Abstract
The p53-MDM2 feedback loop is vital for cell growth control and is subjected to multiple regulations in response to various stress signals. Here we report another regulator of this loop. Using an immunoaffinity method, we purified an MDM2-associated protein complex that contains the ribosomal protein L23. L23 interacted with MDM2, forming a complex independent of the 80S ribosome and polysome. The interaction of L23 with MDM2 was enhanced by treatment with actinomycin D but not by gamma-irradiation, leading to p53 activation. This activation was inhibited by small interfering RNA against L23. Ectopic expression of L23 reduced MDM2-mediated p53 ubiquitination and also induced p53 activity and G1 arrest in p53-proficient U2OS cells but not in p53-deficient Saos-2 cells. These results reveal that L23 is another regulator of the p53-MDM2 feedback regulation.
The tumor suppressor function of p53 is primarily attributed to its ability to activate transcriptional expression of many genes whose protein products induce cell growth arrest, apoptosis, senescence, or inhibition of angiogenesis in response to various stresses, thus protecting cells from transformation and tumorigenesis (54, 63). Cells also develop a negative feedback mechanism to monitor p53 function because p53 activation is toxic to the cells (4, 51, 67). A crucial player in this feedback regulation is an oncoprotein called MDM2 (41). MDM2 specifically binds, through its N-terminal domain, to the N terminus of p53 (10, 47). On one hand, this binding conceals the N-terminal transcription domain of p53, directly blocking its transcriptional activity (10, 41). On the other hand, this binding initiates p53 ubiquitination, thus leading to its degradation by the proteasome system (20, 30), as MDM2 is a Ring-finger-containing E3 ubiquitin ligase (16, 21). Additionally, MDM2 contains a nuclear export signal and induces p53 nuclear export through direct interaction (6, 18, 36, 60), thus preventing p53 from accessing its responsive DNA elements. Consequently, MDM2 suppresses p53-mediated cell growth arrest and apoptosis. This regulation presents an elegant autoregulatory feedback loop because MDM2 is also induced by p53 and in turn inhibits p53 function (4, 51, 67). Indeed, genetic disruption of the p53 gene rescues the lethal phenotype of the mdm2 knockout mouse, firmly validating the notion of the MDM2-p53 feedback loop (27, 43).
This MDM2-p53 feedback loop is subjected to multiple regulations in response to different signals because of its importance in cell growth control and transformation. For instance, DNA damage-induced phosphorylation of p53 or MDM2 suppresses p53-MDM2 binding, thus inhibiting MDM2-mediated p53 suppression (3, 8, 28, 39, 56). Also, viral and cellular oncoproteins, such as Ras and c-Myc, can activate p53 by alleviating the function of MDM2 through induction of an MDM2 inhibitor called p14arf (mouse p19arf) (49, 77). The ARF protein prevents MDM2 from targeting p53, probably through two mechanisms: separating the two proteins from different cellular compartments and inhibiting MDM2-mediated p53 ubiquitination (22, 61, 65, 66, 75). MDM2 is also regulated by its homolog MDMX (55), which assists MDM2 in down-regulating p53 function (19, 24, 32, 57). Furthermore, phosphorylation of MDM2 at serines 166 and 186 by Akt in response to the Her2-mediated cell growth signaling enhances the nuclear localization of MDM2 and, as a result, inactivates p53 (76). Therefore, the MDM2-p53 loop is tightly regulated by distinct proteins in response to different signals.
The p53-MDM2 feedback loop is also regulated by stress on ribosomal biogenesis. Proper ribosome assembly is essential for the health of a cell. Therefore, it is logical that impairment to this function would require cell growth arrest or apoptosis to facilitate repair or to remove affected cells, probably mediated by p53. For example, overexpression of a dominant-negative mutant of Bop1, a nucleolar protein critical for rRNA processing and ribosome assembly, inhibited 28S and 5.8S rRNA formation and led to deficiency of newly synthesized 60S ribosomal subunits in 3T3 fibroblast cells (50). Consequently, the cells underwent p53-dependent G1 arrest (50). Also, actinomycin D, which inhibits RNA polymerase I at a concentration of 5 nM, can stall rRNA synthesis and ribosome assembly. By doing so, this compound activates p53 function without triggering N-terminal phosphorylation of p53 (1, 2). These studies suggest a potential signaling pathway that may mediate p53 activation by sensing stresses on ribosome biogenesis. Indeed, a ribosomal protein, L11, has been recently shown to be a regulator of this pathway (35, 74). Hence, it is important to uncover other regulators in this pathway.
To do so, we have generated a stable human embryonic kidney epithelial 293 cell line that constitutively expresses MDM2. Using this cell line, we purified a cytoplasmic MDM2-associated complex through an immunoaffinity purification followed by a mass spectrometric analysis. Surprisingly, several ribosome proteins, including L23, L11, and L5, were found in the complex. This finding not only confirms the previously reported interaction of MDM2 with L5 (37) and the more recently reported interaction with L11 (35, 74) but also identified L23 as a new regulator of MDM2. Our further study shows that L23, like L5 and L11, bound to MDM2, but unlike L5 and L11, it preferentially interacted with a distinct domain of MDM2, independent of the 80S ribosome and polysome. L23 activated p53 by preventing MDM2 from targeting and ubiquitinating p53. Interestingly, the RNA polymerase I inhibitor actinomycin D, but not gamma-irradiation or translation inhibitors, enhanced the MDM2-L23 binding. Furthermore, removal of L23 by small interfering RNA (siRNA) inhibited actinomycin D-induced p53 activation. Consistently, ectopic expression of L23 induces p53 transcriptional activity and G1 arrest in p53-containing U2OS cells but not in p53-null Saos-2 cells. These results suggest that L23, in response to ribosomal stress, may activate p53 by inhibiting the MDM2-mediated feedback regulation of p53.
MATERIALS AND METHODS
Plasmids and antibodies.
To generate human L23 expression construct pcDNA3-2Flag-L23, the full-length L23 cDNA was amplified by reverse transcriptase PCR (RT-PCR) from HeLa cell mRNA with the following primers (restriction enzyme sites are underlined): P1, 5′-CGCGGATCCATGTCGAAGCGAGGACGTGGTG-3′; P2, 5′-CCGGAATTCTCATGCAATCCTGCCAGCATTG-3′. The PCR product was subcloned into the pcDNA3-2Flag vector. The pcDNA3-2Flag-L23ΔN vector with a deletion of amino acids (aa) 1 to 65 was constructed by PCR amplification with primers P2 and P3 (5′-CGCGGATCCAAGAAAGGCAAACCACAGCTC-3′). The green fluorescent protein (GFP)-L23 expression vector was cloned by inserting a PCR product with the primers P2 and P4 (5′-ACAGAAGATCTATGTCGAAGCGAGGACGTGG-3′) into pEGFP-C1 (Clontech). The glutathione S-transferase (GST)-L23 bacterial expression vector was constructed by subcloning the full-length L23 from pcDNA3-2Flag-L23 into pGEX.4T.1 (Pharmacia Biotech). The GST fusion L23 fragments pGEX.4T.1-L23/1-65, pGEX.4T.1-L23/66-140, pGEX.4T.1-L23/1-105, and pGEX.4T.1-L23/35-105 were subsequently constructed by cloning PCR products into the pGEX.4T.1 vector. GST-MDM2 and GST-MDM2 deletion mutants were described previously (26). The His-MDM2 bacterial expression vector was constructed by inserting a PCR product into the pet24a vector (Novagen); the primers are 5′-CGCGGATCCATGTGCAATACCAACATGTCTG-3′ and 5′-CCGGAATTCGAGGGGGAAATAAGTTAGCAC-3′.
For generation of polyclonal anti-L23 antibody, the full-length L23 was amplified by PCR with primers P1 and P5 (5′-CCGGAATTCCGTGCAATCCTGCCAGCATTG-3′). The PCR product was subcloned into the pet24a-His vector to generate pet24a-His-L23. The His-tagged L23 protein was expressed in Escherichia coli and purified by using Ni-nitrilotriacetic acid (NTA) beads as an antigen to raise rabbit polyclonal anti-L23 antisera. Anti-L11 antibodies were kindly provided by Yangping Zhang (M. D. Anderson Cancer Institute, The University of Texas at Houston, Houston). Anti-Flag (Sigma), anti-p21 (NeeMarkers), and anti-p53 (DO-1; Santa Cruz) were purchased. Anti-MDM2 (2A10) and antihemagglutinin (anti-HA) (12CA5) have been described previously (26).
Buffers and reagents.
Lysis buffer consisted of 50 mM Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 1 mM EDTA, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF). SNNTE buffer contained 50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 1% Nonidet P-40, 500 mM NaCl, and 5% sucrose. Radioimmunoprecipitation assay (RIPA) buffer was comprised of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and 1% (wt/vol) sodium deoxycholate. Buffer C 100 (BC100) included 20 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM PMSF, 1 mM dithiothreitol, and 0.25 μg of pepstatin A/ml.
Cell culture.
Human embryonic kidney epithelial 293 cells, human lung small cell carcinoma H1299 cells, human p53-proficient osteosarcoma U2OS cells, and human p53-null osteosarcoma Saos-2 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 U of penicillin/ml, and 0.1 mg of streptomycin/ml at 37°C in a 5% CO2 humidified atmosphere as previously described (25, 26, 70).
Establishment of HA-MDM2 expression cell lines.
293 cells were transfected with pcDNA3-HA-MDM2 or pcDNA3 vector. Transfected cells expressing HA-MDM2 were selected in the presence of neomycin (0.5 mg/ml) and screened by immunoprecipitation with anti-HA antibodies followed by Western blotting with the monoclonal anti-MDM2 antibody 2A10.
