Abstract
The basic helix-loop-helix transcription factors, E2A and HEB, play important roles in T-cell development at multiple checkpoints. Expression of their inhibitor, Id1, abolishes the function of both transcription factors in a dose-dependent manner. The Id1 transgenic thymus is characterized by an accumulation of CD4− CD8− CD44+ CD25− thymocytes, a dramatic reduction of CD4+ CD8+ thymocytes, and an abundance of apoptotic cells. Here we show that these apoptotic cells carry functional T-cell receptors (TCRs), suggesting that apoptosis occurs during T-cell maturation. In contrast, viable Id1 transgenic CD4 single positive T cells exhibit costimulation-independent proliferation upon treatment with anti-CD3 antibody, probably due to a hyperresponse to TCR signaling. Furthermore, Id1 expression causes apoptosis of CD4 and CD8 double- or single-positive thymocytes in HY- or AND-TCR transgenic mice under conditions that normally support positive selection. Collectively, these results suggest that E2A and HEB proteins are crucial for controlling the threshold for TCR signaling, and Id1 expression lowers the threshold, resulting in apoptosis of developing thymocytes.
T-cell development consists of a series of precisely controlled events involving cell differentiation, proliferation, and survival, which are largely influenced by signals through pre-T-cell receptors (pre-TCRs) and T-cell receptors (TCRs) (27, 32). Several checkpoints are in place to ensure that thymocytes with proper receptors are selected, whereas others with less-than-ideal receptors undergo apoptosis. The β-selection checkpoint at the transition from the CD4 and CD8 double-negative (DN) to double-positive (DP) stage permits only cells with functional pre-TCR to proliferate and differentiate into DP cells (12, 30). DP cells then rearrange the TCRα locus and produce TCRs on their surface. The duration and strength of interaction between TCRs and major histocompatibility complex (MHC)-peptide complexes determine the fate of these DP cells (2, 37, 47). Cells with TCRs mediating appropriate duration and strengths of interaction become positively selected and differentiate into CD4 or CD8 single-positive (SP) cells. However, cells carrying TCRs that interact with MHCs too weakly or too strongly die by neglect and by negative selection, respectively. Therefore, signaling through pre-TCR and TCR must be closely monitored. Otherwise, the default outcome is cell death. Modulation of pre-TCR and TCR signaling occurs at multiple levels from the cell membrane to the nucleus. Although much is known about the positive events transmitting TCR signals, less is understood about the opposing events that balance positive signaling.
The E2A and HEB genes encode basic helix-loop-helix transcription factors, collectively called E proteins, which have redundant functions (15). The function of E proteins can be eliminated by their naturally occurring dominant-negative inhibitors, Id1 to Id4 (44). Complete elimination of the function of these E proteins in the T lineage by expression of various inhibitors arrests T-cell development at early progenitor stages, indicating an essential role for E proteins (8, 19, 23, 24, 33). However, partial inhibition of the function reveals that E proteins also play important roles in pre-TCR and TCR signaling. For example, disruption of the E2A gene or expression of Id1 enables RAG-deficient DN T cells to differentiate into DP cells, suggesting that E2A proteins influence pre-TCR signaling (14, 24). Loss of E2A also moderately facilitates positive selection (5), whereas mutation of the Id3 gene inhibits both positive and negative selection (42). These findings are consistent with the observations that E-protein binding activities are reduced upon pre-TCR and TCR signaling (3, 14, 24).
Furthermore, in Id1 transgenic mice in which E-protein function is more completely abolished than in E2A- or HEB-deficient mice (4, 7, 23), massive apoptosis is observed. We found similar levels of TCRβ and TCRα gene rearrangement in DNA isolated from apoptotic thymocytes of Id1 transgenic mice and viable thymocytes of Id1 transgenic or wild-type mice (23). Thus, the arrest in T-cell development in Id1 transgenic mice is not due to a failure in TCR gene rearrangement. We therefore postulated that these apoptotic cells might have already committed to the T lineage and died during the course of maturation (24). Interestingly, the NF-κB family of transcription factors is dramatically activated in Id1 transgenic thymocytes through activation of IκB kinases. Activation of NF-κB indeed promotes the differentiation of RAG1-deficient DN cells to the DP stage (46). In Id1 transgenic mice, further activation of NF-κB exacerbates the T-cell defects, whereas inhibition of NF-κB alleviates the developmental block (24).
