Abstract
This perspective sets out to critically evaluate the scope of reactive electrophilic small molecules as unique chemical signal carriers in biological information transfer cascades. We consider these electrophilic cues as a new volatile cellular currency and compare them to canonical signaling circulation such as phosphate in terms of chemical properties, biological specificity, sufficiency, and necessity. The fact that nonenzymatic redox sensing properties are found in proteins undertaking varied cellular tasks suggests that electrophile signaling is a moonlighting phenomenon manifested within a privileged set of sensor proteins. The latest interrogations into these on-target electrophilic responses set forth a new horizon in the molecular mechanism of redox signal propagation wherein direct low-occupancy electrophilic modifications on a single sensor target are biologically sufficient to drive functional redox responses with precision timing. We detail how the various mechanisms through which redox signals function could contribute to their interesting phenotypic responses, including hormesis.
Canonical Signaling Small Molecules and Mechanisms
Arguably the most beautiful aspect of cellular design is the intricacy of signaling subsystems.1 These pathways stand as lasting testaments to the wonders of evolution, and our understanding of signaling circuits serve as evidence of the ability of scientists to untie the “Gordian knot”. This perspective evaluates latest developments in our understanding of and methods to study precision redox signaling, a noncanonical chemical signaling paradigm wherein the cell harnesses endogenous reactive chemicals as input signals to precisely control cellular output.
Many canonical signaling pathways involve an external signal, such as a growth factor or hormone that stimulates a downstream signaling cascade starting at the cell surface and relaying information to the nucleus where (a) specific gene or gene(s) is(are) upregulated.1 To propagate the upstream signals, messengers are required that can be handed down a specific pathway. We refer to these small signaling mediators as the currency that can be transferred similar to how cash can be traded between parties. Much like in the global community, cell signaling currency is varied. In biology, currency is encoded in small-molecule messengers, such as phosphate,2 acetate,3 and methyl.4 There are also small-protein signal carriers, including ubiquitin,5 SUMO,6 NEDD,7 and ISG.8 Classical signal transduction operates by three principal methods: (1) turn-on/gain of function (including change of function) in which low-occupancy modification of a specific target elicits signal amplification, such as transcriptional activation through stimulatory low-stoichiometry phosphorylation of an upstream kinase;9−12 (2) turn-off function in which modification turns off the target, such as site-specific histone methylation driving transcription-resistant heterochromatin formation;13,14 and (3) dominant loss-of-function in which one signal modification on a target molecule potentiates inhibition of more than one polypeptide, typically through protein multimerization.15 Signaling carriers can also intersect. Functional trading proceeds at a specific signaling node, leading to sophisticated positive and negative feedback loops all of which serve to help maintain fitness.1
One chemical commonality across these conventional post-translational modifications is that their installation/removal is almost always enzyme-catalyzed.16 In the absence of enzymes assisting the removal, these conventional covalent modifications are largely stable, endowing the cell with exquisite control over the signaling networks while affording a relative ease of detection for these modified states by methods such as mass spectrometry (MS). In addition, the preferred/consensus amino-acid landscape can often direct enzyme-mediated modifications, facilitating bioinformatics prediction. Finally, the signal carriers are not inherently reactive, and these enzyme-catalyzed modifications manifest their influence through either charge/steric/stereoelectronic modulations (e.g., phosphorylation, acetylation, etc.) and/or recruitment of secondary messengers (e.g., ubiquitin).
Demons in Paradise? Redox Signaling Compared to Conventional Enzyme-Orchestrated Signaling Paradigms
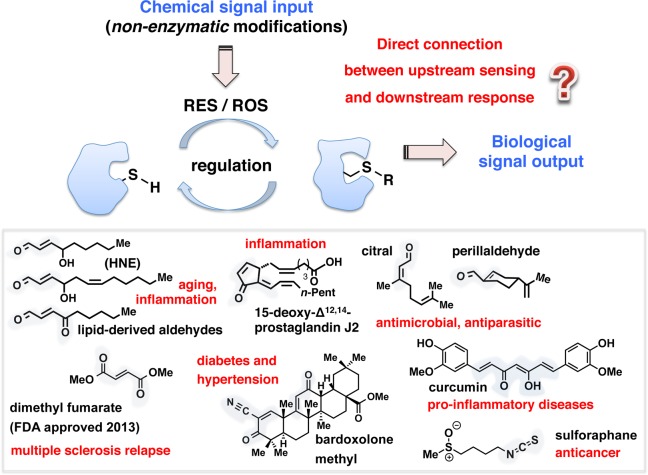
The most recent decade has witnessed the emergence of a distinctive clique of small signaling mediators, reactive electrophilic and oxygen species (RES/ROS), that orchestrate a noncanonical signaling paradigm called redox regulation.17−19 Markedly contrasting precise enzyme-controlled regulation that underlies canonical signaling modalities, spatiotemporal RES/ROS-modification events largely proceed without enzyme catalysis (Figure 1). Precision regulation engendered by these promiscuous and diffusible RES/ROS is intriguing because these chemical agents are deleterious when generated out of control. Yet, the concept that basal, sublethal elevations in endogenous redox signals are beneficial is gaining traction. Regulated reactive signals prime important and possibly essential signaling pathways that promote fitness, bestow longevity, and are indispensable for critical processes such as development.
