Abstract
In this report, we present evidence that R5 human immunodeficiency virus type 1 (HIV-1) replicates more efficiently in primary CD4+ T cells than X4 HIV-1. By comparing CD3/CD28-costimulated CD4+ T-cell cultures infected by several X4 and R5 HIV-1 strains, we determined that R5-infected CD4+ T cells produce more virus over time than X4-infected CD4+ T cells. In the first comparison, we found that more cells were infected by the X4-tropic strain LAI than by the R5-tropic strain JR-CSF and yet that higher levels of viral production were detected in the R5-infected cultures. The differential viral production was partially due to the severe cytopathic effects of the X4 virus. We also compared cultures infected with the isogenic HIV-1 strains NL4-3 (X4) and 49.5 (R5). We found that fewer cells were infected by the R5 strain, and yet similar levels of viral production were detected in both infected cultures. Cell death played less of a role in the differential viral production of these strains, as the cell viability remained comparable in both X4- and R5-infected cultures over time. The final comparison involved the primary R5-tropic isolate KP1 and the primary dual-tropic isolate KP2. Although both strains infected similar numbers of cells and induced comparable levels of cytopathicity, viral production was considerably higher in the R5-infected culture. In summary, these data demonstrate that R5 HIV-1 has an increased capacity to replicate in costimulated CD4+ T cells compared to X4 HIV-1.
Human immunodeficiency virus type 1 (HIV-1) infects cells by binding to the CD4 receptor and to one of several coreceptors expressed on the surface of target cells (2, 13, 15, 16, 18, 26). The chemokine receptors CCR5 and CXCR4 serve as the major coreceptors for HIV-1, although several other chemokine receptors have been linked to minor HIV-1 coreceptor usage (2, 8, 15, 18, 32, 52). Characteristically, non-syncytium-inducing (NSI) isolates utilize CCR5 as a coreceptor and are referred to as R5 strains (2, 14-16). R5 strains often represent the dominant viral population detected during the early stages of clinical HIV-1 infection (9, 14, 41, 43, 46, 53). In contrast, syncytium-inducing (SI) isolates utilize CXCR4 as a coreceptor and are referred to as X4 strains (18, 20, 24, 25, 28, 30, 40, 45, 48). X4 strains are typically detected in the later stages of infection and are associated with rapid CD4+ T-cell loss (11, 14, 24, 25, 46, 48, 49). Despite the link between X4 emergence and disease progression, approximately half of all individuals with AIDS continue to harbor predominantly R5 viruses, suggesting that CXCR4 coreceptor usage alone is not responsible for disease progression (9, 43, 46, 53).
Several mechanisms have been proposed to explain the R5 dominance of early HIV-1 infection. There is evidence that R5 strains may be transmitted at an increased frequency compared to X4 strains. For example, individuals that carry a 32-bp deletion mutation (Δ32) in the CCR5 gene are highly resistant to HIV-1 infection (10, 33, 38, 44). Although these individuals are susceptible to X4 infection, very few cases have been reported, suggesting that X4 transmission occurs with lower frequency (3, 5, 23, 29, 37, 47, 50). Another mechanism leading to R5 dominance in vivo may involve preferential spread of R5 strains. It has been reported that dendritic cells preferentially transport R5 rather than X4 virions, which may lead to selective spread of R5 virus to lymph nodes (19, 41). Also, intestinal epithelial cells have been shown to selectively transport R5 virions to the lamina propria, which may lead to the preferential spread of R5 infection to activated CD4+ T cells (34). Recent evidence suggests that the immune response may also play a role in R5 selection in vivo. Harouse et al. report that both X4 and R5 SHIV replication can be detected in macaques early after coinfection; however, X4 replication is no longer detected after CD8+ T-cell-mediated antiviral immune responses are elicited (22). Furthermore, X4 replication reemerges in coinfected animals following depletion of the antiviral immune response by in vivo infusion with anti-CD8 antibodies. Collectively, these data suggest that X4-infected cells may be more susceptible to immune-mediated killing than R5-infected cells (22).
We propose that an additional mechanism may be involved in R5 dominance. In this report, we present evidence that R5 HIV-1 replicates more efficiently in CD3/CD28-costimulated CD4+ T cells than X4 HIV-1. We tested several viral strains in this system, including the molecular clones LAI (X4) and JR-CSF (R5), the isogenic viral pair NL4-3 (X4) and 49.5 (R5), and two primary isolates that were recovered from the same patient at different time points, KP1 (R5) and KP2 (X4/R5). We found that the R5 HIV-1 JR-CSF strain infected a smaller percentage of costimulated CD4+ T cells than the X4 HIV-1 strain LAI and yet produced more progeny virus over time than the X4-infected culture. It is likely that this result is partially due to the fact that the cell viability of X4-infected cultures decreased rapidly, whereas cell viability remained relatively high in R5-infected cultures over time. Analysis of cultures infected with the isogenic viral pair provided further confirmation that R5-infected CD4+ T cells produce more progeny virus than X4-infected CD4+ T cells. Despite the fact that fewer costimulated CD4+ T cells were infected by the R5-tropic 49.5 strain than by its isogenic X4 counterpart NL4-3, similar amounts of viral production were detected in both infected cultures. Cell death seemed to play less of a role in the differential levels of viral release between these two strains, as the viability characteristics of both infected cultures remained comparable over time. Interestingly, similar results were obtained with the primary R5 isolate KP1 and the primary dual-tropic isolate KP2. Although both KP1 and KP2 infected equivalent numbers of costimulated CD4+ T cells and induced comparable amounts of cell death, viral production was considerably higher in cultures infected by the R5-tropic KP1 strain. In summary, we report that R5 HIV-1 has an increased capacity to replicate in CD4+ T cells compared to X4 HIV-1 and propose that this increased fitness may allow R5 viruses to out-compete X4 viruses in the early stages of HIV-1 infection.
MATERIALS AND METHODS
Antibodies.