We provide here evidence in support of our hypothesis that E proteins play a critical role in controlling the threshold of TCR stimulation and thus prevent apoptosis of developing thymocytes. We found that the frequencies of productive rearrangements in the TCRα and -β loci in apoptotic Id1 transgenic thymocytes are similar to those in wild-type thymocytes, suggesting that these apoptotic cells probably possess functional pre-TCRs or TCRs prior to cell death. Furthermore, Id1 transgenic CD4 SP thymocytes undergo vigorous proliferation in response to anti-CD3 stimulation without costimulation. This result suggests that Id1 transgenic thymocytes are hyperresponsive to TCR stimulation. Consequently, Id1 transgenic thymocytes might be more susceptible to apoptosis through a mechanism analogous to negative selection, which we term pseudo-negative selection. Indeed, we show that Id1 expression turns signals for positive selection into negative selection in HY- or AND-TCR transgenic mice (which carry the AD10 β chain and the AN6.2 α chain [22]), possibly due to a lower threshold for TCR stimulation.
MATERIALS AND METHODS
Mice.
Id1 transgenic mice were described previously (23) and back-crossed to the C57BL/6 background for seven generations. AND-TCR (22) and HY-TCR (25) transgenic mice were also on the C57BL/6 background and bred with Id1 transgenic mice to generate transheterozygotes for each transgene. Id1 transgenic mice used for TCR rearrangement and proliferation assays are on the FVB/N background.
Flow cytometry.
Antibodies used in the present study were purchased from the following sources: Tri-Color-conjugated rat anti-mouse CD4 or CD8, phycoerythrin-conjugated rat anti-mouse CD8 or CD44, and fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD25 were purchased from Caltag Laboratories (Burlingame, Calif.). Monoclonal hamster anti-mouse CD3ɛ (2C11) or CD28 (37.51) and an FITC-conjugated mouse Vβ TCR screening kit were obtained from BD Pharmingen (San Diego, Calif.). Thymocytes were stained with appropriate antibodies and data was collected on a FACScan flow cytometer and analyzed with the CellQuest software (BD Pharmingen).
Analyses of apoptosis.
Detection of apoptotic cells was carried out with Annexin V staining and Apo-Direct kits according to manufacturer's protocols (BD Biosciences, Franklin Lakes, N.J.). Briefly, to stain cells with Annexin V-FITC, 106 thymocytes were washed twice in phosphate-buffered saline (PBS) and resuspended in 1 ml of Annexin V binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2). A total of 100 μl of the cell suspension was mixed with 5 μl of Annexin V-FITC and 250 ng of propidium iodide. The mixture was incubated at room temperature for 15 min, and 300 μl of Annexin V binding buffer was then added. Stained cells were analyzed on a FACScan. To evaluate apoptosis by dUTP labeling, 106 thymocytes were fixed in 4% paraformaldehyde in PBS on ice for 30 min and washed twice with PBS. The fixed cells were resuspended in 75% ice-cold ethanol and kept on ice or at −20°C for at least 1 h. After two washes with washing buffer, the cells were resuspended in 50 μl of the labeling mixture containing 10 μl of the reaction buffer, 1.25 μM FITC-dUTP, and 15 U of terminal deoxynucleotidyl transferase. The labeling reaction was carried out at 37°C for 1 h and washed twice with washing buffer. Cells were resuspended in 300 μl of PBS and analyzed on a FACScan.
Analyses of the rearrangement of TCRβ and -α loci.
Thymocytes were fractionated on a Ficoll cushion to separate viable cells from dead or apoptotic cells. Thymic genomic DNA from the pellet (dead or apoptotic cells) and interface (viable cells) was extracted and gel purified as described previously (23). To amplify rearranged TCRβ fragments, PCRs were carried out with oligonucleotides binding to the Vβ3, Vβ5, or Vβ8 region as a 5′ primer and to Jβ2 as a 3′ primer. Similarly, 5′ primers corresponding to Vα2C, VαF3, or VαH and a 3′ primer from the JαTT11 region were used to amplify rearranged TCRα fragments. Sequences of the primers are as described previously (23). These PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, Wis.). Plasmid DNAs isolated from individual colonies were screened with a Jβ2 or JαTT11 probe by using a dot blot assay. Positive samples were sequenced. The junction sequence of each unique V(D)J recombination event was classified into productive and nonproductive rearrangement, and the percentages of productive rearrangement events were calculated for each DNA sample.
Proliferation assay of CD4+ thymocytes.
Viable thymocytes were purified away from apoptotic cells by collecting cells at the interface after centrifugation at 4°C for 20 min on a Ficoll cushion (Amersham Pharmacia Biotech, Piscataway, N.J.). Cells were then stained with anti-CD4 and anti-CD8 and sorted on a Mo-Flo cell sorter (Cytomation, Ft. Collins, Colo.). Sorted CD4+ thymocytes (2 × 105, >95% purity) were cultured in triplicates in 96-well flat-bottom plates coated with anti-CD3ɛ (clone 2C11, 10 μg/ml) with or without soluble anti-CD28 (clone 37.51, 2 μg/ml) for 48 h in 200 μl of RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and 2 mM l-glutamine at 37°C in 5% CO2. The cells were pulse-labeled with 1 μCi of [3H]thymidine (Perkin-Elmer Life Sciences, Inc., Boston, Mass.) in the last 18 h of the incubation and harvested by using a cell harvester (Cambridge Technology, Inc., Watertown, Mass.). The amount of [3H]thymidine incorporated by proliferating CD4+ thymocytes was measured by liquid scintillation counting. For some experiments, different concentrations of IκB kinase inhibitor (NBD) or its control peptide (NBA) were included (31). These peptides were dissolved in dimethyl sulfoxide and sterilized by passing through a 0.22-μm-pore-size filter.