Figure 1.
Biological inspiration. Ability to directly interrogate on-target redox responses in vivo promises to advance a mechanistic understanding of specific redox pathways and precision therapeutic intervention. RES/ROS, reactive electrophilic/oxygen species. “SH” designates a thiol group of the cysteine residue on the target sensor protein. “R” represents a generic chemical modification. Inset: representative natural and synthetic reactive small electrophilic signaling mediators and their associated bioactivities. Blue shades within the chemical structures highlight electrophilic motifs.
A class of RES known as lipid-derived electrophiles (LDEs) (Figure 1, inset), many of which are endogenously produced from membrane peroxidation events, displays diverse biological roles despite featuring structural simplicity.19 A synthetic RES, Tecfidera, was recently approved for the treatment of multiple sclerosis.20 Other dietary RES are associated with healthy lifestyles, including isothiocyanates, such as sulforaphane currently in clinical trials for diseases such as prostate cancer and diabetes21,22 (Figure 1, inset). These data imply a hormesis wherein priming by low-level exposure to specific RES/ROS is advantageous (eustress), but higher concentrations exert a detrimental effect (distress).23 Indeed, hormesis has been observed for numerous toxins including radiation, RES/ROS, and certain poisons. Many other healthy activities also reportedly function through hormesis, including exercise and calorie restriction.24 Notably, additional fundamental chemical characteristics set RES modifications, the focus of this perspective, apart from ROS signaling. RES modifications oftentimes occur irreversibly, unless modified protein turns over. Furthermore, these LDE signals are often bifunctional; thus once covalently bound to a target typically through Michael addition chemistry to a cysteine, they can undergo secondary reactions at the carbonyl (such as cross-linking via Schiff base formation with a lysine) (Figure 2, inset).
Figure 2.
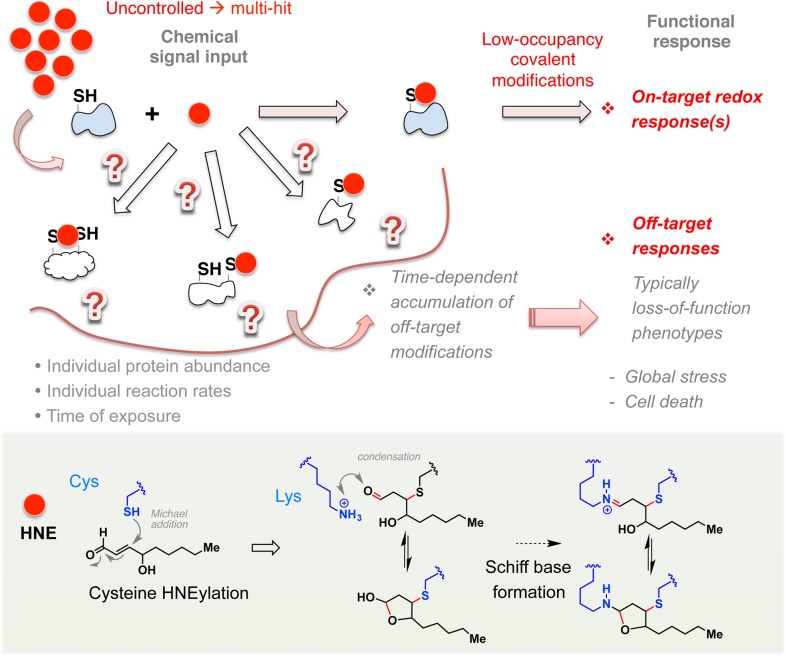
Time is of the essence. Deconvoluting precise impacts of target-specific redox responses remains a challenge because on-target binding and responses are often overwhelmed by off-target mass action of these covalent modifications by highly diffusible and reactive (often bifunctional; see Figure 1, inset) small electrophilic signaling mediators that can react with many targets nonspecifically. The red dot designates a RES signal such as HNE. Inset: dual-reactivity manifested in LDEs such as HNE, potentially resulting in protein cross-links through Schiff base formation (dotted arrow) in addition to conjugate addition (block arrow). Note: Conjugate addition can also occur, but to a lesser extent, with a histdine or lysine (Lys, shown) residue, in addition to with the more nucleophilic residue cysteine (Cys) as shown.
Since these RES/ROS signals are, in terms of the mechanism of conjugation/inherent instability of the modifications, the polar opposite to the canonical functional modifiers discussed above, the mold that was sculpted by early studies into conventional signaling events needs to be reshaped to accommodate the unique signaling properties displayed by these redox-modulatory messengers. However, until recently, it has remained unclear to what extent classical cell signaling concepts/trends are recapitulated by reactive LDEs. Integrating the latest findings in this rapidly moving field, we discuss below how these noncanonical electrophilic modification events function on a specific target. We also highlight relevant methods of mechanistic interrogations into on-target electrophilic responses along with key considerations necessary for investigating the functional impacts of RES regulation.