The following fluorescently labeled monoclonal antibodies were purchased from Becton Dickinson (San Jose, Calif.) and used for flow cytometric analysis: CD3-APC (clone SK7), CD69-FITC (clone FN50), CD38-PE (clone HIT2), CXCR4-APC (clone 12G5), CD4-PerCP (clone SK3), CCR5-APC (clone 2D7), and IFN-γ-PE (clone 25723.11). Monoclonal p24-FITC antibody (clone KC57) was purchased from Coulter Clone (Miami, Fla.), and annexin V-PE was purchased from Caltag Laboratories (Burlingame, Calif.). The following antibodies were used for CD3/CD28 stimulation: CD28 (Leu-28) clone L293 (Becton Dickinson) and CD3 clone SPV-T3b (Zymed, South San Francisco, Calif.).
CD4+ T-cell isolation.
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque (Amersham BioSciences, Uppsala, Sweden) gradient centrifugation of leukopacks (Stanford Blood Bank, Stanford, Calif.) obtained by apheresis of healthy donors. CD4+ T cells were purified by negative selection using Microbeads (Miltenyi Biotec, Auburn, Calif.). Cell purity was determined by staining cells with fluorescently conjugated antibodies directed against CD4, CD3, CD8, and CD14. Cell populations were found to be >95% CD3+ CD4+.
CD4+ T-cell stimulation.
CD4+ T cells were activated by phytohemagglutinin (PHA) stimulation or by CD3/CD28 costimulation. For PHA stimulation, cells were cultured at a density of 2 × 106 cells/ml with 1 μg of PHA (Sigma, St. Louis, Mo.)/ml for 24, 48, or 72 h. Cells were then washed to remove PHA and cultured for 48 h in RPMI 1640 medium (MediaTech, Herndon, Va.) supplemented with 15% fetal bovine serum (FBS) (Gemini, Woodland, Calif.) and 50 U of interleukin-2 (IL-2) (AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH])/ml. For CD3/CD28 costimulation, tissue culture plates were precoated with CD3 antibody. Briefly, wells were washed with 1× phosphate-buffered saline (PBS) and then coated with a 50 μg/ml stock solution of CD3 antibody. Excess liquid was removed, and plates were incubated at 37°C until dry. Cells were then cultured on coated plates at a concentration of 2 × 106 cells/ml in the presence of 1 μg of soluble CD28 antibody (Becton Dickinson)/ml for 24, 48, or 72 h. Cells were removed from the CD3 coated plates, washed to remove soluble CD28, and then cultured in RPMI 1640 medium supplemented with 15% FBS and 50 U of IL-2/ml.
Detection of cell surface protein expression by flow cytometry.
To detect cell surface protein expression, 5 × 105 CD4+ T cells were incubated with appropriate concentrations of fluorescently conjugated monoclonal antibodies diluted in 1× phosphate-buffered saline containing 1% bovine serum albumin (Sigma). Cells were incubated with antibodies for 20 min at 4°C, washed twice, and then fixed in 1% paraformaldehyde. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using CellQuest software (Becton Dickinson).
Measurement of cellular proliferation and IFN-γ expression.
Stimulated CD4+ T cells were incubated with 2 μM CFSE (5- and 6-carboxyfluorescein diacetate, succinimidyl ester) (Molecular Probes, Eugene, Oreg.) for 10 min at 37°C and then washed twice. Stained cells were incubated for 72 h at 37°C. Brefeldin A (Sigma) was added at a final concentration of 10 μg/ml for the last 6 h of culturing. Cells were then washed, fixed in 4% PFA, permeabilized with 1% fluorescence activated cell sorter (FACS) Perm solution (Becton Dickinson), and stained with monoclonal antibodies against CD3, CD4, and gamma interferon (IFN-γ). After antibody staining, cells were washed twice and then stored in 1% paraformaldehyde until acquisition on a FACSCalibur instrument. Subsequent analysis was performed using FlowJo software (Tree Star, Inc).
Virus preparation.
The HIV-1 strains LAI, JR-CSF, NL4-3, and 49.5 were prepared by introducing proviral constructs into 293T cells (American Type Culture Collection, Manassas, Va.) by CaPO4 transfection. The following proviral plasmids were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pLAI.2 (39) from Keith Peden, courtesy of the Medical Research Council AIDS Directed Programme; pYK-JR-CSF (6, 21, 27) from Irvin S. Y. Chen and Yoshio Koyanagi; pNL4-3 (1) from Malcom Martin; and p49.5 (51) from Bruce Chesebro. After the media were changed approximately 16 h after transfection, the virus-containing supernatants were harvested 72 h posttransfection. Viral stocks were centrifuged at 1,000 × g for 10 min to remove cell debris and then passed through a 45-μm-pore-size filter. The infectious titer of each viral preparation was determined by 50% tissue culture infective dose assay. Briefly, PHA-stimulated PBMCs from multiple donors were pooled and infected with serially diluted virus in quadruplicate wells. Cell supernatants were collected 5 days postinfection, and HIV p24 antigen was quantitated by p24 enzyme-linked immunosorbent assay (ELISA). Infections were scored positive for replication when p24 levels were higher than 50 pg/ml. The 50% tissue culture infective dose value represents the virus dilution at which 50% of wells scored positive for infection.
Coreceptor phenotyping assay.
GHOST indicator cells were used to determine coreceptor usage of each viral strain. The GHOST-X4 and GHOST-Hi5 cell lines (35) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Vineet N. Kewalramani and Dan R. Littman. These cell lines were originally derived from human osteosarcoma (HOS) cells. The GHOST-X4 cell line was transduced with a retroviral vector that confers high-level CXCR4 expression, and the GHOST-Hi5 cell line was transduced with a retroviral vector that confers high-level CCR5 expression. The GHOST-Hi5 cells also express low levels of CXCR4 due to endogenous expression in the parental HOS cells. Each cell line was also transduced with a CD4-expressing retroviral vector and a construct that drives expression of green fluorescent protein (GFP) under the control of the HIV long-terminal-repeat (LTR) promoter.
GHOST-X4 and GHOST-Hi5 cells were seeded on 12-well plates at a density of 5 × 105 cells per well. Cells were cultured at 37°C overnight prior to infection with HIV-1 strains. Cells were infected with each viral strain at a multiplicity of infection (MOI) of 0.01 in the presence of 20 μg of Polybrene/ml to enhance infection efficiency. Each infection was performed in a total volume of 300 μl at 37°C for 4 h. After incubation, virus was removed and 1 ml of fresh culture medium was added to each well. Cells were then incubated for an additional 48 h prior to harvest. GFP expression was analyzed by FACS analysis. Samples were acquired on a FACSCalibur instrument, and the resulting data were analyzed using Cell Quest software.
