or a pool of anti-HCV-negative control sera (NC; dilution, 1:50). HCV-LP-antibody complexes were added to HuH-7 cells. After the removal of nonbound HCV-LP-antibody complexes by washing the cells in PBS-2% BSA, binding of HCV-LPs was detected by flow cytometry using mouse monoclonal anti-E2 or human polyclonal anti-HCV antibodies (insets) as described in the legends to Fig. 1 and 2. (A and B) Genotype-dependent inhibition of HCV-LP binding by serum from a patient infected with HCV genotype 1b. (C and D) Genotype-independent inhibition of cellular HCV-LP binding by serum from a patient infected with genotype 1a. Fluorescence intensity (FL2-Height) and relative cell numbers (Counts) are shown on the x and y axes, respectively. The insets show the corresponding results of serum-induced inhibition of HCV-LP binding detected by human polyclonal anti-HCV antibody (HCV-LP binding in the presence of control serum [Control] = 100%).
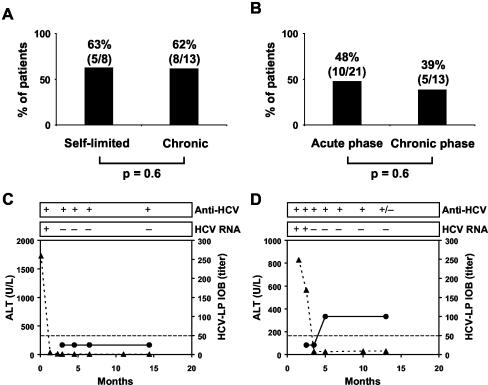
To obtain information on the relationship between the inhibition of binding titers and the outcome of acute or chronic HCV infection, we retrospectively studied serial serum samples from the patients. The presence of antibodies with inhibition of HCV-LP binding activity did not correlate with viral clearance and the outcome of infection (Fig. 5A). Two representative individuals with self-limited HCV infection are shown in Fig. 5. One patient (infected with genotype 3) cleared the viral infection in the absence of measurable antibodies with inhibition of HCV-LP binding activity (Fig. 5C). The other patient (infected with genotype 1b) cleared the infection in parallel with inhibition of binding activity (Fig. 5D). Interestingly, this patient had generated antibodies with cross-strain inhibition of HCV-LP binding. To assess whether the appearance of antibodies with HCV-LP inhibition of binding activity was associated with a defined phase of infection, we compared the presence of inhibitory antibodies in samples in the acute phase of infection (<6 months following the diagnosis of acute infection) with the presence of antibodies in the chronic phase of infection (>6 months following the diagnosis of infection). As shown in Fig. 5B, the frequency of antibodies with HCV-LP inhibition of binding was higher in the acute (48%) than in the chronic (39%) phase of infection, although this difference was not statistically significant (P = 0.6).
FIG. 5.
Inhibition of cellular HCV-LP binding and virus clearance. (A) HCV-LP inhibition of binding activity and outcome of infection in acute hepatitis C. The graph shows the percentages of patients with sera inhibiting cellular HCV-LP binding at a titer of ≥1:50 in relation to the outcome of infection (self-limited infection versus chronic infection). (B) Inhibition of HCV-LP binding and phase of infection. The graph shows the percentages of patients with sera inhibiting cellular HCV-LP binding at a titer of ≥1:50 in relation to the phase of infection (acute versus chronic). (C and D) Serial analyses of antibodies with inhibition of HCV-LP binding activity in two patients with acute self-limited hepatitis C and virus clearance. Alanine aminotransferase (ALT) levels (triangles) are indicated on the left y axis. Titers of antibody-mediated inhibition of HCV-LP binding were determined as described in the legend to Fig. 2 and are indicated on the right y axis (circles). The x axis represents time points after the diagnosis of acute hepatitis C. The presence (+) or absence (−) of HCV RNA and anti-HCV antibodies is shown at the top; +/−, weakly positive. (C) Virus clearance in the absence of antibodies with inhibition of HCV-LP binding activity. (D) Virus clearance in the presence of antibodies with inhibition of HCV-LP binding activity.