RESULTS
Enhanced apoptosis of DP thymocytes in Id1 transgenic mice.
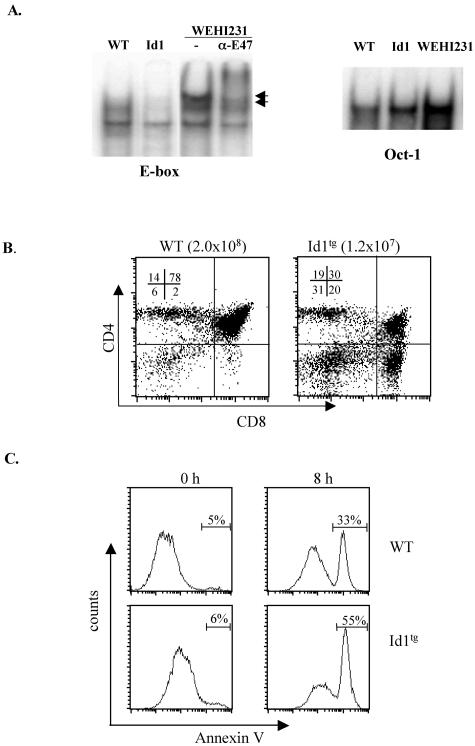
Expression of the Id1 protein in transgenic mice, in which the Id1 cDNA was driven by the proximal promoter of the lck gene, dramatically inhibited E-protein DNA-binding activity in thymocytes. As shown in Fig. 1A, two E-box binding complexes, BCF1 and -2 (34), are usually found in lymphoid cells such as WEHI231 B cells, and both can be supershifted by anti-E47 antibodies. In wild-type total thymocytes, these complexes were readily detectable, but their levels were dramatically reduced in heterozygous Id1 transgenic thymocytes (Fig. 1A). In these Id1 transgenic mice, whereas the total thymic cellularity is significantly decreased, a small number of thymocytes develop to maturity. However, the CD4 and CD8 DP population is disproportionately diminished (Fig. 1B). Since DP cells constitute the majority of total thymocytes in wild-type mice, a loss of DP cells could, at least partially, account for the reduced cellularity. We have previously detected increased apoptosis in the thymuses of Id1 transgenic mice by using scatter analysis, DNA fragmentation, and in situ TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assays (23, 24). Here, we further evaluated apoptosis of freshly isolated viable DP thymocytes in vitro. As determined by Annexin V staining, these cells from Id1 transgenic thymus underwent apoptosis at a rate 70% higher than wild-type cells after 8 h in culture (Fig. 1C), thus confirming the enhanced apoptotic potential of Id1 transgenic thymocytes.
FIG. 1.
Enhanced apoptosis of DP thymocytes in Id1 transgenic mice. (A) EMSA with nuclear extracts prepared with total thymocytes from wild-type and heterozygous Id1 transgenic mice. The E-box and Oct-1 probes were as described previously (24). As a control, EMSA with a nuclear extract from the WEHI231 B-cell line was performed with or without anti-E47 antibodies to supershift the E47 containing complexes as marked with arrows. The amount of Oct-1 binding complex serves as a control for the amount of nuclear extract in each sample. (B). Thymocytes from wild-type and Id1 transgenic mice on the FVB/N background were stained with the indicated antibodies. The number in each quadrant is the percentage of the subset of thymocytes. (C). Sorted viable DP thymocytes were stained with Annexin V-FITC with or without 8 h of culture at 37°C. The percentage of Annexin-positive cells is shown on top of the gate.
Apoptotic cells in Id1 transgenic mice undergo productive TCR rearrangement.
