Abstract
The worldwide outbreak of severe acute respiratory syndrome (SARS) was shown to be associated with a novel coronavirus (CoV) now called SARS CoV. We report here the generation of SARS CoV S protein-pseudotyped murine leukemia virus (MLV) vector particles. The wild-type S protein pseudotyped MLV vectors, although at a low efficiency. Partial deletion of the cytoplasmic tail of S dramatically increased infectivity of pseudotypes, with titers only two- to threefold lower than those of pseudotypes generated in parallel with the vesicular stomatitis virus G protein. S-pseudotyped MLV particles were used to analyze viral tropism. MLV(SARS) pseudotypes and wild-type SARS CoV displayed similar cell types and tissue and host restrictions, indicating that the expression of a functional receptor is the major restraint in permissiveness to SARS CoV infection. Efficient gene transfer could be detected in Vero and CaCo2 cells, whereas the level of gene marking of 293T, HeLa, and HepG2 cells was only slightly above background levels. A cat cell line and a dog cell line were not susceptible. Interestingly, PK-15, a porcine kidney cell line, and primary porcine kidney cells were also highly permissive for SARS S pseudotypes and wild-type SARS CoV. This finding suggests that swine may be susceptible to SARS infection and may be a source for infection of humans. Taken together, these results indicate that MLV(SARS) pseudotypes are highly valuable for functional studies of viral tropism and entry and, in addition, can be a powerful tool for the development of therapeutic entry inhibitors without posing a biohazard to human beings.
A new coronavirus (CoV) was identified as the etiologic agent of severe acute respiratory syndrome (SARS) (8, 12, 22, 28, 35), a life-threatening pulmonary disease which was first reported from Guangdong Province, China. Within a few months, further cases were reported from Vietnam, Canada, and Hong Kong. By 7 August 2003, the SARS epidemic had resulted in 8,422 cases globally, of which 916 were fatal (http://www.who.int/csr/sars/country/2003_08_15/en/).
CoVs comprise a large and diverse family of enveloped, positive-stranded RNA viruses with a genome of 27 to 32 kb. They exhibit a broad host range, infect many mammalian and avian species, and can cause upper respiratory, gastrointestinal, and hepatic disease (14). Although CoVs cause severe diseases in farm animals, in humans they were only known to cause 15 to 30% of mild upper respiratory tract illnesses before SARS emerged (18). Based on genetic and serologic similarities, CoVs are divided into three classes (groups 1, 2, and 3). Sequence analyses of various SARS isolates have indicated that although the virus has many similarities with CoVs, it is genetically distinct from all known previous isolates. Based on such phylogenetic analyses, SARS CoV is classified in a new CoV group (group 4). In addition, sequence data indicate that SARS CoV is a completely new pathogenic strain that evolved neither directly from the known human CoV nor by recombination between different known CoVs (28).
The spike (S) protein, a type I membrane glycoprotein on the viral surface, mediates CoV attachment and entry into host cells. It is synthesized as a 180- to 200-kDa protein which can be cleaved by host-derived proteases into the two similarly sized, noncovalently associated subunits S1 and S2 (13, 40). Receptor binding is mediated by the N-terminal S1 subunit, while the membrane-anchored S2 portion is required for fusion of viral and cellular membranes. Furthermore, S induces neutralizing antibodies (6, 11), and mutations in S can dramatically affect virulence as well as host cell and tissue tropism (24, 25, 36). The overall level of similarity between SARS CoV S and other known CoV S proteins is only 21 to 27% (35).
Currently, murine leukemia virus (MLV)-based retroviral vectors are the main vehicles for stable gene transfer into a variety of cell types. The retroviral envelope protein can be exchanged for envelope proteins from nonrelated viruses, a process called pseudotyping. Many examples of pseudotyping exist in the literature (1, 3, 20, 27, 30, 38, 43). Viral vector pseudotypes comprise an envelope protein of a nonrelated virus and a replication-deficient MLV genome which harbors a transgene (e.g., a reporter gene such as that for enhanced green fluorescent protein [eGFP]). These pseudotypes acquire the host range of the virus from which the heterologous glycoprotein was derived. Early steps in infection, such as receptor binding, membrane fusion, and entry, are determined solely by properties of the nonrelated envelope protein. Pseudotypes can be used as a tool for functional characterization of viral envelope glycoproteins and to study viral tropism and receptor interactions.
Here, the incorporation of SARS S proteins into MLV particles was studied. Our results demonstrate efficient pseudotyping of MLV particles with SARS CoV S protein after partial truncation of the cytoplasmic tail. Moreover, tropism of MLV(SARS) pseudotypes and wild-type (wt) SARS CoV was found to be restricted to the same cell types and host species. This pseudotype system is extremely valuable for further studies on glycoprotein processing or viral entry and may become a key tool in the development new antiviral drugs.
MATERIALS AND METHODS
Cell lines, viruses, and cell culture.
The cell lines Anjou65 (32), 293T, HeLa, TE671, COS7, Vero, PK-15, and PG-4 were maintained in Dulbecco's modified Eagle medium (DMEM). NCI-H292 and HepG2 cells were propagated in RPMI 1640 (Gibco, Karlsruhe, Germany), CaCo2 and D17 cells were propagated in EMEM (Biochrom, Berlin, Germany), and BEAS-2B cells were propagated in F12-DMEM (Gibco). All growth media were supplemented with 4 mM glutamine and 10% fetal calf serum (FCS).