HIV infection.
CD4+ T cells were activated by CD3/CD28 costimulation for 72 h prior to infection. Cells were then washed and incubated with virus at an MOI of 0.01 (for strains LAI, JR-CSF, NL4-3, and 49.5) or 0.001 (for strains KP1 and KP2) for 4 h at 37°C. After infection, cells were washed three times to remove any unbound virions and then cultured in RPMI 1640 medium supplemented with 15% FBS and 50 U of IL-2/ml.
Quantification of viral replication.
Viral replication was assessed by measuring the amount of soluble HIV p24 antigen in culture supernatants. Aliquots (200 μl) of supernatant were removed from infected cell cultures at 3, 5, 7, and 10 days postinfection. Supernatants were stored at −80°C until completion of the experiment. Quantification of p24 was determined using an ELISA (PerkinElmer Life Science, Inc., Boston, Mass.) according to the manufacturer's protocol.
Intracellular p24 staining.
The percentage of infected cells was determined by intracellular staining for the viral p24 antigen. CD4+ T cells (5 × 105) were removed from infected cultures at 3, 5, 7, and 10 days postinfection. Cells were washed, fixed in 4% paraformaldehyde, permeabilized with 1% FACS Perm solution (Becton Dickinson), and then incubated with a fluorescently conjugated monoclonal p24 antibody for 30 min at 4°C. Cells were then washed twice and suspended in 1% paraformaldehyde. Samples were acquired by use of a FACSCalibur instrument, and the resulting data were analyzed using FlowJo software.
RESULTS
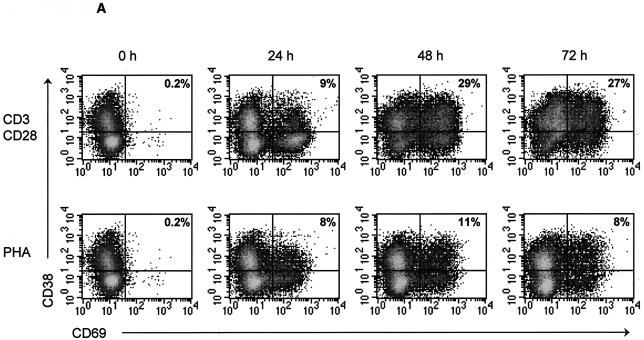
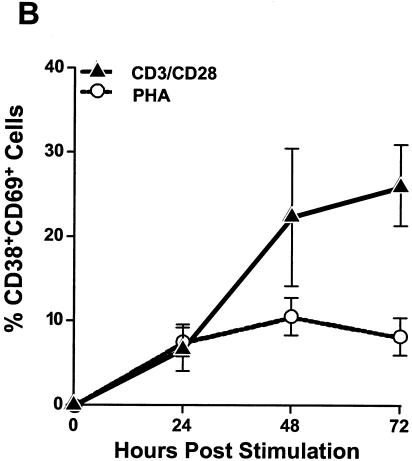
Costimulation with plate-bound CD3 and soluble CD28 antibodies induces high levels of activation in CD4+ T-cell cultures. To study X4 and R5 HIV-1 replication in primary CD4+ T cells, we first optimized in vitro stimulation conditions for these cells. CD4+ T cells were isolated from the PBMC of three donors by magnetic separation. The isolated cells were found to be >95% CD3+ CD4+ (data not shown). In vitro stimulation conditions were optimized by treating cells with increasing concentrations of plate-bound CD3 antibody and increasing concentrations of soluble CD28 antibody for various lengths of time (data not shown). Cellular activation was assessed by monitoring the expression levels of the activation markers CD38 and CD69 by flow cytometry at 0, 24, 48, and 72 h poststimulation. The highest level of activation was achieved following treatment with 50 μg of plate-bound CD3 antibody/ml and 1 μg of soluble CD28 antibody/ml (Fig. 1). CD38 and CD69 expression increased rapidly in these cultures, and by 48 h, an average of 29% of cells coexpressed both activation markers (Fig. 1). Activation marker expression was twofold lower in PHA-stimulated cultures, indicating that CD3/CD28 costimulation is a better method for activating purified CD4+ T cells in vitro (Fig. 1).
FIG. 1.
CD3/CD28 costimulation induces high levels of activation marker expression in CD4+ T cells. CD4+ T cells were isolated from PBMC of three donors and then stimulated with CD3/CD28 antibodies or PHA for 0, 24, 48, or 72 h. At each time point, cells were stained with fluorescently conjugated monoclonal antibodies directed against CD4 and the activation markers CD38 and CD69. Samples were acquired with a FACSCalibur system (Becton Dickinson), and the resulting data were analyzed using CellQuest software (Becton Dickinson). (A) Representative flow cytometric results from one donor. The number in the upper-right hand corner of each FACS plot represents the percentage of CD38+ CD69+-coexpressing cells. (B) The percentage of CD38+ CD69+-coexpressing cells detected in each donor was averaged and plotted as a line graph, with error bars representing the variation among donors.
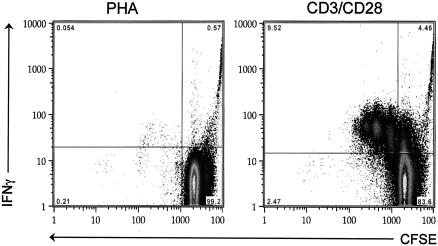
Cellular activation was also assessed by a second, independent assay that measures cellular proliferation and IFN-γ expression. To measure activation-induced proliferation, stimulated CD4+ T cells were stained with the green fluorescent dye CFSE. CFSE is a cytoplasmic dye that is equally divided among daughter cells during cell division. Consequently, the fluorescence of each daughter cell is half as bright as that of the parental cell, allowing for the reduction in fluorescence to be used as a marker of cellular proliferation. After CFSE staining, cells were cultured for 72 h and then fixed, permeabilized, and stained with fluorescently labeled antibodies directed against IFN-γ, CD3, and CD4. A large population of proliferating, IFN-γ-expressing CD4+ T cells was detected in CD3/CD28-costimulated cultures (Fig. 2). This population of cells was not detected in PHA-stimulated cultures, confirming that CD3/CD28 costimulation induces higher levels of activation and proliferation in purified CD4+ T-cell cultures than PHA treatment (Fig. 2).