Time Is of the Essence: Reversible Vs. Irreversible Binders and the “Off-Target Problem”
Many ligands commonly used in biomedical as well as basic research are reversible binders that require a specific folded protein to form a bound complex. Ligand–target interaction equilibria are typically established swiftly and are reversible. By contrast, RES interacts with targets differently. These molecules form a covalent bond to their target through a relatively slow chemical process that can take minutes to hours,25−28 depending on the nature of the target, its subcellular locale, abundance, and potentially, associating partners (Figure 2). Unlike reversible binders, RES interactions with target proteins are often dominated by chemical reactivity (i.e., bond-forming/breakage kinetics) rather than binding affinity. For this reason, assessments of protein modification/inhibition by LDEs using IC50 and Kd’s are not particularly reliable/relevant. Evaluations of interactions in isolated systems for the reactivity of a target or specificity of a residue to a particular RES signal also require caution. For instance, endogenous bioactive signaling LDEs, such as 4-hydroxynonenal (HNE) (Figure 1 and 2, insets), readily conjugate to free cysteine at physiological conditions; thus, the presence of a cysteine within any protein potentially endows susceptibility to HNEylation. This is likely true of many RES/ROS signals. Likewise in a cell, with sufficient quantities of RES/ROS available, most proteins will eventually become labeled. As a consequence, since conditions that lead to endogenous LDE signaling are often unknown, it is no simple task to define to what extent the observed modification is “physiological” and “functional”. Consistent with the above argument, treatment of cells with a bolus of radiolabeled- or alkyne-tagged-HNE leads to hundreds of modified proteins.29 The sheer number of modified targets downplays any profit of “polypharmacology”. Since these modifications are time-dependent, experiments are hard to control, and precise recapitulation of how specific individual modifications directly influence physiologic redox responses in native settings continues to be a formidable challenge in the field. This challenge needs to be addressed since mounting evidence suggests that physiologic redox events proceed with precise timing and spatial/target control.
An additional layer of complexity arises from the fact that the electrophile introduced to the system is not necessarily the active agent. Although strictly true for any ligand, the issue is magnified for reactive LDEs because endogenous small molecules such as glutathione as well as proteins can form adducts with them, changing their chemical and biological properties, while also altering cellular redox state.17 In the case of related ROS treatment of cells, LDE themselves can be generated as secondary products via cellular ROS-induced membrane peroxidation events.19 Furthermore, for dual-reactivity electrophiles such as HNE, the remaining unreacted aldehyde post-Michael conjugation can still react and form inter/intra molecular protein cross-links (Figure 2, inset). Michael addition with other less reactive residues (Lys, His) can also occur19 during uncontrolled treatment or due to proximity.
Deconvoluting the Functional Coupling with Specific Cysteines
Many elegant model systems to study redox signaling exist that allow generation of endogenous RES/ROS in the context of specific (patho)physiological states, such as H2O2 signaling through the Nox-isoenzymes.30 To date, these model systems are yet to be generically applicable. Thus, the most commonly deployed general method to evaluate biochemical mechanisms underlying reactive chemical signal-specific redox pathways of interest involves treating isolated proteins, cells, or animals with excess RES/ROS of interest for minutes to hours. Despite the issues commonly encountered with cumulative off-target effects incurred when bathing with reactive, irreversible binders, the bolus dosing approach is the go-to protocol to identifying large numbers of modified proteins and can be applied to whole organs and model organisms easily. Bolus methods have also shed light on global oxidative stress-related responses. One additional asset of global flooding is the ability to probe phenotypic responses in the context of simultaneous modifications of large numbers of cellular proteins; however, eustress phenotypes are often challenging to be unambiguously linked to modification of specific proteins and to parse from effects of cell death. The reliance on high HNE load continues to muddy the waters limiting our current understanding of physiologic LDE signaling at the molecular level.
Proteome-wide quantitative reactivity profiling has enabled a sizable number of LDE-reactive cysteines (approximately 1% of all cysteines) to be profiled based on their reactivity using MS.31 Addition of a competing reactive ligand (such as HNE) leads to loss of “sensitive” profiled cysteines from the pool identified by MS, allowing modification of that cysteine, or a functionally coupled residue, to be inferred. This method has numerous advantages over bolus dosing: (1) low RES concentrations can be used; (2) around 1000 reactive cysteines can be profiled using a routine protocol; and (3) specific sites (or sites functionally coupled ones) can be identified. Recent years have witnessed exciting extension of this innovation through the development of probes reportedly specific to tyrosines, lysines as well as second-generation cell-permeable probes.32 These interventions have begun to address the initial limitations of the system to profiling serines and cysteines and restriction to perform experiments in lysates, although there is no way to assay multiple reactive groups simultaneously or interrogate direct downstream functional response on a specific modified target. Additional general points of consideration in future proteome-profiling-probe development are to reduce reliance on non-native RES mimics, such as iodoacetamide and N-ethylmaleimide (NEM), as the target spectra of different synthetic electrophiles vary and given the relatively low number of cysteines captured the targets modified by synthetic mimics likely do not recapitulate the bona fide innate sensors that sense native signals; and to possibly sidestep bulky affinity handles such as biotin.