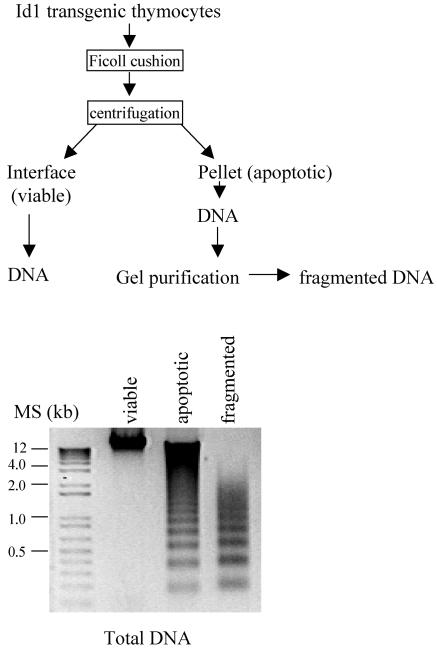
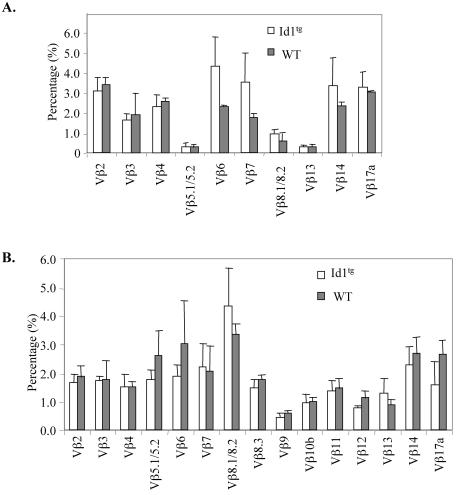
Our previous data indicated that apoptotic cells isolated from thymus of Id1 transgenic mice had substantial levels of TCRβ and -α gene rearrangement compared to viable cells from transgenic or wild-type mice (23), suggesting that apoptosis occurs in developing thymocytes. To examine the nature of these rearrangement events and rule out the possibility that aberrant rearrangement takes place in a few restricted V, D, and J regions of TCRβ and -α genes, we compared the sequences of rearranged V(D)J fragments between viable and apoptotic thymocytes from wild-type and Id1 transgenic mice. As diagrammed in Fig. 2, we isolated viable and apoptotic thymocytes from Id1 transgenic mice by centrifugation on a Ficoll cushion and purified DNA from these cells. Since the genomic DNA from the apoptotic fraction appeared fragmented, we purified the fragmented DNA away from the intact DNA by gel electrophoresis, on the assumption that the fragmented DNA was most likely derived from apoptotic cells. DNA samples isolated from viable and apoptotic cells were examined, along with this preparation of fragmented DNA (Fig. 2). V(D)J and V-J rearrangement products of TCRβ and -α loci were amplified by PCR with the DNA of viable cells and fragmented DNA of apoptotic cells from Id1 transgenic mice, as well as from the DNA of viable wild-type cells as templates. These products were cloned, and the sequences of the individual clones were determined. At least 40 unique recombination events from each DNA sample were analyzed (Table 1). These events represent rearrangements between different V regions and the Jβ2 or JαTT11 regions, respectively (data not shown), suggesting that no skewing of rearrangement of certain V(D)J regions occurred in the apoptotic Id1 transgenic thymocytes. This was further supported by comparison of TCR Vβ usage between wild-type and transgenic viable thymocytes by flow cytometry, where the repertoires of TCRβ in Id1 transgenic mice on both FVB/N and C57BL/6 backgrounds were comparable to those of wild-type mice, even though minor variations exist (Fig. 3).
FIG. 2.
Isolation of genomic DNA from Id1 transgenic mice for TCR gene rearrangement assays. The scheme for isolation of genomic DNA from viable and apoptotic cells is diagrammed. Aliquots of DNA samples prepared were analyzed by agarose gel electrophoresis.
TABLE 1.
Frequencies of productive TCR gene rearrangement
| Locus and cell typec | No. of eventsa
|
% Productive events | ||
|---|---|---|---|---|
| Total | Productive | Nonproductive | ||
| TCRβ locus | ||||
| WT/viable | 55 | 44 | 11 | 80 |
| Id1/viable | 49 | 25 | 24 | 51b |
| Id1/frag. | 67 | 52 | 15 | 78 |
| TCRα locus | ||||
| WT/viable | 43 | 14 | 29 | 33 |
| Id1/viable | 40 | 12 | 28 | 30 |
| Id1/frag. | 42 | 16 | 26 | 38 |
Each event represents a unique VDJ or VJ junction sequence.
χ2 < 0.001.
WT, wild type; frag., fragmented.
FIG. 3.
Normal TCRβ chain usage in Id1 transgenic thymocytes. Thymocytes from wild-type and Id1 transgenic mice on FVB/N (A) and C57BL/6 (B) backgrounds were stained with FITC-conjugated antibodies specific for each of the indicated Vβ chains. The percentage of each Vβ specific population out of total thymocytes is shown.