FIG. 2.
CD3/CD28 costimulation induces high levels of cellular proliferation and IFN-γ expression in CD4+ T-cell cultures. Stimulated CD4+ T cells were stained with CFSE and then cultured for 72 h. Cells were then fixed, permeabilized, and stained with fluorescently labeled monoclonal antibodies directed against CD3, CD4, and IFN-γ. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using FlowJo software (Tree Star, Inc.). The number in the each corner of each FACS plot represents the percentage of cells in that quadrant.
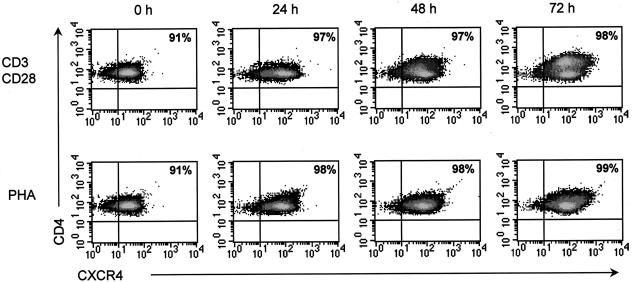
CXCR4 and CCR5, the HIV-1 coreceptors, are expressed on CD3/CD28-costimulated CD4+ T cells.
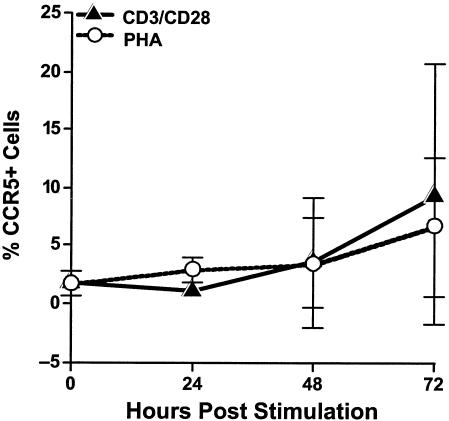
To assess the potential susceptibility of costimulated CD4+ T cells to HIV-1 infection, we examined coreceptor expression on these cells by flow cytometry. Cells from three independent donors were stimulated with either CD3/CD28 antibodies or PHA. At 0, 24, 48, and 72 h poststimulation, cells were stained with fluorescently conjugated monoclonal antibodies directed against CD4, CXCR4, and CCR5. At each time point, nearly all CD4+ T cells from each donor expressed high levels of CXCR4, regardless of the stimulation method (Fig. 3). In contrast, very low levels of CCR5, the R5 coreceptor, were detected in PHA- and CD3/CD28-stimulated CD4+ T-cell culture, and a high degree of variation among donors was detected (Fig. 4). CCR5 expression levels in both PHA- and CD3/CD28-costimulated cultures from each donor were found to increase over time, with expression levels peaking at 72 h poststimulation (Fig. 4).
FIG. 3.
CD3/CD28-costimulated CD4+ T cells express high levels of CXCR4. CD4+ T cells were isolated from PBMC of three donors and then stimulated with CD3/CD28 antibodies or PHA for 0, 24, 48, or 72 h. At each time point, cells were stained with fluorescently conjugated monoclonal antibodies directed against CD4 and CXCR4. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using CellQuest software (Becton Dickinson). Representative results from one donor are shown. The number in the upper-right hand corner of each FACS plot represents the percentage of cells that express both CD4 and CXCR4.
FIG. 4.
CD3/CD28-costimulated CD4+ T cells express low levels of CCR5. CD4+ T cells were isolated from PBMC of 3 donors and then stimulated with CD3/CD28 antibodies or PHA for 0, 24, 48, or 72 h. At each time point, cells were stained with fluorescently conjugated monoclonal antibodies directed against CD4 and CCR5. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using CellQuest software (Becton Dickinson). The average of the CCR5 expression levels of all three donors was plotted as a line graph, with error bars representing the standard deviations among donors.
The coreceptor usage of each HIV-1 strain was determined by GHOST cell assay.
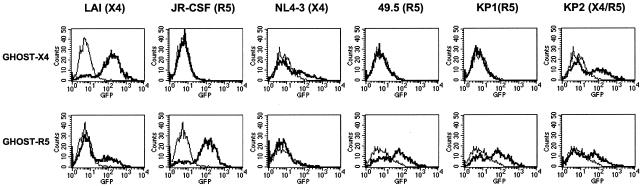
Prior to assessing the susceptibility of costimulated CD4+ T cells to X4 and R5 strains of HIV-1, we first used GHOST-X4 and GHOST-Hi5 indicator cells to examine coreceptor usage of each strain. These cell lines were originally derived from HOS cells and express low levels of endogenous CXCR4. In addition, the GHOST-X4 cells were transduced with retroviral vectors carrying the human genes CD4 and CXCR4 and therefore express high levels of each receptor. The GHOST-Hi5 cells were transduced with vectors carrying the human genes CD4 and CCR5 and express high levels of each receptor as well as low levels of endogenous CXCR4. Each of these cell lines was also stably transfected with a construct carrying the GFP gene under the control of the HIV-1 LTR promoter. This permits HIV infection of these cells to be detected by flow cytometric analysis of GFP expression.
We infected the GHOST-X4 and GHOST-Hi5 cells with the following HIV-1 strains: LAI, JR-CSF, NL4-3, 49.5, KP1, and KP2. GFP expression was detected in GHOST-X4 cells infected with LAI, NL4-3, and KP2, confirming X4 coreceptor usage of these viral strains (Fig. 5). JR-CSF, 49.5, and KP1 did not induce GFP expression in these cells, indicating that these strains do not utilize the X4 coreceptor. High levels of GFP expression were detected in GHOST-Hi5 cells infected with strains JR-CSF, 49.5, KP1, and KP2, indicating R5 coreceptor usage of these strains (Fig. 5). The X4-tropic strain NL4-3 did not infect the GHOST-Hi5 cells; however, the X4-tropic strain LAI did infect a small population of these cells. This low-level infection was likely due to utilization of the endogenous CXCR4 expressed on the GHOST-Hi5 cells. The ability of KP2 to infect both GHOST-X4 and GHOST-Hi5 cells at equivalent levels indicates that this primary isolate has a dual-tropic phenotype.
