Abstract
A post hoc analysis of the ATHENA study was performed to determine whether true HPV‐negative cervical lesions occur and whether they have clinical relevance. The ATHENA database was searched for all CIN2 or worse (CIN2+) cases with cobas HPV‐negative results and comparison was made with Linear Array (LA) and Amplicor to detect true false‐negative HPV results. Immunostaining with p16 was performed on these cases to identify false‐positive histology results. H&E slides were re‐reviewed by the study pathologists with knowledge of patient age, HPV test results and p16 immunostaining. Those with positive p16 immunostaining and/or a positive histopathology review underwent whole tissue section HPV PCR by the SPF10/LiPA/RHA system. Among 46,887 eligible women, 497 cases of CIN2+ were detected, 55 of which tested negative by the cobas® HPV Test (32 CIN2, 23 CIN3/ACIS). By LA and/or Amplicor, 32 CIN2+ (20 CIN2, 12 CIN3/ACIS) were HPV positive and categorized as false‐negatives by cobas HPV; nine of 12 false‐negative CIN3/ACIS cases were p16+. There were 23 cases (12 CIN2, 11 CIN3/ACIS) negative by all HPV tests; seven of 11 CIN3/ACIS cases were p16+. H&E slides were available for six cases for re‐review and all were confirmed as CIN3/ACIS. Tissue PCR was performed on the six confirmed CIN3/ACIS cases (and one without confirmation): four were positive for HPV types not considered oncogenic, two were positive for oncogenic genotypes and one was indeterminate. In summary, subanalysis of a large cervical cancer screening study did not identify any true CIN3/ACIS not attributable to HPV.
Keywords: cervical cancer screening, HPV DNA testing, cervical intraepithelial neoplasia, adenocarcinoma in situ, HPV genotype, histology
Short abstract
What's new?
Human papillomavirus (HPV) testing has a high negative predictive value for detecting histological cervical intraepithelial neoplasia (CIN). False‐negative HPV results can occur, however, though their clinical relevance is little understood. Using data from the U.S.‐based ATHENA study, the authors of the present report show that only a very small percentage of CIN grade 3/adenocarcinoma in situ (ACIS) lesions were missed by the cobas HPV Test, which identifies 14 high‐risk HPV types. False‐negatives by cobas testing were compared with Linear Array and Amplicor testing. Most missed CIN3/ACIS cases were associated with HPV types not included in current tests.
It is well accepted that cervical cancers are caused by high‐risk human papillomavirus (HR‐HPV).1, 2, 3 Nonetheless, a small subset of primary cervical cancers that is not linked to HPV has been described and includes minimal deviation adenocarcinomas, mucinous adenocarcinomas of the gastric type as well as a subset of clear cell adenocarcinoma and serous carcinomas.4, 5 It has also been shown that cervical intraepithelial neoplasia grade 3 (CIN3) lesions are almost exclusively linked to HPV with a negligible proportion associated only with low‐risk HPV types.3 Data from a meta‐analysis of HPV testing in primary screening trials found sensitivity rates of 97–98% for CIN3 with Hybrid Capture 2 and other clinically validated PCR tests.6 As a consequence, the rate of HPV‐test‐negative CIN3 will be perceived as a false‐negative rate and will be used as a major quality indicator for the performance of specific HPV tests. However, some HR‐HPV‐negative cases can be explained by inaccurate histological interpretation or poor sample quality and most reviews were based on very small numbers of HPV‐negative cases. In this analysis, we searched the database of the ATHENA study, a large cervical cancer screening trial, and have performed an in‐depth evaluation of the cobas HPV Test‐negative CIN2+ lesions to determine which ones represent false‐negative HPV test results and which ones are truly HPV negative CIN2 lesions.
Methods
Study population
The ATHENA trial has been previously described in detail.7, 8 Briefly, 47,208 women 21 years or older presenting for routine cervical cancer screening were enrolled at 61 sites across 23 states in the United States. Eligible women (46,887) were ≥21 years, were nonpregnant, had an intact cervix and had not received treatment for CIN within 12 months of enrollment.