Affinity purification of human MDM2-associated protein complexes.
Approximately 109 293 cells were used for the preparation of nuclear extract and cytoplasm (S100) by a previously described method (12). The 12CA5-affinity beads were prepared by conjugating anti-HA monoclonal antibodies (12CA5) to protein A-agarose beads as described previously (31). The beads were washed with phosphate-buffered saline (PBS) and suspended in PBS as a 50% slurry. Fifty milligrams of S100 protein fractions from either 293-HA-MDM2 cells or empty vector expressing 293 cells as a control were incubated with 0.2 ml of 12CA5-affinity beads at 4°C for 4 h. The beads were washed four times in lysis buffer containing protein inhibitors. The bead-bound proteins were eluted in 0.2 ml of lysis buffer containing 4 mg of synthetic HA peptides/ml. Eluted proteins were loaded onto an SDS-5 to 17% gradient polyacrylamide gel electrophoresis gel for colloidal blue staining. Specific bands from 293-HA-MDM2 fractions compared to the 293 control fractions were excised and subjected to mass spectrometric analysis.
Cotransfection, immunoblot, and coimmunoprecipitation analyses.
H1299, U2OS, or Saos-2 cells were transfected with plasmids as indicated in the figure legends with Lipofectin following the manufacturer's protocol (Invitrogen). Cells were harvested at 48 h posttransfection and lysed in lysis buffer. Equal amounts of clear cell lysate were used for immunoblot analysis as described previously (72). Immunoprecipitation was conducted by using antibodies as indicated in the figure legends and described previously (70). Beads were washed twice with lysis buffer, once with SNNTE buffer, and once with RIPA buffer. Bound proteins were detected by immunoblotting with antibodies as indicated in the figure legends.
Transient transfection and luciferase assays.
U2OS, Saos-2, or H1299 cells were transfected with the pCMV-β-galactoside reporter plasmid (0.1 μg) and a luciferase reporter plasmid (0.1 μg) driven by two copies of the p53RE motif derived from the MDM2 promoter (67), together with a combination of different plasmids (total plasmid DNA 1 μg/well) as indicated in the legend to Fig. 6, with Lipofectin (Invitrogen). At 48 h posttransfection, cells were harvested for luciferase assays as described previously (70, 71). Luciferase activity was normalized by a factor of β-galactosidase activity in the same assay.
FIG. 6.
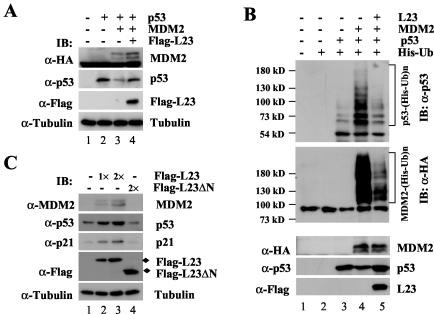
Ectopic expression of L23 stabilizes p53 and inhibits MDM2-mediated p53 ubiquitination. (A) Ectopic expression of L23 reverses MDM2-mediated p53 degradation. H1299 cells were transfected with 1.0 μg of Flag-L23 in the presence of p53 (0.5 μg) with (+) or without (−) MDM2 (1.0 μg) as indicated. Cell lysates (50 μg) were immunoblotted (IB) with anti-MDM2 (α-MDM2), anti-p53 (α-p53), anti-Flag (α-Flag), or antitubulin (α-tubulin) antibodies as indicated to the left. (B) L23 inhibits MDM2-mediated p53 ubiquitination in cells. H1299 cells were transfected with combinations of L23 (2 μg)-, p53 (1 μg)-, or MDM2 (1 μg)-encoding plasmids in the presence of the His-ubiquitin (His-Ub) (2 μg) plasmid as indicated at the top. The cells were treated with MG132 (20 μM) for 8 h before harvesting. The in vivo ubiquitination assay was performed as described in Materials and Methods. Ubiquitinated proteins were detected by immunoblotting with the anti-p53 (DO-1) and anti-HA (α-HA) antibodies. Ubiquitinated p53s [p53-(His-Ub)n] and ubiquitinated MDM2s [MDM2-(His-Ub)n] are indicated to the right of the upper and middle panels. The expression levels of MDM2, p53, and L23 are shown in the lower panels. (C) Ectopic expression of L23 induces endogenous p53. U2OS cells were transfected with 1.0 μg (lane 2) or 2.0 μg (lane 3) of Flag-L23 or 2.0 μg of Flag-L23ΔN (lane 4) plasmids. Cell lysates (50 μg) were used for immunoblot analysis with antibodies as indicated to the right. α-p21, anti-p21.
Glycerol gradient sedimentation centrifugation.
Whole-cell lysates were prepared from 293 cells as described previously (72). Two milligrams of lysates mixed with molecular weight markers was loaded onto the surface of a 12.5 to 25% glycerol gradient solution containing 150 mM NaCl and 10 mM Tris-HCl (pH 7.5) in a 12-ml centrifuge tube. The samples were subjected to centrifugation in a Beckman SW41 rotor at 32,000 rpm at 4°C for 20 h. Two hundred microliters per fraction was collected from each tube. Thirty microliters of each fraction was loaded onto an SDS gel for electrophoresis followed by immunoblot analysis.
Polysome and mRNP distribution analysis.
Postmitochondrial supernatant extractions, sucrose gradient sedimentation of polysomes, and analysis of the polysome and mRNP distribution of proteins and RNAs were carried out as previously described with minor modifications (17, 69). Briefly, cells were incubated with 100 μg of cycloheximide/ml for 15 min, which arrests polysome migration prior to the isolation of postmitochondrial supernatant. The cells were homogenized in polysome lysis buffer containing 30 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 100 mM KCl, 0.3% Nonidet P-40, 50 μg of cycloheximide/ml, 30 U of RNasin inhibitor/ml, 1 mM dithiothreitol, 1 mM PMSF, 1 mM pepstatin, and 1 mM leupeptin. After incubation on ice for 5 min, the lysates were centrifuged at 12,000 × g at 4°C for 8 min. Supernatants were subjected to sedimentation centrifugation in a 15 to 47% sucrose gradient solution containing 30 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 100 mM KCl in a Beckman SW41 rotor at 37,000 rpm for 2 h. Fourteen fractions were collected from each tube. RNAs were extracted from the fractions by phenol-chloroform extraction.
Cell cycle analysis.
U2OS or Saos-2 cells were transfected with plasmids encoding GFP, GFP-L23, or GFP-L23ΔC. At 32 h posttransfection, cells were treated with 200 μg of nocodazole/ml for an additional 16 h. Cells were harvested, suspended in 100 μl of PBS, and transferred to a polystyrene tube. Cells were stained in 500 μl of propidium iodide (PI; Sigma) stain buffer (50 μg of PI/ml, 30 μg of polyethylene glycol 8000/ml, 200 μg of RNase A/ml, 0.1% Triton X-100, and 0.38 M NaCl [pH 7.2]) at 37°C for 30 min and then analyzed for DNA content by using a Becton Dickinson FACScan flow cytometer. Data were collected by using the Modfit software program. GFP-positive cells were gated for cell cycle analysis.
GST fusion protein association assays.
His-tagged L23 and MDM2 proteins were expressed in E. coli, purified through an Ni-NTA (QIAGEN) column, and eluted with 0.5 M imidazole. Protein-protein interaction assays were conducted as described previously by using fusion protein-containing glutathione beads (26). Purified L23 proteins were incubated with the glutathione-Sepharose 4B beads (Sigma) containing 200 ng of GST-MDM2/1-491, GST-MDM2/1-301, GST-MDM2/1-150, GST-MDM2/151-301, GST-MDM2/294-491, GST-MDM2/384-491, GST-MDM2/425-491, or GST. Purified MDM2 proteins were incubated with GST-L23/1-140, GST-L23/1-105, GST-L23/1-65, GST-L23/66-140, GST-L23/36-105, or GST. Thirty minutes after incubation at room temperature, the mixtures were washed once in BC100 containing 0.1% Nonidet P-40, twice in SNNTE buffer, and once in RIPA buffer. Bound proteins were analyzed on a 10 or 15% SDS gel and detected by immunoblotting with anti-L23 and anti-MDM2 (2A10) monoclonal antibodies.
In vivo ubiquitination assay.
The in vivo ubiquitination assay was conducted as previously described with minor modifications (68). H1299 cells (60% confluence, 100-mm-diameter plate) were transfected with His6-ubiquitin (2 μg), p53 (2 μg), L23 (2 μg), or HA-MDM2 (2 μg) expression plasmids with Lipofectin. At 48 h after transfection, cells from each plate were harvested and split into two aliquots, one for Western blotting and the other for ubiquitination assays. Cell pellets were lysed in buffer I (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol) and incubated with Ni-NTA beads (QIAGEN) at room temperature for 4 h. Beads were washed once with buffer I, buffer II (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol), and buffer III (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 6.3], 10 mM β-mercaptoethanol). Proteins were eluted from beads in buffer IV (200 mM imidazole, 0.15 M Tris-HCl [pH 6.7], 30% glycerol, 0.72 M β-mercaptoethanol, and 5% SDS). Eluted proteins were analyzed by Western blotting with monoclonal anti-p53 (DO-1; Santa Cruz) or anti-HA antibodies.
Immunofluorescent staining.