FIG. 5.
Coreceptor usage of each HIV-1 strain was determined by GHOST cell assays. The GHOST-X4 and GHOST-Hi5 cells were infected with virus for 48 h prior to measurement of GFP expression by flow cytometry. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using CellQuest software (Becton Dickinson). The top row of FACS plots represents GHOST-X4 infections, and the bottom row represents GHOST-Hi5 infections. The thin-lined peak in each plot represents the background fluorescence of uninfected cells, and the bold-lined peak represents the GFP fluorescence detected in cells infected with the indicated HIV-1 strain. The HIV-1 strains LAI and NL4-3 utilize the X4 coreceptor, the strains JR-CSF, 49.5, and KP1 utilize the R5 coreceptor, and the KP2 strain utilizes both the X4 and R5 coreceptors.
R5-infected CD4+ T cells produce more progeny virus over time than X4-infected CD4+ T cells.
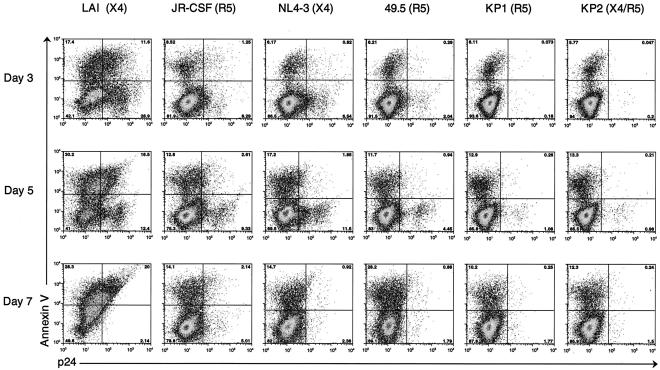
CD4+ T cells were isolated from PBMC and then stimulated for 72 h with CD3/CD28 antibodies. Costimulated cultures were infected with several strains of X4 and R5 HIV-1. Strains included the HIV-1 molecular clones; LAI (X4) and JR-CSF (R5), an isogenic viral pair, which differ only in the V1-V3 loop of gp120; NL4-3 (X4) and 49.5 (R5), two primary viral isolates recovered from the same patient at different stages of clinical infection; and KP1 (R5, early infection) and KP2 (X4/R5, late infection). Cells were infected with strains LAI, JR-CSF, NL4-3, and 49.5 at an MOI of 0.01 and with strains KP1 and KP2 at an MOI of 0.001. At several time points postinfection, the percentage of apoptotic cells was determined by surface staining with fluorescently labeled annexin V, and the percentage of infected cells was determined by intracellular p24 staining with a fluorescently conjugated antibody directed against HIV-1 p24 antigen (Fig. 6 and 7). The FACS plots from the flow cytometric analysis of the annexin V and intracellular p24 staining are shown in Fig. 6. Percentages of infected cells detected in each culture over time are summarized graphically in Fig. 7. In addition, the amount of viral production released by each infected culture was determined by p24 ELISA by measuring the amount of HIV-1 p24 antigen in the culture supernatants. Results of these assays are shown in Fig. 7, with error bars representing the standard deviations among triplicate infections. Also, the cell viability of each infected culture over time was monitored using the trypan blue exclusion assay. Results of these assays are depicted in Fig. 7.
FIG. 6.
The percentage of costimulated CD4+ T cells infected by each viral strain was determined by intracellular p24 staining. CD3/CD28-costimulated CD4+ cells were infected with LAI, JR-CSF, NL4-3, and 49.5 at an MOI of 0.01 and with KP1 and KP2 at an MOI of 0.001. Apoptotic cells were detected by surface staining with fluorescently conjugated annexin V, and infected cells were detected by intracellular staining for HIV-1 p24 antigen. Samples were acquired on a FACSCalibur instrument (Becton Dickinson), and the resulting data were analyzed using FlowJo software (Tree Star, Inc.). Results from days 3, 5, and 7 postinfection are shown. The number in each corner of each FACS plot represents the percentage of cells in that quadrant.
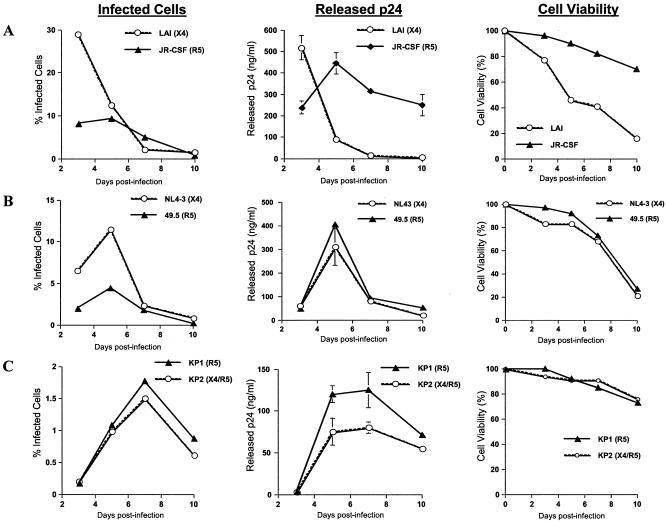
FIG. 7.
R5-infected CD4+ T cells produce more progeny virus over time than X4-infected CD4+ T cells. CD4+ T cells were isolated from PBMC and then stimulated for 72 h with CD3/CD28 antibodies prior to HIV-1 infection. At 3, 5, 7, and 10 days postinfection, the percentage of infected cells in each culture was determined by intracellular p24 staining and flow cytometry, the amount of virus released from each culture was measured by p24 ELISA, and the cell viability of each culture was determined by trypan blue exclusion assay. (A) Comparison of percentages of infected cells, viral release characteristics, and percentages of cell viability in costimulated CD4+ T-cell cultures infected with the HIV-1 molecular clones LAI (X4) and JR-CSF (R5). (B) Comparison of percentages of infected cells, viral release characteristics, and percentages of cell viability in costimulated CD4+ T-cell cultures infected with the isogenic HIV-1 strains NL4-3 (X4) and 49.5 (R5). (C) Comparison of percentages of infected cells, viral release characteristics, and percentages of cell viability in costimulated CD4+ T-cell cultures infected with the primary HIV-1 isolates KP1 (R5) and KP2 (X4/R5).