Study procedures
At enrollment, a liquid‐based cervical cytology (LBC) sample (ThinPrep, Hologic, Bedford, MA) was obtained from all women and was used for LBC and HPV testing. HPV testing was performed with three HPV assays (Roche Molecular Systems, Pleasanton, CA): the Amplicor HPV test that reports out results for 13 high‐risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68), the Linear Array HPV Genotyping Test (LA) modified to detect 16 types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 73 and 82) and the cobas HPV Test that reports out 12 pooled high‐risk genotypes (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) and simultaneously individual results for HPV 16 and HPV 18.
Cytology was performed at four certified clinical laboratories in the United States and results reported using the Bethesda terminology. HPV testing was performed according to the manufacturer's instructions at the same four laboratories and at Roche Molecular Systems. Histologic diagnoses of cervical biopsies were determined by an expert panel of three pathologists using standard CIN terminology. To provide adjudication for those cases with CIN2+ histology results that were cobas HPV Test negative (cobas negative), immunohistochemistry for p16INK4a (p16) was performed on cervical biopsies using the CINtec® Histology Kit (Roche mtm Laboratories AG, Mannheim, Germany) according to the manufacturer's instructions.
For this post hoc analysis, the cobas HPV Test results of all samples were first compared with Amplicor and LA results to identify false‐negative cobas results and to evaluate if there was a particular pattern of HPV types that may be missed by the cobas HPV Test.
In a second phase of the analysis, all cobas‐negative CIN2+ cases underwent p16 immunostaining.9 As overexpression of p16 in cervical lesions is a consequence of a transforming HPV infection, p16‐positive cobas‐negative cases could be best explained by false‐negative HPV or false‐positive histology results.
To determine further whether false‐positive histology results occurred, in the third phase all cobas‐negative CIN2+ cases underwent a second histopathology review. H&E slides were re‐read by the study pathologists unmasked to patient age and all HPV test results and with knowledge of the p16 staining results.
In the final phase of review, cobas‐negative CIN2 cases with positive p16 immunostaining and CIN3+ cases with either positive p16 or negative p16 plus a positive histopathology review underwent analysis by whole tissue section polymerase chain reaction (WTS‐PCR) using a second broad spectrum SPF10LiPA/RHA PCR system, SPF10 PCR/LiPA25 version 1 with 25 HPV genotypes (Labo Bio‐medical Products, Rijswijk, The Netherlands).10 The same amplimers were analyzed with an additional in house RHA strip+ to determine the presence of 17 HPV genotypes, as previously described.6 In brief, hematoxylin and eosin (H&E) slides were prepared with sections from a biopsy block and were scanned in an image system. Specimens were tested with the HPV SPF10 PCR/LiPA25 (version 1) system that identifies the following HPV genotypes: 6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70, 74. An additional strip identifies HPV 26, 30, 55, 61, 62, 64, 67, 69, 71, 82, 83, 84, 85, 87, 89, 90 and 91. Using this SPF10 DEIA/LiPA/RHA methodology, some cases had “undetermined” results which occurred when the DNA enzyme immunoassay (DEIA) used to detect a broad spectrum of HPV amplification products of at least 68 different mucosal HPV genotypes was positive but no HPV genotypes were detected by the LiPA 25 or the RHA strip.
The WTS‐PCR was performed blinded to all other results and a control group of five cobas HPV‐positive CIN2+ cases was randomly included with cobas‐negative CIN2+ cases.
Our a priori definitions of findings in cobas‐negative CIN2+ case were:
| Result | Final evaluation |
|---|---|
| HPV‐HR types detected with other tests | 1: cobas HPV false negative |
| HPV types detected that are not considered as HR | 2: cobas HPV true negative |
| Review and p16 review negative | 3: cobas HPV true negative; CIN2+ false positive |
| Discrepancy between review and p16 review | 4: Indeterminate |
| HPV negative with all tests and reviews positive | 5: HPV‐negative CIN2+ |
We considered a false‐positive diagnosis of CIN2+ the strongest outcome of final evaluation. Therefore, cases with negative review and negative p16 review were classified as final evaluation Group 3 even when high‐risk HR or other HPV genotypes were detected with any of the other tests.