Because of the random addition and deletion of nucleotides at Vβ-Dβ, Dβ-Jβ, and Vα-Jα junctions at the TCRβ and -α loci, only a fraction of recombination events results in in-frame gene rearrangements, allowing production of functional TCRβ or -α chains. Thymocytes without productive TCRβ gene rearrangements normally undergo apoptosis due to failure of passing β-selection and those without productive TCRα rearrangements die by neglect. It is therefore necessary to determine whether the apoptotic cells in Id1 transgenic mice are simply those that have failed the selection processes. As summarized in Table 1, the frequency of productive TCRβ rearrangement in apoptotic thymocytes of Id1 transgenic mice was comparable to that of viable thymocytes of wild-type mice (78% versus 80%). These values are consistent with the theoretical frequency of productive TCRβ rearrangement in viable cells following β selection (71%), taking into account allelic exclusion at the TCRβ locus (45). Thus, we conclude that the majority of Id1 transgenic thymocytes undergo apoptosis after passing the β-selection checkpoint. Interestingly, the frequency of productive TCRβ rearrangements in viable thymocytes of Id1 transgenic mice was significantly lower (51%) than that in wild-type thymocytes or apoptotic Id1 thymocytes. Indeed, we found a higher percentage of low or no TCRβ-expressing DP cells in the Id1 transgenic mice compared to wild-type mice (data not shown). This is in agreement with our previous finding that Id1 expression enables RAG1-deficient thymocytes to differentiate to the double positive stage without a functional TCRβ (24). Since allelic exclusion does not occur during the rearrangement of the TCRα locus (10, 17), thymocytes with productive gene rearrangements at the TCRα locus are not enriched. Multiple attempts of rearrangement of each TCRα allele also occurs and the excision circles persist in thymocytes. Therefore, the probability of detecting in-frame TCRα rearrangement is expected to be ca. 33%, which is what we found for all cell populations from Id1 transgenic and wild-type mice (Table 1). Taken together, these results thus support the notion that the Id1 transgenic apoptotic cells possess intact T-cell receptors.
Id1 transgenic CD4+ cells proliferate independently of costimulation.
To evaluate TCR signaling in Id1 transgenic thymocytes, the proliferation potential of activated CD4+ cells was determined. Activation of naive CD4+ T cells leads to proliferation and differentiation to effector cells. This usually requires two signals: the signal originating from the TCR after the recognition of MHC class II-peptide complexes on antigen-presenting cells (APC) and the signal initiating from the costimulatory receptors of T cells by binding to costimulatory molecules on APC (1, 28, 29). One well-characterized costimulatory signal comes from the interaction between CD28 on CD4+ T cells and CD80/CD86 on APC. Activation of CD4+ cells can be mimicked by treatment with antibodies against CD3 and CD28 in vitro.
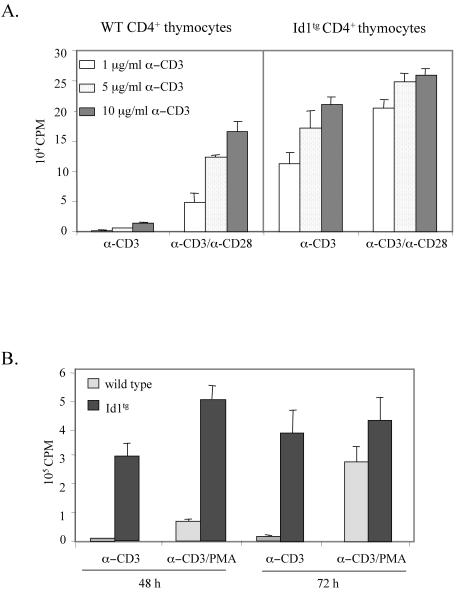
Because Id1 transgenic mice have very few T cells in the periphery, and the CD8+ thymocyte population contains a mixture of immature and mature CD8+ cells, we sorted CD4+ thymocytes from wild-type and Id1 transgenic mice. Their ability to proliferate was assayed by [3H]thymidine incorporation 48 h after stimulation with different concentrations of plate-bound anti-CD3 antibodies in the presence or absence of soluble anti-CD28 (Fig. 4A). Wild-type CD4+ cells barely proliferated when treated with anti-CD3ɛ antibodies alone. In contrast, CD4+ thymocytes from Id1 transgenic mice treated with anti-CD3ɛ antibodies underwent substantial proliferation in the absence of exogenous costimulation by the anti-CD28 antibody (Fig. 4A). These results suggested that Id1 expression in CD4+ thymocytes created effects similar to those generated by costimulation. Furthermore, in the presence of both anti-CD3ɛ and anti-CD28 antibodies, Id1 transgenic CD4+ cells also displayed up to fourfold-higher response than wild-type cells, indicating that Id1 expression may potentiate the cellular response to TCR stimulation and costimulation (Fig. 4A). Incidentally, anti-CD28 costimulation of Id1 transgenic CD4+ cells also triggered a more vigorous proliferation than the same cells treated with anti-CD3 antibodies alone. This is probably due to quantitatively or qualitatively different costimulatory signals delivered through the CD28 coreceptor and that mediated by Id1 expression. Furthermore, similar results were also obtained by using phorbol myristate acetate (PMA) as a costimulus (Fig. 4B). At 48 h after stimulation, Id1 transgenic CD4+ thymocytes treated with anti-CD3 alone or with both anti-CD3 and PMA already exhibited high levels of thymidine uptake, whereas wild-type cells stimulated with both anti-CD3 and PMA only incorporated a low level of [3H]thymidine. Furthermore, at 72 h Id1 transgenic cells cultured in the presence or absence of PMA continued to proliferate more vigorously than wild-type cells.