Our data demonstrate that R5-infected costimulated CD4+ T-cell cultures produce more progeny virus than X4-infected CD4+ T-cell cultures. In comparing cultures infected by the molecular clones LAI (X4) and JR-CSF (R5), we found that a much larger percentage of cells was infected by LAI than by JR-CSF at the early time points postinfection (Fig. 6 and 7A). For example, 29% of CD4+ T cells were infected by clone LAI at day 3 postinfection, but only 8% were infected by clone JR-CSF (Fig. 6 and 7A). This was likely due to the high expression level of CXCR4 and low expression level of CCR5 on these cells (Fig. 3 and 4). At the later time points postinfection, however, the percentage of apoptotic cells increased in the LAI-infected cultures, and the number of infected cells decreased (Fig. 6 and 7A). By day 7 postinfection, more than half of the cells in the LAI-infected culture were dead, obscuring the FACS analysis due to the large amount of background autofluorescence caused by the dead and dying cells (Fig. 6 and 7A). Despite the small number of JR-CSF-infected cells, high levels of viral production were detected in these cultures (Fig. 7A). The differential in viral production levels between the R5-tropic JR-CSF and the X4-tropic LAI clones was partially due to the fact that the cell viability in the LAI-infected cultures decreased rapidly whereas the cell viability remained relatively high in the JR-CSF-infected cultures over time (Fig. 7A). Analysis of costimulated CD4+ T-cell cultures infected with the isogenic pair NL4-3 (X4) and 49.5 (R5) provided further confirmation that R5-infected CD4+ T cells produce more progeny virus than X4-infected CD4+ T cells. A smaller percentage of costimulated CD4+ T cells was infected by the R5-tropic 49.5 at the early time points postinfection compared to the results seen with its isogenic X4 counterpart, NL4-3 (Fig. 6 and 7B). At day 5, 11.5% of CD4+ T cells were infected by the X4 strain whereas only 4.45% were infected by the R5 strain (Fig. 6 and 7B). Despite the presence of fewer R5-infected cells, similar levels of viral production were detected in the R5- and X4-infected cultures (Fig. 7B). Cell death seemed to play less of a role in the differential viral production levels between these two strains, as the viability characteristics of both infected cultures remained comparable over time (Fig. 7B). Similar results were obtained in comparisons of cultures infected with the primary R5 isolate KP1 and the primary dual-tropic isolate KP2. Although KP1 and KP2 were found to infect similar numbers of cells and induce comparable amounts of cell death, viral production was considerably higher in cultures infected by the R5-tropic KP1 strain compared to the results seen with the dual-tropic KP2 strain (Fig. 6 and 7C). In summary, these data demonstrate that R5 HIV-1 has an increased capacity to replicate in costimulated CD4+ T cells compared to X4 HIV-1.
DISCUSSION
Our data provide evidence that R5 HIV-1 replicates more efficiently in primary CD4+ T cells than X4 HIV-1. Our first experiments focused on optimizing the in vitro stimulation conditions of primary CD4+ T cells. We found that costimulation with plate-bound anti-CD3 antibodies and soluble anti-CD28 antibodies induced high levels of activation and rendered cells permissible to X4 and R5 HIV-1 infection. This contradicts earlier reports that stimulation with CD3 and CD28 antibodies induced an R5-resistant state in CD4+ T-cell cultures (7, 31, 42). Reports from subsequent studies have indicated that the R5 resistance only occurred when CD4+ T cells were stimulated with CD3 and CD28 antibodies immobilized on magnetic beads. This phenotype was thought to be mediated by down-regulation of the CCR5 receptor and increased expression levels of β-chemokines (4, 12). Following costimulation with our protocol, CD4+ T cells were found to express low levels of CCR5 but were still able to replicate R5 virus efficiently. This may be due in part to the high activation state of the CD4+ T cells following CD3/CD28 costimulation. In addition, costimulated CD4+ T cells were found to express high levels of CXCR4 and to be highly susceptible to infection by X4 HIV-1.
We present evidence that R5-infected CD4+ T cells produce more progeny virus than X4-infected cells. Direct comparisons were made between costimulated CD4+ T-cell cultures infected with X4 and R5 HIV-1 strains. We first compared cultures infected with two molecular clones of HIV-1, LAI (X4) and JR-CSF (R5). The R5-tropic JR-CSF clone infected far fewer cells than the X4-tropic LAI clone and yet produced greater amounts of progeny virus over time. The striking difference in the replication capacity characteristics of these two strains can partly be explained by the variation in virus-induced cell death in these infected cultures. Cell viability decreased rapidly in LAI-infected cultures and yet remained relatively high in JR-CSF-infected cultures during the course of the experiment. These data demonstrate that X4 replication can be limited in CD4+ T-cell cultures by extensive virus-induced cell death, whereas viral production remains high in cultures infected by less-cytopathic viruses. We next compared cultures infected with the isogenic HIV-1 strains NL4-3 (X4) and 49.5 (R5). Because these viruses differ only in the V3 region of the envelope gene, differences in viral replication kinetics are likely mediated by coreceptor usage. We found that a smaller percentage of costimulated CD4+ T cells was infected by the R5-tropic 49.5 strain than by its isogenic X4 counterpart strain NL4-3, and yet similar levels of viral production were detected in both infected cultures. Virus-induced cell death played less of a role in the differential viral production of these two viruses, since the viability characteristics of the two infected cultures remained comparable over time. The final comparison between the R5-tropic primary isolate KP1 and the dual-tropic primary isolate KP2 yielded similar results. Although KP1 and KP2 infected similar percentages of costimulated CD4+ T cells and induced comparable amounts of cell death, viral production was considerably higher in KP1-infected cultures. Taken together, these data provide evidence that R5-infected CD4+ T cells produce more virus over time than X4-infected CD4+ T cells.
