Abstract
Neuropeptides that act as muscle relaxants have been identified in chordates and protostomian invertebrates but little is known about the molecular identity of neuropeptides that act as muscle relaxants in deuterostomian invertebrates (e.g. echinoderms) that are ‘evolutionary intermediates’ of chordates and protostomes. Here, we have used the apical muscle of the starfish Patiria pectinifera to assay for myorelaxants in extracts of this species. A hexadecapeptide with the amino acid sequence Phe‐Gly‐Lys‐Gly‐Gly‐Ala‐Tyr‐Asp‐Pro‐Leu‐Ser‐Ala‐Gly‐Phe‐Thr‐Asp was identified and designated starfish myorelaxant peptide (SMP). Cloning and sequencing of a cDNA encoding the SMP precursor protein revealed that it comprises 12 copies of SMP as well as 3 peptides (7 copies in total) that are structurally related to SMP. Analysis of the expression of SMP precursor transcripts in P. pectinifera using qPCR revealed the highest expression in the radial nerve cords and lower expression levels in a range of neuromuscular tissues, including the apical muscle, tube feet and cardiac stomach. Consistent with these findings, SMP also caused relaxation of tube foot and cardiac stomach preparations. Furthermore, SMP caused relaxation of apical muscle preparations from another starfish species – Asterias amurensis. Collectively, these data indicate that SMP has a general physiological role as a muscle relaxant in starfish. Interestingly, comparison of the sequence of the SMP precursor with known neuropeptide precursors revealed that SMP belongs to a bilaterian family of neuropeptides that include molluscan pedal peptides (PP) and arthropodan orcokinins (OK). This is the first study to determine the function of a PP/OK‐type peptide in a deuterostome.
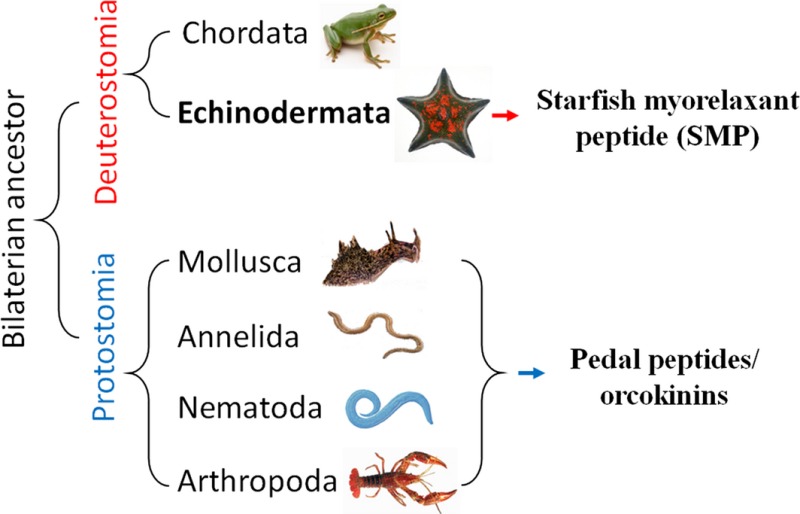
Pedal peptide/orcokinin (PP/OK)‐type peptides are a family of structurally related neuropeptides that were first identified and functionally characterised in protostomian invertebrates. Here, we report the discovery of starfish myorelaxant peptide (SMP), a novel member of the PP/OK‐type neuropeptide identified in the starfish Patiria pectinifera (phylum Echinodermata). SMP is the first PP/OK‐type neuropeptide to be functionally characterised in a deuterostome.
Keywords: muscle, neuropeptide, orcokinin, pedal peptide, relaxation, starfish
Abbreviations used
- ACh
acetylcholine
- RP
reversed‐phase
- RT‐qPCR
real‐time quantitative polymerase chain reaction
- SMP
starfish myorelaxant peptide
- TFA
trifluoroacetic acid
A variety of neuropeptides that act as smooth muscle relaxants in vertebrates have been identified, including calcitoningene‐related peptide (CGRP), adrenomedullin, corticotropin‐releasing hormone (CRH), urocortin, vasoactive intestinal peptide (VIP), pituitary adenylyl cyclase‐activating peptide (PACAP) and peptide histidine isoleucine (PHI) (Robberecht et al. 1982; Brain et al. 1985; Grider and Makhlouf 1986; Williams et al. 1987; Miyata et al. 1989; Kitamura et al. 1993; Schilling et al. 1998). Likewise, studies on protostomian invertebrates such as insects have identified a number of myoinhibitory neuropeptides; for example type‐A allatostatins, type‐B allatostatins or myoinhibitory peptides (MIPs) and myosupressins (Holman et al. 1986; Blackburn et al. 1995; Bendena et al. 1997). In the context of our understanding of the evolutionary history and relationships of neuropeptides in the animal kingdom (Mirabeau and Joly 2013), it appears that neuropeptides belonging to different families have been recruited to act as muscle relaxants in vertebrates and protostomes. It is of interest, therefore, to identify neuropeptides that act as muscle relaxants in animals that occupy an ‘intermediate’ position with respect to the vertebrates and protostomes in animal phylogeny – the deuterostomian invertebrates, which include two chordate subphyla that are closely related to vertebrates (Urochordata and Cephalochordata) and the Ambulacraria (Hemichordata and Echinodermata) (Adoutte et al. 2000).
Nothing is known about the molecular identity of neuropeptides that act as muscle relaxants in hemichordates but neuropeptides that act as muscle relaxants have been identified in echinoderms – the SALMFamides. The prototypes for this family of neuropeptides were both identified in the starfish Asterias rubens and Asterias forbesi – S1 (GFNSALMF‐NH2) and S2 (SGPYSFNSGLTF‐NH2) (Elphick et al. 1991a,b). In vitro pharmacological tests with S1 and S2 revealed that both peptides cause relaxation of neuromuscular preparations from A. rubens – the cardiac stomach, tube feet and apical muscle – but with S2 more potent/effective than S1 (Elphick et al. 1995; Melarange et al. 1999; Elphick and Melarange 2001; Melarange and Elphick 2003). Subsequently, other members of the SALMFamide neuropeptide family were identified in other echinoderms (e.g. sea cucumbers) and these peptides were also found to act as muscle relaxants (Diaz‐Miranda and Garcia‐Arraras 1995; Ohtani et al. 2002).
It is unlikely that SALMFamides are the only family of neuropeptides that act as muscle relaxants in echinoderms, given the multitude of neuropeptides that have been found to act as muscle relaxants in vertebrates and protostomian invertebrates (see above). Therefore, here we set out to employ use of an in vitro muscle bioassay to screen for muscle relaxants in an echinoderm. The starfish species Patiria pectinifera was selected as a model system because it is widely distributed in the northern Pacific Ocean, and can be easily collected and transported as it is found in shallow coastal waters. This species adapts well to artificial conditions in the laboratory and as a non‐specialized predator and/or scavenger it can be fed on algae, detritus and small invertebrates. For these reasons, this species has been used in many scientific studies as a model organism for studying starfish physiology, and it is also of interest from both economic and environmental perspectives (Ikegami et al. 1967; Dan‐Sohkawa et al. 1986; Davydov et al. 1990; Mita et al. 2009; Jo et al. 2013; Haraguchi et al. 2015). The apical muscle of P. pectinifera was selected as a bioassay because it can be easily dissected from the aboral body wall of the arms in this species. Furthermore, as highlighted above, previous studies have revealed that SALMFamides cause relaxation of the apical muscle from the starfish A. rubens (Melarange and Elphick 2003).
Here, we report the isolation of a novel neuropeptide from P. pectinifera that causes relaxation of the apical muscle from this species – starfish myorelaxant peptide (SMP). A cDNA encoding the SMP precursor protein was cloned and sequenced, enabling investigation of its expression pattern in P. pectinifera and investigation of relationships with neuropeptides that have been identified in other echinoderms and other phyla.
Methods
Animals
Live specimens of the starfish species Patiria pectinifera (Fig. 1a and b) and Asterias amurensis were collected at Cheongsapo of Busan, Korea, and maintained in a recirculating seawater system at 15°C until use. The animals were fed once every 3 days with live manila clam, Ruditapes philippinarum. Live specimens of the starfish species Asterias rubens were collected at low tide from the Thanet coast of Kent in the UK, and maintained in a recirculating seawater system at 12°C until use. The animals were fed weekly with live mussels (Mytilus edulis). Approval by the local institution/ethics committee was not required for this work because experimental work on starfish is not subject to regulation.
Figure 1.