293-HA-MDM2 cells were transfected with Flag-L23 expression plasmid. Forty-eight hours after transfection, cells were fixed for immunofluorescent staining with monoclonal anti-Flag antibodies and polyclonal anti-MDM2 antibodies and for DNA staining with 4′,6′diamidino-2-phenylindole (DAPI). The Alexa Fluor 488 (green) goat anti-mouse antibody and the Alexa Fluor 546 (red) goat anti-rabbit antibody (Molecular Probes, Eugene, Oreg.) were used for Flag-L23 and MDM2, respectively. Stained cells were analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Inhibition of L23 by siRNA and treatment of cells with actinomycin D.
U2OS cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. RNA interference-mediated removal of endogenous L23 was performed essentially as previously described (46). A 21-nucleotide siRNA duplex with a 3′ dTdT overhang, corresponding to L23 mRNA (AATTCCGGATTTCCTTGGGTC) or the scramble II RNA duplex (AAGCGCGCTTTGTAGGATTC) as a control, were synthesized (Dhamacon). These siRNA duplexes (0.2 μM) were introduced into cells by using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Cells were then treated with or without 5 nM actinomycin D for 8 h before harvesting. Cells were harvested 72 h after transfection for immunoblot, RT-PCR, cell cycle, and luciferase activity analyses. For cell cycle analysis, cells were treated with 200 μg of nocodazole/ml for an additional 16 h before harvesting. For luciferase assay, cells were transfected with the pCMV-β-galactoside reporter plasmid (0.2 μg) and a luciferase reporter plasmid (0.2 μg) driven by two copies of the p53RE motif derived from the MDM2 promoter as mentioned above before siRNA transfections.
RT-PCR analysis.
U2OS cells were transfected with or without L23 siRNA and treated with or without 5 nM actinomycin D as described above. RNA was isolated from cells by using RNeasy mini kits (QIAGEN). Reverse transcriptions were performed as described previously (70). PCRs were performed in a 20-μl mixture containing 1× PCR buffer, 60 μmol of deoxynucleoside triphosphates/liter, 1 U of Taq polymerase (Boehringer, Mannheim, Germany), 0.5 μmol of each primer/liter, and 0.2 μCi of [32P]dCTP for 18 to 20 cycles as described previously (70). PCR products were resolved onto a 6% polyacrylamide gel. The gel was dried, followed by autoradiography. The primers for amplifying p21waf1/cip1, MDM2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were described previously (70). The other primers were for p53 (5′-TACAGTCAGAGCCAACCTCAG-3′ and 5′-AGATGAAGCTCCCAGAATGCC-3′) and L23 (5′-ATGTCGAAGCGAGGACGTGGTG-3′ and 5′-TCATGCAATCCTGCCAGCATTG-3′).
RESULTS
Isolation of a human MDM2-associated protein complex.
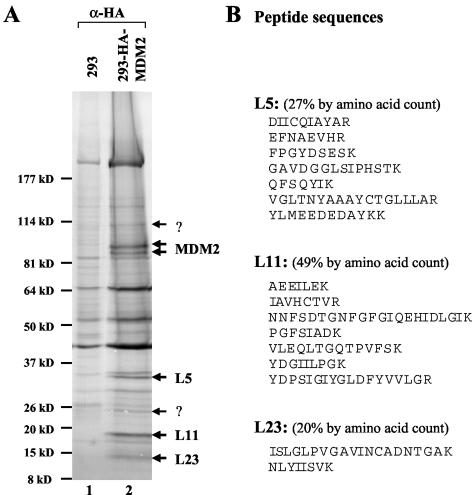
To understand how nuclear and cytoplasmic proteins regulate the MDM2-p53 pathway, our laboratory previously purified a nuclear complex containing MDM2, p300, and p53 from HeLa nuclear extracts and identified a cytoplasmic MDM2-associated protein complex free of p53 and p300 from HeLa cytoplasmic extracts via biochemical fractionation (29). To further illustrate the identity of the cytoplasmic MDM2-associated proteins, we have tried to purify the complex by using conventional chromatography. However, due to the unstable nature of the endogenous MDM2 (data not shown), this purification failed to yield sufficient amounts of protein for further analysis. Thus, to surmount this obstacle, we have generated a cell line stably overexpressing MDM2 (293-HA-MDM2) by using human 293 cells and neomycin selection. We used an affinity purification to isolate the MDM2-associated proteins in this cell line. Immunoprecipitation was performed with the cytoplasmic fractions (S100) from 293-HA-MDM2 cells as well as 293 cells expressing the empty vector pcDNA3. MDM2 and associated proteins were eluted with HA peptides and visualized on an SDS-polyacrylamide gel electrophoresis gel by colloidal blue staining (Fig. 1A). Several bands appeared specifically in the HA-MDM2-expressing sample but not in the control sample. Three proteins were revealed by mass spectrometric analysis to be ribosomal proteins L5, L11, and L23, respectively (Fig. 1). Two doublet bands were MDM2. This result not only identifies L23 as another potential regulator of MDM2 but also suggests a possible MDM2-ribosomal protein complex.
FIG. 1.
Isolation of a human MDM2-associated protein complex by immunoaffinity purification. (A) Colloidal blue staining analysis of proteins eluted from 12CA5 beads loaded with either empty vector expressing 293 cytoplasmic extracts (293) or the cytoplasmic extracts from the 293-HA-MDM2 cell line (lane 2). MDM2-associated polypeptides were digested and subjected to sequence analysis by mass spectrometry. The MDM2, L5, L11, and L23 bands are indicated. The question marks denote unidentified polypeptides. α-HA, anti-HA. (B) Peptide sequences for L5, L11, and L23 bands obtained from mass spectrometry analysis.
MDM2 binds to L23 in cells.
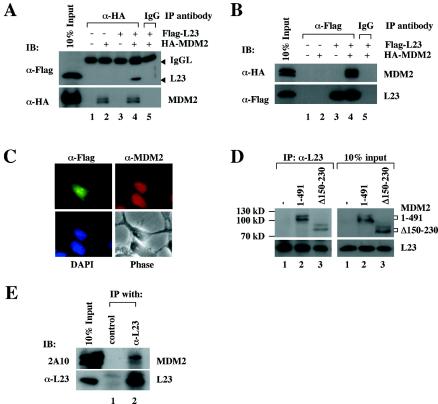
To verify the association of MDM2 with L23, Flag-L23 was expressed in 293-HA-MDM2 cells. Whole-cell lysates were prepared and subjected to coimmunoprecipitation with either anti-HA or anti-Flag antibodies, followed by immunoblot analysis. Immunoblotting results revealed that Flag-L23 and HA-MDM2 were specifically coimmunoprecipitated by either anti-HA (Fig. 2A) or anti-Flag antibodies (Fig. 2B) but not by control antibodies, indicating that MDM2 binds to L23 in cells. Further, to determine whether this interaction is also true with endogenous MDM2 and L23 proteins, we generated rabbit polyclonal antisera against the full-length L23 and employed them in coimmunoprecipitation assays with U2OS cells. Indeed, the endogenous MDM2 and L23 proteins were specifically coimmunoprecipitated by anti-L23 antisera but not by preimmune sera (Fig. 2E). Using the same approaches, we also verified the previously reported interaction between L5 and MDM2 (37) and the recently reported interaction between L11 and MDM2 (35, 74) (data not shown).
FIG. 2.
L23 interacts with MDM2 in cells. (A) Exogenous MDM2 and L23 interact with each other in 293 cells. HA-MDM2 (1.5 μg), Flag-L23 (1.5 μg), or both vectors (1.5 μg each) were used for transfection, as indicated at the top. Whole-cell lysates (500 μg) were subjected to immunoprecipitation (IP) with anti-HA (α-HA) or a control antibody followed by immunoblotting (IB) with anti-Flag (α-Flag) (upper) or anti-HA (lower) antibodies. IgG, immunoglobulin G. (B) The same transfections as shown in panel A were conducted except that anti-Flag antibodies were used for immunoprecipitation. (C) MDM2 colocalized with L23 in both the nucleus and the cytoplasm but not in the nucleolus. 293-HA-MDM2 cells were transfected with Flag-L23 and immunostained with both polyclonal anti-MDM2 (red) and monoclonal anti-Flag (green) antibodies. (D) L23 binds to MDM2 deletion mutants in cells. 293 cells were transfected with 6 μg of wild-type MDM2, MDM2Δ150-230, or empty (−) plasmids, as indicated at the top. Whole-cell lysates (500 μg) were immunoprecipitated with anti-L23 (α-L23) antibodies followed by immunoblotting with anti-MDM2 (2A10) and anti-L23 antibodies. Ten percent of the lysates loaded as input are shown in the panels on the right side. (E) Endogenous L23 interacts with endogenous MDM2 in U2OS cells. Whole-cell lysate (500 μg) was used for immunoprecipitation with either rabbit polyclonal anti-L23 antibody or preimmune serum (control), followed by immunoblotting with anti-MDM2 (2A10) (top panel) or anti-L23 (bottom panel) antibody.
To determine where the L23-MDM2 binding may occur in cells, several experiments were conducted. First, 293-HA-MDM2 cells were transfected with Flag-L23 and immunostained with anti-Flag (green) and anti-MDM2 (red) antibodies. As shown in Fig. 2C, stably expressed MDM2 in 293 cells was localized in both the cytoplasm and the nucleus but not in the nucleolus, whereas Flag-L23 was detected in the cytoplasm, the nucleoplasma, and the nucleolus. Second, we introduced wild-type MDM2 and its deletion mutant that lacks the nuclear localization sequence (Δ150-230) into 293 cells and carried out a coimmunoprecipitation assay with an anti-L23 antibody followed by an immunoblot assay. As shown in Fig. 2D, either wild-type or mutant MDM2 was coimmunoprecipitated by the anti-L23 antibody. This result indicates that L23 can bind to MDM2 in the cytoplasm because L23 bound to the Δ150-320 mutant of MDM2 that has been shown to locate only in the cytoplasm (25). Also, by using both cytoplasmic and nuclear fractions from 293-HA-MDM2 cells transfected with Flag-L23, we were able to immunoprecipitate MDM2 from both fractions with anti-Flag antibody (data not shown). Taken together, L23 may bind to MDM2 in both the nucleoplasm and the cytoplasm.
