Assignment of 16S rRNA gene sequences to operational taxonomic units (OTUs) allows microbial ecologists to overcome the inconsistencies and biases within bacterial taxonomy and provides a strategy for clustering similar sequences that do not have representatives in a reference database.
KEYWORDS: 16S rRNA gene sequences, environmental microbiology, OTU, QIIME, bioinformatics, metagenomics, microbial ecology, microbiome, mothur
ABSTRACT
Assignment of 16S rRNA gene sequences to operational taxonomic units (OTUs) allows microbial ecologists to overcome the inconsistencies and biases within bacterial taxonomy and provides a strategy for clustering similar sequences that do not have representatives in a reference database. I have applied the Matthews correlation coefficient to assess the ability of 15 reference-independent and -dependent clustering algorithms to assign sequences to OTUs. This metric quantifies the ability of an algorithm to reflect the relationships between sequences without the use of a reference and can be applied to any data set or method. The most consistently robust method was the average neighbor algorithm; however, for some data sets, other algorithms matched its performance.
COMMENTARY
Numerous algorithms have been developed for solving the seemingly simple problem of assigning 16S rRNA gene sequences to operational taxonomic units (OTUs). These algorithms were recently the subject of benchmarking studies performed by Westcott and myself (1, 2), He et al. (3), and Kopylova et al. (4). These studies provide a thorough review of the sequencing clustering landscape, which can be divided into three general approaches: (i) de novo clustering, where sequences are clustered without first mapping sequences to a reference database; (ii) closed-reference clustering, where sequences are clustered based on the references that the sequences map to; and (iii) open-reference clustering, where sequences that do not map adequately to the reference are then clustered using a de novo approach. Assessment of the quality of the clustering assignments has been a persistent problem in the development of clustering algorithms.
The recent analysis by Kopylova et al. (4) repeated many of the benchmarking strategies employed by previous researchers. Many algorithm developers have clustered sequences from simulated communities or sequencing data from synthetic communities of cultured organisms and quantified how well the OTU assignments matched the organisms’ taxonomy (5–16). Although an OTU definition would ideally match bacterial taxonomy, bacterial taxonomy has proven itself to be fluid and to reflect the biases of various research interests. Furthermore, it is unclear how the methods scale to sequences from the novel organisms we are likely to encounter in deep sequencing surveys. In a second approach, developers have compared the time and memory required to cluster sequences in a data set (6, 13, 17, 18). These are valid parameters to assess when judging a clustering method but indicate little regarding the quality of the OTU assignments. For example, reference-based methods are very efficient but do a poor job of reflecting the genetic diversity within the community when novel sequences are encountered (2). In a third approach, developers have compared the number of OTUs generated by various methods for a common data set (4, 5). Although methods need to guard against excessive splitting of sequences across OTUs, by focusing on minimizing the number of OTUs in a community, developers risk excessively lumping sequences together that are not similar. In a fourth approach, a metric of OTU stability has been proposed as a way to assess algorithms (3). Although it is important that the methods generate reproducible OTU assignments when the initial order of the sequences is randomized, this metric ignores the possibility that the variation in assignments may be equally robust or that the assignments by a highly reproducible algorithm may be quite poor. In a final approach, some developers have assessed the quality of clustering based on the method’s ability to generate the same OTUs generated by other methods (18, 19). Unfortunately, without the ability to ground truth by any method, such comparisons are tenuous. There is a need for an objective metric to assess the quality of OTU assignments.
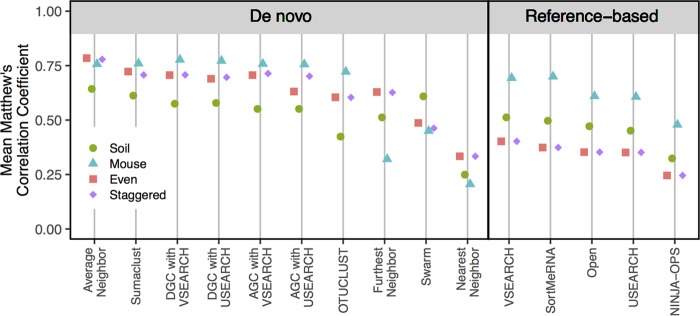
Westcott and I have proposed an unbiased and objective method for assessment of the quality of OTU assignments that can be applied to any collection of sequences (1, 2). Our approach uses the observed dissimilarity between pairs of sequences and information about whether sequences were clustered together to quantify how well similar sequences are clustered together and dissimilar sequences are clustered apart. To quantify the correlation between the observed and expected OTU assignments, we synthesize the relationship between OTU assignments and the distances between sequences using the Matthews correlation coefficient (MCC) (20). I have expanded our previous analysis to evaluate three hierarchical and seven “greedy” de novo algorithms, one open-reference clustering algorithm, and four closed-reference algorithms (Fig. 1). To test these approaches, I applied each of them to data sets from soil (21) and mouse feces (22), as well as two simulated data sets. The simulated communities were generated by randomly selecting 10,000 16S rRNA sequences that were unique within the V4 region from the SILVA nonredundant database (4, 23). Next, an even community was generated by specifying that each sequence had a frequency of 100 reads, and a staggered community was generated by specifying that the abundance of each sequence was a randomly drawn uniform distribution between 1 and 200.