It is in principle possible to interrogate functional and pathway relevance of LDE signaling using siRNA/targeted knockout in combination with bolus dosing. However, because of the promiscuity of LDEs coupled with our inability to recapitulate/define/predict a priori “redox signaling”, this approach is much less informative than in traditional genetics experiments that probe canonical enzyme-mediated signaling mechanisms. Thus, from both biochemical and genetic grounds, a major limitation in understanding precision redox signaling had been the dearth of methods to directly and precisely link individual targets captured to downstream phenotypes with specificity in timing and target (Figure 1).
Biological Sufficiency within Single-Protein-Specific Redox Signaling
Given the promiscuity and reactivity of diffusible electrophilic small molecules, LDE signaling had been largely considered to operate in a “multi-hit” model in which cumulative modifications of several localized sensor proteins yield a phenotype (Figure 3). At the other extreme, a model in which modification of a specific sensor is sufficient for response has recently been considered, although until recently it remained an untestable possibility. However, such mechanistic knowledge is of fundamental importance; for instance, if redox signaling pathways are triggered through specific low-stoichiometry modifications leading to a new biological state of a specific target protein, it is critical that these new state(s) and the mechanism whereby this state can elicit functional redox response be interrogated in cells and whole organisms.
Figure 3.
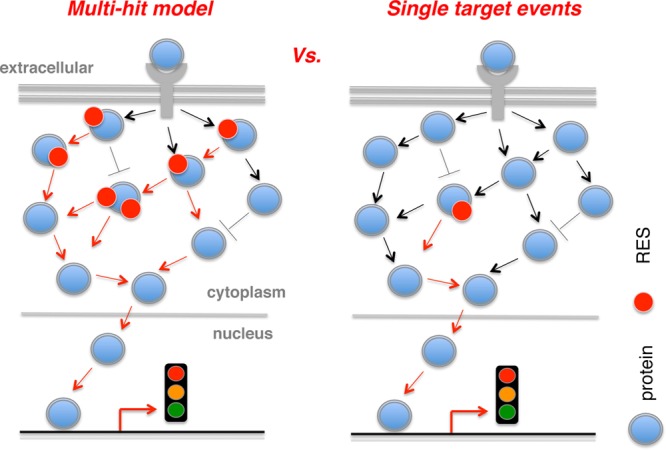
Evolution of RES signaling concepts. Multihit paradigm necessitates modifications of many localized sensor targets in order to elicit a response downstream, such as modulation of transcriptional response (this illustration). By contrast, the latest findings in the field initially suggest and subsequently provide direct evidence that redox responses operate similarly to canonical signal transduction wherein low-stoichiometry modest modifications of a single target are capable of driving a functional response. See text for details. The double-head and blunt-end arrows, respectively, indicate direct/indirect activation and inhibition. Arrows highlighted in red designate the nuclear signaling trajectory of interest.
On-Target Redox Events Are Sufficient Drivers of Functional Signaling Response
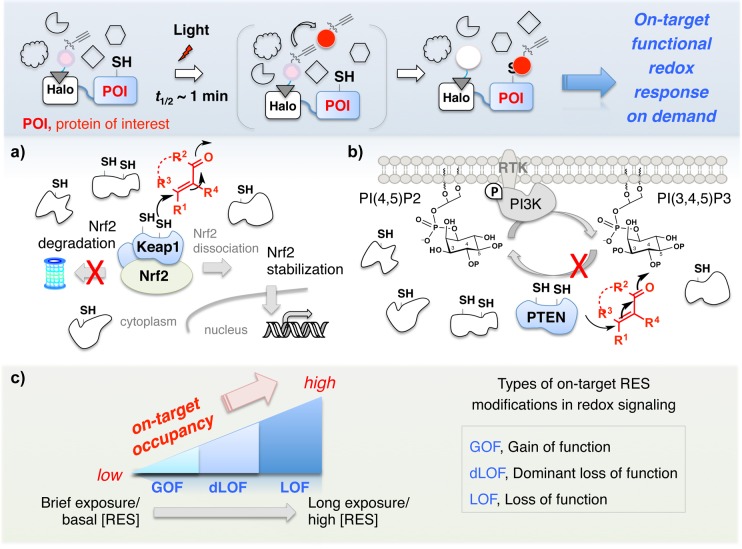
A recently developed chemistry-driven platform, “targetable reactive electrophiles and oxidants (T-REX)”, ultimately aimed at on-target on-demand tripping of specific redox signaling nodes, addresses precision redox responses in physiologic settings.33−36 T-REX takes advantage of proximity-assisted targeting to enable controlled delivery of a reactive chemical signal to a specific protein of interest upon photoactivation (Figure 4, inset) (Class II proximity enhancement concept37). The nuts and bolts of the platform are detailed elsewhere.33 Briefly, a protein of interest (POI) is expressed as a genetic fusion to HaloTag (an engineered protein that reacts stoichiometrically and irreversibly with chloroalkanes). Cells expressing the HaloTag-POI construct are treated with a specific chloroalkane-decorated inert probe capable of releasing a single HNE molecule in the presence of light. After washing away excess probe, the HaloTag is stoichiometrically and irreversibly labeled with the photocaged-precursor to HNE. Light then releases HNE in the vacinity of the POI. If the POI is HNE sensitive, low-occupancy modification (up to ∼60% of released HNE has been delivered to the POI; whereas as low as ∼15% HNEylation efficiency has been scored as a positive sensor) of the POI by HNE will occur. HNE that does not bind to the POI is either intercepted by GSH or averaged over the whole proteome, leading to essentially no background labeling of off-target proteins. This system has significant potential to influence how redox signaling is studied/considered because if single-target HNEylation events are able to elicit gain-of-function or dominant loss-of-function, this would prove that these states are single, biologically relevant entities that have defined signaling capacity and must be studied separately. Such states would ideally be examined in the context of a healthy cell, mandating study through single-protein on-demand modifications, like T-REX. Our early findings using T-REX have indeed unveiled fundamental nuances of electrophile signaling summarized below.