Results
From the 46,887 eligible women in the ATHENA study, 497 cases of CIN2+ were detected (192 CIN2 and 305 CIN3+; CIN3+ to CIN2 ratio 1.59), 55 of which tested negative for HPV with the cobas HPV Test on LBC samples. There were 32 cobas‐negative CIN2 and 23 cobas‐negative CIN3+ [including two adenocarcinoma in situ (ACIS) cases; CIN3+ to CIN2 ratio 0.72]. Of note, there were no cobas‐negative invasive cancers. All six squamous carcinomas or adenocarcinomas in ATHENA tested positive with the cobas HPV Test; CIN3+ cases will therefore here be referred to here as CIN3/ACIS.
The cobas‐negative cohort underwent three different reviews to further characterize these lesions. The first review identified 32 cobas‐negative CIN2+ as false HPV‐negative cases, that is positive for HPV by Amplicor (the more analytically sensitive test) and/or LA. HPV types could be detected with LA genotyping in 22 of the 32 cobas false‐negative CIN2+ cases (Table 1). HPV 82, which is not considered to be a high‐risk HPV genotype, was detected in three CIN3 (including one co‐infection with HPV 51) and four CIN2 lesions (including one co‐infection with HPV 51). Overall, according to our a priori definitions ten cases associated with HPV 73 or 82 were classified as CIN2+ with non‐HR‐HPV, and 22 cases as cobas false‐negative CIN2+.
Table 1.
Linear Array Genotyping results of cobas HPV Test‐negative CIN2+ casesa
| Genotype by Linear Array | CIN2 | CIN3/ACIS |
|---|---|---|
| HPV 16 | 1 | |
| HPV 33 | 1 | |
| HPV 39 | 1 | |
| HPV 51 | 2 | 1 |
| HPV 52 | 3 | 3 |
| HPV 58 | 2 | 1 |
| HPV 66 | 1 | |
| HPV 73 | 3 | |
| HPV 82 | 4 | 3 |
Including four cases with co‐infections.
Table 2 summarizes the LA/Amplicor, p16 immunostaining and unblinded histology review findings as well as the final classification categories among all 55 cobas‐negative CIN2+ cases. Overall, 20 of 32 cobas‐negative CIN2 (62.5%) tested positive for HPV with at least one HPV test compared to 12 of 23 cobas‐negative CIN3/ACIS (52.2%). Twelve LA and/or Amplicor‐positive CIN3/ACIS cases underwent p16 staining and histology review, and ten of these 12 cases were confirmed as CIN3/ACIS on re‐review and p16. One LA/Amplicor‐negative case had HPV 39 identified but was p16 negative and was considered not to represent CIN2+ on re‐review. One LA‐negative/Amplicor‐positive case was also negative and not classified as CIN2+ on re‐review (Table 2).
Table 2.