Other groups have also observed a connection between R5 coreceptor usage and increased viral replication. In these studies, X4 and R5 isogenic strains were used to infect human lymphoid tissue cultures (17, 20; A. M. Roy, D. A. Eckstein, and M. A. Goldsmith, Abstr. 2002 Keystone Symposia: HIV Pathogenesis, abstr. 329, 2002). These cultures contain various cell types in addition to CD4+ T cells and require no exogenous stimulation to confer susceptibility to HIV-1 infection. Infection with R5 and X4 HIV-1 isogenic strains resulted in similar amounts of viral production in the lymphoid cultures despite the presence of fewer R5-infected cells (17, 20; Roy et al., 2002 Keystone Symposia). The results of these studies support the conclusion that R5 HIV-1 has a higher replication capacity than X4 HIV-1.
We propose that the increased replication capacity of R5 strains may contribute to the R5 dominance of early HIV-1 infection. R5 viruses have the selective advantage of targeting the more activated CD4+ T cells that express higher levels of transcription factors, such as NFκB, which have been linked to increased HIV LTR promoter activity (36). This may lead to higher viral production levels and preferential spread of R5 viruses in vivo. In addition, X4 viral strains are often associated with increased cytopathicity, which may lead to differential life spans of X4- and R5-infected CD4+ T cells. As a result, R5-infected CD4+ T cells may live longer and release more virus over time than X4-infected CD4+ T cells.
It has been suggested that R5-infected CD4+ T cells may be less susceptible to immune-mediated killing than X4-infected CD4+ T cells. Experiments performed in the macaque model have shown that both X4 and R5 SHIV replication can be detected early after coinfection; however, R5 dominance develops within weeks of the initial infection (22). Interestingly, experimental depletion of CD8+ T cells in these animals results in reemergence of X4 replication, suggesting that the CD8-mediated immune response is more effective at eliminating X4-infected cells (22). This may partially be due to compartmentalization of X4- and R5-infected cells. X4 viruses are more likely to infect circulating CD4+ T cells, which may be more accessible to CD8+ T-cell surveillance than R5-infected activated CD4+ T cells and macrophages that are located deep within tissues. The idea that X4-infected cells are more susceptible to immune-mediated elimination suggests that the emergence of X4 strains in the later stages of disease may be the result of immune exhaustion.
In summary, we present evidence that R5 HIV-1 strains replicate more efficiently in CD3/CD28-costimulated CD4+ T cells than X4 HIV-1 strains. We found that noncytopathic R5 HIV-1 has a greater capacity to replicate in CD4+ T cells than cytopathic X4 strains. In addition, we further analyzed the impact of coreceptor usage on viral production by comparing X4 and R5 viruses that share greater homology and induce similar cytopathic effects. These experiments have provided evidence that R5-infected CD4+ T cells produce more virus over time than X4-infected CD4+ T cells. We suggest that this replication advantage may contribute to the preferential spread of R5 viruses during the early stages of HIV-1 infection.
Acknowledgments
We thank Joseph M. McCune and Einar M. Aandahl for valuable discussions, John Carroll for assistance with manuscript preparation, and Andreas Jekle, Christian Callebaut, Marielle Cavrois, and Jason Kreisberg for helpful discussions and critical reading of the manuscript. We also thank Jay A. Levy, Christopher P. Locher, and Kyle Bonneau for providing the KP1 and KP2 primary HIV-1 isolates. In addition, we thank the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, for providing proviral plasmids.
This research was supported by funds provided by the Universitywide AIDS Research Program (grant F02-GI-212) awarded to B.S. and NIH grant AI44595 awarded to D.F.N. Partial funding was also obtained from the Gladstone Institute of Virology and Immunology. D.F.N. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Balotta, C., P. Bagnarelli, M. Violin, A. L. Ridolfo, D. Zhou, A. Berlusconi, S. Corvasce, M. Corbellino, M. Clementi, M. Clerici, M. Moroni, and M. Galli. 1997. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS 11:F67-F71. [DOI] [PubMed] [Google Scholar]
- 4.Barker, E., K. N. Bossart, and J. A. Levy. 1998. Differential effects of CD28 costimulation on HIV production by CD4+ cells. J. Immunol. 161:6223-6227. [PubMed] [Google Scholar]
- 5.Biti, R., R. Ffrench, J. Young, B. Bennetts, G. Stewart, and T. Liang. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 3:252-253. [DOI] [PubMed] [Google Scholar]
- 6.Cann, A. J., J. A. Zack, A. S. Go, S. J. Arrigo, Y. Koyanagi, P. L. Green, S. Pang, and I. S. Chen. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, R. G., J. L. Riley, B. L. Levine, Y. Feng, S. Kaushal, D. W. Ritchey, W. Bernstein, O. S. Weislow, C. R. Brown, E. A. Berger, C. H. June, and D. C. St. Louis. 1997. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science 276:273-276. [DOI] [PubMed] [Google Scholar]
- 8.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 9.Connor, R. I., H. Mohri, Y. Cao, and D. D. Ho. 1993. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 67:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. I., W. A. Paxton, K. E. Sheridan, and R. A. Koup. 1996. Macrophages and CD4+ T lymphocytes from two multiply exposed, uninfected individuals resist infection with primary non-syncytium-inducing isolates of human immunodeficiency virus type 1. J. Virol. 70:8758-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creson, J. R., A. A. Lin, Q. Li, D. F. Broad, M. R. Roberts, and S. J. Anderson. 1999. The mode and duration of anti-CD28 costimulation determine resistance to infection by macrophage-tropic strains of human immunodeficiency virus type 1 in vitro. J. Virol. 73:9337-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 14.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 17.Eckstein, D. A., M. P. Sherman, M. L. Penn, P. S. Chin, C. M. De Noronha, W. C. Greene, and M. A. Goldsmith. 2001. HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J. Exp. Med. 194:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 19.Granelli-Piperno, A., E. Delgado, V. Finkel, W. Paxton, and R. M. Steinman. 