MDM2 binds to L23 in vitro.
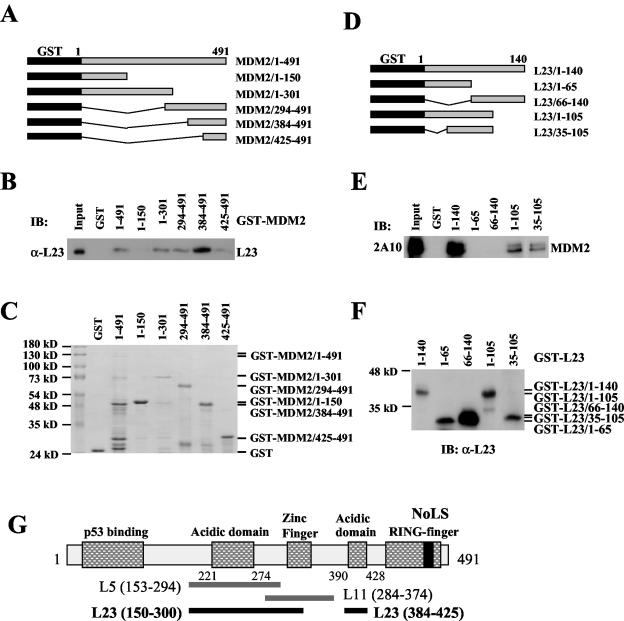
Next, we wanted to determine whether the interaction between MDM2 and L23 is direct or indirect through other ribosome proteins, L11 (35, 74) or L5 (37). To do so, GST fusion protein-protein association assays were conducted by using His-L23 and GST-MDM2 deletion fusion proteins purified from bacteria (Fig. 3A to C). As shown in Fig. 3B, GST-MDM2, but not GST alone, interacted directly with His-L23. It was shown previously that L5 binds to a central acidic domain of MDM2 (residues 153 to 294) (15), and it was recently reported that L11 binds to the residues 212 to 296 of MDM2 (35). Different from L5 and L11, L23 appeared to bind to the aa 150 to 301 and aa 384 to 425 regions of MDM2 with a strong preference for the latter in vitro (Fig. 3C). A close examination of the MDM2 sequence revealed a second acidic domain in this region, which contains 35% acidic amino acids. These residues may be important for L23 binding. Thus, the central region of MDM2 may possess two L23-binding sites.
FIG. 3.
Ribosomal protein L23 interacts with MDM2 in vitro. (A) Schematic presentation of recombinant full-length MDM2 and its fragments fused to GST. Black rectangles indicate GST. Gray rectangles indicate MDM2 or its fragments. (B) L23 preferentially binds to the acidic domains of MDM2. About 200 ng of purified GST alone, full-length GST-MDM2, or GST-MDM2 deletion mutants including MDM2/1-150, MDM2/1-301, MDM2/294-491, MDM2/384-491, and MDM2/425-491 immobilized on glutathione beads were used in GST pull-down assays with 200 ng of His-L23 purified from bacteria. Bound L23 was detected by immunoblotting (IB) with anti-L23 (α-L23) antibodies. (C) Coomassie blue staining of GST-MDM2 fusion proteins used in panel B. (D) Schematic presentation of recombinant full-length L23 and its fragments fused to GST. (E) MDM2 binds to the middle domain of L23 in vitro. The same GST fusion protein pull-down assay as that described for panel B was conducted except that purified 200 ng of GST-L23 and 200 ng of GST-L23 deletion mutants were incubated with 200 ng of His-MDM2 purified from bacteria, as indicated. Bound MDM2 was detected by immunoblotting with anti-MDM2 antibodies (2A10). (F) Immunoblot of GST-L23 fusion proteins used in panel E with anti-L23 antibodies. (G) Schematic presentation of MDM2 domains that bind to L23, L5, and L11.
To map the MDM2 binding site in L23, we performed a similar GST pull-down assay with His-MDM2 and GST-L23 fusion proteins purified from bacteria (Fig. 3D and F). As shown in Fig. 3E, His-MDM2 bound to the GST full-length L23 protein but not to its N-terminal or C-terminal half fused with GST nor GST alone, indicating that the central portion of L23 may be essential for the binding. Indeed, His-MDM2 bound to the GST-L23/1-105 or GST-L23/35-105 fragment, although less efficiently. These results demonstrate that MDM2 can physically bind to L23 in vitro. The observation that L23, L11, and L5 bind to different regions of MDM2 (Fig. 3G) suggests that MDM2 may form a complex with these ribosome proteins.
MDM2 forms a complex with L5, L11, and L23 in cells.
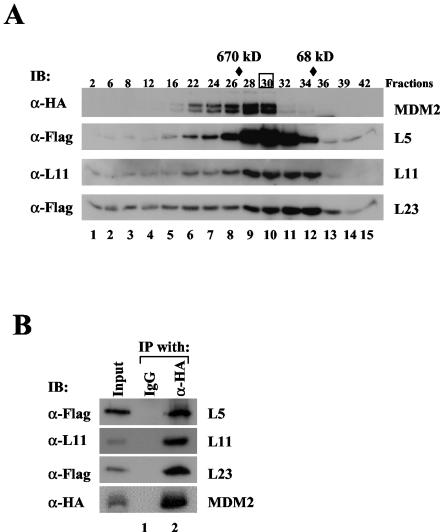
To determine whether the three ribosomal proteins identified from our immunoprecipitation purification interact with MDM2 in the same complex in cells, we fractionated 293-HA-MDM2 cell lysates by using glycerol gradient sedimentation centrifugation and analyzed the fractions with an immunoblot assay. As shown in Fig. 4A, some L5, L11, and L23 proteins were coeluted with HA-MDM2 (lanes 6 to 10). MDM2 may form a complex with the ribosomal proteins in fractions 28 to 30 (lanes 9 to 10) because these fractions eluted around where a 670-kDa molecular mass marker eluted. To test this idea, we performed a coimmunoprecipitation assay with fraction 30. As shown in Fig. 4B, indeed, L5, L11, and L23 were all specifically coimmunoprecipitated with HA-MDM2 by the anti-HA antibody but not by a control antibody. Noticeably, there were some fractions, such as fractions 22 to 26, which also possessed all the tested proteins and were eluted before the 670-kDa marker. Two possibilities may account for this observation. First, the protein samples may be overloaded. Alternatively, MDM2 may form a larger complex that contains not only the identified ribosomal proteins but also other yet unidentified proteins (Fig. 1). In summary, these results indicate that MDM2 may form a complex at least with L5, L11, and L23 in cells.
FIG. 4.
MDM2 forms a complex with L23, L5, and L11 in cells. (A) MDM2 cosedimented with L23, L5, and L11. Whole-cell lysates (2 mg) prepared from HA-MDM2-expressing 293 cells were subjected to a 12.5 to 25% glycerol gradient sedimentation centrifugation. Fractions were collected for immunoblot (IB) analysis with antibodies indicated to the left of each panel. Molecular markers coeluted with fractions are indicated at the top. Fraction 30, boxed, was used for immunoprecipitation and immunoblot analysis shown in panel B. (B) MDM2 is coimmunoprecipitated with L23, L5, and L11 in fraction 30. Fraction 30 (200 μl) was used for immunoprecipitation (IP) with the anti-HA (α-HA) or control antibody as indicated at the top, followed by immunoblotting with the antibodies indicated to the left. IgG, immunoglobulin G; α-Flag, anti-Flag; α-L11, anti-L11.
MDM2 is not associated with 80S ribosomes or polysomes.
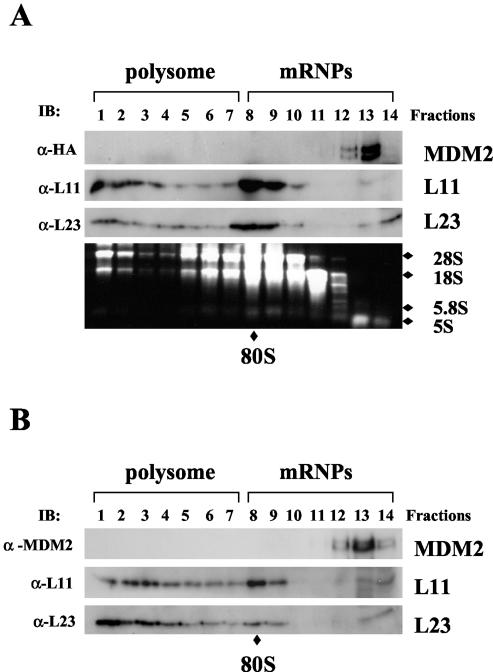
The observation that MDM2 associated with three ribosomal proteins, L5, L11, and L23, in one complex raises the question of whether MDM2 may associate with 80S ribosomes or polysomes through these proteins. To address this issue, we performed a polysome profile analysis. Cytoplasmic extracts prepared from 293-HA-MDM2 cells were subjected to a linear sucrose gradient sedimentation centrifugation. Fourteen fractions were collected and subjected to immunoblot assays for detection of MDM2, L11, and L23 as well as rRNA analysis with ethidium bromide (Fig. 5A). The result showed that MDM2 was not coeluted with either polysomes or 80S ribosomes, both of which contain L11 and L23 (fractions 1 to 7 and 8 to 10), and instead stayed near the top of the gradient where the ribosome-free ribosomal proteins L11 and L23 were also detected by blotting with anti-L11 and anti-L23 antibodies. The polysome and 80S ribosome profile was verified by determining the distribution of rRNAs (Fig. 5A, lower panel). The distribution of rRNAs together with L11 and L23 coordinated well with polysome, ribosome, and mRNP (small ribonuclear protein) profiles, as expected (17, 69). Consistent with the result shown in Fig. 5A, endogenous MDM2 proteins associated with free L11 and L23 but not with intact 80S ribosomes and polysomes (Fig. 5B). Thus, we conclude that MDM2 does not associate with the 80S ribosomes or polyribosomes.