Details of cases with negative cobas HPV Test results and CIN2+ histology
| Case | Initial consensus histology diagnosis | AMPLICOR | LA | LA genotype | p16 result | Unblinded histology review | Whole tissue section‐PCR HPV | Final evaluationa |
|---|---|---|---|---|---|---|---|---|
| 1 | CIN2 | Positive | Positive | 52 | Positive | Positive | 1 | |
| 2 | CIN2 | Positive | Positive | 52 | Negative | Negative | 1 | |
| 3 | CIN2 | Positive | Positive | 52, 73 | Positive | Positive | 1 | |
| 4 | CIN2 | Positive | Positive | 51 | Negative | Negative | 3 | |
| 5 | CIN2 | Positive | Positive | 16 | Positive | Positive | 1 | |
| 6 | CIN2 | Positive | Positive | 58, 66 | Negative | Negative | 3 | |
| 7 | CIN2 | Positive | Positive | 51, 82 | Positive | Positive | 1 | |
| 8 | CIN2 | Positive | Positive | 33 | Positive | Negative | 4 | |
| 9 | CIN2 | Positive | Positive | 58 | Positive | Positive | 1 | |
| 10 | CIN2 | Negative | Positive | 73 | Negative | Negative | 3 | |
| 11 | CIN2 | Negative | Positive | 73 | Negative | Negative | 3 | |
| 12 | CIN2 | Negative | Positive | 82 | Positive | Positive | 2 | |
| 13 | CIN2 | Negative | Positive | 82 | Positive | Positive | 2 | |
| 14 | CIN2 | Negative | Positive | 82 | Positive | Positive | 2 | |
| 15 | CIN2 | Positive | Negative | Negative | Negative | 3 | ||
| 16 | CIN2 | Positive | Negative | Negative | Negative | 3 | ||
| 17 | CIN2 | Positive | Negative | − | No p16 slide | N/Ab | ||
| 18 | CIN2 | Positive | Negative | Positive | Indeterminatec | N/Ab | ||
| 19 | CIN2 | Positive | Negative | Negative | Negative | 3 | ||
| 20 | CIN2 | Positive | Negative | Positive | Positive | 1 | ||
| 21 | CIN2 | Negative | Negative | Positive | No review | Negative | 4 | |
| 22 | CIN2 | Negative | Negative | Negative | Negative | 3 | ||
| 23 | CIN2 | Negative | Negative | Negative | Negative | 3 | ||
| 24 | CIN2 | Negative | Negative | Negative | Negative | 3 | ||
| 25 | CIN2 | Negative | Negative | Negative | Negative | 3 | ||
| 26 | CIN2 | Negative | Negative | Negative | Positive | 4 | ||
| 27 | CIN2 | Negative | Negative | Negative | Positive | 4 | ||
| 28 | CIN2 | Negative | Negative | Negative | Negative | 3 | ||
| 29 | CIN2 | Negative | Negative | Positive | Positive | 30, 68, 89 | 1 | |
| 30 | CIN2 | Negative | Negative | Negative | Positive | 4 | ||
| 31 | CIN2 | Negative | Negative | Positive | Positive | 70 | 2 | |
| 32 | CIN2 | Negative | Negative | Positive | Positive | Negative | 5 | |
| 33 | CIN3 | Positive | Positive | 52 | Negative | Positive | 4 | |
| 34 | CIN3 | Positive | Positive | 52 | Positive | Positive | 1 | |
| 35 | CIN3 | Positive | Positive | 82 | Positive | Positive | 2 | |
| 36 | CIN3 | Positive | Positive | 58 | Positive | Positive | 1 | |
| 37 | CIN3 | Positive | Positive | 82 | Positive | Positive | 2 | |
| 38 | CIN3 | Positive | Positive | 52 | Positive | Positive | 1 | |
| 39 | CIN3 | Negative | Positive | 39 | Negative | Negative | 3 | |
| 40 | CIN3 | Negative | Positive | 51, 82 | Positive | Positive | 1 | |
| 41 | CIN3 | Positive | Negative | Positive | Positive | 1 | ||
| 42 | CIN3 | Positive | Negative | Positive | Positive | 1 | ||
| 43 | CIN3 | Positive | Negative | Positive | Positive | 1 | ||
| 44 | CIN3 | Positive | Negative | Negative | Negative | 3 | ||
| 45 | CIN3 | Negative | Negative | Negative | Negative | 3 | ||
| 46 | CIN3 | Negative | Negative | Negative | Positive | 4 | ||
| 47 | CIN3 | Negative | Negative | Positive | Positive | 6 | 2 | |
| 48 | CIN3 | Negative | Negative | Negative | Negative | 3 | ||
| 49 | CIN3 | Negative | Negative | Positive | Positive | Undetermined | N/Ab | |
| 50 | CIN3 | Negative | Negative | Positive | Positive | 26, 54, 67 | 2 | |
| 51 | CIN3 | Negative | Negative | Positive | Positive | 67 | 2 | |
| 52 | CIN3 | Negative | Negative | Positive | No slide | 16, 51 (33, 58 in sections without dysplasia) | 1 | |
| 53 | CIN3 | Negative | Negative | Positive | Positive | 53 | 2 | |
| 54 | Adenocarcinoma in situ (ACIS) | Negative | Negative | Positive | Positive | 16 | 1 | |
| 55 | Adenocarcinoma in situ (ACIS) | Negative | Negative | Negative | Negative | 3 |
1 = cobas HPV false negative; 2 = cobas HPV true negative; 3 = cobas HPV true negative; CIN2+ false positive; 4 = indeterminate; 5 = HPV negative CIN2+.