FIG 1 .
Comparison of OTU quality generated by multiple algorithms applied to four data sets. The nearest, average, and furthest neighbor clustering algorithms were used as implemented in mothur (v.1.37) (25). Abundance-based greedy clustering (AGC) and distance-based greedy clustering (DGC) were implemented using USEARCH (v.6.1) and VSEARCH (v.1.5.0) (3, 5, 26). Other de novo clustering algorithms included Swarm (v.2.1.1) (6, 7), OTUCLUST (v.0.1) (27), and Sumaclust (v.1.0.20). The MCC values for swarm were determined by selecting the distance threshold that generated the maximum MCC value for each data set. The USEARCH and SortMeRNA (v.2.0) closed-reference clusterings were performed using QIIME (v.1.9.1) (28, 29). Closed-reference clustering was also performed using VSEARCH (v.1.5.0) and NINJA-OPS (v.1.5.0) (16). The order of the sequences in each data set was randomized 30 times, and the intramethod range in MCC values was smaller than the plotting symbol. MCC values were calculated using mothur.
I replicated the benchmarking approach that I have used previously to assess the ability of an algorithm to correctly group sequences that are similar to each other and split sequences that are dissimilar to each other using the MCC (1, 2). When I compared the MCC values calculated using the 10 de novo algorithms with the four data sets, the average neighbor algorithm reliably performed as well or better than the other methods (Fig. 1). For the murine data set, the MCC values for the VSEARCH (abundance-based greedy clustering [AGC], 0.76; distance-based greedy clustering [DGC], 0.78) and USEARCH-based (AGC, 0.76; DGC; 0.77) algorithms, Sumaclust (0.76), and average neighbor (0.76) were similarly high. For each of the other data sets, the MCC value for the average neighbor algorithm was at least 5% higher than the next best method. Swarm does not use a traditional distance-based criterion to cluster sequences into OTUs and instead looks for natural subnetworks in the data. When I used the distance threshold that gave the best MCC value for the Swarm data, the MCC values were generally not as high as they were using the average neighbor algorithm. The one exception was for the soil data set. Among the reference-based methods, all of the MCC values suffer because when sequences that are at least 97% similar to a reference are pooled, the sequences within an OTU could be as much as 6% different from each other. The effect of this is observed in the MCC values that were calculated for the OTUs assigned by these methods generally being lower than those observed using the de novo approaches (Fig. 1). It is also important to note that the MCC values for the closed-reference OTUs are inflated because sequences were removed from the analysis if there was not a reference sequence that was more than 97% similar to the sequence. By choosing to focus on the ability to regenerate taxonomic clusterings, minimizing the number of OTUs, and computational performance, Kopylova et al. (4) concluded that Swarm and Sumaclust had the most consistent performance among the de novo methods. My objective MCC-based approach found that Sumaclust performed well but was matched or outperformed by the average neighbor algorithm; using a 3% threshold, Swarm was actually one of the worst methods. Given the consistent quality of the clusterings formed by the average neighbor algorithm, these results confirm the conclusion from the previous analysis that researchers should use the average neighbor algorithm or calculate MCC values for several methods and use the clustering that gives the best MCC value (2).
Next, I investigated the ability of the reference-based methods to properly assign sequences to OTUs. The full-length 16S rRNA gene sequences in the default reference taxonomy that accompanies QIIME are less than 97% similar to each other. Within the V4 region, however, many of the sequences were more similar to each other and even identical to each other. As a result, we previously found that there was dependence between the ordering of sequences in the reference database and the OTU assignments with USEARCH and VSEARCH (2). To explore this further, we analyzed the 32,106 unique sequences from the murine data set with randomized databases. VSEARCH always found matches for 27,737 murine sequences; the reference matched to those sequences differed between randomizations. For USEARCH, there were between 28,007 and 28,111 matches, depending on the order of the reference. In the updated analysis, we found that SortMeRNA resulted in between 23,912 and 28,464 matches. Using NINJA-OPS with different orderings of the reference sequences generated the same 28,499 matches. These results point to an additional problem with closed-reference clustering, which is the inability of the method to assign sequences to OTUs when a similar reference sequence does not exist in the database. For the well-characterized murine microbiota, NINJA-OPS did the best by finding relatives for 88.8% of the unique murine sequences. As indicated by the variation in the number of sequences that matched a reference sequence, these methods varied in their sensitivity and specificity to find the best reference sequence. Of the closed-reference methods, NINJA-OPS had the best sensitivity (99.7%) and specificity (79.7%), while SortMeRNA had the worst sensitivity (95.7%), and VSEARCH had the worst specificity (60.3%). Reference-based clustering algorithms are much faster than de novo approaches, but they do not generate OTUs that are as robust.