FIG. 5.
MDM2 does not associate with the 80S ribosome and polysomes. (A) Ectopically expressed MDM2 does not associate with polysomes. Cytoplasmic extracts (5 mg) containing polysomes from HA-MDM2-expressing 293 cells were subjected to a 15 to 47% linear sucrose gradient sedimentation centrifugation. Fourteen fractions were collected, and 30 μl of each fraction was used for immunoblotting (IB) with anti-HA (α-HA), anti-L11 (α-L11), or anti-L23 (α-L23) antibodies as indicated to the left. Total RNAs were isolated from each fraction and subjected to electrophoresis on a 1% agarose gel and stained with ethidium bromide as shown in the bottom panel. 28S, 18S, 5.8S, and 5S rRNAs are indicated to the right. The fractions containing polysomes and mRNPs are indicated on the top. The 80S ribosome is indicated at the bottom. (B) Endogenous MDM2 does not associate with polysomes. The same fractionation as that shown in panel A was performed with U2OS cell extracts. The distributions of polysomes and mRNPs are indicated. Thirty microliters of each fraction was subjected to immunoblot analysis with anti-MDM2 (2A10, α-MDM2), anti-L11, or anti-L23 antibodies as indicated to the left.
L23 activates p53 by overcoming MDM2-mediated suppression.
The finding that L23 associates with MDM2 in cells suggests that L23 may regulate the MDM2-p53 feedback loop. We first examined whether overexpression of L23 affects MDM2-mediated p53 degradation by introducing exogenous proteins into p53-deficient human non-small-cell carcinoma H1299 cells because MDM2 mediates ubiquitination and proteasome-mediated degradation of p53 (16, 20, 21, 30). As expected, overexpression of MDM2 remarkably reduced p53 levels (Fig. 6A, lane 3). By contrast, further overexpression of L23 partially rescued MDM2-mediated p53 degradation (Fig. 6A, lane 4). This rescue appeared to be dependent on the interaction of L23 with MDM2, as the C-terminal domain of L23, which did not bind to MDM2 (Fig. 3), was unable to stabilize p53 (data not shown). Overexpression of L23 also slightly stabilized HA-MDM2 (Fig. 6A, top panel, lane 4). Consistent with these results, overexpression of L23 led to marked inhibition of MDM2-mediated p53 ubiquitination and MDM2 ubiquitination (Fig. 6B). Thus, L23 can stabilize p53 by alleviating MDM2-mediated p53 ubiquitination and degradation.
Next, we examined the effect of L23 on endogenous p53 by introducing Flag-L23 into human osteosarcoma U2OS cells that contain wild-type p53. Interestingly, overexpression of Flag-L23, but not a Flag-L23 deletion mutant that does not bind to MDM2 (Flag-L23ΔN), markedly induced p53 in a dose-dependent fashion (Fig. 6C, second panel from top). Correspondingly, the levels of the p53 targets p21cip1 and MDM2 were also induced (Fig. 6C, middle and top panels). This result together with the results above suggests that ectopic expression of L23 induces the level of the endogenous p53 as well as its targets p21cip1 and MDM2 by blocking MDM2-mediated p53 degradation.
L23 stimulates p53-dependent transcription and G1 arrest.
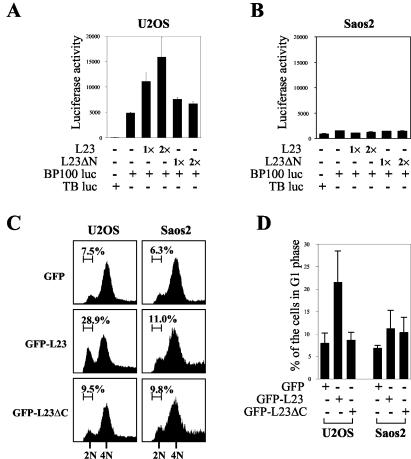
The finding that overexpression of L23 led to the induction of p53 and p21cip1 levels suggests that the high level of cellular L23 may enhance p53-dependent transcription and cell growth arrest. To test this concept, we transfected U2OS cells with Flag-L23 or Flag-L23ΔN together with a luciferase reporter plasmid driven by the p53RE derived from the MDM2 promoter (67) and then carried out luciferase assays. Consistent with the result shown in Fig. 6C, ectopic expression of full-length L23 markedly stimulated p53RE-driven transcription, as presented as luciferase activity, in a dose-dependent manner (Fig. 7A). This stimulation was dependent on p53, as no significant change in luciferase activity was detected in human osteosarcoma Saos-2 cells that are deficient in p53 (Fig. 7B). This stimulation may also require the interaction of L23 with MDM2, as the L23 mutant Flag-L23ΔN (aa 66 to 140), which was unable to bind to MDM2 (Fig. 3E; data not shown), failed to enhance p53-dependent transcription activity (Fig. 7A) and induction of p21cip1 (Fig. 6C). Hence, these results suggest that L23 may stimulate p53 activity and that this stimulation requires the MDM2-L23 interaction.
FIG. 7.
Ectopic expression of L23 stimulates p53-dependent transcription and G1 arrest. (A) Ectopic expression of L23 increases p53RE-dependent luciferase activity in p53-proficient U2OS cells. U2OS cells were transfected with increasing amounts of Flag-L23 (0.4 μg [1×] and 0.8 μg [2×]) or Flag-L23ΔN (0.4 μg [1×] and 0.8 μg [2×]) in the presence of a luciferase reporter plasmid driven by p53RE (BP100 luc, 0.1 μg) or a control luciferase reporter plasmid (TB luc, 0.1 μg). Luciferase activity is presented in arbitrary units. (B) L23 does not affect p53RE-dependent luciferase activity in p53-deficient Saos-2 cells. The same transfection followed by a luciferase assay as that described for panel A was conducted except Saos-2 cells were used here. (C) Ectopic expression of L23 leads to p53-dependent G1 cell cycle arrest. U2OS or Saos-2 cells were transfected with GFP (2 μg), GFP-L23 (2 μg), or GFP-L23ΔC (2 μg) plasmids and treated with nocodazole as described in Materials and Methods. GFP-expressing cells were then gated for cell cycle analysis. The histograms of PI staining from one representative experiment are shown. Percentages indicate the cells that were arrested in G1 phase. (D) The mean percentage of cells arrested in the G1 phase obtained from four separate experiments is presented. Bars indicate standard deviations. +, present; −, absent.
Next, we determined whether induction of p21cip1 by L23 through p53 activation could result in G1 arrest because activated p53 triggers p21cip1-dependent cell cycle arrest (63). To do so, p53-proficient U2OS or p53-deficient Saos-2 cells were transiently transfected with either the GFP-fused L23 (GFP-L23) or the GFP-L23 C-terminal deletion (GFP-L23ΔC) (retaining only aa 1 to 65). Cells were then treated with the mitotic inhibitor nocodazole before fluorescence-activated cell sorter analysis, thus leading to G2/M arrest (5). Therefore, cells found in G1 phase are previously arrested and do not reach G2/M phase. GFP-positive cells were then gated for cell cycle analysis. As shown in a representative result in Fig. 7C, 28.9% of GFP-L23 expressing U2OS cells were arrested in the G1 phase while only 7.5 and 9.5% of U2OS cells expressing GFP and GFP-L23ΔC, respectively, were detected in the G1 phase. The G1 arrest induced by L23 was dependent on p53 because only a marginal change was observed in p53-null Saos-2 cells (Fig. 7C and D). These results were reproducible, as summarized in Fig. 7D. Therefore, ectopic expression of L23 induced p53-dependent G1 arrest, and this induction is also dependent on the MDM2-L23 interaction because the L23 N-terminal domain (aa 1 to 65) was unable to exert such an effect (Fig. 7C and D).
Actinomycin D, but not pactamycin or gamma irradiation, enhances L23-MDM2 interaction and activates p53.