Evaluation not available.
Very limited tissue.
Of the 55 cobas‐negative CIN2+ cases, 23 tested negative for HPV with all tests (12 CIN2 and 11 CIN3/ACIS lesions, Table 2 and Figs. 1 and 2). Of the CIN3/ACIS cases, seven tested p16 positive and four showed negative p16 staining. On review, H&E slides were available for six of the seven p16‐positive cases and all were confirmed as CIN3. The one CIN3 case for which the H&E slide could not be located did however have material available for WTS‐PCR. Of the p16‐negative CIN3/ACIS cases, three of four were downgraded to atrophy or metaplasia (Fig. 2). The two ACIS cases tested negative with both Amplicor and LA; one also tested p16 negative and on review was reclassified as squamous metaplasia. Six cobas‐negative CIN3/ACIS cases with positive p16 staining and/or positive reviews and the one cobas‐negative CIN3 without any review were available for WTS‐PCR. Among the seven CIN3/ACIS cases that underwent WTS‐PCR, four were positive for HPV types not considered oncogenic, one was positive for HPV 16, one was “undetermined” and the case that had no slide available for histopathology review was positive for HPV 16 and 51 (Table 3).
Figure 1.
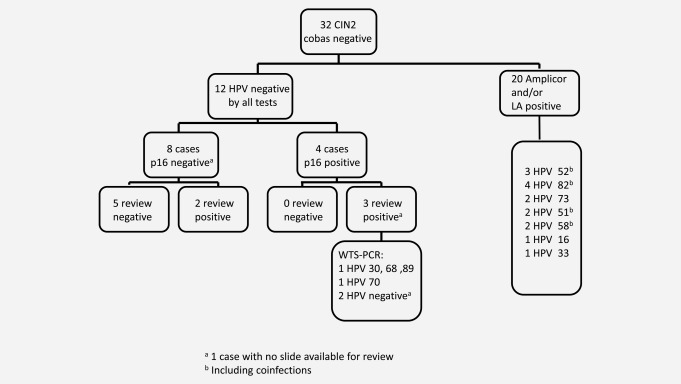
Overview of cobas‐negative CIN2 cases. aOne case with no slide available for review; bincluding coinfections.
Figure 2.
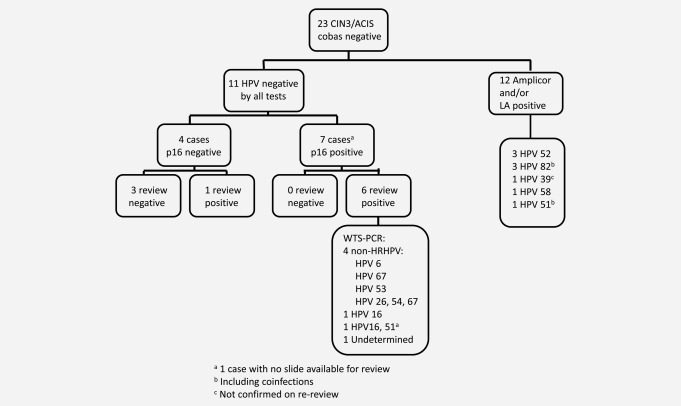
Overview of cobas‐negative CIN3/ACIS cases. aOne case with no slide available for review; bincluding coinfections; cnot confirmed on re‐review.
Table 3.