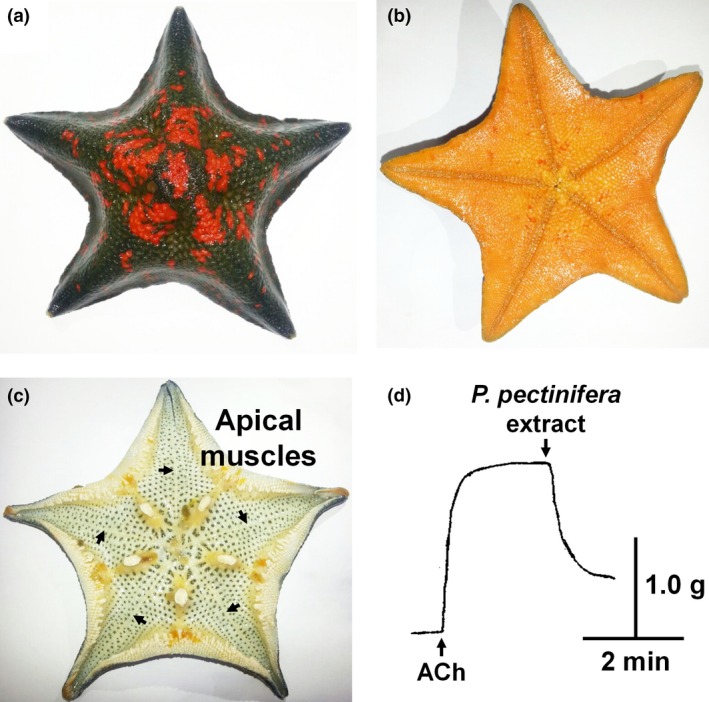
The starfish Patiria pectinifera. The aboral side (a) and oral side (b) of an intact animal are illustrated. The position of the apical muscles on the inner surface of the aboral body wall of a dissected animal is shown in (c) marked by arrows. An extract of P. pectinifera containing peptidic materials relaxed apical muscle that was pre‐contracted with 1 μM acetylcholine (ACh); up and down arrows represent application of ACh and the extract, respectively (d).
Peptide extraction
Starfish (P. pectinifera) were cut into pieces using scissors, soaked in 70% methanol and then heated in a double boiler for 5 min to denature proteins and inhibit proteolytic enzyme activity. The boiled sample was cooled on ice and then homogenized (PT10‐35; Kinematica AG, Luzern, Switzerland), followed by addition of glacial acetic acid to yield a final concentration of 5% acetic acid. The homogenate was then centrifuged (10, 000 g, 40 min, 4°C). The pellet was re‐extracted in 5% acetic acid with same extraction method. The supernatant was pooled and concentrated using a rotary evaporator. The concentrated solution was diluted with 10 volumes of ethanol and then the suspension was centrifuged (10, 000 g, 40 min, 4°C) to remove the precipitate. The supernatant was evaporated to 100 mL, and then 100 mL of ethanol with 1.1 g sodium chloride was added to it. After centrifugation to remove precipitate, the supernatant was concentrated by evaporation, and 0.1 volume of 1 N hydrochloric acid was added. The precipitate was removed again by centrifugation (20, 000 g, 50 min, 4°C) and the supernatant was applied to a C18 cartridge (Sep‐pak C18; Waters Corp., Miford, Massachusetts, USA). The column was washed with 10% methanol/0.1% trifluoroacetic acid (TFA) and retained materials were then eluted with 60% methanol/0.1% TFA. The eluate was evaporated and its biological activity on the apical muscle of P. pectinifera was investigated, as described below in the methods section for in vitro bioassay and pharmacology.
Peptide purification
The 60% methanol eluate was applied to a cation‐exchange column (CM‐52, 2.5 × 30 cm; Whatman, Maidstone, UK), and eluted with a linear gradient of 0.02–1.5 M ammonium acetate (pH 5.0) for 6 h at a flow rate of 2.75 mL/min. Absorbance peaks were monitored at 254 nm (ISCO Model UA‐6 detector; Lincoln, NE, USA) and fractions were collected every 4 min. The bioactive fractions, which eluted between fraction numbers 40–45, were pooled and then subjected to reversed phase (RP)‐HPLC (Vydac 218TP510 Protein & Peptide C18, 9.2 × 250 mm; The Separation Group. Inc., Hesperia, CA, USA). Elution was performed with a linear gradient of 0–60% acetonitrile/0.1% TFA at a flow rate of 3.0 mL/min for 120 min, and fractions were collected every 2 min.
Bioactive fractions eluted between 50 and 54 min with RP‐HPLC and these were subjected to further purification steps using an anion‐exchange column (TSKgel DEAE‐5PW, 7.5 × 75 mm; Tosho Corp., Minato‐ku, Tokyo, Japan) with a linear gradient of 0–0.5 M sodium chloride in 10 mM Tris‐HCl (pH 9.2) at a flow rate of 0.5 mL/min for 100 min. A fraction that eluted with a concentration of about 0.1 M sodium chloride from the anion‐exchange column caused relaxation of the apical muscle from P. pectinifera. This eluate was subjected to further RP‐HPLC (Capcellpak C18, 4.6 × 250 mm; Shisheido CO. LTD., Chuo‐ku, Tokyo, Japan). The absorbance peaks were recovered with a linear gradient of 15–30% acetonitrile/0.1% TFA at flow rate of 1 mL/min for 60 min. The bioactive peak was then subjected again to RP‐HPLC using the same solvent gradient as in the previous RP‐HPLC step but with a different column (Hypersil‐BDS C18, 2 × 125 mm; HP, Waldbronn, Germany). Finally, the active peak was applied to the same column as in the previous step but with an isocratic elution of 20% acetonitrile/0.1% TFA at a flow rate of 0.5 mL/min.
Structure determination and synthesis of peptides
To determine the molecular mass and amino acid sequence of the purified SMP, it was analysed using an automated N‐terminal amino acid gas‐phase sequencer (PPSQ‐1; Shimadzu Corp. Nakagyo‐ku, Kyoto, Japan) and a MALDI‐TOF mass spectrometer (Voyager‐DE™ PRO spectrometer; Perseptive Biosystem, Framingham, MA, USA). On the basis of the structural determination results, two peptides, with or without the carboxyl‐terminus amidated, were automatically synthesized by a conventional solid‐phase method with Fmoc‐protected amino acids and coupling reagents, 1‐hydroxybenzotriazole and N,N‐diisopropylcarbodimide, using a peptide synthesizer (PSSM‐8; Shimadzu) as described previously (Kim et al. 2015). Other neuropeptides, S1 (GFNSALMFamide), S2 (SGPYSFNSGLTFamide), FMRFamide and FLRFamide were synthesized to enable comparison of their activities with that of the identified peptide.
In vitro bioassay and pharmacology
Three neuromuscular preparations, apical muscle, cardiac stomach and tube feet, were dissected from P. pectinifera for in vitro bioassay and pharmacology according to a slightly modified version of previously reported methods (Elphick et al. 1995; Melarange and Elphick 2003). Synthetic neuropeptides were also tested for bioactivity on apical muscle preparations from a different starfish species – A. amurensis. Briefly, the apical muscle was cut from the aboral body wall of an arm, where the apical muscle forms a thickening of longitudinally orientated muscle that runs along the mid‐line of the inner side (Fig. 1c). A piece of cardiac stomach between the oral opening and extrinsic retractor strand was obtained by removing the aboral body wall from the central disc and the proximal region. An individual whole tube foot was dissected from the arm ambulacra but without the ampulla. All muscle preparations were cut to approximately 10 mm, and both ends of the muscle preparations were tied with cotton threads. The preparations were then suspended vertically in a 2 mL polypropylene chamber containing artificial seawater (ASW) with aeration, one end being connected to silver hook on the bottom of the chamber and the other to a force displacement transducer (Type 45196A; NEC‐Sanei Instrument Ltd., Tokyo, Japan). Output from the force displacement transducer was monitored by a recorder (WR7300; GRAPHTEC CORP., Yokohama, Japan) via an amplifier (AS1302; NEC‐Sanei Instrument Ltd.), which recorded the mechanical responses of the device. Prior to testing, the muscle preparations were allowed to stabilize for about 90 min. The resting tension was then adjusted to 1.0 g for apical muscle and 0.5 g for cardiac stomach and tube foot. Muscles in the chamber were allowed to equilibrate for about 30 min in ASW, during which time the ASW in the chamber was freshly replaced every 15 min. Pre‐contraction of apical muscle, cardiac stomach or tube foot preparations was induced by applying 1 μM acetylcholine (ACh), 10 μM carbachol or 30 mM high‐potassium ASW respectively. Then immediately after equilibration, the muscles were treated with test samples to measure relaxation responses.
The bioassay system adopted for monitoring purification of the bioactive peptide was a system that measures relaxation of apical muscle from P. pectinifera pre‐contracted for 2 min at 20 min intervals with 1 μM ACh. An aliquot of each test fraction was evaporated to dryness, dissolved with 50 μL of phosphate‐buffered saline, and added into the chamber.