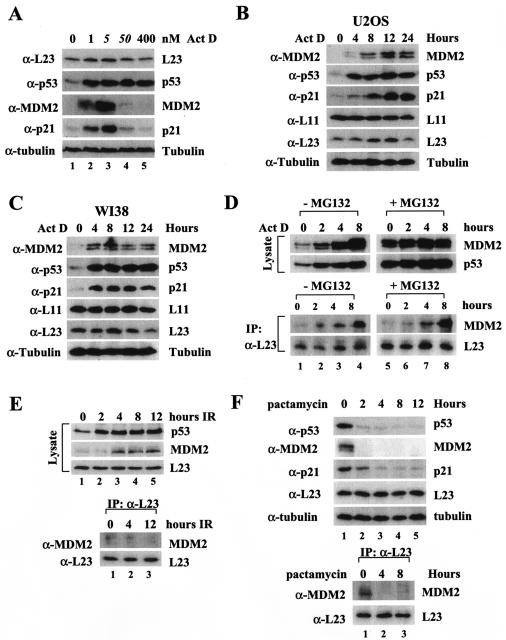
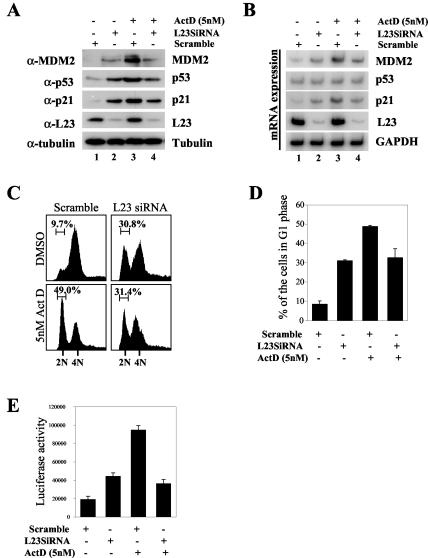
It has been shown that a low dose of actinomycin D (5 nM) specifically inhibits RNA polymerase I and consequently reduces rRNA synthesis, leading to perturbation of ribosomal biogenesis (2). Intriguingly, this biogenesis perturbation activates p53 without inducing phosphorylation at its N-terminal domain in cells, suggesting a previously uncharacterized p53 signaling pathway (2). The identification of the MDM2-L23 interaction led us to determine whether L23 may play a role in this pathway. Consistent with previous studies (2), our result from the experiment with U2OS cells treated with different doses of actinomycin D showed that low doses of actinomycin D (1 and 5 nM) markedly induced p53 as well as MDM2 and p21cip1 (Fig. 8A). However, higher doses (50 and 400 nM) of the drug, which inhibit all RNA polymerases including RNA polymerase II (33), only induced p53 but not MDM2 or p21 levels (Fig. 8A). We wanted to test whether actinomycin D inhibition of ribosomal biogenesis may affect the interaction between L23 and MDM2. The activation of p53 by a low dose (5 nM) of actinomycin D is time dependent. It was induced as early as 2 h after treatment followed by MDM2 and p21 induction in U2OS cells (Fig. 8B). Of note, the L23 level slightly increased 8 h after treatment of U2OS or WI38 cells with 5 nM actinomycin D and then decreased 24 h after the treatment. By contrast, the L11 level was not affected by actinomycin D treatment of both cell lines (Fig. 8B and C). Next, we wanted to determine whether actinomycin D could affect the interaction between MDM2 and L23. U2OS cells were treated with 5 nM actinomycin D and harvested at different time points posttreatment for immunoprecipitation and immunoblot assays. As shown in Fig. 8D, the MDM2-L23 interaction was drastically increased after actinomycin D treatment in a time-dependent manner; this increase began from 2 h and peaked at 8 h after this treatment (Fig. 8D, lower panels), consistent with p53 induction in response to this stress (Fig. 8D, upper left panels). The enhancement of the MDM2-L23 interaction was not merely due to the increase of MDM2 levels, as this interaction was not increased when MDM2 was stabilized by the proteasome inhibitor MG132 (Fig. 8D, compare lane 1 with lane 5). The decrease of L23 24 h after the drug treatment does not underscore the importance of L23 in regulating MDM2 function in response to this stress, as the induction of the L23-MDM2 interaction and p53 level was detected much earlier (approximately 2 h after actinomycin D treatment) (Fig. 8D). In contrast to actinomycin D, the enhancement of the L23-MDM2 binding was not observed when U2OS cells were treated with gamma irradiation, regardless of high or low levels of MDM2 (Fig. 8E). These results suggest that the L23-MDM2 binding is highly related to p53 induction by 5 nM actinomycin D but not by gamma irradiation.
FIG. 8.
A low dose of actinomycin D enhances MDM2-L23 interaction and p53 activation. (A) Low doses of actinomycin D induce p53 and its function, whereas high doses of actinomycin D induce p53 but not its function. U2OS cells were treated with increasing amounts of actinomycin D (Act D) as indicated at the top. Cell lysates (50 μg) were used for an immunoblot analysis with antibodies indicated to the right. (B) Time-dependent effect of actinomycin D on p53 and L23 levels in U2OS cells. U2OS cells were treated with 5 nM actinomycin D and harvested at different time points as indicated at the top. Cell lysates (50 μg) were used for an immunoblot analysis with antibodies indicated to the left of each panel. (C) Time-dependent effect of actinomycin D on p53 and L23 levels in WI38 cells. WI38 cells were treated with 5 nM actinomycin D, harvested, and blotted with antibodies as described for panel B. (D) Five nanomolar actinomycin D enhances MDM2-L23 interaction. U2OS cells were treated with 5 nM actinomycin D and harvested at different time points as indicated at the top. Cells were incubated with (+) or without (−) MG132 (20 μM) for 6 h before harvesting. Cell lysates (500 μg) were subjected to an immunoprecipitation (IP) with anti-L23 antibodies and immunoblotting with anti-MDM2 or anti-L23 antibodies (lower panels). The lysates were also directly loaded onto an SDS gel for an immunoblot analysis with anti-MDM2, anti-p53, or anti-L23 antibodies (upper panels). (E) Ionizing irradiation does not affect L23-MDM2 interaction. U2OS cells were treated with gamma irradiation (10 Gy) and harvested at different time points as indicated. Cell lysates (50 μg) were subjected to an immunoblot analysis with anti-p53, anti-MDM2, or anti-L23 antibodies (upper panels). The cell lysates (500 μg) were subjected to an immunoprecipitation analysis with anti-L23 antibodies, followed by an immunoblot analysis with anti-MDM2 or anti-L23 antibodies (bottom panels). (F) Pactamycin treatment does not induce p53 and the MDM2-L23 interaction. U2OS cells were treated with 0.2 μg of pactamycin/ml for different numbers of hours as indicated at the top. The cells were harvested for immunoblotting with the indicated antibodies (top panels). The cell lysates were also subjected to immunoprecipitation with anti-L23 followed by immunoblotting with anti-MDM2 and anti-L23 antibodies (bottom panels). α-L23, anti-L23; α-p53, anti-p53; α-MDM2, anti-MDM2; α-p21, anti-p21; α-tubulin, antitubulin; α-L11, anti-L11.
To test whether the induction of p53 and MDM2-L23 interaction by actinomycin D is due to general inhibition of translation machinery, we also assayed the consequences of translation inhibition by protein synthesis inhibitors. Pactamycin, a translation initiation inhibitor acting on inhibition of Met-tRNA binding (23), did not activate p53 and induce p21 and MDM2. Instead, these proteins decreased after pactamycin treatment as shown in Fig. 8F. Consequently, MDM2-L23 interaction was reduced after pactamycin treatment (Fig. 8F). The same result was also obtained from cells treated with cycloheximide, which inhibits ribosomal translocation and thus translation elongation (data not shown). These data demonstrated that the L23-MDM-p53 pathway is specifically activated in response to the ribosomal stress only but not to translation inhibition.
Removal of L23 by siRNA activates p53 but inhibits actinomycin D-induced p53 activation.
To demonstrate the physiological relevance of L23 in this signaling pathway, we employed siRNA against L23 and determined whether removal of L23 by its siRNA could affect p53 induction by actinomycin D. Indeed, as shown in Fig. 9, this was the case, as reduction of L23 levels by its siRNA, but not by the scrambled siRNA duplex, correlated well with a decrease of actinomycin-induced p53 levels (Fig. 9A, compare lane 3 with lane 4). Consistently, actinomycin D-induced p53-dependent transcription, as represented by induction of its targets, p21, MDM2, and luciferase activity, driven by the p53RE-containing MDM2 promoter (Fig. 9A, B, and E), and G1 arrest (Fig. 9C and D) were also reduced by L23 siRNA. These results were reproducible and suggest that L23 may mediate p53 activation in response to this drug. Noticeably, in the absence of actinomycin D, siRNA against L23 (Fig. 9A, lane 2), but not the scrambled siRNA (lane 1), also induced p53 as well as the protein and mRNA levels of MDM2 and p21cip1 (Fig. 9A and B). Consistently, p53 transcriptional activity and p53-dependent G1 arrest were also induced by L23 siRNA (Fig. 9C, D, and E). The reason for p53 activation by reduction of endogenous L23 is presently unknown. One possibility would be that lowering the L23 level may cause another ribosomal biogenesis stress that in turn induces p53, probably through a yet unidentified pathway. In summary, the results, as described above, demonstrate that ribosomal biogenesis stress caused by the low dose of actinomycin D, but not by gamma-irradiation or direct inhibition of translation, can activate p53, possibly by inducing the association of L23 with MDM2 and inhibiting MDM2-mediated p53 degradation.
FIG. 9.