Epitope mapping of antibodies inhibiting cellular binding of HCV-LPs.
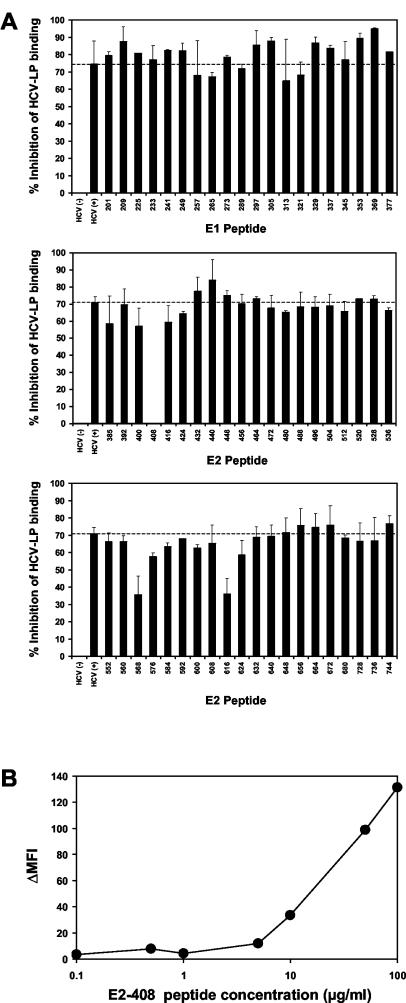
To identify HCV-LP envelope epitopes targeted by human antibodies inhibiting cellular HCV-LP binding, an anti-HCV-positive serum with marked inhibition of binding activity was preincubated with overlapping 15-mer peptides of the HCV envelope glycoproteins (comprising amino acid positions 201 to 758) derived from the HCV-J strain (26) prior to the addition of HCV-LPs. As shown in Fig. 6, three E2 peptides were able to reverse inhibition of binding activity (peptides 408 to 422, 568 to 582, and 616 to 630). Whereas preincubation with E2 peptides 568 to 582 and 616 to 630 resulted in a partial reversion of antibody-mediated inhibition of cellular HCV-LP binding, E2 peptide 408 to 422 (amino acid sequence, SQKIQLVNTNGSWHI) completely reversed inhibition of the binding activity of human anti-HCV antibodies. Reversion of antibody-mediated inhibition of binding was concentration dependent, as shown in Fig. 6B. These data indicate that the viral envelope E2 epitope SQKIQLVNTNGSWHI is targeted by human anti-envelope antibodies, resulting in blocking of HCV-LP binding to the target cell surface.
FIG. 6.
Epitope mapping of antibodies inhibiting cellular binding of HCV-LPs. (A) To identify HCV-LP envelope epitopes targeted by human antibodies inhibiting cellular HCV-LP binding, an anti-HCV-positive serum (HCV+) with marked inhibition of binding activity (Fig. 3) was incubated with overlapping 15-mer peptides of the HCV envelope glycoproteins (comprising amino acid positions 201 to 758) derived from the HCV-J strain (26) or PBS (as a control) for 1 h at RT (peptide concentration, 100 μg/ml; serum dilution, 1:50). HCV-LPs (derived from the homologous isolate as the peptides) were then added to the peptide-antibody complexes and incubated for 1 h at 37°C. The HCV-LPs, antibodies, and peptides were added to HuH-7 cells and incubated for 1 h at 4°C. Following the removal of nonbound HCV-LPs, antibodies, and peptides by washing the cells in PBS, binding of HCV-LPs was detected by flow cytometry as described above. Inhibition of HCV-LP binding (as indicated on the y axis) was calculated as described in the legend to Fig. 2. The error bars indicate standard deviations of results of a representative experiment performed in triplicate. (B) Concentration-dependent reversion of HCV-LP inhibition of binding activity by E2 peptide 408. Anti-HCV-positive serum (dilution, 1:50) was preincubated with E2 peptide 408 (SQKIQLVNTNGSWHI; corresponding to amino acid positions 408 to 422) at the peptide concentrations indicated on the x axis. Cellular HCV-LP binding (corresponding to ΔMFI, indicated on the y axis) in the presence of the antiviral antibody-peptide complex was determined as described in Materials and Methods.