At least four separate experiments to test the pharmacological activities of synthetic SMP, C‐terminally amidated SMP (SMPamide) and other neuropeptides were performed, using a concentration range of 10−10 M to 10−5 or 10−4 M at 25°C. EC50 values represent the concentration of peptide required to cause a response 50% of the maximum. The maximal response (E max) was expressed as the percentage of the maximal relaxation induced by 10−4 or 10−5 M of each peptide compared to the maximal contraction of apical muscle by 1 μM ACh, of cardiac stomach by 10 μM carbacol or of tube foot by 30 mM high‐potassium ASW. The relative activity was calculated as the ratio of the concentration of SMP or other peptides required to produce responses equivalent to a half‐maximal response.
cDNA cloning and sequence analysis
Total RNA was extracted using RNeasy Mini kit (Qiagen, Valencia, CA, USA) from total tissues (except body wall) of P. pectinifera, and then mRNA was purified using Oligotex mRNA mini kit (Qiagen) following the manufacturer's instructions. The synthesis rapid amplification cDNA end (RACE)‐ready cDNA template was performed with SMARTer™ RACE cDNA amplification Kit (Clontech, Mountain view, CA, USA) according to manufacturer's instructions.
Based on the amino acid sequence of the purified peptide, two degenerate primers were designed for 3′ RACE PCR, and then 5′ RACE PCRs were conducted with sequence‐specific primers designed from the sequencing result of the 3′ RACE product. The sequences of primers used in RACE are listed in Table S1. The first PCR conditions for 3′ RACE included initial denaturation at 94°C for 3 min followed by: 5 cycles of 94°C for 1 min, 59°C for 1 min and 72°C for 1 min; 5 cycles of 94°C for 1 min, 57°C for 1 min and 72°C for 1 min; 20 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min. Nested PCR for 3′RACE was performed with the same conditions as the first PCR. The first PCR product of 5′ RACE was obtained by the following thermal cycle profile: 5 cycles of 94°C for 30 s, 67°C for 30 s and 72°C for 1 min; 5 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 1 min; 25 cycles of 94°C for 30 s, 63°C for 30 s and 72°C for 1 min. Nested PCR for 5′ RACE was as follows: 5 cycles of 94°C for 30 s, 69°C for 30 s and 72°C for 1 min; 5 cycles of 94°C for 30 s, 67°C for 30 s and 72°C for 1 min; 25 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 1 min.
PCR products in the last step of 3′ and 5′ RACE were introduced into the pGEM‐Teasy vector system (Promega, Madison, WI, USA) and sequenced. The full‐length translated sequence of the SMP precursor, based on the cloned cDNA nucleotide sequence, was aligned by BLAST (http://blast.ncbi.nlm.nih.gov/blast.cgi) and the sequence was submitted to the GenBank database (Accession number: KT870152). Multiple sequence alignment of the full‐length P. pectinifera SMP precursor and related proteins in other species was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Asterias rubens radial nerve cord transcriptome sequence data obtained by Illumina HiSeq sequencing, as reported previously (Semmens et al. 2013), were analysed using BLAST to identify a homologue of the P. pectinifera SMP precursor. Using the sequence of the P. pectinifera SMP precursor as a query, a 444 bp A. rubens contig (1025452) comprising a partial sequence corresponding to the 3′ region of the P. pectinifera SMP precursor cDNA was identified. Then ovarian transcriptome sequence data obtained from multiple echinoderm species [(Reich et al. 2015); http://www.echinobase.org/Echinobase/Blasts] was analysed and non‐overlapping contigs encoding the 5′ region (GAUS01027726.1) and the 3′ region (GAUS01027727.1) of a SMP‐type precursor transcript was identified from the starfish species Asterias forbesi. Combining these partial sequence data from A. rubens and A. forbesi, primers were designed to enable PCR amplification of the full‐length SMP precursor coding sequence from A. rubens, as described below.
Total RNA was extracted from radial nerve cords of A. rubens using the SV Total RNA Isolation System according to the manufacturer's instructions (Promega). Then cDNA was synthesized using the QuantiTect Rev. Transcription Kit in accordance with the manufacturer's instructions (Qiagen). A cDNA containing the coding sequence of the A. rubens SMP‐type precursor was amplified by PCR using Phusion high‐fidelity PCR master mix (NEB, Ipswich, MA, USA) and the oligo primers 5′‐ATGCGGCTCATCATGCAC‐3′ and 5′‐TACACACCAAGCAGTGACA‐3′. The conditions for PCR included initial denaturation at 98°C for 2 min followed by: 30 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 1 min, 72°C for 8 min and hold at 4°C. 1% gel electrophoresis was performed to analyse the PCR products and then the PCR product was gel‐extracted and purified using a QIAquick gel extraction kit (Qiagen). Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA, USA) was used to ligate the PCR product into the pCR‐Blunt II with TOPO vector for sequencing. The sequence obtained (GenBank accession number KT870153) was translated into protein sequence using ExPASy (http://web.expasy.org/translate/) and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was used to predict the signal peptide of the translated protein sequence.
Real‐time quantitative PCR (RT‐qPCR) analysis
To quantitatively analyse expression of SMP precursor transcripts in different starfish tissues/organs, RT‐qPCR was employed using a LightCycler 480 Real‐Time PCR System (Roche, Mannheim, Germany) with LightCycler 480 SYBR green master I (Roche). Total RNA extracted from the apical muscle, radial nerve cord, cardiac stomach, pyloric stomach, coelomic lining containing transverse muscles, tube feet, pyloric caecae, testis and ovary were obtained from five specimens of P. pectinifera. cDNA was synthesized using the TOPscript cDNA synthesis Kit with oligo dT (dT18) (Enzynomics, Daejeon, Korea) according to the manufacturer's instructions. The primer pairs used for amplifying SMP precursor cDNA and elongation factor 1α (EF1α) cDNA as a control for normalization were SMP RT‐F and SMP RT‐R, and EF1α RT‐F and EF1α RT‐R respectively (see Table S1 for sequences). Based on the standard curves for both SMP and EF1α, the relative expression levels of SMP transcripts in each tissue were normalized against the level of the EF1α control using the following formula: relative expression = [(1 + E SMP)CP_SMP]−1/[(1 + E EF1α)CP_EF1α]−1, in which E is PCR efficiency (E = 10−1/slope − 1) and CP is the threshold cycle number. Triplicate amplifications were carried out independently, and the relative quantification results were expressed as the fold levels of SMP precursor transcripts.
Statistical analysis
All data are presented as means ± standard deviation. The statistical analysis for pharmacological data were performed using two‐way analysis of variance (anova) supported by Bonferroni's multiple comparisons test when carrying out pair wise comparison between the same doses of different peptides. For RT‐qPCR data, comparison between tissues was carried out by one‐way anova, followed by Duncan's Multiple Range test.
Statistical analyses were performed using spss 21 program (SPSS, Chicago, IL, USA) and graphs were generated using GraphPad Prism software version 6.0 for Windows (GraphPad Software, San Diego, CA, USA). p values with p < 0.05 were considered statistically significant.
Results
Purification of a novel hexadecapeptide that relaxes the apical muscle of P. pectinifera
A whole‐body extract of P. pectinifera induced relaxation of apical muscle pre‐contracted with ACh (Fig. 1d), indicating that it was an appropriate source to isolate myorelaxants. A single absorbance peak (peak A) containing a myorelaxant was successfully purified from the whole‐body extract through six steps of column purification which sequentially were cation, repeated RP and anion HPLC. Finally, peak A was isocratically eluted with 20% acetonitrile/0.1% TFA at 16.1 min of the retention time (Fig. 2a). An aliquot of peak A relaxed the apical muscle of P. pectinifera that was pre‐contracted by ACh (Fig. 2b). Purified peak A was identified as a sixteen‐residue peptide with the sequence Phe‐Gly‐Lys‐Gly‐Gly‐Ala‐Tyr‐Asp‐Pro‐Leu‐Ser‐Ala‐Gly‐Phe‐Thr‐Asp with a free carboxy terminus based on N‐terminal amino acid sequencing and molecular mass analysis (Fig. 2c and Figure S1a). The purified hexadecapeptide was designated SMP. To confirm the primary structure and chemical properties of SMP under RP‐HPLC, SMP with a free carboxy terminus (SMP) and SMP with an amidated carboxy terminus (SMPamide) were synthesized. The synthetic SMP and native SMP eluted with an identical retention time, and a mixture of the two peptides eluted as a single peak under RP‐HPLC (Fig. 2d). Moreover, SMP and SMPamide did not have an identical retention time on RP‐HPLC (Figure S1b). Collectively, the results demonstrate that the purified SMP is the hexdecapeptide Phe‐Gly‐Lys‐Gly‐Gly‐Ala‐Tyr‐Asp‐Pro‐Leu‐Ser‐Ala‐Gly‐Phe‐Thr‐Asp‐OH, with a free carboxy terminus and without any post‐translational modifications.