SARS CoV isolate FFM-1 (8) was obtained from the sputum of a patient hospitalized with a diagnosis of probable SARS in the isolation unit of Frankfurt University Hospital, Frankfurt, Germany. SARS CoV was grown in Vero cell cultures (African green monkey kidney, ATCC CCL-81). The maintenance medium consisted of minimum essential medium without FCS and containing 100 IU of penicillin per ml and 100 μg of streptomycin per ml. Virus stocks were stored at −80°C. Infectious virus titers were determined as 50% tissue culture infective doses in confluent cells in 96-well microtiter plates as described previously (34). In accordance with World Health Organization recommendations, all work involving infectious SARS CoV was performed under biosafety level 3 conditions in a biosafety level 3 facility.
Cell culture infectivity.
To assess virus infectivity in different cell cultures, monolayers were inoculated with SARS CoV at a multiplicity of infection of 5 in 12.5-cm2 flasks and incubated at 37°C. After 24 and 48 h of incubation, cells were fixed with acetone-methanol (40:60). SARS CoV infectivity was detected by recognition of a cytopathic effect and additionally by immunostaining of infected cells with reconvalescent-phase serum from a SARS patient as described before (5).
Construction of S gene expression plasmids.
The S gene of SARS CoV strain FFM-1 (GenBank accession no. AY291315.1) was amplified by reverse transcription-PCR from viral RNA isolated from supernatants of infected Vero cells. Reverse transcription was performed with Superscript II (Invitrogen, Karlsruhe, Germany) and random primers, as described by the manufacturer. The cDNA was amplified with the Expand long-template PCR system (Roche, Mannheim, Germany) with the primers 5′-GCGGATCCACCATGTTTATTTTCTTATTATTTCTTACTCTCACTAGTGG-3′ and 5′-GGGGTACCTTATGTGTAATGTAATTTGACACCCTTGAG-3′ as recommended by the manufacturer. The S gene was then inserted between the BamHI and KpnI sites downstream of the human cytomegalovirus immediate-early promoter and rabbit beta-globin intron into the pHCMV vector (44) by using the primer-introduced restriction sites. DNA sequencing of the resulting plasmid pHCMV-SARS-S revealed three mute point mutations (nucleotide [nt] 834, T→C; nt 1386, T→C; and nt 1866, T→C) and one point mutation (nt 1921, C→T) which was present in all sequenced clones and lead to an amino acid exchange at position 641 (H→Y). Hemagglutinin (HA)-tagged and truncated S expression constructs were generated not by PCR amplification of the whole S gene but by amplification of an approximately 250-bp fragment by using the forward primer 5′-CAGGCATTAACGCTTCTGTCGTCAAC-3′ in combination with different reverse primers given below. The resulting PCR fragments were treated with SwaI (present in the S gene) and KpnI (introduced by the primers) and inserted into pHCMV-SARS-S vector. S protein expression plasmids were amplified by using the following reverse primers: pHCMV-SARS-SHA, 5′-GGTACCTTAAGCGTAATCTGGAACATCGTATGGGTATGTGTAATGTAATTTGACACCCTTG-3′; pHCMV-SARS-SCΔ19HA, 5′-GGTACCTTAAGCGTAATCTGGAACATCGTATGGGTAGCAGCAAGAACCACAAGAGC-3′; pHCMV-SARS-SCΔ8, 5′-GGGGTACCTTAGAGAACTGGCTCAGAGTCATCC-3′; pHCMV-SARS-SCΔ19, 5′-GGGGTACCTTAG-CAGCAAGAACCACAAGAGC-3′; pHCMV-SARS-SCΔ26, 5′-GGGGTACCTTATGCACCCTTGAGGCAACTGC-3′; pHCMV-SARS-SCΔ34, 5′-GGGGTACCTTAACTAGTCATGCAACAAAGCAAG-3′; and pHCMV-SARS-SCΔ39, 5′-GGGGTACCTTAAAGCAAGATTGTAACCATGACG-3′.
Transient production and purification of vector pseudotypes.
Oncoretroviral and lentiviral pseudotypes were prepared essentially as described previously (3), with slight modifications. Briefly, one 10-cm-diameter dish of subconfluently grown 293T or Anjou65 cells was transfected with 7.5 μg of pMP71-eGFP-pre (37), 12.5 μg of pSV-Mo-MLVgagpol, and 1 to 4 μg of env expression plasmids by using a calcium phosphate transfection kit (Sigma, Taufkirchen, Germany). The medium was replaced 6 to 8 h after transfection. Pseudotype vector-containing supernatants were harvested after an additional 24, 39, and 48 h; filtered through a 0.45 μm-pore-size Millex-HV filter (Millipore, Schwalbach, Germany); and used for transduction. For the generation of lentiviral pseudotypes, the second-generation packaging system was used. Lentiviral vectors were produced likewise by transfecting 293T cells with 7.5 μg of packaging plasmid pCMVΔR8.91 (encoding human immunodeficiency virus [HIV] Gag/Pol), 12.5 μg of eGFP harboring transfer vector pHR′SIN.cPPT-SEW (7), and 1 to 4 μg of env plasmids. The lentiviral transfer vector is a self-inactivating HIV type 1 (HIV-1)-derived vector (46) which contains the posttranscriptional regulatory element of woodchuck hepatitis virus to enhance transgene expression as well as a central polypurine tract to improve gene transfer efficiency. The expression of eGFP is driven by the spleen focus-forming virus promoter.