DISCUSSION
In this study, we demonstrate for the first time that cellular binding of HCV-LPs to target cells represents a novel, convenient model system for the functional characterization of antiviral antibodies present in sera from patients with acute or chronic HCV infection. The system is characterized by a fast, simple, and highly standardizable flow cytometry-based method for antibody characterization. Our model system allowed us, by studying a large panel of serial serum samples from HCV-infected patients, to define the roles of antibodies inhibiting cellular binding of the viral envelope to target cells in virus clearance in HCV infection.
A major obstacle in assessing the relevance of the antibody responses in HCV infection is the lack of a convenient in vitro or in vivo neutralization assay for HCV. Various cell lines and primary hepatocytes or biliary epithelial cells have been shown to be susceptible to HCV infection (3, 9, 32, 39, 43, 52). However, variability in infection efficiency and the limited availability of primary cells hamper the use of these assays for the analysis of large serial serum panels. Other models for the study of the HCV-host interaction include binding of unpurified or sucrose gradient-purified virions to human cell lines (1, 49). These systems use partially purified virions but require either the use of ultrasensitive detection methods, such as reverse transcriptase PCR and in situ hybridization, or flow cytometry-based systems using less standardized reagents, such as polyclonal anti-HCV antibodies derived from human serum, for the detection of virus binding.
Binding of individually expressed recombinant glycoprotein E2 to human cell lines has been used as a surrogate model for the binding of HCV to host cells, allowing the study of antibody-mediated neutralization of binding (25, 38). However, it is still unclear whether individually expressed recombinant E2 protein reflects the natural interaction of the viral envelope with target cells. The use of C-terminally truncated E2 protein as a surrogate ligand for virus binding is limited by the fact that the proper conformation of the envelope protein requires the coexpression of both the E1 and E2 proteins (36). The signal sequences of the C termini of the E1 and E2 transmembrane domains are important for membrane anchoring, heterodimerization, and membrane retention (14). The relevance of this finding is reflected by the recently described antigenic differences between C-terminally truncated E2, full-length E1/E2 complexes, and HCV-LPs, pointing to the existence of structural differences with important functional implications (13, 34). In contrast to recombinant C-terminally truncated E2, HCV-LP E2 is present as an E1/E2 heterodimer expressed from a full-length E1/E2 cDNA. Several studies have demonstrated that HCV-LPs contain E2 in a native conformation, which may resemble properly folded E2 in the virion (13, 45, 48). HCV-LPs and virions share distinct features in their cellular binding profiles, suggesting that cellular binding of HCV-LPs represents an appropriate model system for the study of HCV-host cell membrane interaction (44, 48, 50). In contrast to previous model systems for the study of virus-host interaction (1, 49), our model system uses a clearly defined ligand and highly standardized conditions to quantify cellular binding (monoclonal antibodies and flow cytometry). Binding could easily be detected and quantified by mouse monoclonal anti-E2 antibodies, whereas human polyclonal anti-HCV antibody served as a reagent to confirm the specificity of the inhibition of binding.
In this study, we demonstrate for the first time that cellular HCV-LP binding is inhibited by anti-HCV-positive sera from HCV-infected patients. Specific inhibition of HCV-LP binding by antiviral antibodies is demonstrated by the following experimental findings: (i) only sera from HCV-infected patients, but not from uninfected control subjects, inhibited cellular HCV-LP binding in a concentration-dependent manner (inhibitor-specific inhibition of binding [Fig. 2 and 3]); (ii) only purified IgG from HCV-infected patients, but not from uninfected control subjects, inhibited cellular HCV-LP binding in a concentration-dependent manner (antibody-specific inhibition of binding [Fig. 3]); (iv) antibody-mediated inhibition of binding is reverted by preincubation with a defined viral envelope peptide (Fig. 6).