Figure 2.
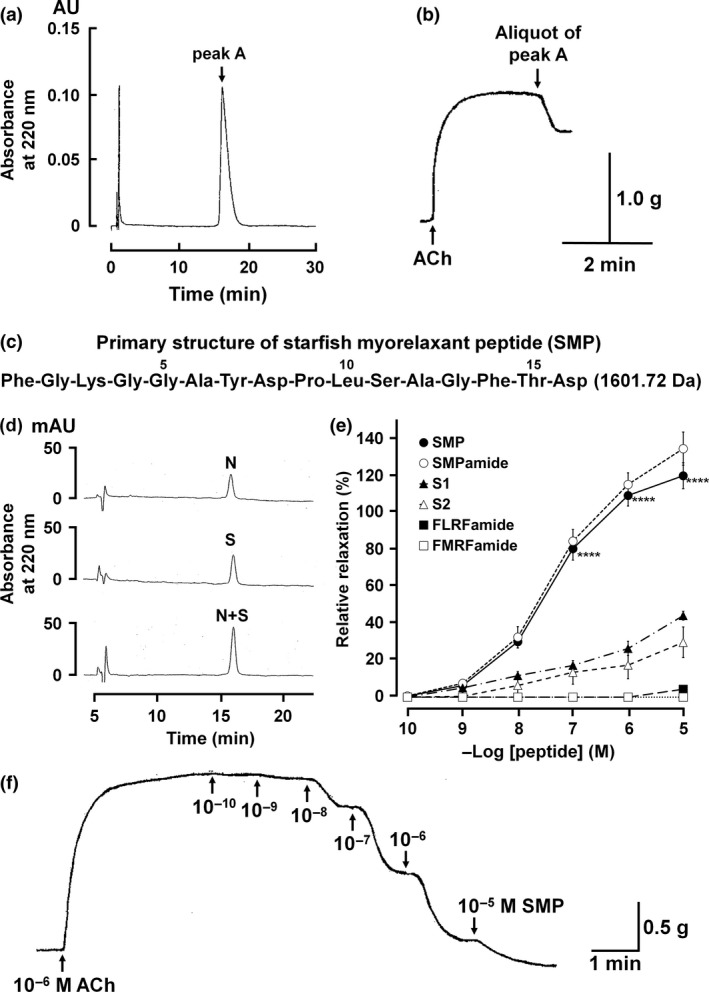
Isolation, structure determination and pharmacology of purified myorelaxant peptide. Peak A was isocratically eluted with 20% acetonitrile/0.1% trifluoroacetic acid on RP‐HPLC (a), and an aliquot of purified peak A caused relaxation of the apical muscle (b). Purified peak A was identified as a peptide comprised of sixteen amino acid residues with a molecular mass of 1601.72 Da, which we have named starfish myorelaxant peptide or SMP (c). Comparison of chromatographic properties of native SMP (N) and synthetic SMP (S) on RP‐HPLC showed that native SMP and synthetic SMP with a free carboxy terminal have identical retention times on RP‐HPLC (d). The concentration‐dependent relaxing activity of SMP on the apical muscle of P. pectinifera. SMP with free carboxyl terminus and amidated carboxy terminus is SMP (●) and SMPamide (○), respectively. The effects of S1 (▲) and S2 (▵) from the starfish A. rubens and the molluscan neuropeptides FLRFamide (■) and FMRFamide (□) are shown to compare their activity with SMP. Each point represents the mean ± standard deviation determined from four separate experiments. ****p < 0.0001 for SMP (●) compared with S1/S2. The percentage relaxing activity was calculated by comparing each relaxation effect to the maximal contraction of the apical muscle by 1 μM ACh (e). Representative recording of the concentration‐dependent relaxing effect of SMP on P. pectinifera apical muscle pre‐contracted with 1 μM ACh (f).
SMP is a potent relaxant of P. pectinifera apical muscle in vitro
SMP and a C‐terminally amidated analogue of SMP (SMPamide) both caused dose‐dependent relaxation of in vitro apical muscle preparations from P. pectinifera (Fig. 2e). The threshold response, ED50 and E max for SMP were 10−10 M, 6.0 × 10−8 M and 120.02 ± 7.00% and for SMPamide were 10−10 M and 4.0 × 10−8 M and 134.69 ± 9.57% respectively (Fig. 2e). These results corroborate the structural determination that SMP is not C‐terminally amidated and does not require C‐terminal modification for its bioactivity. Comparison of the bioactivity of SMP with the starfish SALMFamide neuropeptides S1 and S2 revealed that SMP was more potent/efficacious than these peptides as a relaxant of the apical muscle from P. pectinifera (Fig. 2e). Thus, the E max for S1 and S2 at a concentration of 10−5 M were only 44.15 ± 2.41 and 29.72 ± 8.29 respectively. Furthermore, the molluscan neuropeptides FLRFamide and FMRFamide, which share some sequence similarity with SALMFamides, exhibited little or no bioactivity as relaxants of apical muscle preparations, even at concentrations as high as 10−5 M (Fig. 2e).
The P. pectinifera SMP precursor protein comprises twelve copies of SMP and seven copies of other SMP‐like peptides
A cDNA encoding the P. pectinifera SMP precursor was cloned and sequenced (GenBank accession number: KT870152) and the nucleotide sequence and the deduced protein sequence are shown in Fig. 3. The cDNA sequence comprised 1682 bp, starting with a 5′ untranslated region of 148 bp, followed by an open reading frame of 1281 bp, a 3′ untranslated region of 253 bp including a poly‐A tail. The open reading frame of the SMP precursor encodes a 426 amino acid residue protein that contains four regions: a signal peptide (Met1‐Ala19), as predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), a N‐terminal spacer peptide (Ser20‐Arg56) containing several acidic amino acids, a region containing twelve copies of SMP and seven copies of SMP‐like peptides with each peptide copy bounded by dibasic cleavage sites (Phe57‐Arg402) and a C‐terminal region (Thr403‐Arg426).
Figure 3.
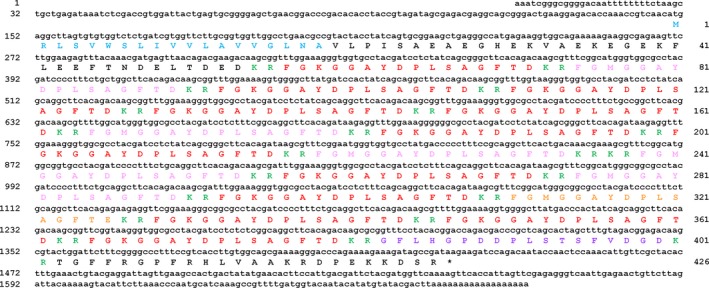
Precursor of starfish myorelaxant peptide (SMP) in Patiria pectinifera. The DNA sequence of a transcript (lowercase, 1682 bases) encoding the P. pectinifera SMP precursor (uppercase, 426 amino acid residues) is shown. The predicted signal peptide, the purified mature SMP (SMP a) and three other variants (SMP b, [Met3]‐SMP a; SMP c, [Met3, Glu16]‐SMP a; SMP d, SMP a‐related octadecapeptide) are shown in blue, red, pink, orange and purple, respectively, and putative dibasic cleavage sites (KR) are shown in green. The asterisk shows the position of the stop codon. The SMP precursor protein comprises twelve copies of SMP and seven copies of SMP‐like peptides.
SMP precursor transcripts are widely expressed in P. pectinifera and SMP causes in vitro relaxation of other muscle preparations from P. pectinifera
The relative expression levels of the SMP precursor mRNA in different tissues (apical muscle, radial nerve cord, cardiac stomach, pyloric stomach, coelomic lining, tube feet, pyloric caecae, testis and ovary) were determined by RT‐qPCR (Fig. 4a). The highest expression of SMP precursor transcripts was detected in radial nerve cords, which are the major components of the nervous system in starfish. In addition, relatively high expression levels of SMP precursor transcripts were observed, in descending order, in apical muscle, tube feet, coelomic lining, cardiac stomach, pyloric stomach and pyloric caecae. However, the expression of SMP precursor transcripts in reproductive organs (ovary and testis) was barely detectable. These findings indicate that SMP is a neuropeptide and suggest that SMP may have widespread roles as a regulator of muscle activity in P. pectinifera. To address this issue, SMP was tested in vitro on two other neuromuscular preparations in which SMP precursor transcripts are detected – cardiac stomach and tube feet. SMP caused dose‐dependent relaxation of both preparations and, as with apical muscle preparations, SMP was more potent/effective as a muscle relaxant than the SALMFamides S1 and S2 (Fig. 4b and c).
Figure 4.