Figure 4.
Single-target RES modifications are individual events that drive functional redox response. Inset: ability to directly and precisely flip a single redox switch in living systems by T-REX offers a lens to understand functional on-target redox responses on demand (see text for details). Red dot, alkyne-functionalized HNE photouncaged from its photocaged precursor (pink dot) covalently bound to HaloTag. T-REX shows that (a) Keap-1-alone electrophilic modification is sufficient to stabilize Nrf2 and activate transcriptional antioxidant response in a way similar to canonical gain-of-function signaling; and (b) Pten-specific electrophilic modification modulates cellular phosphoinositide levels through a conventional mechanism of dominant loss-of-function inhibitory cell signaling. (RTK, receptor tyrosine kinase; PI3K, phosphoinositide 3-kinase; PI[(3,)4,5]P2(3), phosphatidylinositol (3,)4,5-bis(tris)phosphate. (c) A potential model subcategorizing the types of RES modifications that drive on-target redox signal propagation as a gradient of occupancy (e.g., “LDEylation stoichiometry”) and time/dose of RES. See text for details.
Gain-of-Function Redox Signaling through Keap1-Specific Electrophilic Modification.33,35,36
Keap1 is an E3-ligase adaptor that facilitates polyubiquitination and degradation of the transcription factor Nrf2, the master regulator of cellular antioxidant response (AR).38 In the prevailing model, AR activation results when RES-modified Keap1 releases Nrf2, which subsequently activates downstream genes. However, this pathway is modulated by multiple upstream redox-sensor proteins.38 Many existing data argued for requirements of LDE-modification on other coregulators of the pathway to observe AR activation38,39 (a multihit model, Figure 3). We thus considered this pathway a good test case for T-REX because it was at the time impossible to define to what extent Keap1-specific redox modifications control AR signaling under bolus multihit conditions that modify hundreds of sensors simultaneously. Under T-REX-enabled targeted LDEylation of Keap1 in low stoichiometry (20–60% modification efficiency depending on cell lines/context, RES chemotype, etc.),33−36 with no background labeling detected, selective Nrf2 stabilization and gain-of-function Nrf2-driven AR upregulation were observed. No AR activation was observed under otherwise identical conditions when Keap1 was not specifically targeted.35,36 Interestingly, the magnitude as well as latency of AR pathway activation showed subtle differences compared to global flooding with various LDEs.36 Hundreds of proteins are typically modified under bolus methods, and these discrepancies may be attributable to secondary, synergistic, compensatory, and/or off-target effects or permeability of LDEs versus temporally controlled targeted delivery of a specific LDE from a photocaged probe in T-REX. The key result of pathway activation demonstrated that on-target HNEylation could function in a similar way to canonical signaling pathways: a single target-alone modification can trigger downstream response (Figure 4a).
Dominant Loss-of-Function Response through Pten-Specific Electrophilic Modulation33,34
Pten is a key tumor suppressor phosphatase frequently mutated in cancer cells.40 Importantly, there is evidence that heterozygous individuals with one loss-of-function allele are more prone to disease than hemizygotes.41 This finding has led to the postulate that hypomorphic/loss-of-function alleles are dominant negative. Consistent with prior data suggesting that alkylation of this enzyme leads to loss of protein function, T-REX-enabled selective HNEylation of PTEN in cells resulted in accumulation of endogenous PIP3 phosphoinositide.33 These data reinforce that inhibitory electrophilic modification on a single target is a functionally relevant event that can intercept canonical currency transfer processes such as phosphosignaling pathways (Figure 4b). Together with the Keap1 example above, these systems provide direct evidence supporting the single-hit model (Figure 3).