Using antibody-mediated inhibition of HCV-LP binding as a marker for virus neutralization at the level of envelope binding to the host cell membrane, our study provides for the first time a detailed functional characterization of anti-HCV antibodies in acute and chronic HCV infections. The key findings of our study are the following: (i) antibodies inhibiting HCV-LP binding are present in acute or chronic HCV infection; (ii) antibodies that inhibit HCV-LP binding are of low titer (≤1:200); (iii) in the majority of studied individuals, antibody-mediated inhibition of cellular HCV-LP binding is subtype or isolate specific; (iv) the presence of antibodies with inhibition of HCV-LP binding activity may not predict virus clearance.
Using antibody-mediated inhibition of HCV-LP binding as a surrogate marker for virus neutralization, we conclude that virus-neutralizing antibodies are present in acute and chronic HCV infections. Since the presence of such antibodies did not correlate with the outcome of infection, we conclude that neutralizing antibodies presumably do not play a major role in virus clearance. Our data indicate that limited induction of high-titer antibodies with neutralization capacity, as well as isolate-specific restriction of neutralization, most likely contributes to the failure of antibody-mediated virus clearance in HCV infection.
The absence of a high-titer humoral immune response with neutralizing properties in patients with self-limited acute hepatitis C is in line with the concept that a robust and multispecific antiviral cellular immune response is central for sustained virus clearance in acutely infected individuals (22, 23, 30, 46). Several studies with humans have shown that virus clearance is associated with a strong HCV-specific CD4+ T helper and cytotoxic-T-cell response (for a review, see references 23, 30, and 46).
Recently, pseudotyped retroviral particles containing HCV glycoproteins have been suggested as a model system for HCV entry (5, 5a, 24). Whether the entry of retroviral HCV pseudoparticles containing heterologous retroviral proteins represents HCV entry leading to viral infection is not yet known. In a previously published pilot study, Bartosch et al. analyzed sera from four HCV-infected chimpanzees and one HCV-infected human for the presence of antibodies neutralizing pseudoparticle entry (5a). Interestingly, the authors could detect antibodies (titer, >1:320) neutralizing pseudoparticle entry in the single human individual and in two out of four chimpanzees that progressed to chronic HCV infection. Cross-neutralization of pseudoparticle entry was observed in one of the two animals and the single human (5a). In contrast, no antibodies with neutralizing properties were observed in two chimpanzees with self-limited infections (5a). Since only one human individual and four experimentally infected chimpanzees were assessed in this study (5a), it is difficult to compare the results obtained in the HCV pseudoparticle and HCV-LP models at the present time. Interestingly, both systems indicate the presence of cross-neutralizing antibodies in chronic HCV infection. A prospective study assessing serum samples from a large number of HCV-infected humans side by side is needed to compare findings obtained using the two systems. Studies are under way to address this important question.
We cannot exclude the possibility that anti-envelope antibodies with binding-inhibitory properties are present in antigen-antibody immune complexes. In this case, these antibodies are not detected by this assay or by ELISAs based on HCV-LPs (8) or individual envelope proteins (12, 21) or by other neutralization-of-binding assays (25, 38). Virus neutralization can also occur at stages of the viral life cycle following virus binding to the cell membrane (16). These stages include fusion of the viral envelope with the cell membrane, blocking of viral uncoating, and replication and would not be detected by our assay.