FIG. 4.
Costimulation-independent proliferation of CD4+ thymocytes from Id1 transgenic mice. (A) Sorted CD4+ thymocytes were plated in triplicates (2 × 105 cells per well) in a 96-well plate coated with indicated concentrations of anti-CD3ɛ antibody with or without soluble anti-CD28 MAb (2 μg/ml) and incubated for 48 h. Cells were pulsed with 1 μCi of [3H]thymidine per well for the last 18 h of incubation. The amount of [3H]thymidine incorporated by proliferating thymocytes was measured by scintillation counting. (B) Sorted CD4+ thymocytes from wild-type and Id1 transgenic mice were stimulated with plate-bound anti-CD3ɛ antibody (10 μg/ml) in a 96-well plate with or without PMA (2.5 ng/ml) for 48 or 72 h. [3H]thymidine incorporation was measured as described for panel A.
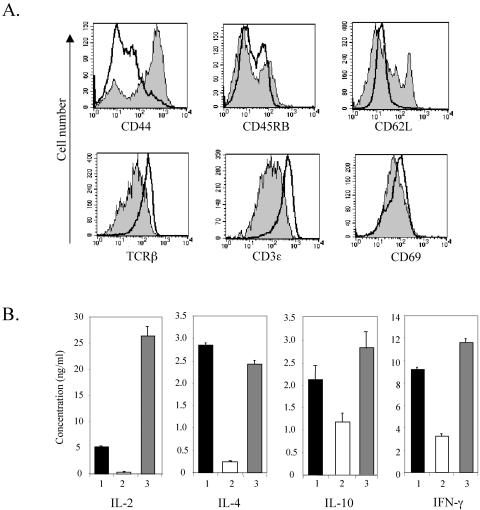
The ability of Id1 transgenic CD4+ cells to proliferate independently of costimulation resembles the behavior of memory T cells. Memory T cells usually proliferate and secrete a broad spectrum of cytokines in response to lower doses of peptides recognized by TCR and in the absence of costimulation (13). Murine memory T cells are identified by surface markers as CD44+ CD45RBlo CD62L− CD69lo. To determine whether the CD4+ thymocytes in Id1 transgenic mice represent a population of memory T cells, we analyzed the surface phenotype of these cells by flow cytometry. Gated CD4+ thymocytes from wild-type and transgenic mice were examined for expression of each of the memory T-cell markers individually (Fig. 5A). Although the majority of CD4+ cells were CD44+, no distinct subpopulation of these cells displayed lower levels of CD45RB, CD62L, and CD69. Therefore, it is unlikely that memory T cells predominate in the Id1 transgenic CD4+ population. Interestingly, levels of TCRβ and CD3ɛ were significantly lower in Id1 transgenic cells than wild-type cells (Fig. 5A), even though the transgenic cells responded more avidly to TCR stimulation (Fig. 4). The lower levels of TCR and CD3 on Id1 transgenic CD4+ cells may be explained by an increase in receptor internalization or by selection against higher level of TCR expression, both of which could result from a stronger response to TCR signaling due to Id1 expression.
FIG. 5.
CD4+ thymocytes from Id1 transgenic mice do not display the phenotype of memory T cells. (A) Surface marker expression in CD4+ thymocytes. Thymocytes from wild-type and Id1 transgenic mice were stained with anti-CD4 and anti-CD8 plus one of the antibodies specific for the indicated antigens. The levels of the indicated surface antigens on gated CD4 SP thymocytes are shown in histograms. Solid lines represent wild-type thymocytes, and shaded areas designate Id1 transgenic thymocytes. (B) Cytokine secretion by activated CD4+ thymocytes. CD4+ thymocytes (2 × 105 cells per well) were stimulated for 48 h with plate-bound anti-CD3ɛ (10 μg/ml) with or without soluble anti-CD28 monoclonal antibody (2 μg/ml). Culture media were collected and analyzed for the indicated cytokines by enzyme-linked immunosorbent assay. Bars: 1, wild-type CD4+ thymocytes stimulated with anti-CD3ɛ and anti-CD28; 2, Id1 transgenic CD4+ thymocytes stimulated with anti-CD3ɛ alone; 3, Id1 transgenic CD4+ thymocytes stimulated with anti-CD3ɛ and anti-CD28.