Redox Sensing and Response as a Moonlighting Function
Since LDEs function as bona fide signaling molecules, the cell must have evolved many sensing hubs that enable transmutation of reactive LDE signals to precise molecular events, including transcription regulation and intersection with known signaling subsystems. These privileged nodes are likely to be excellent LDE-sensors such that one could consider this behavior to be a moonlighting activity. We predict that these sensors will mainly function through the typical signal amplification mechanisms, namely, gain-of-function and (dominant) loss-of-function. We anticipate many new sensor proteins that use these signal transduction methods will be unveiled through continued novel applications and future developments of innovative tools. The foundational knowledge that precision RES targeting and global RES proteomics tools have collectively established thus promises to ultimately address unmet therapeutic needs through precision perturbation of these moonlighting events on diverse sensor proteins.
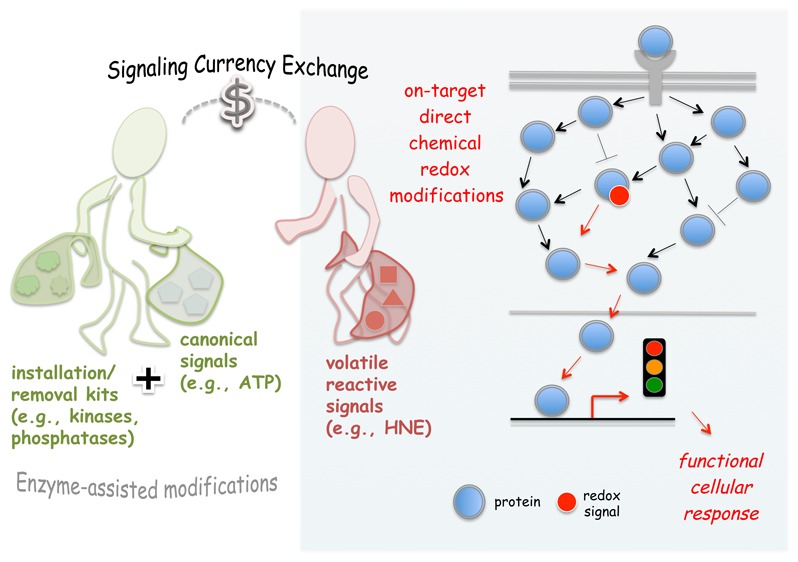
HNE Is Part of the Information Currency of the Cell
Interrogations of Nrf2/AR pathway and PI3K/Pten phospholipid signaling subsystems have shown that HNE is part of the information currency of the cell. The changing of hands of information carriers we show above constitute the first examples of RES-to-canonical-signaling “information currency exchange”, from the volatility of the primary LDE signal, to the measured response of phosphate-signaling (such as in PI3K/Pten), or transcriptional modulation (such as Nrf2/AR), a process akin to going to the “Dollar standard”. These currencies have their own idiosyncrasies that the cell uses to its advantage. LDEs as small signaling mediators are short-lived and unpredictable but act rapidly and under specific conditions act highly selectively. It is likely that these attributes are beneficial under times of stress where resources can be limited, and some enzymes function at suboptimal capacity. Although single-protein-specific signaling proves that these pathways do not require that the cell be stressed in order for them to function. (De)Phosphorylation/gene activation is much more able to respond to global market fluxes and has multiple checks and balances built in. Thus, it makes sense that redox signaling should ultimately intersect with traditional pathways, at least in part as a “reality check”. Thus, specific proteins appear to behave as “brokers” in this critical information transfer system, that is, ultimately “the eyes and ears” of the cell.
We finally propose that the mechanism by which a specific LDE effects downstream signaling may help to control threshold trigger points and in part be responsible for hormesis. For similarly reactive proteins, at low-concentration/brief HNE exposure, phenotypes likely stem from gain-of-function, such as Keap1-initiated Nrf2/AR-activation. As HNE-concentration or exposure-time increases, HNE-occupancy of sensor proteins increases, and dominant-loss-of-function signaling pathways can influence phenotypes (Figure 4c). Pten-inhibitory signaling for instance promotes growth-stimulating signaling. Finally, at high/prolonged/chronic HNE-exposure loss-of-function phenotypes come to light. Using this yardstick, it is no coincidence that beneficial antioxidant signals are ushered through Keap1 gain-of-function. At the other extreme, cell death42 is likely elicited by excess (bolus) HNE as a result of loss-of-function (high-occupancy) of sensor proteins.
Acknowledgments
We thank Professor Carolyn S. Sevier (Cornell University) for helpful discussions and comments on the manuscript.