Interestingly, antibody-mediated inhibition of HCV-LP binding did not correlate with the immunoreactivity of anti-HCV antibodies against HCV-LPs or recombinant E1/E2 proteins as capture antigens in an ELISA (8). Several sera from patients with chronic HCV infection containing high-titer antibodies against HCV-LPs (>1:128,000) (8) or recombinant E1/E2 proteins (>1:64,000) did not result in inhibition of HCV-LP binding (data not shown). In contrast, several sera from patients with acute self-limited hepatitis C and low-titer anti-HCV-LP or anti-E1/E2 antibodies exhibited marked inhibition of HCV-LP binding (data not shown). The difference between the presence (as measured by HCV-LP [8] or E1/E2 [48, 29] ELISAs) and function (as measured by inhibition of HCV-LP binding [this study]) of anti-envelope antibodies is also reflected by the interaction of HCV-LPs with anti-HCV antibodies from patients infected with different HCV genotypes. Using HCV-LPs as capture antigens in an ELISA format, HCV-LPs of genotype 1b cross-interacted with antibodies of patients infected with HCV genotypes 1a, 2, 3, 4, 5, and 6 (8). In contrast, cross-reactivity between the same HCV subtypes (1a or 1b) was a less frequent event (4 of 13 patients) in antibody-mediated inhibition of HCV-LP binding. Although we cannot exclude the possibility that cross-reactivity observed in studies using the HCV-LP ELISA may be partly due to the interaction of antibodies with partly deenveloped nucleocapsids present in the HCV-LP preparation, our data indicate that HCV elicits abundant antibodies directed against the HCV structural proteins (as demonstrated in HCV-LP ELISA) (8) that have little or no ability to block envelope docking to the target cell (this study).
Several mechanisms may explain this finding. First, the majority of anti-HCV antibodies are targeted against epitopes of the HCV structural proteins that are not required for virus neutralization. Envelope epitopes presented on the surfaces of HCV-LPs have been mapped recently (13, 45). Our data are consistent with the hypothesis that many epitopes exposed on the HCV-LP surface and recognized by human antiviral antibodies are most likely not target epitopes for virus neutralization. Epitopes mediating viral envelope binding and targeted by virus-neutralizing antibodies are still poorly defined. So far, two epitopes in envelope glycoproteins E1 and E2 (amino acids 197 to 207 and 640 to 653) have been proposed to play a potential role in mediating cellular HCV-LP binding (44). In chimpanzees, the E2 HVR-1 has been inferred to represent a potential B-cell epitope associated with virus neutralization (18). In contrast, antibodies directed against several epitopes of the HCV core protein were not able to block HCV-LP binding to target cells, suggesting that the nucleocapsid is not involved in mediating HCV-LP cellular binding (M. Triyatni and T. J. Liang, unpublished observations). Using a panel of overlapping envelope peptides covering the entire HCV envelope, we identified an epitope of the N-terminal E2 region (SQKIQLVNTNGSWHI; amino acid positions 408 to 422) as the target of human antiviral antibodies inhibiting cellular particle binding. This epitope overlaps with the E2 hypervariable region and corroborates the importance of this region for the binding of the viral envelope to the cell surface. Interestingly, the same region has been shown to be involved in E2-CD81 interaction (34, 35) and in mediating cellular binding and entry of HCV pseudoparticles (5, 24).
Second, the presence of HCV as a highly variable pool of rapidly mutating viral quasispecies in an individual patient may contribute to virus escape from antibody-mediated virus neutralization. A previous study elegantly demonstrated that an antiserum against the E2 hypervariable region from HCV recovered from a single patient induced protection against homologous HCV infection in chimpanzees but not against the emergence of neutralization escape mutants that were found to be already present in the complex viral quasispecies of the inoculum (18). This study, performed with serum samples from a single patient, suggested that neutralizing antibodies can be induced but isolate-specific virus neutralization and the rapid emergence of viral escape mutants may represent a major limitation for antibody-mediated virus neutralization in HCV infection. Our data identifying antibodies with strain-specific neutralization of viral envelope binding in a large serum panel of 21 well-characterized patients indicates that the proposed mechanism may indeed contribute to the failure of antibody-mediated virus neutralization in patients with HCV infection.
Third, the virus has developed highly specific escape strategies to evade antibody-mediated neutralization. These strategies may include conformational masking of receptor binding sites following envelope-antibody interaction (27) or alteration in envelope N-glycosylation motifs (11, 47), as described recently for human immunodeficiency virus. The HCV-LP-based model system described in this study may be helpful in defining similar mechanisms for HCV.