Further distinction of the Id1 transgenic CD4+ cells from memory T cells came from the analysis of cytokine production by proliferating CD4+ cells. Treatment of Id1 transgenic thymocytes with anti-CD3 alone did not produce significant amounts of interleukin-2 (IL-2) and IL-4 cytokines typically secreted by memory T cells or cells treated with both anti-CD3 and anti-CD28 antibodies (Fig. 5B). There were low levels of IL-10 and gamma interferon production in Id1 transgenic CD4+ cells treated with anti-CD3, but these cytokines are not known to stimulate proliferation (41).
NF-κB activation contributes to costimulation-independent proliferation of Id1 transgenic thymocytes.
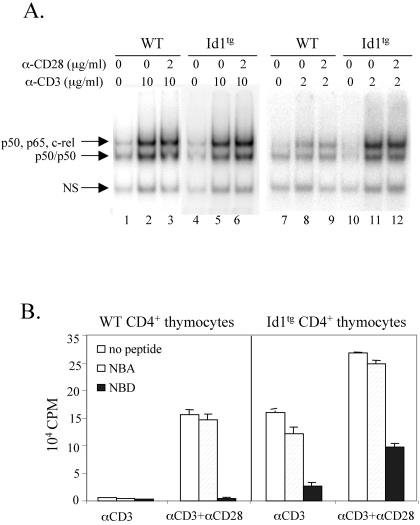
Freshly isolated total thymocytes from Id1 transgenic mice have dramatically increased NF-κB activities (24). Since CD28-mediated signaling is thought to cause activation of NF-κB (21), we examined NF-κB DNA-binding activity in these CD4+ thymocytes cultured in vitro. At 24 h after treatment with or without anti-CD3 or anti-CD3 plus anti-CD28, nuclear extracts were prepared and electrophoretic mobility shift assays (EMSAs) were performed. NF-κB activities in both wild-type and Id1 transgenic cells were very low without any treatment but increased significantly upon stimulation with 10 μg of anti-CD3/ml alone and further increased slightly with anti-CD3 plus anti-CD28 (Fig. 6A, lanes 1 to 6). The transcriptionally active forms of NF-κB complexes (slower-migrating band) predominantly consist of c-rel or p65 homodimers or heterodimers with p50 as determined by using supershift assays (data not shown). Furthermore, to distinguish the responses of wild-type and Id1 transgenic CD4+ thymocytes to these stimulatory signals, we lowered the concentration of anti-CD3 antibody to 2 μg/ml in these experiments. Indeed, NF-κB activation in wild-type cells was much less dramatic than that in Id1 transgenic cells (Fig. 6A, lanes 7 to 12), suggesting that Id1 expression potentiates NF-κB activation by TCR signaling.
FIG. 6.
NF-κB is necessary but not sufficient for costimulation-independent proliferation. (A) EMSA for NF-κB DNA-binding activities. CD4+ thymocytes (2 × 105 cells per well) were stimulated with indicated concentrations of plate-bound anti-CD3ɛ with or without soluble anti-CD28 for 24 h. Nuclear extracts were prepared from the stimulated and unstimulated thymocytes and used in EMSAs. The indicated subunits present in each of the specific NF-κB binding complexes were identified by supershift assays (data not shown). A ubiquitously expressed nonspecific binding complex (NS) served as a loading control. (B) Inhibition of CD4+ thymocyte proliferation by an inhibitor of NF-κB activation. Proliferation cultures were set up as described for Fig. 5B, except the inhibitor peptide (NBD) or control peptide (NBA) were included at the concentration of 200 μM. [3H]thymidine incorporation was measured as described for Fig. 4A.
We next tested whether proliferation of these CD4+ cells depends on NF-κB activation. A cell-permeable peptide, NBD, was shown to inhibit NF-κB activation by competing with the NEMO-binding domains of IKKα and IKKβ for interacting with NEMO (31). Consequently, IKK complexes cannot be activated upon stimulation, and IκB molecules cannot be phosphorylated and signaled for degradation in the presence of NBD. The addition of NBD peptides completely abolished the proliferation of wild-type CD4+ cells stimulated with anti-CD3 and anti-CD28 and inhibited the proliferation of Id1 transgenic cells by about 80 and 60% when treated with anti-CD3 alone or both anti-CD3 and anti-CD28, respectively (Fig. 6B). In contrast, a control peptide, NBA, which carries mutations in the residues critical for NEMO binding (31), had no significant effect on the proliferation of wild-type or transgenic cells. This result suggests that NF-κB activity is necessary for T-cell proliferation. It remains to be determined whether the partial inhibition of proliferation in Id1 transgenic cells is due to an incomplete inhibition of NF-κB activity or to the action of additional factors.