The expression levels for the starfish myorelaxant peptide (SMP) precursor transcript in various organs/tissues from P. pectinifera and the pharmacological effects of SMP on cardiac stomach and tube foot from P. pectinifera. Relative expression levels of SMP transcripts in each organ/tissue were normalized against the level of the EF1α gene as an internal control. Mean ± standard deviation (n = 3) are shown. Means denoted by the same letter did not differ significantly (p > 0.05) whilst different letters (a, b, c, d, e) at the top of the bars indicate statistically significant differences (p < 0.05) between tissues determined by one‐way anova followed by Duncan's Multiple Range test (a). SMP caused concentration‐dependent relaxation of the cardiac stomach (b) and tube foot (c) from P. pectinifera. The relaxing activity of SMP (●) was compared with S1 (▲) and S2 (▵). Each point represents the mean ± standard deviation determined from four separate experiments. Statistically significant difference between SMP and S1/S2 represents with ****p < 0.0001. The percentage relaxing activity was calculated by comparing each relaxation effect to the maximal contraction of cardiac stomach caused by 10 μM carbacol and of tube foot caused by 30 mM high‐potassium artificial seawater respectively. Representative recordings of the effects of SMP on cardiac stomach (d) and tube foot (e) preparations are shown.
SMP causes relaxation of apical muscle preparations from the starfish Asterias amurensis and identification of an SMP‐type precursor in Asterias rubens
Having identified SMP as a muscle relaxant in P. pectinifera, we then investigated if this peptide also acts as a muscle relaxant in other starfish species. To address this issue we tested synthetic SMP on apical muscle preparations from A. amurensis. SMP caused dose‐dependent relaxation and the E max was 82.1 ± 1.94% at a concentration of 10−5 M (Fig. 5a). Previous studies have shown that the SALMFamide neuropeptides S1 and S2 cause relaxation of apical muscle preparations from Asterias rubens, which is closely related to A. amurensis (Melarange and Elphick 2003). Therefore, we compared the bioactivity of SMP with S1 and S2 and found that the E max for S1 and S2 at a concentration of 10−5 M were less than for SMP, 62.3 ± 4.4 and 38.7 ± 1.21, respectively (Fig. 5a). However, by comparison with S1 and S2, SMP was less effective as a relaxant of the apical muscle from A. amurensis (Fig. 5a) than from P. pectinifera (Fig. 2e). These findings indicate that SMP or a related peptide(s) exists in A. amurensis and that SMP‐type peptides are likely to act as muscle relaxants throughout the Asteroidea. Accordingly, a cDNA encoding an SMP‐type precursor was identified in A. rubens, comprising a 224‐residue protein with a predicted 20‐residue signal peptide and eight copies of putative SMP‐like peptides: four copies of the peptide FGGKGAFDPLSAGFTD, two copies of the peptide FGGSRGAFDPLSAGFTD and one copy each of GFGMGAYDPLSAGFTD and SFVHGDFDPLSTGFVDGD (Fig. 5b and Figure S2, GenBank Accession number: KT870153). It is noteworthy that the C‐terminal region of three of these peptides (DPLSAGFTD) is identical to the corresponding region of P. pectinifera SMP.
Figure 5.
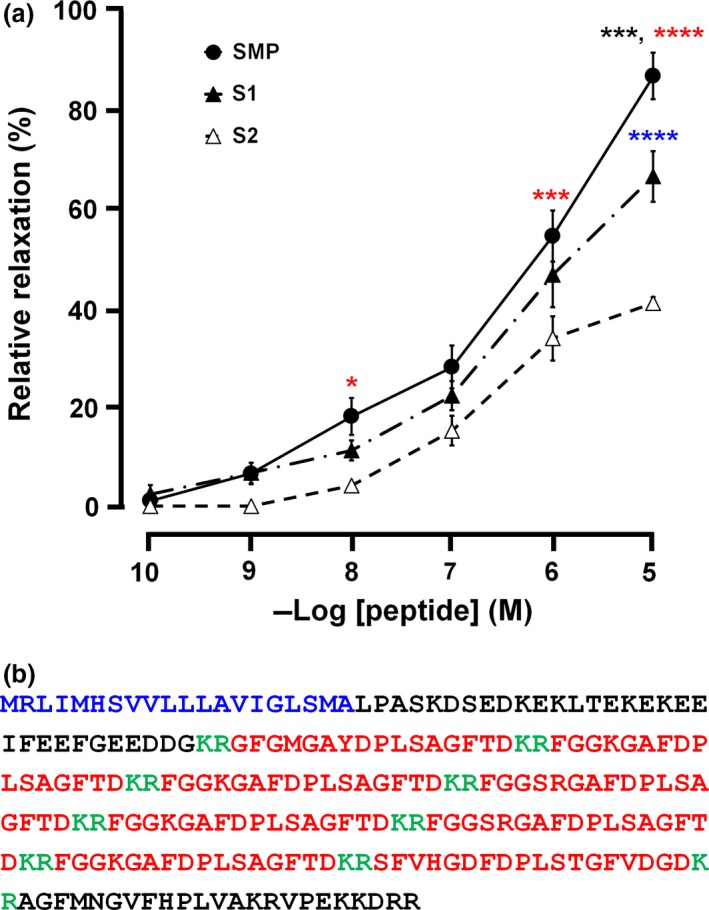
Pharmacological effect of starfish myorelaxant peptide (SMP) on apical muscle from Asterias amurensis and identification of an SMP‐type precursor in Asterias rubens. (a). The concentration‐dependent relaxing activity of SMP (●) compared with S1 (▲) and S2 (▵) on the apical muscle of A. amurensis. Each point represents the mean ± standard deviation determined from four separate experiments. Statistically significant differences between the effects of SMP and S1, SMP and S2, and S1 and S2 are represented by black, red and blue asterisks (*p < 0.05, ***p < 0.001 and ****p < 0.0001), respectively. The percentage relaxing activity was calculated by comparing each relaxation effect to the maximal contraction of apical muscle caused by 1 μM ACh. (b) Amino acid sequence of a 224‐residue SMP‐type precursor protein identified in A. rubens, which comprises a predicted 20‐residue signal peptide (blue) and eight copies of putative SMP‐like peptides (red) and putative dibasic cleavage sites (KR, green). The sequence of the cDNA encoding this protein is shown in Figure S2.
Starfish SMP precursors are homologues of neuropeptide precursors that have been identified in other echinoderms
To investigate relationships with neuropeptide precursors that have been in other animals, the P. pectinifera and A. rubens SMP precursor proteins were submitted as queries against the GenBank nr database using BLAST. The top two hits (XP_785647.1 and XP_003727926) were identified as neuropeptide precursor proteins that have been described previously from the sea urchin Strongylocentrotus purpuratus and designated as Spnp6 and Spnp7, respectively (Rowe and Elphick 2012). In Fig. 6, we show a multiple sequence alignment of the P. pectinifera SMP precursor, the A. rubens SMP‐type precursor, Spnp6, Spnp7 and a homologue of Spnp7 that has been identified in the sea cucumber Apostichopus japonicus [Ajnp7 (Rowe et al. 2014)]. Furthermore, alignment of the putative neuropeptides derived from the P. pectinifera SMP precursor, the A. rubens SMP‐type precursor, Spnp6, Spnp7 and Ajnp7 (Fig. 7) reveals that the peptides have a number of features in common. These include two phenylalanine residues located at or near the N‐ and C‐termini of the peptides as well as a conserved core region with the motif (D/E)‐(P)‐(L/M), structural characteristics that may be important for the bioactivity of these peptides.
Figure 6.
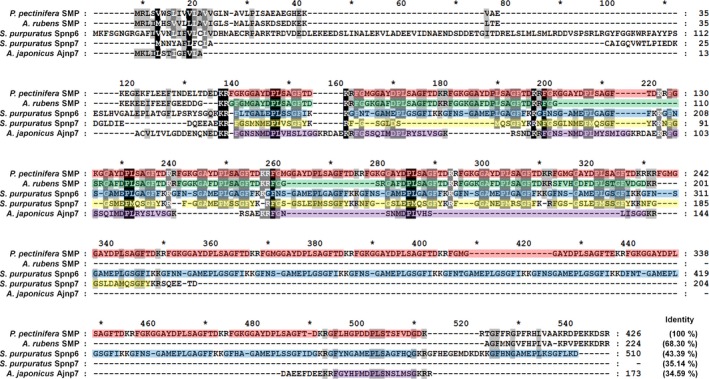
Multiple sequence alignment of the P. pectinifera starfish myorelaxant peptide (SMP) precursor with related neuropeptide precursors in other echinoderms. Highlighted red, green, blue, yellow and purple boxes represent multiple copies of neuropeptides separated by putative cleavage sites (KR or KK). All of the precursors contain multiple copies of related peptides: P. pectinifera SMP precursor contains twelve copies of SMP and seven copies of SMP‐like peptides; A. rubens SMP precursor contains eight copies of SMP‐like peptides; S. purpuratus neuropeptide precursor 6 (Spnp6) contains twenty‐one copies of nine SMP‐like peptides; Spnp7 precursor contains ten copies of nine SMP‐like peptides; A. japonicus neuropeptide precursor 7 (Ajnp7) contains six copies of five SMP‐like peptides. The sequences of Spnp6, Spnp7 and Ajnp7 are from (Rowe and Elphick 2012; Rowe et al. 2014).