Although the goal of Kopylova et al. (4) was to compare various clustering algorithms, they also studied these algorithms in the broader context of raw sequence processing, screening for chimeras, and removal of singletons. Each of these is a critical decision in a comprehensive pipeline. By including these steps, they confounded their analysis of how best to cluster sequences into OTUs. The effect of differences in MCC values on one’s ability to draw inferences is unclear and admittedly may be relatively minor for some data sets. Because of this uncertainty, researchers should use the most reliable methods available in case the differences in clustering do affect the conclusions that can be drawn from a particular data set. Through the use of objective criteria that measure the quality of the clusterings, independent of taxonomy or database, researchers will be able to evaluate which clustering algorithm is the best fit for their data.
(A reproducible version of this article and analysis has been submitted to an online repository [24].)
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
For the article discussed, see 10.1128/mSystems.00003-15.
REFERENCES
- 1.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/aem.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westcott SL, Schloss PD. 2015. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ 3:e1487. doi: 10.7717/peerj.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Caporaso JG, Jiang X-T, Sheng H-F, Huse SM, Rideout JR, Edgar RC, Kopylova E, Walters WA, Knight R, Zhou H-W. 2015. Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome 3:20. doi: 10.1186/s40168-015-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopylova E, Navas-Molina JA, Mercier C, Xu ZZ, Mahé F, He Y, Zhou H-W, Rognes T, Caporaso JG, Knight R. 2016. Open-source sequence clustering methods improve the state of the art. mSystems 1:e00003-15. doi: 10.1128/msystems.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 6.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. 2014. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. 2015. Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3:e1420. doi: 10.7717/peerj.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barriuso J, Valverde JR, Mellado RP. 2011. Estimation of bacterial diversity using next generation sequencing of 16S rDNA: a comparison of different workflows. BMC Bioinformatics 12:473. doi: 10.1186/1471-2105-12-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonder MJ, Abeln S, Zaura E, Brandt BW. 2012. Comparing clustering and pre-processing in taxonomy analysis. Bioinformatics 28:2891–2897. doi: 10.1093/bioinformatics/bts552. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Zhang CK, Cheng Y, Zhang S, Zhao H. 2013. A comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS One 8:e70837. doi: 10.1371/journal.pone.0070837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May A, Abeln S, Crielaard W, Heringa J, Brandt BW. 2014. Unraveling the outcome of 16S rDNA-based taxonomy analysis through mock data and simulations. Bioinformatics 30:1530–1538. doi: 10.1093/bioinformatics/btu085. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Sun Y. 2011. ESPRIT-tree: hierarchical clustering analysis of millions of 16S rRNA pyrosequences in quasilinear computational time. Nucleic Acids Res 39:e95. doi: 10.1093/nar/gkr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Cai Y, Huse SM, Knight R, Farmerie WG, Wang X, Mai V. 2012. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief Bioinform 13:107–121. doi: 10.1093/bib/bbr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White JR, Navlakha S, Nagarajan N, Ghodsi M-R, Kingsford C, Pop M. 2010. Alignment and clustering of phylogenetic markers—implications for microbial diversity studies. BMC Bioinformatics 11:152. doi: 10.1186/1471-2105-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Ghalith GA, Montassier E, Ward HN, Knights D. 2016. NINJA-OPS: fast accurate marker gene alignment using concatenated ribosomes. PLoS Comput Biol 12:e1004658. doi: 10.1371/journal.pcbi.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W, Farmerie W. 2009. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res 37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou H-W, Knight R, Caporaso JG. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt TS, Matias Rodrigues JF, von Mering C. 2015. Limits to robustness and reproducibility in the demarcation of operational taxonomic units. Environ Microbiol 17:1689–1706. doi: 10.1111/1462-2920.12610. [DOI] [PubMed] [Google Scholar]
- 20.Matthews B. 1975. Comparison of the predicted and observed secondary structure of t4 phage lysozyme. Biochim Biophys Acta 405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 21.Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. 2012. Stabilization of the murine gut microbiome following weaning. Gut Microbes 3:383–393. doi: 10.4161/gmic.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD. 2016. Application of database-independent approach to assess the quality of OTU picking methods. GitHub https://github.com/SchlossLab/Schloss_Cluster_PeerJ_2015. [DOI] [PMC free article] [PubMed]
- 25.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/aem.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rognes T, Mahé F, Flouri T, McDonald D. 2015. Vsearch: VSEARCH 1.4.0. Zenodo, European Organization for Nuclear Research, Geneva, Switzerland. [Google Scholar]
- 27.Albanese D, Fontana P, De Filippo C, Cavalieri D, Donati C. 2015. Micca: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5:9743. doi: 10.1038/srep09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]