Purified vector particles were obtained by ultracentrifugation of 32 ml of viral supernatant through 5 ml of a 20% sucrose cushion in an SW28 Beckman rotor (2 h, 25,000 rpm, 4°C). Viral pellets were resuspended over night in 500 μl of phosphate-buffered saline (PBS) or DMEM. For further purification, equilibrium density centrifugation was performed. Two milliliters of concentrated particles was layered on the top of a 20 to 60% continuous sucrose gradient and centrifuged overnight in an SW41 Beckman rotor (35,000 rpm, 4°C). Fractions of 1 ml were collected, of which 0.6 ml was precipitated with trichloroacetic acid, resolved in 50 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (60 mM Tris-HCl, 5% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.05% bromphenol blue), and analyzed by Western blotting.
Western blot analysis.
To detect SARS S protein expression and putative cleavage, 293T cells were transfected with pHCMV SARS SCΔ19HA. Cells were grown in 24-well plates and transfected with 800 ng of plasmid DNA per well by using Lipofectamine 2000 (Invitrogen). At 48 h posttransfection, cells were lysed in 1× SDS-PAGE loading buffer, either directly in the well or after they were trypsinized (Gibco BRL) and then heated for 5 min at 95°C. Protein samples were separated by SDS-7% PAGE and transferred to nitrocellulose. The Western blot analyses were performed according to standard methods (41). HA-tagged S proteins were detected by chemiluminescence with the ECL system (Amersham, Freiburg, Germany) after incubation of the blot with a rat monoclonal anti-HA antibody (clone 3F10; Roche), followed by incubation with horseradish peroxidase-conjugated goat anti-rat immunoglobulin G (IgG) (Dianova, Hamburg, Germany).
For detection of gag-pol-encoded proteins in the individual gradient fractions, blots which were used to detect SARS S SCΔ19HA were incubated in stripping buffer (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris HCl [pH 6.7]) for 3 h at 65°C and reprobed with polyclonal rabbit antiserum against MLV gag (a generous gift from B. Schnierle) at a dilution of 1:3,000. After being washed, blots were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Dianova).
Protease treatment of viral particles.
Viral particles were purified by ultracentrifugation through a 20% sucrose cushion and were treated with a 10-μg/ml concentration of trypsin treated with l-(tosylamido-2-phenylethyl)chloromethyl ketone (TPCK trypsin; Pierce, Rockford, Ill.) for 30 min at room temperature in PBS. After digestion was stopped by adding SDS-PAGE loading buffer, proteins were analyzed by SDS-PAGE.
Flow cytometry.
For cell surface staining of SARS S proteins, transfected and mock-transfected 293T cells were resuspended in PBS-3% FCS and incubated for 1 h at 4°C with antiserum from a SARS patient (1:50). After being washed three times with PBS-3% FCS, cells were incubated with phycoerythrin-conjugated goat anti-human IgG (Dianova) for 30 min at 4°C, washed intensively, and subsequently analyzed by flow cytometry on a FACSCalibur (Becton Dickinson).
Vector titration.
Pseudotype titers were determined by fluorescence-activated cell sorter (FACS) analysis of transduced cells. Target cells were seeded at a density of 5 × 104 cells per well in 24-well plates and incubated overnight. Dilutions of supernatants were added to the cells, and the plates were centrifuged for 1 h at 1,000 × g. One to 24 hours later, vector supernatants were removed and the cells were cultivated in regular medium. The percentage of eGFP-positive cells was determined at 72 h posttransduction by FACS analysis.
Neutralization assay.
Neutralization assays were performed by using sera from four convalescent SARS patients. Two of the patients (patients 1 and 3) were hospitalized in Frankfurt, Germany, and one (patient 4) was hospitalized in Hemer, Germany. This antiserum was obtained from the Bernhard Nocht Institute for Tropical Medicine (BNI). The forth antiserum was derived from a SARS patient (patient 2) from Hong Kong. Viral supernatants were pelleted by ultracentrifugation as described above. Fifty microliters of concentrated pseudotypes was incubated with serial dilutions of either SARS patient antisera or unspecific human control serum (Biowhittaker) for 1 h at room temperature, and then 450 μl of DMEM was added and transduction of Vero cells was performed.
Isolation of primary cells from porcine lung and kidney.
Representative portions of freshly removed lung and kidney were minced by using a scalpel and digested with dispase (10 U/ml in FCS; Roche) for 1 h at 37°C. In order to separate undesired fibrous material and erythrocytes from single cells, the suspension was mixed with an equal amount of 40% Percoll (Biochrom) and centrifuged for 20 min in a Heraeus Minifuge RF centrifuge (4,000 rpm, 4°C). Cells immediately above the pelleted erythrocytes were transferred into a new 50-ml Falcon tube, washed twice with DMEM containing kanamycin, and resuspended in DMEM supplemented with antibiotics and 15% heat-inactivated FCS. Cells were grown to confluency on six-well plates.
Nucleotide sequence accession number.
The sequence of pHCMV-SARS-S can be found under GenBank accession no. AY569693.
RESULTS
Cytoplasmically truncated SARS CoV S protein pseudotypes oncoretroviral vectors.