In summary, this model system may be useful in providing a detailed map of epitopes targeted by human antiviral antibodies on the level of envelope-cell surface receptor interaction. Furthermore, the HCV-LP-based model system may allow the elucidation of viral escape mechanisms for the evasion of antibody-mediated neutralization. Understanding these mechanisms may ultimately provide important clues for the development of new antivirals inhibiting virus-cell surface interaction, as well as define strategies for the efficient induction of virus cross-neutralizing antibodies for vaccine development.
Acknowledgments
We thank F. von Weizsäcker and P. Hafkemeyer (Department of Medicine II, University of Freiburg) for providing HuH-7 and HepG2 cell lines, C. Grüllich and J. Finke (Department of Medicine I, University of Freiburg) for support in flow cytometry analysis, M. Follo (Core Facility, Department of Medicine I, University of Freiburg) for performing confocal laser scanning microscopy, M. Nauck (Division of Clinical Chemistry, Department of Medicine, University of Freiburg) for determination of serum IgG concentrations, and J. Baumert (National Research Centre for Environment and Health [GSF], Institute of Epidemiology, Neuherberg, Germany) for support in statistical analysis.
This work was supported by grants from the European Union, Brussels, Belgium (QLK-2-1999-00356 and QLK-2-2002-01329); the Wilhelm Sander Foundation, Munich, Germany (99041.1); the Deutsche Forschungsgemeinschaft (Ba1417/6-1 and 7-1); the Alexander von Humboldt Foundation; and the Flemish Government, Brussels, Belgium (IWT grant 000405).
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 4.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397-1407. [DOI] [PubMed] [Google Scholar]
- 8.Baumert, T. F., S. Wellnitz, S. Aono, J. Satoi, D. Herion, J. Tilman Gerlach, G. R. Pape, J. Y. Lau, J. H. Hoofnagle, H. E. Blum, and T. J. Liang. 2000. Antibodies against hepatitis C virus-like particles and viral clearance in acute and chronic hepatitis C. Hepatology 32:610-617. [DOI] [PubMed] [Google Scholar]
- 9.Bichr, S., R. Rende-Fournier, G. Vona, A. M. Yamamoto, E. Depla, G. Maertens, and C. Brechot. 2002. Detection of neutralizing antibodies to hepatitis C virus using a biliary cell infection model. J. Gen. Virol. 83:1673-1678. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, I. A. Wilson, R. Pantophlet, X. Wei, J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 12.Chen, M., M. Sallberg, A. Sonnerborg, O. Weiland, L. Mattsson, L. Jin, A. Birkett, D. Peterson, and D. R. Milich. 1999. Limited humoral immunity in hepatitis C virus infection. Gastroenterology 116:135-143. [DOI] [PubMed] [Google Scholar]
- 13.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocquerel, L., A. Op de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 16.Dimmock, N. J. 1993. Neutralization of animal viruses. Curr. Top. Microbiol. Immunol. 183:1-149. [DOI] [PubMed] [Google Scholar]
- 17.Ezelle, H. J., D. Markovic, and G. N. Barber. 2002. Generation of hepatitis C virus-like particles by use of a recombinant vesicular stomatitis virus vector. J. Virol. 76:12325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feray, C., M. Gigou, D. Samuel, B. Ducot, P. Maisonneuve, M. Reynes, A. Bismuth, and H. Bismuth. 1998. Incidence of hepatitis C in patients receiving different preparations of hepatitis B immunoglobulins after liver transplantation. Ann. Intern. Med. 128:810-816. [DOI] [PubMed] [Google Scholar]
- 20.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournillier-Jacob, A., F. Lunel, A. Cahour, P. Cresta, L. Frangeul, M. Perrin, M. Girard, and C. Wychowski. 1996. Antibody responses to hepatitis C envelope proteins in patients with acute or chronic hepatitis C. J. Med. Virol. 50:159-167. [DOI] [PubMed] [Google Scholar]