Figure 7.
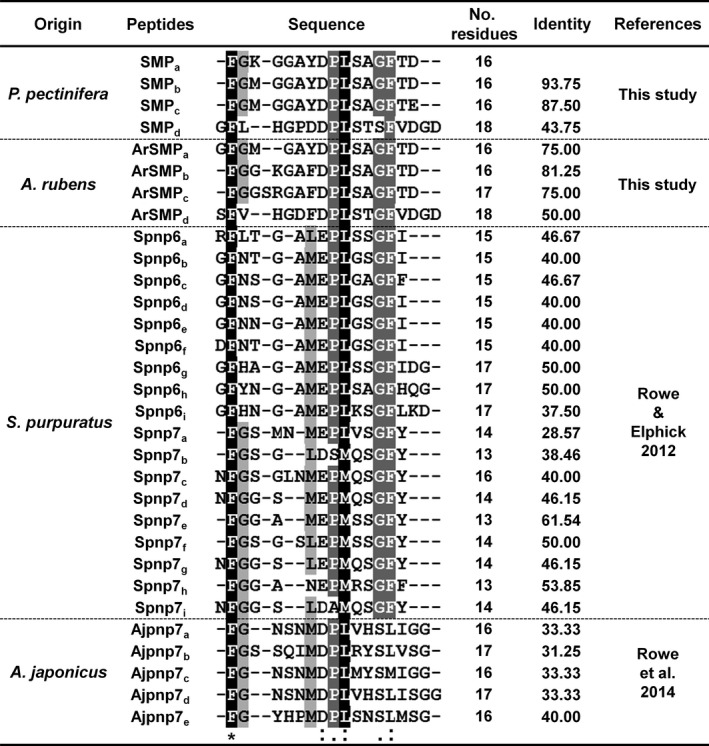
Alignment of starfish myorelaxant peptide (SMP a) with putative SMP‐like neuropeptides derived from echinoderm SMP‐type precursors: the starfish P. pectinifera and A. rubens; sea urchin S. purpuratus; sea cucumber A. japonicus. Conserved residues are highlighted in black and grey.
Discussion
Here, we have isolated a novel hexadecapeptide (FGKGGAYDPLSAGFTD) from starfish that acts as a muscle relaxant and which we have designated as SMP. Previous studies have identified the SALMFamide neuropeptides S1 and S2 as muscle relaxants in starfish (Melarange et al. 1999; Melarange and Elphick 2003) and here the bioactivity of SMP, S1 and S2 as muscle relaxants were compared. When tested on three preparations from P. pectinifera (apical muscle, cardiac stomach and tube feet), SMP was more effective/potent than S1 or S2. This finding is likely to be physiologically relevant with respect to S1 because we know that S1 occurs in the closely related species Patiria miniata. However, P. miniata does not contain S2 and this species has instead an S2‐like peptide (Elphick et al. 2013, 2015). Therefore, the inferior bioactivity of S2 as a myorelaxant in P. pectinifera may in part be attributable to differences in peptide structure. Furthermore, analysis of the sequences of the two SALMFamide precursor proteins in P. miniata reveals that they comprise S1, the S2‐like peptide and fourteen other SALMFamide‐type peptides (Elphick et al. 2013, 2015). So comparison of the effects of SMP with S1 or S2 tested in isolation does not reflect the physiological occurrence of ‘cocktails’ of SALMFamides. Nevertheless, the superior bioactivity of SMP as a myorelaxant, compared to S1 and S2, in tests on muscle preparations from both P. pectinifera and Asterias amurensis clearly indicates that SMP is a physiologically important regulator of muscle relaxation in starfish.
Analysis of the distribution of the expression of the SMP precursor in P. pectinifera using qPCR revealed a widespread pattern of expression, including all three neuromuscular preparations that SMP causes relaxation of in vitro – the apical muscle, cardiac stomach and tube feet. Likewise, immunocytochemical‐ and radioimmunoassay‐based analysis of the distribution of S1 and S2 in A. rubens reveals a widespread pattern of expression (Moore and Thorndyke 1993; Elphick et al. 1995; Newman et al. 1995a,b). Therefore, it is likely that SMP and SALMFamide neuropeptides act in concert as muscle relaxants to regulate a variety of physiological processes in starfish. For example, relaxing effects on the apical muscle in vivo may be associated with neural mechanisms that control changes in body posture, whereas relaxing effects on tube feet in vivo may be associated with locomotor activity. The relaxing action of SALMFamides on the cardiac stomach is thought be relevant to neural mechanisms controlling stomach eversion during feeding in starfish (Melarange et al. 1999) and this role may equally apply to the novel SMP neuropeptide identified here. Further insights into the physiological roles of SMP and other SMP‐like peptides derived from the same precursor protein may be obtained by analysis of the distribution of these peptides at the cellular level. As highlighted above, detailed immunocytochemical analyses of the distribution of S1 and S2 in A. rubens have been reported previously (Moore and Thorndyke 1993; Elphick et al. 1995; Newman et al. 1995a,b) and it would be interesting to compare the distribution of SMP and SALMFamides using this approach.
Comparative analysis of the sequence of SMP and the SMP precursor with neuropeptides and neuropeptide precursors that have been identified in other animals reveals that SMP belongs to a bilaterian family of neuropeptides that includes molluscan pedal peptides (PP) and arthropodan orcokinins (OK) (Fig. 8, Figure S3 and Table S2). The occurrence of PP/OK‐type peptides in echinoderms has been reported previously based on analysis of genome/transcriptome sequence data (Rowe and Elphick 2012; Rowe et al. 2014) and in Figs 6 and 7, respectively, we show alignments of SMP and the SMP precursor with related PP/OK‐type neuropeptides and precursor proteins that have been identified in the sea urchin S. purpuratus and the sea cucumber A. japonicus. These alignments reveal conserved residues that may be important for the bioactivity of PP/OK‐type peptides in echinoderms. In Fig. 8 we show an alignment of SMP and other representative echinoderm PP/OK‐type peptides with molluscan pedal peptides and arthropodan orcokinins. What this reveals is the conservation of hydrophobic residues, which are typically phenylalanine, proximal to or at the N‐ and C‐termini of the peptides. This suggests that these evolutionarily conserved structural features are important for the bioactivity of PP/OK‐type peptides in the Bilateria. However, the motif (D/E)‐(P)‐(L/M) that is a conserved feature of the core of echinoderm PP/OK‐type peptides, including SMP (Fig. 7), is not seen in molluscan and arthropodan PP/OK‐type peptides and therefore this may be a unique characteristic of echinoderm representatives of this neuropeptide family. Investigation of the structure‐activity relationships of orcokinin in Orconectes limosus has revealed that C‐terminal amidation results in a reduction of bioactivity (Bungart et al. 1995), whereas here we found that C‐terminal amidation of SMP neither reduces nor enhances its bioactivity. These findings contrast with neuropeptides that are naturally C‐terminally amidated in vivo, where loss of the C‐terminal amide typically results in a dramatic loss of bioactivity (Greenberg and Price 1979).
Figure 8.
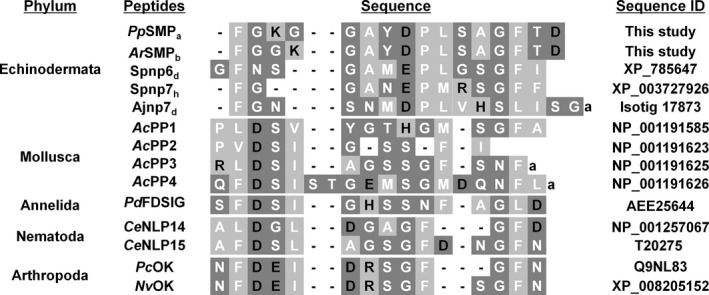
Alignment of echinoderm starfish myorelaxant peptide (SMP)‐type peptides with protostomian pedal peptide (PP)/orcokinin(OK)‐type peptides. The basic amino acids Lys, Arg and His are shown in the black with light grey highlighting, and the acidic residues Glu and Asp are shown in black with dark grey highlighting. All other amino acids are classified as hydrophobic (white with light grey highlighting) or hydrophilic (white with dark grey highlighting). Lower case ‘a’ denotes a C‐terminal amide group. Species abbreviations and references: Pp, P. pectinifera; Ar, A. rubens; Sp, S. purpuratus (Reich et al. 2015); Aj, A. japonicus (Du et al. 2012; Rowe and Elphick 2012); Ac, Aplysia californica (Moroz et al. 2006); Pd, Platynereis dumerilii (Conzelmann et al. 2011); Ce, Caenorhabditis elegans (Nathoo et al. 2001); Pc, Procambrus clarkii (Yasuda‐Kamatani and Yasuda 2000); Nv, Nasonia vitripennis (Hauser et al. 2010).