Inhibition of endogenous L23 by siRNA induces p53 but inhibits actinomycin D (Act D)-induced p53 activation and G1 arrest. (A) Inhibition of endogenous L23 by siRNA induces the p53 level but inhibits actinomycin D-induced p53 induction. U2OS cells were transfected with L23 siRNA oligonucleotides (0.2 μM, lanes 2 and 4) or a scrambled RNA duplex (0.2 μM, lanes 1 and 3). Cells were then incubated with (+) (lanes 3 and 4) or without (−) (lanes 1 and 2) 5 nM actinomycin D for 8 h before harvesting. Cell lysates (50 μg) were then immunoblotted with anti-MDM2, anti-p53, anti-p21, anti-L23, or antitubulin antibodies. (B) Inhibition of endogenous L23 by siRNA inhibits actinomycin D-induced p53 transcriptional activity. Cells were prepared as described for panel A, and total RNA was extracted. RT-PCR analysis was performed to detect MDM2, p53, p21, L23, and GAPDH mRNA levels as indicated. (C) Actinomycin-induced G1 arrest is inhibited by elimination of endogenous L23 by siRNA. U2OS cells were treated as described for panel A except that cells were treated with nocodazole for 16 h before harvesting. Cells were then stained with PI for fluorescence-activated cell sorter analysis. The histograms of PI staining from the results of one representative experiment are shown. (D) Summary of the results from three independent experiments as described for panel C. The mean percentages of cells arrested in G1 phase are presented. Bars indicate standard deviations. (E) Elimination of endogenous L23 by siRNA inhibits actinomycin D-induced p53RE-dependent luciferase activity. U2OS cells were transfected with a luciferase reporter plasmid driven by the p53RE (BP100 luc, 0.2 μg) and β-galactosidase (0.2 μg) plasmids followed by treatment with L23 siRNA and actinomycin D as described for panel A. Luciferase activity is presented in arbitrary units. α-L23, anti-L23; α-p53, anti-p53; α-MDM2, anti-MDM2; α-p21, anti-p21; α-tubulin, antitubulin; DMSO, dimethyl sulfoxide.
DISCUSSION
The p53-MDM2 feedback loop is regulated by distinct pathways in response to different stress signals (48). Here we have described a ribosomal protein that regulates this loop in response to ribosomal biogenesis stress. First, we have purified an MDM2-associated cytoplasmic complex by immunoaffinity purification from human 293 cells. This complex contains multiple ribosomal proteins, including the previously reported L5 (37), the recently reported L11 (35, 74), and a new component—the ribosomal protein L23 (Fig. 1). Our further characterization of this complex indicates that L23, unlike L5, which binds to the first acidic region (aa 221 to 274) of MDM2 (15), and L11, which binds to aa 284 to 374 of MDM2 (35, 74), preferentially binds to the second acidic domain (aa 384 to 425) of MDM2 with a minor binding site at the first acidic domain (Fig. 3). Interestingly, L23 appears to interact with MDM2 in both the nucleus and the cytoplasm when they are overexpressed (Fig. 2). In response to actinomycin D, this interaction may mostly occur in the nuclei of U2OS cells (data not shown). Functionally, ectopic expression of L23 inhibits MDM2-mediated p53 degradation and thus induces p53 levels as well as its activity (Fig. 6). L23 also leads to p53-dependent G1 arrest (Fig. 7) by inducing p53-dependent p21cip1 production (Fig. 6). Finally, the interaction between L23 and MDM2 is dramatically enhanced by a low dose of actinomycin D that only inhibits rRNA synthesis. Further, removal of endogenous L23 molecules by siRNA alleviates p53 induction by this drug (Fig. 9). Hence, our study documents L23 as another possible regulator of the p53-MDM2 feedback pathway in response to ribosomal biogenesis perturbation.
Association of MDM2 with multiple ribosomal proteins.
Another finding from our study is that the three ribosomal proteins associate with MDM2 in one complex, although MDM2-L5 and MDM2-L11 interactions were individually reported (35, 37). Several lines of evidence support this notion. First, an MDM2-associated protein complex has been purified and has been found to contain all three ribosomal proteins from 293 cells (Fig. 1), whose native mass is similar to our previously identified MDM2-associated complex free of p53 and p300 from HeLa cytoplasmic extracts (29). Second, MDM2 cosediments with L5, L11, and L23 by glycerol gradient sedimentation centrifugation with a molecular mass ranging from ∼200 to ∼800 kDa (Fig. 4A). The simultaneous association of MDM2 with L23, L5, and L11 in one of the peak fractions (fraction 30) is confirmed by a coimmunoprecipitation analysis with anti-HA antibodies (Fig. 4B). Consistent with this result is that MDM2 directly interacts with L23 (Fig. 3), L11, and L5 in vitro through its different domains (15) (data not shown). Although whether these ribosomal proteins could work in concert to inhibit the function of MDM2 remains to be studied, overexpression of L11 (35, 74), L23 (this study), and L5 (unpublished data) alone is able to activate p53 by preventing MDM2-mediated p53 ubiquitination and degradation. The MDM2-associated complex may contain other yet unidentified proteins in addition to these ribosomal proteins because of its large native molecular mass (Fig. 4), and some MDM2-associated polypeptides remain to be identified (Fig. 1). Despite its interaction with three ribosomal proteins, MDM2 does not appear to bind to the 80S ribosome and polysomes, as our sucrose gradient sedimentation centrifugation separates MDM2 from the 80S ribosome and polysome (Fig. 5). Hence, MDM2 forms a complex with ribosomal proteins L5, L11, and L23 independently of the 80S ribosome and polysome.
L23 activates p53 by blocking MDM2 feedback regulation.
Our study suggests that L23 induces p53 through direct binding to MDM2 (Fig. 1 to 4) and inhibition of MDM2-mediated p53 ubiquitination and MDM2 ubiquitination (Fig. 6). This mode of action seems similar to that mediated by the ARF protein (22, 34, 40, 61, 65, 66, 75) or by L11 (35, 74). Both ARF and L11 could relocalize MDM2 into the nucleolus. However, our data show that the interaction of L23 with MDM2 appears to occur in the nucleus as well as the cytoplasm but not in the nucleolus. Four lines of evidence support this assumption. First, the ribosomal proteins containing the MDM2 complex were isolated from the cytoplasmic fraction of the cells. Second, the MDM2 deletion mutation which lacks the nuclear localization signal sequence (Δ150-230) and stays in the cytoplasm (25) can interact with L23 as well as wild-type MDM2 (Fig. 2E). Third, MDM2 was coimmunoprecipitated by anti-L23 in both the cytoplasm and the nucleus (data not shown). Finally, in our 293-HA-MDM2 cells transfected with Flag-L23, MDM2 was expressed in both the cytoplasm and the nucleus, but not in the nucleolus, while L23 was expressed in all three compartments (Fig. 2D). Therefore, MDM2 is probably retained in the cytoplasm and the nucleus as a complex, and this complex formation may represent one mechanism by which L23 suppresses the MDM2 feedback regulation. Alternatively, L23 inhibits MDM2-mediated p53 ubiquitination and thus stabilizes p53 most likely by simply binding to MDM2, while the L23 deletion, which is defective in MDM2 binding (Fig. 3), is unable to induce p53 (Fig. 6) and its activity (Fig. 7). Although these possibilities need to be investigated further, it seems that L23 does not directly bind to p53 (data not shown) and the cytoplasmic MDM2-associated complex is free of p53 (29), suggesting that it is less likely that L23 directly targets p53. Therefore, L23 may activate p53 by preventing the MDM2 feedback suppression.
Role of L23 in the ribosome biogenesis-p53 pathway.
In mammals, ribosomal biogenesis is well coordinated by cell growth signals (14, 53). Although the mechanism underlying this precise coordination remains obscure, several studies suggest that some cell cycle regulators and tumor suppressors may be involved in this regulation. For example, p53 and Rb have been shown to inhibit rRNA synthesis, thus lessening ribosome assembly (7, 9, 11, 64, 73). Also, the other regulator of the MDM2-p53 loop, p19arf, has recently been reported to inhibit rRNA processing (59). These studies suggest that overproduction of these tumor suppressors would inhibit protein synthesis while stopping cell growth in response to stress signals. Remarkably, p53 can also sense the perturbation of ribosomal biogenesis, such as inhibition of rRNA synthesis and processing or disruption of ribosome assembly (1, 2, 50, 58). For instance, overexpression of dominant-negative mutants of a nucleolar protein Bop1, an inhibitor of rRNA processing, induced p53-dependent G1 arrest (50, 58). Another newly identified nucleolar protein called nucleostemin prevented cells from entering mitosis and caused p53-dependent apoptosis (62), though its exact function on ribosomal biogenesis is unknown. Also, as aforementioned, a low dose (5 nM) of actinomycin D rapidly induced p53 activity independently of the p53 N-terminal phosphorylation (1, 2), which is critical for p53 activation by gamma irradiation (3, 8, 28, 56). These studies suggest an elaborate coordination between p53-mediated cell growth regulation and cellular responses to malfunction of ribosomal biogenesis through a novel signaling pathway.
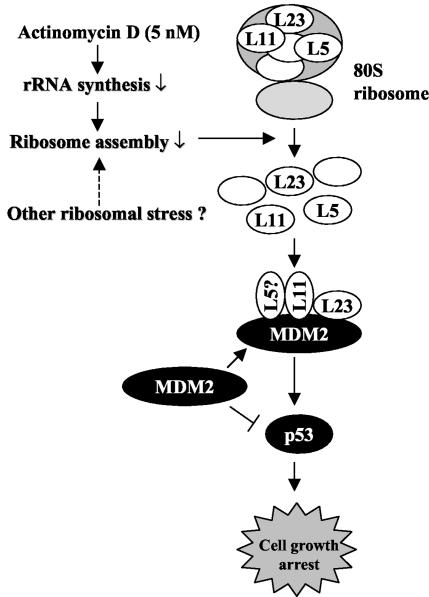
The findings that L23 and L11 activate p53 by binding to MDM2 and inhibiting the MDM2-mediated p53 degradation process, as described here and by others (35), suggest a plausible model for the role of these ribosomal proteins in the ribosomal biogenesis-p53 pathway (Fig. 10). Inhibition of ribosomal biogenesis, such as that caused by actinomycin D, but not direct inhibition of translation by protein synthesis inhibitors such as pactamycin or cycloheximide, would interfere with the assembly of the 80S ribosome complex and hence release free ribosomal proteins such as L23, L11, or L5. These free ribosomal proteins could then bind to MDM2, probably forming a multiple-subunit complex in the cytoplasm or the nucleus. In doing so, they could either prevent MDM2 from being transported into the nucleus to target p53 or inhibit MDM2-mediated p53 ubiquitination. Consequently, p53 would become stabilized and activated to induce p21cip1-dependent G1 arrest and to inhibit protein synthesis. Strongly supporting this model are two lines of evidence. For instance, a low dose of actinomycin D induces the interaction of L23 with MDM2, leading to p53 activation (Fig. 8B). Also, removal of L23 by siRNA markedly reduced actinomycin D-caused p53 induction (Fig. 9). Hence, L23 may play a role in the ribosomal biogenesis-p53 pathway, although other pathological signals that turn on this pathway remain to be uncovered.