- 22.Gerlach, J. T., H. M. Diepolder, M. C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii, K., D. Rosa, Y. Watanabe, T. Katayama, H. Harada, C. Wyatt, K. Kiyosawa, H. Aizaki, Y. Matsuura, M. Houghton, S. Abrignani, and T. Miyamura. 1998. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology 28:1117-1120. [DOI] [PubMed] [Google Scholar]
- 26.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 28.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 29.Lechmann, M., K. Murata, J. Satoi, J. Vergalla, T. F. Baumert, and T. J. Liang. 2001. Hepatitis C virus-like particles induce virus-specific humoral and cellular immune responses in mice. Hepatology 34:417-423. [DOI] [PubMed] [Google Scholar]
- 30.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1161. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 32.Mizutani, T., N. Kato, S. Saito, M. Ikeda, K. Sugiyama, and K. Shimotohno. 1996. Characterization of hepatitis C virus replication in cloned cells from a human T-cell leukemia virus type 1-infected cell line, MT-2. J. Virol. 70:7219-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murata, K., M. Lechmann, M. Qiao, T. Gunji, H. J. Alter, and T. J. Liang. 2003. Immunization with hepatitis C virus-like particles protects mice from recombinant hepatitis C virus-vaccinia infection. Proc. Natl. Acad. Sci. USA 100:6753-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 35.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 36.Patel, J., A. H. Patel, and J. McLauchlan. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58-68. [DOI] [PubMed] [Google Scholar]
- 37.Qiao, M., K. Murata, A. R. Davis, S. H. Jeong, and T. J. Liang. 2003. Hepatitis C virus-like particles combined with novel adjuvant systems enhance virus-specific immune responses. Hepatology 37:52-59. [DOI] [PubMed] [Google Scholar]
- 38.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. Weiner, J. Y. N. Lau, Q.-L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rumin, S., P. Berthillon, E. Tanaka, K. Kiyosawa, M. A. Trabaud, T. Bizollon, C. Gouillat, P. Gripon, C. Guguen-Guillouzo, G. Inchauspe, and C. Trepo. 1999. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. J. Gen. Virol. 80:3007-3018. [DOI] [PubMed] [Google Scholar]
- 40.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiff, E. R., M. de Medina, and R. S. Kahn. 1999. New perspectives in the diagnosis of hepatitis C. Semin. Liver Dis. 19:3-15. [PubMed] [Google Scholar]
- 42.Seipp, S., H. M. Mueller, E. Pfaff, W. Stremmel, L. Theilmann, and T. Goeser. 1997. Establishment of persistent hepatitis C virus infection and replication in vitro. J. Gen. Virol. 78:2467-2476. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, Y., A. Iwamoto, M. Hijikata, R. H. Purcell, and H. Yoshikura. 1992. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc. Natl. Acad. Sci. USA 89:5477-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Triyatni, M., B. Saunier, P. Maruvada, A. R. Davis, L. Ulianich, T. Heller, A. Patel, L. D. Kohn, and T. J. Liang. 2002. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J. Virol. 76:9335-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 46.Ward, S., G. Lauer, R. Isba, B. Walker, and P. Klenerman. 2002. Cellular immune responses against hepatitis C virus: the evidence base 2002. Clin. Exp. Immunol. 128:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 48.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang, J., S. Wunschmann, S. L. George, D. Klinzman, W. N. Schmidt, D. R. LaBrecque, and J. T. Stapleton. 2002. Recombinant hepatitis C virus-like particles expressed by baculovirus: utility in cell-binding and antibody detection assays. J. Med. Virol. 68:537-543. [DOI] [PubMed] [Google Scholar]
- 51.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, X., Z. Y. Tang, B. Klumpp, G. Wolff-Vorbeck, H. Barth, S. Levy, F. von Weizsacker, H. E. Blum, and T. F. Baumert. 2002. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J. Clin. Investig. 109:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]