For CoV, receptor binding and fusion of the viral and cell membrane are thought to be mediated exclusively by the S protein (14, 19). In this study, we analyzed whether packaging of the S protein into the envelope of a retroviral vector generates infectious pseudotype virions. The S protein gene of the SARS CoV strain FFM-1 was amplified by reverse transcription-PCR and cloned into a cytomegalovirus promoter-driven expression plasmid. As MLV assembles at the plasma membrane, whereas CoVs bud in the endoplasmic reticulum (ER), FACS analysis was performed to determine whether sufficient amounts of S glycoprotein are expressed on the cell surface after transfection of 293T cells with the envelope expression constructs. SARS S protein was expressed at the cell surface (see Fig. 2A), most probably due to leakiness of the ER retention upon overexpression. Initial experiments (see Fig. 3A) using wt S protein indicated that MLV particles can be pseudotyped with SARS S protein and are able to infect Vero cells, albeit at low efficiency.
FIG. 2.
Expression of S protein constructs. (A) Cell surface expression of S mutants in 293T cells. Cells were transfected with DNA encoding either full-length S (wt) or cytoplasmically truncated S protein (CΔ8 to CΔ39). Mock-transfected 293T cells (thin line) were used as a negative control. FACS analysis was performed with SARS antiserum (1:50) and anti-human IgG antibody conjugated with phycoerythrin. (B) Immunoblot of transfected 293T cells (left panel) and purified viral particles (right panel). At 48 h after transfection with HA-tagged CΔ19 S expression construct, cells were lysed either directly (CΔ19-HA) or after being detached from the wells with trypsin-EDTA (CΔ19-HA/Trypsin) and subjected to Western blot analysis with monoclonal anti-HA antibody. Lysates of mock-transfected 293T cells were used as a control (mock). Viral particles with or without trypsin treatment were analyzed likewise. Supernatants produced in the absence of a viral envelope (no env) served as a control. Molecular size standards (in kilodaltons) are indicated.
FIG. 3.
Infectivities of S pseudotypes. (A) Pseudotype titers for different S expression constructs. Vector titers were calculated from end point dilutions on Vero cells and are displayed as TU per milliliter of supernatant (mean ± standard deviation from up to four independent experiments). Envelope proteins of VSV and LCMV were used as positive controls. (B) Infectivities of CΔ19 pseudotypes in the presence of 1, 10, and 25 μM AZT, with HIV-1 core proteins, and with an HA tag without concentration and after 60× concentration by ultracentrifugation. Infectious titers were determined on Vero cells.
We assumed that putative ER retention signals at the cytoplasmic part of the glycoprotein might lead to the retention of the S protein in the ER and thus reduce pseudotyping efficiency. Furthermore, it has been shown for several other pseudotypes that a long cytoplasmic domain of viral glycoproteins interferes with particle formation. Although the length of the S protein cytoplasmic tail is only moderate, we tested whether pseudotype titers could be increased by truncating the S protein C terminus. Five serially truncated S proteins were generated (Fig. 1), and their surface expression was analyzed by flow cytometry. All mutants were expressed at the cell surface (Fig. 2A), even when cytoplasmic deletion reached into the transmembrane domain as in construct CΔ39. However, this construct and also the CΔ8 mutant displayed decreased surface expression. None of the constructs showed an increase in cell surface expression in comparison to the wt S protein. Interestingly, fusion of the cytoplasmic domain of amhotrophic MLV to the truncated protein constructs completely abolished surface expression of these constructs (data not shown).
FIG. 1.
Schematic illustration of SARS S expression constructs. The extracellular, membrane-spanning, and cytoplasmic domains of the SARS S expression constructs as determined by computational prediction (www.cbs.dtu.dk) are shown. Deletions of the cytoplasmic domains were performed via PCR. Amino acid residues of the cytoplasmic domains of truncated S proteins are indicated.
In general, CoV S protein is expressed as a glycoprotein precursor that is cleaved by cellular proteases into two subunits, S1 and S2. The extent of cleavage into S1 and S2 varies among CoVs and also depends on the host cell types (18). Amino acid comparison of SARS S with known cleavage sites of other CoVs failed to predict a specific cleavage site in SARS S protein (35). To analyze the potential cleavage of the SARS CoV S protein, we generated C-terminal HA-tagged S protein of the CΔ19 construct. Expression mediated by this construct was examined by transient transfection of 293T cells. At 48 h after transfection, cells were lysed either directly in the well or after trypsin treatment and expression and putative cleavage were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody (Fig. 2B). One distinct band migrating with an apparent molecular mass of about >175 kDa could be detected in lysates of transfected cells, whereas a further band with a molecular mass of about 90 kDa appeared in cells that were detached from the well by trypsin-EDTA treatment prior to lysis. The protein sizes corresponded to the previously described migration pattern of glycosylated unprocessed S protein (180 kDa) and cleaved S protein (90 kDa). These data indicate that the SARS CoV S protein is not endoproteolytically cleaved but is sensitive to treatment with trypsin. Furthermore, uncleaved SARS S protein on viral particles was cleaved upon trypsin digestion. These particles exhibited a twofold decrease in virus infectivity (data not shown).
SARS S protein constructs are efficiently incorporated into MLV particles.
The ability of the different S protein mutants to form pseudotypes with MLV vectors was then tested. Pseudotypes were produced by transfection of 293T cells with plasmids encoding MLV Gag/Pol, with an MLV genome containing the eGFP marker gene, and with either wt or C-terminally truncated S protein expression plasmids. Supernatants were collected and used in different dilutions for transduction of Vero cells. Parallel infections were carried out with supernatants produced in absence of a viral envelope and with the vesicular stomatitis (VSV) G and the lymphocytic choriomeningitis virus (LCMV) GP proteins, both of which are known to efficiently pseudotype MLV. Transducing units (TU) per milliliter were calculated from transduction levels determined by flow cytometry. Deletion mutant CΔ19 (with 19 amino acids deleted from the C terminus) yielded about 100-fold-higher pseudotype titers than the wt S protein. Independently prepared pseudotype particles showed variations in titers, ranging from 4 × 103 to 1 × 104 TU/ml. Further truncations involved highly conserved cysteine residues and resulted in decreased pseudotype titers (Fig. 3A). Particles without envelope protein failed to infect Vero cells (not shown), whereas VSV and LCMV glycoprotein pseudotypes yielded two- to sevenfold-higher titers than the SARS S CΔ19 construct.