Glossary
Abbreviations
- AR
antioxidant response
- ATP
adenosine triphosphate
- Cys
cysteine
- dLOF
dominant loss of function
- GOF
gain of function
- HaloTag
an engineered domain that reacts irreversibly with chloroalkanes
- His
histidine
- HNE
4-hydroxy-2-nonenal
- IC50
inhibitor constant (inhibitor concentration required for 50% inhibition)
- ISG
interferon-stimulated gene
- Kd
dissociation constant
- LDE
lipid-derived electrophile
- LOF
loss of function
- Lys
lysine
- Me
methyl
- MS
mass spectrometry
- NEDD
neural precursor cell expressed developmentally down-regulated protein
- Nrf2
nuclear factor (erythroid-derived 2)-like 2, also known as NFE2L2
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- PI[(3,)4,5]P2(3)
phosphatidylinositol (3,)4,5-bis(tris)phosphate
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- n-Pent
normal pentyl
- POI
protein of interest
- PTEN
phosphatase and tensin homologue
- R
generic chemical modification
- RES
reactive electrophilic species
- ROS
reactive oxygen species
- RTK
receptor tyrosine kinase
- T-REX
targetable reactive electrophiles and oxidants
- SUMO
small ubiquitin-like modifier
- vs
versus
Biographies
Marcus Long was born in Beverley, East Yorkshire, UK. He studied chemistry at Oxford University, UK, graduating in 2003. Having completed his Ph.D. in biochemistry in Brandeis University, USA, he now focuses on understanding the small-molecule regulation of cell signaling pathways, including redox signaling.
Yimon Aye was born in Burma. She moved to UK, and studied chemistry at Oxford University, UK, graduating in 2004. She moved to Harvard University, USA, achieving a Ph.D. in organic chemistry under the supervision of Prof. Dave Evans. She then moved to Massachusetts Institute of Technology to research the cellular and biochemical regulatory mechanisms of the enzyme ribonucleotide reductase with Prof. JoAnne Stubbe. In her independent career at Cornell University that began in mid-2012, she set out to understand the detailed mechanisms of redox signaling. This impetus culminated in the development of T-REX.
NIH-New-Innovator (1DP2GM114850), NSF CAREER (CHE-1351400), Beckman Young Investigator, Burroughs Wellcome CRTG, Cornell Ithaca-Weill intercampus seed funding, and the Sloan fellowship programs (to Y.A.) are acknowledged for supporting the RES signaling research program in the Aye lab.
The authors declare no competing financial interest.
Notes
We are honored to contribute this Perspective as part of the 2016 ACS CRT young investigator award (to Y.A.).
References
- Lim W., Mayer B., and Pawson T. (2015) Cell Signaling: Principles and Mechanisms, Garland Science Taylor & Francis Group, New York. [Google Scholar]
- Chen Z.; Cole P. A. (2015) Synthetic approaches to protein phosphorylation. Curr. Opin. Chem. Biol. 28, 115–122. 10.1016/j.cbpa.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E.; Ott M. (2015) 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264. 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Paik W. K.; Paik D. C.; Kim S. (2007) Historical review: the field of protein methylation. Trends Biochem. Sci. 32, 146–152. 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- (2005) Protein Degradation, Wiley-VCH, Weinheim, Germany. [Google Scholar]
- Flotho A.; Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385. 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Enchev R. I.; Schulman B. A.; Peter M. (2015) Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44. 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. M.; Chevillotte M. D.; Rice C. M. (2014) Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. F.; Wang Y. T.; Yen H. Y.; Tsou C. C.; Ku W. C.; Lin P. Y.; Chen H. Y.; Nesvizhskii A. I.; Ishihama Y.; Chen Y. J. (2015) Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat. Commun. 6, 6622. 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.; Haas W.; Dephoure N.; Huttlin E. L.; Zhai B.; Sowa M. E.; Gygi S. P. (2011) A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683. 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V.; Vermeulen M.; Santamaria A.; Kumar C.; Miller M. L.; Jensen L. J.; Gnad F.; Cox J.; Jensen T. S.; Nigg E. A.; Brunak S.; Mann M. (2010) Quantitative Phosphoproteomics Reveals Widespread Full Phosphorylation Site Occupancy During Mitosis. Sci. Signaling 3, ra3–ra3. 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Dar A. C.; Shokat K. M. (2011) The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu. Rev. Biochem. 80, 769–795. 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- Martin C.; Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849. 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Holt M.; Muir T. (2015) Application of the protein semisynthesis strategy to the generation of modified chromatin. Annu. Rev. Biochem. 84, 265–290. 10.1146/annurev-biochem-060614-034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich S. G.; Garbarino J. E.; Wei P.; Lim W. A. (2010) Rapid Diversification of Cell Signaling Phenotypes by Modular Domain Recombination. Science 328, 368–372. 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. (2005) Posttranslational Modification of Proteins: Expanding Nature’s Inventory, W. H. Freeman. [Google Scholar]
- Banerjee R., Becker D. F., Dickman M. B., Gladyshev V. N., and Ragsdale S. W. (2008) Redox Biochemistry. John Wiley & Sons, Inc., New York. [Google Scholar]