FIG. 10.
A model for p53 activation by interfering with the MDM2-p53 feedback loop in response to perturbation of ribosomal biogenesis. Bars indicate inhibition, and arrows indicate activation.
Emerging evidence implies that alterations of the ribosomal biogenesis pathway may contribute to tumorigenesis (53). For instance, mutations in a ribosomal gene encoding S19 lead to a cancer susceptibility syndrome called Diamond-Blackfan anemia (13). Also, mutations of the dyskerin gene encoding a crucial pseudouridine synthase that mediates posttranscriptional modification of rRNA are frequent in the dyskeratosis congenital disease characterized by premature aging and an increased susceptibility to cancer (52). Moreover, experimentally generated tumor mice harbor characteristic mutations in the ribosomal protein L11 (38) or L9 (42). Inversely, overexpression of some ribosomal proteins has been shown to induce cell cycle arrest (44, 45). Therefore, it would be interesting and worthwhile to investigate whether L23 is altered in human cancers.
Acknowledgments
We thank Yanping Zhang and Karen Vousden for providing L11 plasmids and antibodies, Hunjoo Lee for plasmid preparation, David M. Keller and Jayme Gallegos for critically reading the manuscript, and other members in our laboratory for active discussion. We thank Yanping Zhang and his colleagues for communicating their work prior to publication.
L.D. was supported by an NIH grant (EY10572). This work is supported by grants to H.L. from NIH/NCI (CA095441, CA93614, and CA079721).
REFERENCES
- 1.Ashcroft, M., M. H. Kubbutat, and K. H. Vousden. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 4.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, M. T., N. Vlatkovic, and D. S. Haines. 2000. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J. Biol. Chem. 275:31883-31890. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 7.Budde, A., and I. Grummt. 1999. p53 represses ribosomal gene transcription. Oncogene 18:1119-1124. [DOI] [PubMed] [Google Scholar]
- 8.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciarmatori, S., P. H. Scott, J. E. Sutcliffe, A. McLees, H. M. Alzuherri, J. H. Dannenberg, H. te Riele, I. Grummt, R. Voit, and R. J. White. 2001. Overlapping functions of the pRb family in the regulation of rRNA synthesis. Mol. Cell. Biol. 21:5806-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draptchinskaia, N., P. Gustavsson, B. Andersson, M. Pettersson, T. N. Willig, I. Dianzani, S. Ball, G. Tchernia, J. Klar, H. Matsson, D. Tentler, N. Mohandas, B. Carlsson, and N. Dahl. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21:169-175. [DOI] [PubMed] [Google Scholar]
- 14.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197-239. [DOI] [PubMed] [Google Scholar]
- 15.Elenbaas, B., M. Dobbelstein, J. Roth, T. Shenk, and A. J. Levine. 1996. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol. Med. 2:439-451. [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Y., D. Absher, D. E. Eberhart, V. Brown, H. E. Malter, and S. T. Warren. 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1:109-118. [DOI] [PubMed] [Google Scholar]
- 18.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 19.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277:19251-19254. [DOI] [PubMed] [Google Scholar]
- 20.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 21.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 22.Honda, R., and H. Yasuda. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iordanov, M. S., D. Pribnow, J. L. Magun, T. H. Dinh, J. A. Pearson, S. L. Chen, and B. E. Magun. 1997. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 17:3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22:6365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, Y., S. X. Zeng, M. S. Dai, X. J. Yang, and H. Lu. 2002. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J. Biol. Chem. 277:30838-30843. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 28.Khanna, K. K., K. E. Keating, S. Kozlov, S. Scott, M. Gatei, K. Hobson, Y. Taya, B. Gabrielli, D. Chan, S. P. Lees-Miller, and M. F. Lavin. 1998. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat. Genet. 20:398-400. [DOI] [PubMed] [Google Scholar]
- 29.Kobet, E., X. Zeng, Y. Zhu, D. Keller, and H. Lu. 2000. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc. Natl. Acad. Sci. USA 97:12547-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 31.Lane, D., and E. Harlow. 1982. Two different viral transforming proteins bind the same host tumour antigen. Nature 298:517. [DOI] [PubMed] [Google Scholar]
- 32.Little, N. A., and A. G. Jochemsen. 2001. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene 20:4576-4580. [DOI] [PubMed] [Google Scholar]
- 33.Ljungman, M., F. Zhang, F. Chen, A. J. Rainbow, and B. C. McKay. 1999. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene 18:583-592. [DOI] [PubMed] [Google Scholar]
- 34.Lohrum, M. A., M. Ashcroft, M. H. Kubbutat, and K. H. Vousden. 2000. Contribution of two independent MDM2-binding domains in p14(ARF) to p53 stabilization. Curr. Biol. 10:539-542. [DOI] [PubMed] [Google Scholar]
- 35.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577-587. [DOI] [PubMed] [Google Scholar]
- 36.Lohrum, M. A., D. B. Woods, R. L. Ludwig, E. Balint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marechal, V., B. Elenbaas, J. Piette, J. C. Nicolas, and A. J. Levine. 1994. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 14:7414-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsutake, T., and P. K. Srivastava. 2001. The immunoprotective MHC II epitope of a chemically induced tumor harbors a unique mutation in a ribosomal protein. Proc. Natl. Acad. Sci. USA 98:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 41.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 42.Monach, P. A., S. C. Meredith, C. T. Siegel, and H. Schreiber. 1995. A unique tumor antigen produced by a single amino acid substitution. Immunity 2:45-59. [DOI] [PubMed] [Google Scholar]
- 43.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 44.Naora, H. 1999. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol. Cell Biol. 77:197-205. [DOI] [PubMed] [Google Scholar]
- 45.Neumann, F., and U. Krawinkel. 1997. Constitutive expression of human ribosomal protein L7 arrests the cell cycle in G1 and induces apoptosis in Jurkat T-lymphoma cells. Exp. Cell Res. 230:252-261. [DOI] [PubMed] [Google Scholar]
- 46.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. Parc: a cytoplasmic anchor for p53. Cell 112:29-40. [DOI] [PubMed] [Google Scholar]
- 47.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 48.Oren, M., A. Damalas, T. Gottlieb, D. Michael, J. Taplick, J. F. Leal, R. Maya, M. Moas, R. Seger, Y. Taya, and A. Ben-Ze'Ev. 2002. Regulation of p53: intricate loops and delicate balances. Ann. N. Y. Acad. Sci. 973:374-383. [DOI] [PubMed] [Google Scholar]
- 49.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125-126. [DOI] [PubMed] [Google Scholar]
- 50.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picksley, S. M., and D. P. Lane. 1993. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays 15:689-690. [DOI] [PubMed] [Google Scholar]
- 52.Ruggero, D., S. Grisendi, F. Piazza, E. Rego, F. Mari, P. H. Rao, C. Cordon-Cardo, and P. P. Pandolfi. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299:259-262. [DOI] [PubMed] [Google Scholar]
- 53.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 54.Sharpless, N. E., and R. A. DePinho. 2002. p53: good cop/bad cop. Cell 110:9-12. [DOI] [PubMed] [Google Scholar]
- 55.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Laar, P. Dekker, M. Bazuine, R. C. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 56.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stad, R., N. A. Little, D. P. Xirodimas, R. Frenk, A. J. van der Eb, D. P. Lane, M. K. Saville, and A. G. Jochemsen. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277:29617-29625. [DOI] [PubMed] [Google Scholar]
- 59.Sugimoto, M., M. L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 60.Tao, W., and A. J. Levine. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA 96:3077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao, W., and A. J. Levine. 1999. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai, R. Y., and R. D. McKay. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 64.Voit, R., K. Schafer, and I. Grummt. 1997. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 17:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber, J. D., M. L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 67.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 68.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20:4972-4983. [DOI] [PubMed] [Google Scholar]
- 69.Zalfa, F., M. Giorgi, B. Primerano, A. Moro, A. Di Penta, S. Reis, B. Oostra, and C. Bagni. 2003. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112:317-327. [DOI] [PubMed] [Google Scholar]
- 70.Zeng, S. X., M. S. Dai, D. M. Keller, and H. Lu. 2002. SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 21:5487-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng, X., H. Lee, Q. Zhang, and H. Lu. 2001. p300 does not require its acetylase activity to stimulate p73 function. J. Biol. Chem. 276:48-52. [DOI] [PubMed] [Google Scholar]
- 72.Zeng, X., X. Li, A. Miller, Z. Yuan, W. Yuan, R. P. Kwok, R. Goodman, and H. Lu. 2000. The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell. Biol. 20:1299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhai, W., and L. Comai. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, Y., G. W. Wolf, K. Bhat, A. Jin, T. Allio, W. A. Burkhart, and Y. Xiong. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Y., and Y. Xiong. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, B. P., Y. Liao, W. Xia, Y. Zou, B. Spohn, and M. C. Hung. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 3:973-982. [DOI] [PubMed] [Google Scholar]
- 77.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]