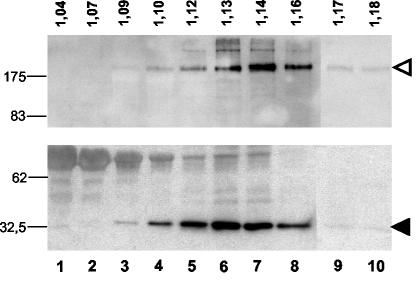
To further characterize the SARS S pseudotypes, we performed equilibrium density centrifugation of particles derived from HA-tagged CΔ19, which generated viral titers in the same range as untagged CΔ19 (Fig. 3B). Immunoblots of viral particles, which were purified through sucrose density gradient centrifugation (20 to 60% sucrose), demonstrated comigration of MLV Gag and CΔ19-HA, indicating correct incorporation of SARS envelope protein into MLV particles (Fig. 4). MLV(SARS) pseudotypes were detected in fractions corresponding to densities from 1.12 to 1.16 g/ml, similar to the previously reported density of MLV (39). The infectivities of individual fractions correlated with S protein expression levels, although the overall infectivity of sucrose-purified pseudotypes was rather low (data not shown). Further evidence that the SARS glycoprotein was properly incorporated into particles was provided by experiments using zidovudine (3′-azido-3′-deoxythymidine) (AZT), a reverse transcriptase inhibitor that prevents conversion of the retroviral genomic RNA into proviral DNA. The infectivity of SARS pseudotypes was reduced when Vero cells were transduced in the presence of AZT. Higher concentrations of AZT even abolished infection completely (Fig. 3B).
FIG. 4.
Incorporation of SARS S protein into MLV particles. Pelleted virus particles were layered on top of a 20 to 60% sucrose gradient and centrifuged to equilibrium. The gradient was fractionated from the top, and individual aliquots were trichloroacetic acid pelleted and then analyzed for the presence of HA-tagged CΔ19 (arrowhead in upper panel) and MLV Gag/p30 (arrowhead in lower panel) by Western blotting with monoclonal anti-HA or polyclonal anti-Gag antibody. The density of each fraction, in grams per milliliter, is shown. Molecular sizes of marker proteins (in kilodaltons) are indicated on the left.
In conclusion, cotransfection of plasmids encoding C-terminally truncated SARS S protein and MLV proteins produced pseudotypes that incorporated SARS S proteins and were infectious for Vero cells, which are known to be permissive for SARS CoV.
Lentivirus-based vectors are attractive gene delivery vectors as they are able to transduce both quiescent and dividing cells, while oncoretroviruses infect only dividing cells. We therefore were interested in whether CΔ19 could also efficiently assemble on lentiviral cores. As depicted in Fig. 3B, pseudotype formation on HIV-derived cores was less efficient.
Concentration by ultracentrifugation was also tested. An approximately 20-fold increase in viral titers could be detected after a 60-fold reduction in volume (Fig. 3B).
Pseudotype virus infection is neutralized by SARS-specific patient serum.
To confirm the specificity of SARS pseudotype infection, antisera from four different SARS patients were tested for their ability to neutralize infection of Vero cells. All antisera were previously shown to neutralize SARS CoV infection, although with various efficiencies (H.W. Doerr, unpublished data). Nonspecific human antiserum served as a control. SARS pseudotypes were preincubated either with the patient serum or with a SARS-negative control serum at the indicated dilutions for 1 h at room temperature. The pseudotype-antiserum mix was then used for infection of Vero cells (Fig. 5). Infectivity was reduced only when pseudotypes were preincubated with SARS-specific antisera. Neutralization was dose dependent. The antiserum from patient 1 was most efficient in neutralizing transduction of Vero cells. With a 1:50 dilution, transduction was reduced by >90% in comparison to the control, whereas neutralization was not found at a dilution of 1:1,000.
FIG. 5.
Neutralization of SARS pseudotypes. Prior to infection of Vero cells, pseudotype particles were incubated with antisera from four different SARS patients or with a control serum for 1 h at room temperature at the indicated dilutions. The infectivity of pseudotypes in absence of antiserum was set as 100%. Patient antisera were named according to their origin: BNI, Bernhard Nocht Institute, Hamburg, Germany; FFM, Frankfurt, Germany; HK, Hong Kong.
Analysis of SARS S protein-mediated host range.