With the identification of SMP as a member of the PP/OK‐type family of neuropeptides, it is of interest to consider what is known about the physiological roles of these neuropeptides in other phyla. PP was originally discovered in the mollusc Aplysia californica as a peptide that causes contraction of pedal muscles (Lloyd and Connolly 1989; Hall and Lloyd 1990); it also stimulates beating of cilia associated with the foot (Longley and Peterman 2013). OK was first isolated from neural extracts of the crayfish Orconectus limosus on account of its stimulatory effect on hindgut activity (Stangier et al. 1992). Subsequently, OK‐type peptides have been identified in several arthropod species and found to have a variety of effects, including stimulation of the prothoracic gland and regulation of ecdysteroidogenesis in the silk moth Bombyx mori (Yamanaka et al. 2011) and regulation of circadian activity in the cockroach Leucophaea maderae (Hofer and Homberg 2006; Soehler et al. 2011; Wei and Stengl 2011). Thus, in both molluscs and arthropods, PP/OK‐type neuropeptides have stimulatory effects on the activity of muscle and other tissues. This contrasts with the inhibitory effect that SMP has in causing relaxation of muscle in starfish, as reported here. It will be interesting, therefore, to investigate in future studies if PP/OK‐type peptides also act as muscle relaxants in other echinoderms or if this is a unique characteristic of PP/OK‐type peptides in starfish.
Thus far, PP/OK‐type peptides have not been identified in other deuterostomian phyla such as hemichordates, which are a sister clade to the echinoderms, or chordates. One possibility is that PP/OK‐type peptides have been lost in hemichordates and chordates and the echinoderms are unique amongst the deuterostomes in retaining peptides belonging to this bilaterian neuropeptide family. Alternatively, the possibility remains that members of this neuropeptide family exist in hemichordates and chordates but their relationship with PP/OK‐type peptides has not been observed because of sequence divergence. Addressing this issue would be facilitated if the receptors that mediate the effects of PP/OK‐type peptides in echinoderms or in protostomes were identified, and therefore this represents an important objective for future research on PP/OK‐type peptides.
Supporting information
Figure S1. MALDI‐TOF mass spectrum of purified peak A (A). Comparison of the chromatographic properties of synthetic SMP with a free carboxyl terminus (SMP) and SMP with an amidated carboxyl terminus (SMPamide) on RP‐HPLC reveals that the two peptides elute at different retention times with isocratic 20% acetonitrile/0.1% TFA (B).
Figure S2. Precursor of starfish myorelaxant peptide (SMP)‐type neuropeptides in Asterias rubens.
Figure S3. Multiple sequence alignment showing that starfish SMP precursors share sequence similarity with other echinoderm pedal peptide‐type precursors (Spnp6, Spnp7, Ajnp7) and with protostomian pedal peptide (PP)/orcokinin(OK)‐type peptide precursors.
Table S1. Primers used for RACE and RT‐qPCR analysis of SMP precursor expression in P. Pectinifera.
Table S2. Identity and similarity grid showing the identity/similarity percentages for the amino acid sequences of SMP precursors and PP/OK‐type peptide precursors.
Acknowledgements and conflict of interest disclosure
This work was supported by Korea Ministry of Environment (MOE) as ‘Eco‐innovation Program (201300030002)’, the China Scholarship Council (ML) and Queen Mary University of London (MRE). The authors declare that they have no conflicts of interest with the contents of this article.
All experiments were conducted in compliance with the ARRIVE guidelines.
References
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud'homme B. and de Rosa R. (2000) The new animal phylogeny: reliability and implications. Proc. Natl Acad. Sci. USA 97, 4453–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendena W. G., Garside C. S., Yu C. G. and Tobe S. S. (1997) Allatostatins: diversity in structure and function of an insect neuropeptide family. Ann. N. Y. Acad. Sci. 814, 53–66. [DOI] [PubMed] [Google Scholar]
- Blackburn M. B., Wagner R. M., Kochansky J. P., Harrison D. J., Thomas‐Laemont P. and Raina A. K. (1995) The identification of two myoinhibitory peptides, with sequence similarities to the galanins, isolated from the ventral nerve cord of Manduca sexta . Regul. Pept. 57, 213–219. [DOI] [PubMed] [Google Scholar]
- Brain S. D., Williams T. J., Tippins J. R., Morris H. R. and MacIntyre I. (1985) Calcitonin gene‐related peptide is a potent vasodilator. Nature 313, 54–56. [DOI] [PubMed] [Google Scholar]
- Bungart D., Kegel G., Burdzik S. and Keller R. (1995) Structure‐activity relationships of the crustacean myotropic neuropeptide orkinin. Peptides 16, 199–204. [DOI] [PubMed] [Google Scholar]
- Conzelmann M., Offenburger S. L., Asadulina A., Keller T., Munch T. A. and Jekely G. (2011) Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl Acad. Sci. USA 108, E1174–E1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan‐Sohkawa M., Yamanaka H. and Watanabe K. (1986) Reconstruction of bipinnaria larvae from dissociated embryonic cells of the starfish, Asterina pectinifera . J. Embryol. Exp. Morphol. 94, 47–60. [PubMed] [Google Scholar]
- Davydov P. V., Shubravyi O. I. and Vassetzky S. G. (1990) The starfish Asterina pectinifera, in Animal Species for Developmental Studies, (Dettlaff T. A. and Vassetzky S. G., eds.), pp. 287–311. Springer, US. [Google Scholar]
- Diaz‐Miranda L. and Garcia‐Arraras J. E. (1995) Pharmacological action of the heptapeptide GFSKLYFamide in the muscle of the sea cucumber Holothuria glaberrima (Echinodermata). Comp. Biochem. Physiol. C 110, 171–176. [DOI] [PubMed] [Google Scholar]
- Du H., Bao Z., Hou R. et al (2012) Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867). PLoS ONE 7, e33311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick M. R. and Melarange R. (2001) Neural control of muscle relaxation in echinoderms. J. Exp. Biol. 204, 875–885. [DOI] [PubMed] [Google Scholar]
- Elphick M. R., Price D. A., Lee T. D. and Thorndyke M. C. (1991a) The SALMFamides: a new family of neuropeptides isolated from an echinoderm. Proc. Biol. Sci. 243, 121–127. [DOI] [PubMed] [Google Scholar]
- Elphick M. R., Reeve J. R., Jr , Burke R. D. and Thorndyke M. C. (1991b) Isolation of the neuropeptide SALMFamide‐1 from starfish using a new antiserum. Peptides 12, 455–459. [DOI] [PubMed] [Google Scholar]
- Elphick M. R., Newman S. J. and Thorndyke M. C. (1995) Distribution and action of SALMFamide neuropeptides in the starfish Asterias rubens . J. Exp. Biol. 198, 2519–2525. [DOI] [PubMed] [Google Scholar]
- Elphick M. R., Achhala S. and Martynyuk N. (2013) The evolution and diversity of SALMFamide neuropeptides. PLoS ONE 8, e59076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick M. R., Semmens D. C., Blowes L. M., Levine J., Lowe C. J., Arnone M. I. and Clark M. S. (2015) Reconstructing SALMFamide neuropeptide precursor evolution in the phylum echinodermata: ophiuroid and crinoid sequence data provide new insights. Front. Endocrinol. (Lausanne) 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. J. and Price D. A. (1979) FMRFamide, a cardioexcitatory neuropeptide of molluscs: an agent in search of a mission. Am. Zool. 19, 163–174. [Google Scholar]
- Grider J. R. and Makhlouf G. M. (1986) Colonic peristaltic reflex: identification of vasoactive intestinal peptide as mediator of descending relaxation. Am. J. Physiol. 251, G40–G45. [DOI] [PubMed] [Google Scholar]
- Hall J. D. and Lloyd P. E. (1990) Involvement of pedal peptide in locomotion in Aplysia: modulation of foot muscle contractions. J. Neurobiol. 21, 858–868. [DOI] [PubMed] [Google Scholar]
- Haraguchi S., Ikeda N., Abe M., Tsutsui K. and Mita M. (2015) Nucleotide sequence and expression of relaxin‐like gonad‐stimulating peptide gene in starfish Asterina pectinifera . Gen. Comp. Endocrinol. pii: S0016‐6480(15)00185‐9. doi: 10.1016/j.ygcen.2015.06.017. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hauser H., Neupert S., Williamson M., Predel R., Tanaka Y. and Grimmelikhuijzen C. J. (2010) Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J. Proteome Res. 9, 5296–5310. [DOI] [PubMed] [Google Scholar]