Results of the WTS‐PCR
| Initial diagnosis | p16 staining | Review CIN2+ | WTS‐PCR HPV types |
|---|---|---|---|
| CIN2a | No review | No review | Negative |
| CIN2 | Positive | Positive | HPV 30, 68, 89 |
| CIN2 | Positive | Positive | HPV 70 |
| CIN2 | Positive | Positive | Negative |
| CIN3 | Negative | Positive | HPV 6 |
| CIN3 | Positive | Positive | HPV X |
| CIN3 | Positive | Positive | HPV 26, 54, 67 |
| CIN3 | Positive | Positive | HPV 67 |
| CIN3 | Positive | Positive | HPV 53 |
| CIN3 | Positive | No reviewb | HPV 16, 51 |
| ACIS | Positive | Positive | HPV 16 |
Case excluded due to suboptimal tissue quality for p16 staining and WTS‐PCR.
H&E slide not available.
Among the 12 CIN2 cases, four tested positive for p16 and eight showed negative staining. Five of seven p16‐negative cases with slides available for review were downgraded on review (Table 2 and Fig. 1), while the four p16‐positive cases were confirmed as CIN2 and underwent WTS‐PCR; one case had no slide available for p16 review but underwent WTS‐PCR. Of these four p16‐positive cases, one tested negative for HPV by WTS‐PCR, one was a co‐infection (HPV 30, 68, 89), one was HPV 70 positive and the case without review tested HPV negative (Table 3).
Of note, the relatively high percentage of p16‐negative CIN2+ cases reported here should be viewed in the context of the unique cohort of disease‐positive/cobas‐negative discordant cases. For reference, the overall concordance between the adjudicated histology review of a random subset of ATHENA study cases and the p16 histology immunostaining results was very high: 86.8 and 94.5% for CIN2+ and CIN3/ACIS, respectively (Table 4).
Table 4.
Concordance between adjudicated histology review and p16 histology results in a random subset of ATHENA study CIN2+/CIN3+ cases
| ATHENA histological diagnosis | p16+ | ||
|---|---|---|---|
| n | n | % (95% CI) | |
| CIN2+ | 189 | 164 | 86.8 (81.1–91.3%) |
| CIN3+ | 109 | 103 | 94.5 (88.4–98.0) |
95% CI, exact binomial 95% confidence interval.
Discussion
Our analysis showed that a false‐negative HPV test result was the most frequent reason for a cobas‐negative CIN2+ lesion. Overall, 62.5% of all cobas‐negative CIN2 and 52.2% of all cobas‐negative CIN3 tested positive with other HPV tests. A reasonable proportion of cobas‐negative CIN2 and to a lesser extent cobas‐negative CIN3 was explained by non–high‐risk HPV types 73 and 82. The high‐risk types missed were only found each in a single case with Linear Array, with the exception of HPV type 52; improving the limit of detection for this genotype should be solved easily by the manufacturer. HPV 82, which is not considered to be a Group 1 carcinogen by IARC but is a member of the high‐risk Alpha 5 clade, was detected as single infection in three CIN2 and two CIN3 lesions. The fact that p16 was positive in four CIN2 cases (including one as a coinfection) and three CIN3 cases (including one as a coinfection) that were associated with HPV 82, as well as reports on the detection of HPV 82 as a single HPV type in anal cancers,11 indicates that HPV 82 might occasionally be associated with high‐grade neoplasia. Recently, researchers from Barcelona and Heidelberg demonstrated that eight potential or probable high‐risk HPV types including HPV 82 are rarely but consistently found as single HPV‐type infections in invasive cervical cancers. The biological activities in cancer cells with single HPV‐type infections of any of these eight types did not differ from HPV 16‐associated cancers.12 An accompanying editorial pointed out that HPV 82 and five other HPV types occur significantly more often in invasive cancers than in the normal population but are so rare that they should not be included in HPV screening tests.13 Likewise, a single CIN3 case that was confirmed by p16 was found to be positive for HPV 6 on WTS‐PCR. Given the rarity of the contribution of HPV 6 to invasive cancer,14 this represents a lesion of unknown clinical significance.