When we started our studies, little was known about the SARS CoV receptor. SARS CoV was propagated mostly in Vero cells. In order to investigate the potential cellular tropism mediated by the SARS S protein, we examined transduction of a variety of cell lines derived from different species and tissues with S pseudotypes (Table 1). Since VSV G protein has been shown to confer a broad host range to MLV, we used VSV G pseudotypes as a control to distinguish a block in entry from postentry restrictions. Only a few of the tested cell lines displayed detectable peudotype titers. Viral titers were highest in Vero cells (5.2 × 103 TU/ml), followed by CaCo2 cells (2.4 × 103 TU/ml) and, unexpectedly, by porcine PK-15 cells (7.7 × 102 TU/ml). Low levels of transduction could be detected in 293T and HeLa cells, indicating basal infection of these cells. The other target cell types used in the infection assay displayed very low (HepG2) or undetectable (COS7 and TE671) levels of transduction with SARS S pseudotypes. Even the two human lung cell lines BEAS-2B and NCI-H292 were refractory to transduction with SARS S pseudotypes. The canine cell line D17 and the feline cell line PG-4 were also resistant to transduction with SARS S pseudotypes. Similar results were obtained when SARS CoV strain Frankfurt was used to determine susceptibilities of the indicated cell lines. Vero, CaCo2, and PK-15 cells were highly permissive for SARS CoV. Low levels of replication were detected in COS7, HepG2, and NCI-H929 cells, whereas the remaining cell lines were not infected.
TABLE 1.
Host ranges of MLV(SARS) pseudotypes and SARS CoV strain FFM-1
| Cell line | Species tissue of origin | SARS S pseudotype titer (TU/ml)a | SARS CoV susceptibilityb |
|---|---|---|---|
| Vero | Simian, kidney | 5.2 (±1.6) × 103 | ++++ |
| COS7 | Simian, kidney | <101 | + |
| CaCo2 | Human, colon | 2.4 (±0.5) × 103 | ++++ |
| HeLa | Human, cervix | 7.2 (±1.4) × 101 | − |
| 293T | Human, kidney | 9.3 (±6.8) × 101 | − |
| HepG2 | Human, liver | 1.5 (±0.4) × 101 | + |
| TE671 | Human, muscle | <101 | − |
| BEAS-2B | Human, lung | <101 | − |
| NCI-H929 | Human, lung | <101 | + |
| PG-4 | Feline, brain | <101 | − |
| PK-15 | Swine, kidney | 7.7 (±1.6) × 102 | +++ |
| D17 | Canine, bone | <101 | − |
All cells were susceptible to transduction with VSV G pseudotypes, yielding titers that ranged from 6 × 103 to 9 × 104 TU/ml.
Virus infectivity was determined by peroxidase staining with antiserum from a SARS patient. ++++, 75 to 100% infected cells; +++, 50 to 75% infected cells; +, <10% infected cells; −, <1% infected cells.
Susceptibility of primary porcine cells to SARS S pseudotypes and SARS CoV strain FFM-1.
The tropism for porcine cells was analyzed further. Since we had noticed that kidney cells were particularly permissive to infection, we isolated primary porcine kidney and lung cells and performed transduction assays with oncoretroviral and lentiviral S pseudotypes. Control titers were determined on Vero cells. Whereas primary lung cells were refractory to HIV/MLV(SARS) pseudotype transduction (not shown), primary porcine kidney cells were readily transduced with both SARS S pseudotyped oncoretroviral and lentiviral vectors (Fig. 6). In comparison to Vero cells, transduction of primary kidney cells with SARS pseudotypes was less efficient. VSV G-pseudotyped viral vectors displayed only marginally decreased transduction efficiencies in primary cells. We then studied the susceptibility of primary porcine cells to infection with wt SARS CoV. As shown in Fig. 6C, approximately 50 to 75% of the primary porcine kidney cells stained positive for SARS antigens after inoculation with SARS CoV, indicating that SARS CoV can infect porcine kidney cells. In contrast, primary porcine lung cells were not infected with SARS CoV.
FIG. 6.
Susceptibility of primary porcine cells to MLV/HIV(SARS) pseudotypes and to SARS CoV. (A) Primary cells of porcine kidney were grown on 48-well plates and transduced with 5.4 × 102 TU of HIV(SARS) pseudotype per ml (upper panel) and with 1.6 × 104 TU of MLV(SARS) pseudotype per ml (lower panel). The phase-contrast and eGFP fluorescences are shown (magnification, ×200). (B) Supernatants containing either HIV(SARS) or MLV(SARS) pseudotypes were used for transduction of Vero cells and primary porcine kidney cells. Pseudotype titers on each cell type were determined by FACS analysis as described in Materials and Methods. Means and standard deviations from duplicates are shown. VSV G-pseudotyped lentiviral and oncoretroviral vectors served as controls. (C) Primary cells of porcine lung and kidney were grown on six-well plates and infected with SARS CoV strain Frankfurt at a multiplicity of infection of 5. Virus replication was assessed by immunostaining with reconvalescent-phase serum from a SARS patient.
DISCUSSION
In this report, we demonstrate that the SARS envelope glycoprotein can be efficiently incorporated into MLV particles after truncation of the cytoplasmic tail. MLV(SARS) pseudotypes displayed properties of MLV particles, such as the characteristic buoyant density and the susceptibility to AZT. On the other hand, cell and tissue tropism was mediated solely by the SARS S envelope protein. SARS pseudotypes and authentic SARS CoV showed very similar tropisms.
Cleavage of viral glycoproteins was found to be a prerequisite for membrane fusion and thus infectivity in a variety of retroviruses, orthomyxoviruses, paramyxoviruses, and arenaviruses (2, 21). We found that most SARS CoV S protein was not cleaved when cells were lysed directly in the culture vessels, although we observed a weak band which migrates at the same position as trypsin-cleaved S protein. This may be due to random digestion by proteases. On the other hand, cleavage of S protein was detected in lysates from cells which were trypsinized. This suggests that the S protein of SARS CoV is sensitive to exogenous trypsin treatment rather than being endoproteolytically cleaved. In virions, only full-length S protein was detected, suggesting that cleavage is not required for incorporation into virions and for viral infectivity. For the mouse hepatitis virus, trypsin cleavage of S into S1 and S2 may enhance fusion activity or viral infectivity (40). Trypsin treatment of SARS pseudotypes led to loss of infectivity (not shown); however, fusion assays have to be performed to clarify the role of trypsin digestion in the fusogenicity of S protein.