- Hofer S. and Homberg U. (2006) Evidence for a role of orcokinin‐related peptides in the circadian clock controlling locomotor activity of the cockroach Leucophaea maderae . J. Exp. Biol. 209, 2794–2803. [DOI] [PubMed] [Google Scholar]
- Holman G. M., Cook B. J. and Nachman R. J. (1986) Isolation, primary structure and synthesis of leucomyosuppressin, an insect neuropeptide that inhibits spontaneous contractions of the cockroach hindgut. Comp. Biochem. Physiol. C 85, 329–333. [DOI] [PubMed] [Google Scholar]
- Ikegami S., Tamura S. and Kanatani H. (1967) Starfish gonad: action and chemical identification of spawning inhibitor. Science 158, 1052–1053. [DOI] [PubMed] [Google Scholar]
- Jo Y. B., Park S. H., Jeon J. K., Ko C. H., Ryu C. and Park Y. K. (2013) Biodiesel production via the transesterification of soybean oil using waste starfish (Asterina pectinifera). Appl. Biochem. Biotechnol. 170, 1426–1436. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Go H. J. and Park N. G. (2015) Two myomodulins isolated from central nervous system of Northwest Pacific Sea Hare, Aplysia kurodai, and their activities on other mollusks. Protein Pept. Lett. 22, 341–347. [DOI] [PubMed] [Google Scholar]
- Kitamura K., Kangawa K., Kawamoto M., Ichiki Y., Nakamura S., Matsuo H. and Eto T. (1993) Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem. Biophys. Res. Commun. 192, 553–560. [DOI] [PubMed] [Google Scholar]
- Lloyd P. E. and Connolly C. M. (1989) Sequence of pedal peptide: a novel neuropeptide from the central nervous system of Aplysia . J. Neurosci. 9, 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley R. D. and Peterman M. (2013) Neuronal control of pedal sole cilia in the pond snail Lymnaea stagnalis appressa . J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 199, 71–86. [DOI] [PubMed] [Google Scholar]
- Melarange R. and Elphick M. R. (2003) Comparative analysis of nitric oxide and SALMFamide neuropeptides as general muscle relaxants in starfish. J. Exp. Biol. 206, 893–899. [DOI] [PubMed] [Google Scholar]
- Melarange R., Potton D. J., Thorndyke M. C. and Elphick M. R. (1999) SALMFamide neuropeptides cause relaxation and eversion of the cardiac stomach in starfish. Proc. Biol. Sci. 266, 1785–1785. [Google Scholar]
- Mirabeau O. and Joly J. S. (2013) Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl Acad. Sci. USA 110, E2028–E2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita M., Yoshikuni M., Ohno K., Shibata Y., Paul‐Prasanth B., Pitchayawasin S., Isobe M. and Nagahama Y. (2009) A relaxin‐like peptide purified from radial nerves induces oocyte maturation and ovulation in the starfish, Asterina pectinifera . Proc. Natl Acad. Sci. USA 106, 9507–9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata A., Arimura A., Dahl R. R., Minamino N., Uehara A., Jiang L., Culler M. D. and Coy D. H. (1989) Isolation of a novel 38 residue‐hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem. Biophys. Res. Commun. 164, 567–574. [DOI] [PubMed] [Google Scholar]
- Moore S. J. and Thorndyke M. C. (1993) Immunocytochemical mapping of the novel echinoderm neuropeptide SALMFamide 1 (S1) in the starfish Asterias rubens . Cell Tissue Res. 274, 605–618. [DOI] [PubMed] [Google Scholar]
- Moroz L. L., Edwards J. R., Puthanveettil S. V. et al (2006) Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127, 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo A. N., Moeller R. A., Westlund B. A. and Hart A. C. (2001) Identification of neuropeptide‐like protein gene families in Caenorhabditis elegans and other species. Proc. Natl Acad. Sci. USA 98, 14000–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S. J., Elphick M. R. and Thorndyke M. C. (1995a) Tissue distribution of the SALMF amide neuropeptides S1 and S2 in the starfish Asterias rubens using novel monoclonal and polyclonal antibodies. II. digestive system. Proc. Biol. Sci. 261, 187–192. [DOI] [PubMed] [Google Scholar]
- Newman S. J., Elphick M. R. and Thorndyke M. C. (1995b) Tissue distribution of the SALMFamide neuropeptides S1 and S2 in the starfish Asterias rubens using novel monoclonal and polyclonal antibodies. I. Nervous and locomotory systems. Proc. Biol. Sci. 261, 139–145. [DOI] [PubMed] [Google Scholar]
- Ohtani M., Iwakoshi E., Muneoka Y., Minakata H. and Nomoto K. (2002) Isolation and characterization of bioactive peptides from the sea cucumber, Stichopus japonicus, in Peptide Science ‐ Present and Future, (Shimonishi Y., ed.), pp. 419–420. Springer, Netherlands. [Google Scholar]
- Reich A., Dunn C., Akasaka K. and Wessel G. (2015) Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa. PLoS ONE 10, e0119627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht P., Tatemoto K., Chatelain P. et al (1982) Effects of PHI on vasoactive intestinal peptide receptors and adenylate cyclase activity in lung membranes. A comparison in man, rat, mouse and guinea pig. Regul. Pept. 4, 241–250. [DOI] [PubMed] [Google Scholar]
- Rowe M. L. and Elphick M. R. (2012) The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus . Gen. Comp. Endocrinol. 179, 331–344. [DOI] [PubMed] [Google Scholar]
- Rowe M. L., Achhala S. and Elphick M. R. (2014) Neuropeptides and polypeptide hormones in echinoderms: new insights from analysis of the transcriptome of the sea cucumber Apostichopus japonicus . Gen. Comp. Endocrinol. 197, 43–55. [DOI] [PubMed] [Google Scholar]
- Schilling L., Kanzler C., Schmiedek P. and Ehrenreich H. (1998) Characterization of the relaxant action of urocortin, a new peptide related to corticotropin‐releasing factor in the rat isolated basilar artery. Br. J. Pharmacol. 125, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmens D. C., Dane R. E., Pancholi M. R., Slade S. E., Scrivens J. H. and Elphick M. R. (2013) Discovery of a novel neurophysin‐associated neuropeptide that triggers cardiac stomach contraction and retraction in starfish. J. Exp. Biol. 216, 4047–4053. [DOI] [PubMed] [Google Scholar]
- Soehler S., Stengl M. and Reischig T. (2011) Circadian pacemaker coupling by multi‐peptidergic neurons in the cockroach Leucophaea maderae . Cell Tissue Res. 343, 559–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangier J., Hilbich C., Burdzik S. and Keller R. (1992) Orcokinin: a novel myotropic peptide from the nervous system of the crayfish, Orconectes limosus . Peptides 13, 859–864. [DOI] [PubMed] [Google Scholar]
- Wei H. and Stengl M. (2011) Light affects the branching pattern of peptidergic circadian pacemaker neurons in the brain of the cockroach Leucophaea maderae . J. Biol. Rhythms 26, 507–517. [DOI] [PubMed] [Google Scholar]
- Williams C. L., Peterson J. M., Villar R. G. and Burks T. F. (1987) Corticotropin‐releasing factor directly mediates colonic responses to stress. Am. J. Physiol. 253, G582–G586. [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Roller L., Zitnan D., Satake H., Mizoguchi A., Kataoka H. and Tanaka Y. (2011) Bombyx orcokinins are brain‐gut peptides involved in the neuronal regulation of ecdysteroidogenesis. J. Comp. Neurol. 519, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda‐Kamatani Y. and Yasuda A. (2000) Identification of orcokinin gene‐related peptides in the brain of the crayfish Procambarus clarkii by the combination of MALDI‐TOF and on‐line capillary HPLC/Q‐Tof mass spectrometries and molecular cloning. Gen. Comp. Endocrinol. 118, 161–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. MALDI‐TOF mass spectrum of purified peak A (A). Comparison of the chromatographic properties of synthetic SMP with a free carboxyl terminus (SMP) and SMP with an amidated carboxyl terminus (SMPamide) on RP‐HPLC reveals that the two peptides elute at different retention times with isocratic 20% acetonitrile/0.1% TFA (B).
Figure S2. Precursor of starfish myorelaxant peptide (SMP)‐type neuropeptides in Asterias rubens.
Figure S3. Multiple sequence alignment showing that starfish SMP precursors share sequence similarity with other echinoderm pedal peptide‐type precursors (Spnp6, Spnp7, Ajnp7) and with protostomian pedal peptide (PP)/orcokinin(OK)‐type peptide precursors.
Table S1. Primers used for RACE and RT‐qPCR analysis of SMP precursor expression in P. Pectinifera.
Table S2. Identity and similarity grid showing the identity/similarity percentages for the amino acid sequences of SMP precursors and PP/OK‐type peptide precursors.