Of 23 CIN2+ cases that tested negative with all HPV tests, a total of 12 were not confirmed as CIN2+ by p16 staining. Of the remaining 11 CIN2+ that were p16 positive, nine were confirmed by review and two had no review. Of the confirmed CIN2+ cases, eight of nine were characterized by WTS‐SPF10LiPA/RHA analysis either as associated with HPV genotypes not considered to be Group 1 carcinogens by IARC (n = 5), undetermined (n = 1) or as CIN2+ associated with HPV types 16 (n = 1) or 68 (n = 1). Only a single confirmed CIN2 lesion tested negative for HPV with all tests and fulfilled our a priori definition of HPV‐negative CIN2. Analogous to HPV 82 which was found in several CIN2 and CIN3 lesions, most of the other HPV genotypes identified by WTS‐PCR analysis were members of high‐risk clades such as HPV 26 (Alpha clade 5), HPV 30 and 53 (Alpha clade 6), HPV 67 (Alpha clade 9) and HPV 70 (Alpha clade 7).15 Although these HPV genotypes are not classified as Group 1 carcinogens, our results and the above mentioned Barcelona–Heidelberg evaluation indicate that they can occasionally be found in CIN2 as well as in CIN3 lesions and cancers. The single CIN3 case that tested negative for p16 but considered positive on histopathology review unfortunately did not have additional tissue available for WTS‐PCR testing.
As the natural history of a substantial proportion of CIN2 resembles CIN1 rather than CIN3 and because only CIN3 and ACIS are recognized as true precursors of cervical cancer, we focused on cobas‐negative CIN3/ACIS rather than on cobas‐negative CIN2 lesions. Of the 11 CIN3/ACIS lesions that tested HPV negative with all tests (cobas, LA and Amplicor) seven cases were confirmed as CIN3 by p16 immunostaining and/or pathology review. Four of these seven cases were associated with low‐risk or other HPV types not generally considered to be high‐risk, and only two cases were found to be CIN3 associated with HPV 16.
Overall, just six of 23 cobas‐negative CIN3/ACIS cases were confirmed by all review criteria as CIN3 associated with high‐risk HPV types missed by cobas [2× HPV 16 (1× with HPV 51 co‐infection), 3×HPV 52, 1×HPV 58]. Out of the total of 305 CIN3+ lesions detected within the ATHENA trial, only 1.97% (six of 305) CIN3/ACIS lesions were therefore confirmed as CIN3/ACIS associated with high‐risk HPV types and missed by the cobas HPV Test. Only two of these six cases were missed by all HPV tests (Amplicor, LA and the cobas HPV Test), probably because of low viral load. Of the review‐positive CIN3/ACIS cases, seven additional cases were associated with low‐risk HPV types or other HPV types that are not identified by current HPV tests and that are not considered to be a useful inclusion in population‐based HPV screening. However, we cannot exclude the possibility that additional HPV types that are found to be associated with high‐grade disease, but have low prevalence in the general population, may be included in future HPV tests.
Our findings are in accordance to observations of others that not all HPV‐negative CIN3/ACIS cases are clinically meaningful.3 Immunostaining for p16 of all HPV‐negative CIN2+ and additional histopathology review should help exclude false diagnoses. It seems more difficult to resolve false‐negative HPV results explained by HPV 52 or other high‐risk HPV types with low viral load and similarly CIN2+ cases associated with non–high‐risk types. Any increase in sensitivity of HPV testing for such cases will result in a loss of specificity but a high specificity is crucial for a screening test.
Conclusion
Out of a large screening population with >47,000 women with 497 CIN2+ and 305 CIN3+ cases we did not find any sign for the existence of a true CIN3+ lesion that is not linked to HPV.
In the management of HPV‐negative CIN2+, clinicians should be aware that approximately two thirds of these cases may be explained by false‐positive histopathology diagnoses and/or infection with non–high‐risk HPV types and therefore can be considered almost meaningless. However, even very good HPV tests may miss CIN3 associated with high‐risk HPV types in 1–3% of cases.
Acknowledgements
The authors dedicate this publication to co‐author Mario Sideri, Istituto Europeo di Oncologia, Milano, Italy, who was very much involved in the development of the manuscript but died unexpectedly and tragically in an accident before we could finish our collaboration completely.