- Murphy M. P.; Holmgren A.; Larsson N. G.; Halliwell B.; Chang C. J.; Kalyanaraman B.; Rhee S. G.; Thornalley P. J.; Partridge L.; Gems D.; Nystrom T.; Belousov V.; Schumacker P. T.; Winterbourn C. C. (2011) Unraveling the biological roles of reactive oxygen species. Cell Metab. 13, 361–366. 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer F. J.; Cipollina C.; Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021. 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D.; Kieseier B. C.; Hartung H. P.; Hemmer B.; Warnke C.; Menge T.; Miller-Little W. A.; Stuve O. (2015) Dimethyl fumarate in relapsing-remitting multiple sclerosis: rationale, mechanisms of action, pharmacokinetics, efficacy and safety. Expert Rev. Neurother. 15, 339–346. 10.1586/14737175.2015.1025755. [DOI] [PubMed] [Google Scholar]
- Traka M. H.; Melchini A.; Mithen R. F. (2014) Sulforaphane and prostate cancer interception. Drug Discovery Today 19, 1488–1492. 10.1016/j.drudis.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova A. T.; Kostov R. V. (2012) Glucosinolates and isothiocyanates in health and disease. Trends Mol. Med. 18, 337–347. 10.1016/j.molmed.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Yun J.; Finkel T. (2014) Mitohormesis. Cell Metab. 19, 757–766. 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E. J.; Baldwin L. A. (2003) Hormesis: the dose-response revolution. Annu. Rev. Pharmacol. Toxicol. 43, 175–197. 10.1146/annurev.pharmtox.43.100901.140223. [DOI] [PubMed] [Google Scholar]
- Singh J.; Petter R. C.; Baillie T. A.; Whitty A. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discovery 10, 307–317. 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Connor R. E.; Marnett L. J.; Liebler D. C. (2011) Protein-selective capture to analyze electrophile adduction of hsp90 by 4-hydroxynonenal. Chem. Res. Toxicol. 24, 1275–1282. 10.1021/tx200157t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C. E.; Tallman K. A.; Porter N. A.; Marnett L. J. (2011) Structure-activity analysis of diffusible lipid electrophiles associated with phospholipid peroxidation: 4-hydroxynonenal and 4-oxononenal analogues. Chem. Res. Toxicol. 24, 357–370. 10.1021/tx100323m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J. T.; Zhang X.; Leach A. G.; Houk K. N. (2009) Beyond Picomolar Affinities: Quantitative Aspects of Noncovalent and Covalent Binding of Drugs to Proteins. J. Med. Chem. 52, 225–233. 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Tallman K. A.; Porter N. A.; Liebler D. C. (2015) Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal. Chem. 87, 2535–2541. 10.1021/ac504685y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer T. F.; Garcia F. J.; Onak C. S.; Carroll K. S.; Chang C. J. (2015) Chemical approaches to discovery and study of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins. Annu. Rev. Biochem. 84, 765–790. 10.1146/annurev-biochem-060614-034018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Weerapana E.; Blewett M. M.; Cravatt B. F. (2014) A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85. 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon D. A.; Weerapana E. (2015) Covalent protein modification: the current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol. 24, 18–26. 10.1016/j.cbpa.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Parvez S., Long M. J. C., Lin H.-Y., Zhao Y., Haegele J. A., Pham V. N., Lee D. K., and Aye Y. (2016) T-REX on-demand redox targeting: a toolset for functional discoveries and validations. Nat. Protoc., in press, DOI: 10.1038/nprot.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Fu Y.; Long M. J.; Haegele J. A.; Ge E. J.; Parvez S.; Aye Y. (2013) Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. J. Am. Chem. Soc. 135, 14496–14499. 10.1021/ja405400k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S.; Fu Y.; Li J.; Long M. J.; Lin H. Y.; Lee D. K.; Hu G. S.; Aye Y. (2015) Substoichiometric hydroxynonenylation of a single protein recapitulates whole-cell-stimulated antioxidant response. J. Am. Chem. Soc. 137, 10–13. 10.1021/ja5084249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. Y.; Haegele J. A.; Disare M. T.; Lin Q.; Aye Y. (2015) A generalizable platform for interrogating target- and signal-specific consequences of electrophilic modifications in redox-dependent cell signaling. J. Am. Chem. Soc. 137, 6232–6244. 10.1021/ja5132648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Poganik J. R.; Aye Y. (2016) On-Demand Targeting: Investigating Biology with Proximity-Directed Chemistry. J. Am. Chem. Soc. 138, 3610–3622. 10.1021/jacs.5b12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D.; Dinkova-Kostova A. T. (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218. 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Bryan H. K.; Olayanju A.; Goldring C. E.; Park B. K. (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 85, 705–717. 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Worby C. A.; Dixon J. E. (2014) Pten. Annu. Rev. Biochem. 83, 641–669. 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- Leslie N. R.; den Hertog J. (2014) Mutant PTEN in Cancer: Worse Than Nothing. Cell 157, 527–529. 10.1016/j.cell.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Yang W. S.; Stockwell B. R. (2016) Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 26, 165–176. 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]