1998. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J. Virol. 72:2733-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grivel, J. C., M. L. Penn, D. A. Eckstein, B. Schramm, R. F. Speck, N. W. Abbey, B. Herndier, L. Margolis, and M. A. Goldsmith. 2000. Human immunodeficiency virus type 1 coreceptor preferences determine target T-cell depletion and cellular tropism in human lymphoid tissue. J. Virol. 74:5347-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haltiner, M., T. Kempe, and R. Tjian. 1985. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 13:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harouse, J. M., C. Buckner, A. Gettie, R. Fuller, R. Bohm, J. Blanchard, and C. Cheng-Mayer. 2003. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc. Natl. Acad. Sci. USA 100:10977-10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heiken, H., S. Becker, I. Bastisch, and R. E. Schmidt. 1999. HIV-1 infection in a heterosexual man homozygous for CCR-5 delta32. AIDS 13:529-530. [DOI] [PubMed] [Google Scholar]
- 24.Jekle, A., O. T. Keppler, E. De Clercq, D. Schols, M. Weinstein, and M. A. Goldsmith. 2003. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 77:5846-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jekle, A., B. Schramm, P. Jayakumar, V. Trautner, D. Schols, E. De Clercq, J. Mills, S. M. Crowe, and M. A. Goldsmith. 2002. Coreceptor phenotype of natural human immunodeficiency virus with Nef deleted evolves in vivo, leading to increased virulence. J. Virol. 76:6966-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 27.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 28.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, J. I. Mullins, A. B. van't Wout, and M. A. Goldsmith. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75:8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuipers, H., C. Workman, W. Dyer, A. Geczy, J. Sullivan, and R. Oelrichs. 1999. An HIV-1-infected individual homozygous for the CCR-5 delta32 allele and the SDF-1 3′A allele. AIDS 13:433-434. [DOI] [PubMed] [Google Scholar]
- 30.Kwa, D., J. Vingerhoed, B. Boeser-Nunnink, S. Broersen, and H. Schuitemaker. 2001. Cytopathic effects of non-syncytium-inducing and syncytium-inducing human immunodeficiency virus type 1 variants on different CD4+-T-cell subsets are determined only by coreceptor expression. J. Virol. 75:10455-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine, B. L., J. D. Mosca, J. L. Riley, R. G. Carroll, M. T. Vahey, L. L. Jagodzinski, K. F. Wagner, D. L. Mayers, D. S. Burke, O. S. Weislow, D. C. St Louis, and C. H. June. 1996. Antiviral effect and ex vivo CD4+ T cell proliferation in HIV-positive patients as a result of CD28 costimulation. Science 272:1939-1943. [DOI] [PubMed] [Google Scholar]
- 32.Littman, D. R. 1998. Chemokine receptors: keys to AIDS pathogenesis? Cell 93:677-680. [DOI] [PubMed] [Google Scholar]
- 33.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 34.Meng, G., X. Wei, X. Wu, M. T. Sellers, J. M. Decker, Z. Moldoveanu, J. M. Orenstein, M. F. Graham, J. C. Kappes, J. Mestecky, G. M. Shaw, and P. D. Smith. 2002. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat. Med. 8:150-156. [DOI] [PubMed] [Google Scholar]
- 35.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien, T. R., C. Winkler, M. Dean, J. A. Nelson, M. Carrington, N. L. Michael, and G. C. White II. 1997. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet 349:1219-1224. [DOI] [PubMed] [Google Scholar]
- 38.Paxton, W. A., S. R. Martin, D. Tse, T. R. O'Brien, J. Skurnick, N. L. VanDevanter, N. Padian, J. F. Braun, D. P. Kotler, S. M. Wolinsky, and R. A. Koup. 1996. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat. Med. 2:412-417. [DOI] [PubMed] [Google Scholar]
- 39.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 40.Penn, M. L., J. C. Grivel, B. Schramm, M. A. Goldsmith, and L. Margolis. 1999. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc. Natl. Acad. Sci. USA 96:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reece, J. C., A. J. Handley, E. J. Anstee, W. A. Morrison, S. M. Crowe, and P. U. Cameron. 1998. HIV-1 selection by epidermal dendritic cells during transmission across human skin. J. Exp. Med. 187:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riley, J. L., R. G. Carroll, B. L. Levine, W. Bernstein, D. C. St. Louis, O. S. Weislow, and C. H. June. 1997. Intrinsic resistance to T cell infection with HIV type 1 induced by CD28 costimulation. J. Immunol. 158:5545-5553. [PubMed] [Google Scholar]
- 43.Roos, M. T., J. M. Lange, R. E. de Goede, R. A. Coutinho, P. T. Schellekens, F. Miedema, and M. Tersmette. 1992. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J. Infect. Dis. 165:427-432. [DOI] [PubMed] [Google Scholar]
- 44.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 45.Schramm, B., M. L. Penn, R. F. Speck, S. Y. Chan, E. De Clercq, D. Schols, R. I. Connor, and M. A. Goldsmith. 2000. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J. Virol. 74:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheppard, H. W., C. Celum, N. L. Michael, S. O'Brien, M. Dean, M. Carrington, D. Dondero, and S. P. Buchbinder. 2002. HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion. J. Acquir. Immune Defic. Syndr. 29:307-313. [DOI] [PubMed] [Google Scholar]
- 48.Tersmette, M., R. E. de Goede, B. J. Al, I. N. Winkel, R. A. Gruters, H. T. Cuypers, H. G. Huisman, and F. Miedema. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62:2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tersmette, M., R. A. Gruters, F. de Wolf, R. E. de Goede, J. M. Lange, P. T. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodorou, I., L. Meyer, M. Magierowska, C. Katlama, and C. Rouzioux, et al. 1997. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet 349:1219-1220. [PubMed] [Google Scholar]
- 51.Toohey, K., K. Wehrly, J. Nishio, S. Perryman, and B. Chesebro. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70-79. [DOI] [PubMed] [Google Scholar]
- 52.Verrier, F., A. M. Borman, D. Brand, and M. Girard. 1999. Role of the HIV type 1 glycoprotein 120 V3 loop in determining coreceptor usage. AIDS Res. Hum. Retrovir. 15:731-743. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]