Efficient incorporation of foreign viral envelope proteins into MLV particles has been demonstrated for several viruses. The VSV (9), Ebola virus (43), LCMV (30), hepatitis C virus (1, 20), and gibbon ape leukemia virus (31) envelope glycoproteins all efficiently pseudotype retroviral vectors without any modifications. However, some virus envelope glycoproteins pseudotype MLV particles only if modifications are introduced into the transmembrane and/or cytoplasmic domain. SARS S protein allows efficient formation of infectious MLV particles only after truncation of the cytoplasmic tail. Similar findings have been described for MLV (HIV-1 and HIV-2) pseudotypes, which form only with truncated HIV envelope proteins (17, 38). Interestingly, although all truncated SARS CoV S proteins were expressed at the cell surface, the infectivity of pseudotypes was severely impaired when the highly conserved cysteines were deleted. We cannot rule out the possibility that truncations in the cytoplasmic domain influence the structure and function of the ectodomain, as was shown previously for other viral glycoproteins. Alterations in the cytoplasmic domains of measles virus and simian virus glycoproteins, for example, had various effects on receptor binding, fusogenicity, and viral infectivity (4, 45). Therefore, the increased or decreased infectivity of the cytoplasmic domain-truncated S protein pseudotypes may be due to altered (i) incorporation of S protein into the virus particles, (ii) receptor binding, or (iii) fusion/entry process. Further experiments may clarify the role of the SARS S protein cytoplasmic domain in these processes. Chimeric envelopes containing the extracellular domain of a foreign viral glycoprotein fused to the transmembrane and cytoplasmic domains of the MLV glycoproteins have also been shown to enhance pseudotype formation. An example is the incorporation of human foamy virus/MLV chimeric envelope proteins into MLV particles (27). However, we found that the chimeric glycoproteins of SARS S were not efficiently expressed on the cell surface, possibly due to misfolding.
Comigration of MLV protein and S protein in a linear sucrose gradient provided further evidence for the proper incorporation of S protein into MLV particles. Whether the observed slightly offset peaks of MLV Gag and S protein result from empty Gag particles, which may be abundant in our supernatants, or whether this is a technical artifact has to be addressed by use of more-appropriate sucrose gradients.
The tissue tropisms of MLV(SARS) pseudotypes and wt SARS CoV were analyzed by using different cell lines. We found that SARS pseudotypes and the SARS CoV preferentially transduced cell lines of renal origin, such as Vero, MA104 (not shown), PK-15 (porcine), and, to some extent, 293T cells. In previous studies, SARS CoV has also been found in the kidneys of infected patients (22) and could be isolated from the kidney of an experimentally infected macaque (23), suggesting that this organ is a site of viral replication. Furthermore, these findings are consistent with efficient replication of SARS CoV in the primate kidney cell lines Vero (8, 22), MA104 (Doerr, unpublished data), and FRhK-4 (23), as described previously. Basal replication has also been observed in the human kidney cell line 293T (26). We also found that CaCo2 cells, which are derived from a human colon carcinoma, were highly permissive for SARS pseudotypes and SARS CoV (5). This is consistent with the detection of SARS CoV in feces (8), suggesting that the gut may be a further site of virus replication. Other cell types, even those of lung origin, were resistant to both transduction and infection. Interestingly, our data demonstrate that primary porcine lung cells, in contrast to primary kidney cells, were permissive neither for MLV(SARS) pseudotypes nor for native SARS CoV. Similarly, SARS CoV also does not grow on primary human bronchial epithelial cells (Doerr, unpublished data). This is surprising, as the lung is the organ affected most severely during SARS. Moreover, mRNA expression profiling of the SARS virus receptor ACE2 (16) showed expression in the lung and the gastrointestinal tract. ACE2 (26) is also found in the endothelia of coronary and intrarenal vessels and in renal tubular epithelium (10, 42). The lack of virus replication in lung cells ex vivo may reflect the loss of susceptible cells during the preparation and isolation of cells from lung tissue. On the other hand, the ACE2 could be lost during ex vivo culture. Finally, lung pathology could be caused by indirect mechanisms rather than by local virus replication. Taken together, our observations are consistent with previous studies showing that SARS CoV does not grow in most of the commonly used cell lines (8, 22, 33). The observation that pseudotypes and wt virus had very similar tropisms indicates that susceptibility to infection is determined primarily by the availability of a functional cellular receptor and is expected to correlate with expression levels of ACE2 in the different cell types in culture. This issue will be addressed in future studies.
The natural reservoir of SARS CoV is still unknown, although SARS CoV-like viruses were isolated from different wild animals which were found in live-animal markets in Guangdong in southern China. Guan and coworkers performed serological and virological studies on 25 animals from a live-animal retail market (15). Their study indicates that masked palm civets, raccoon dogs, and Chinese ferret badgers can be infected with a SARS CoV-like virus that has an extra stretch of 29 nt, which is not found in most human isolates. Virus could not be detected in any of the samples taken from four domestic cats. On the other hand, Martina and coworkers demonstrated that ferrets and domestic cats are susceptible to experimental infection with human SARS CoV (29). In our hands, the feline cell line PG-4 can be neither transduced with SARS pseudotypes nor infected with SARS CoV. These observations may not conflict, considering that PG-4 cells are from glial origin and thus probably are not the appropriate target cell type for SARS CoV. Further feline cell lines from different tissues must be studied in the future.