Conflicts of interest: KUP has served as a speaker and advisor for Roche Diagnostics (including Ventana) and as a speaker for Seegene. JTC has served as an advisor to Roche, and has served as a speaker for Roche. KJ and CB are employed by Roche Molecular Systems. WQ is employed by DDL Diagnostic Laboratory, is a minority shareholder in Diassay BV and has received a grant from GSK. RR is employed by Ventana Medical Systems, Inc, a Member of the Roche group. TCW Jr. is a consultant and speaker for Roche Diagnostics
References
- 1. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–19. [DOI] [PubMed] [Google Scholar]
- 2. Liebrich C, Brummer O, Von Wasielewski R, et al. Primary cervical cancer truly negative for high‐risk human papillomavirus is a rare but distinct entity that can affect virgins and young adolescents. Eur J Gynaecol Oncol 2009;30:45–8. [PubMed] [Google Scholar]
- 3. Böhmer G, van den Brule AJ, Brummer O, et al. No confirmed case of human papillomavirus DNA‐negative cervical intraepithelial neoplasia grade 3 or invasive primary cancer of the uterine cervix among 511 patients. Am J Obstet Gynecol 2003;189:118–20. [DOI] [PubMed] [Google Scholar]
- 4. Holl K, Nowakowski AM, Powell N, et al. Human papillomavirus prevalence and type‐distribution in cervical glandular neoplasias: results from a European multinational epidemiological study. Int J Cancer 2015;137:2858–68. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kurman RJ, Carcangiu ML, Herrington CS, et al., eds. WHO classification of tumors of female reproductive organs, 4th edn., vol. 6 Geneva: World Health Organization, 2014. [Google Scholar]
- 6. Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012;30:F88–F99. [DOI] [PubMed] [Google Scholar]
- 7. Stoler MH, Wright TC, Jr , Sharma A, et al.; ATHENA (Addressing the Need for Advanced HPV Diagnostics) HPV Study Group. High‐risk human papillomavirus testing in women with ASC‐US cytology: results from the ATHENA HPV study. Am J Clin Pathol 2011;135:468–75. [DOI] [PubMed] [Google Scholar]
- 8. Wright TC, Jr, Stoler MH, Behrens CM, et al. The ATHENA human papillomavirus study: design, methods, and baseline results. Am J Obstet Gynecol 2012;206:46.e1–46.e11. [DOI] [PubMed] [Google Scholar]
- 9. Darragh TM, Colgan TJ, Thomas Cox J, et al.; Members of the LAST Project Work Groups . The Lower Anogenital Squamous Terminology Standardization project for HPV‐associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology [published errata appear in Int J Gynecol Pathol 2013;32:241; Int J Gynecol Pathol 2013;32:432]. Int J Gynecol Pathol 2013;32:76–115. [DOI] [PubMed] [Google Scholar]
- 10. Quint W, Jenkins D, Molijn A, et al. One virus, one lesion–individual components of CIN lesions contain a specific HPV type. J Pathol 2012;227:62–71. [DOI] [PubMed] [Google Scholar]
- 11. Ouhoummane N, Steben M, Coutlée F, et al. Squamous anal cancer: patient characteristics and HPV type distribution. Cancer Epidemiol 2013;37:807–12. [DOI] [PubMed] [Google Scholar]
- 12. Halec G, Alemany L, Lloveras B, et al.; Retrospective International Survey and HPV Time Trends Study Group . Pathogenic role of the eight probably/possibly carcinogenic HPV types 26, 53, 66, 67, 68, 70, 73 and 82 in cervical cancer. J Pathol 2014;234:441–51. [DOI] [PubMed] [Google Scholar]
- 13. Arbyn M, Tommasino M, Depuydt C, et al. Are 20 human papillomavirus types causing cervical cancer? J Pathol 2014;234:431–5. [DOI] [PubMed] [Google Scholar]
- 14. Munoz N, Bosch X, Casellsague X, et al. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer 2004;111:278–85. [DOI] [PubMed] [Google Scholar]
- 15. International Agency for Cancer Research Human papillomaviruses. In: IARC monographs on the evaluation of carcinogenic risks to humans. Biological agents: a review of human carcinogens, vol. 100 B. Lyon, France: International Agency for Research on Cancer, 2012. 255–313. [Google Scholar]