So far, only primate or human cell lines have been reported to efficiently replicate SARS CoV. Here, we report that PK-15, a cell line of porcine origin, and primary porcine kidney cells were efficiently transduced with MLV(SARS) pseudotypes and were highly susceptible to SARS CoV infection. This observation may indicate that domestic pigs play a crucial role in the interspecies transmission of SARS. To our knowledge, no data on the susceptibility of pigs or porcine cells to SARS CoV have been published. This is surprising, as pigs can transmit several different viruses to humans. The most prominent example is influenza virus. In contrast to influenza virus, however, SARS CoV does not seem to be merely due to recombination between known CoV species. However, pigs may well serve as an intermediate host that facilitates infection of humans with SARS, while it seems highly unlikely to us that pigs are a major reservoir for SARS CoV. Although not published, it must be assumed that the major farm animal species, including pigs, have been extensively tested for SARS CoV infection in China.
Safety issues are a major restraint to research on SARS CoV, and human infections acquired during laboratory work have been reported from Singapore and Taiwan. For a broad range of research topics, this restraint can be overcome by the use of the MLV(SARS) pseudotypes described in this study as relatively safe surrogates for native SARS CoV. Examples are the investigation of S protein-mediated cell binding and entry, the specific and fast screening of antiviral compounds that target SARS CoV entry, and the development of an effective SARS vaccine.
Acknowledgments
We thank C. Münk for the cell lines PG-4, PK-15, and D17 and T. Merovci, G. Bauer, and P. Schult-Dietrich for excellent technical assistance.
REFERENCES
- 1.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer, W. R., M. Westphal, W. Ostertag, and D. von Laer. 2002. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J. Virol. 76:1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Treatment of SARS with human interferons. Lancet 362:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demaison, C., K. Parsley, G. Brouns, M. Scherr, K. Battmer, C. Kinnon, M. Grez, and A. J. Thrasher. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803-813. [DOI] [PubMed] [Google Scholar]
- 8.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 9.Emi, N., T. Friedmann, and J. K. Yee. 1991. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 65:1202-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson, U., U. Danilczyk, and J. M. Penninger. 2002. Just the beginning: novel functions for angiotensin-converting enzymes. Curr. Biol. 12:1745-1752. [DOI] [PubMed] [Google Scholar]
- 11.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen., G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, K. F. Lim, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 16.Harmer, D., M. Gilbert, R. Borman, and K. L. Clark. 2002. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 532:107-110. [DOI] [PubMed] [Google Scholar]
- 17.Hohne, M., S. Thaler, J. C. Dudda, B. Groner, and B. S. Schnierle. 1999. Truncation of the human immunodeficiency virus-type-2 envelope glycoprotein allows efficient pseudotyping of murine leukemia virus retroviral vector particles. Virology 261:70-78. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, K.V. 2001. Coronaviruses, p. 1187-1203. In D. Knipe et al. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 19.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klenk, H. D., and W. Garten. 1994. Activation cleavage of viral spike proteins, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Monograph 28. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Ksiazek, T. G., D. Erdman, C. C. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 23.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leparc-Goffart, I., S. T. Hingley, M. M. Chua, J. Phillips, E. Lavi, and S. R. Weiss. 1998. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J. Virol. 72:9628-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 29.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miletic, H., M. Bruns, K. Tsiakas, B. Vogt, R. Rezai, C. Baum, K. Kuhlke, F. L. Cosset, W. Ostertag, H. Lother, and D. von Laer. 1999. Retroviral vectors pseudotyped with lymphocytic choriomeningitis virus. J. Virol. 73:6114-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Movassagh, M., C. Desmyter, C. Baillou, S. Chapel-Fernandes, M. Guigon, D. Klatzmann, and F. M. Lemoine. 1998. High-level gene transfer to cord blood progenitors using gibbon ape leukemia virus pseudotype retroviral vectors and an improved clinically applicable protocol. Hum. Gene Ther. 9:225-234. [DOI] [PubMed] [Google Scholar]
- 32.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, et al.. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed, J. L., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-498. [Google Scholar]
- 35.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schambach, A., H. Wodrich, M. Hildinger, J. Bohne, H. G. Krausslich, and C. Baum. 2000. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol. Ther. 2:435-445. [DOI] [PubMed] [Google Scholar]
- 38.Schnierle, B. S., J. Stitz, V. Bosch, F. Nocken, H. Merget-Millitzer, M. Engelstadter, R. Kurth, B. Groner, and K. Cichutek. 1997. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc. Natl. Acad. Sci. USA 94:8640-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma, S., F. Murai, A. Miyanohara, and T. Friedmann. 1997. Noninfectious virus-like particles produced by Moloney murine leukemia virus-based retrovirus packaging cells deficient in viral envelope become infectious in the presence of lipofection reagents. Proc. Natl. Acad. Sci. USA 94:10803-10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 56:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner, A. J., and N. M. Hooper. 2002. The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 2:177-183. [DOI] [PubMed] [Google Scholar]
- 43.Wool-Lewis, R. J., and P. Bates. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 72:3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]
- 45.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]