Effect of Id1 expression on thymocyte positive selection.
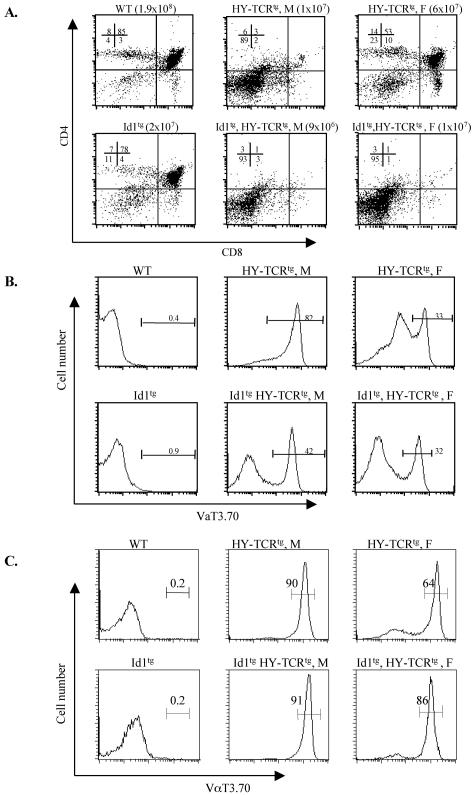
As another measure of TCR signaling in Id1 transgenic mice, we examined the effect of Id1 expression on the negative and positive selection of DP thymocytes. Id1 transgenic mice were crossed with HY-TCR transgenic mice. HY-TCR is specific for a male-specific antigen in the context of class I MHC, H-2Db (25). DP thymocytes are positively selected to become CD8 SP cells in female mice expressing H-2Db and eliminated after negative selection in male mice. In female HY-TCR transgenic mice, CD8+ thymocytes constituted 10% of the total thymocytes compared to 3% of that in wild-type littermates (Fig. 7A). When thymocytes of female HY-TCR transgenic mice were gated on HY-TCR expression by staining with a monoclonal antibody against the α chain of the HY-TCR (Fig. 7B), the percentage of CD8+ cells increased to 20% (data not shown). In contrast, male HY-TCR transgenic mice had few thymocytes beyond the DN stage due to negative selection against the majority of cells bearing the HY-TCR (Fig. 7A and B). Their total cellularity was consequently reduced to ca. 10% of that in wild-type littermates. Interestingly, the thymuses of either male or female HY-TCR transgenic mice carrying the Id1 transgene had an appearance reminiscent of that seen in the male HY-TCR mice, i.e., few thymocytes beyond the DN stage (Fig. 7A). The total number of thymocytes in female transheterozygotes of Id1 and HY-TCR transgenic mice was similar to male HY-TCR transgenic mice. Thus, thymocytes in female Id1tg, HY-TCRtg transheterozygous mice appeared to have undergone negative selection.
FIG. 7.
Effects of Id1 on thymocyte positive selection in H-Y TCR transgenic mice. (A) Total thymocytes from littermates with indicated genotypes were analyzed by fluorescence-activated cell sorting. The numbers shown indicate the percentages of the subsets. Total thymic cellularity is shown in parenthesis. Male (M) and female (F) mice are as labeled. Eleven litters of mice were analyzed and identical results were obtained. The data shown are representative of analyses of one such litter. Histograms of anti-VαT3.70 staining of total thymocytes (B) or the CD3ɛhi population (C) represent HY-TCR expression. The percentage of HY-TCR-positive cells is shown on top of the gate.
Examination of HY-TCR expression in these mice revealed that 82% of male HY-TCR transgenic thymocytes express the HY TCR but only 33% of female transgenic cells carry this TCR (Fig. 7B). This is perhaps due to opportunities of rearranging the endogenous TCRα locus in female HY-TCR transgenic mice, thus resulting in the expression of nontransgenic TCR. Cells expressing a nontransgenic TCR would not be subjected to the same selection as HY-TCR. Because DP thymocytes with HY-TCR in male mice die quickly by negative selection, they do not have the opportunity to rearrange their endogenous TCRα locus. Similarly, Id1 transgenic thymocytes with HY-TCR also undergo apoptosis before they are able to rearrange endogenous TCRα alleles. Although only 42 and 32% of the thymocytes in male and female Id1 and HY-TCR trans-heterozygotes bore the HY-TCR, respectively (Fig. 7B), the remainder of thymocytes was arrested at DN stages prior to the expression of HY-TCR transgenes (data not shown). When the CD3ɛhi population, which consists of TCR-bearing cells, was analyzed for HY-TCR expression with the anti-VαT3.70 antibody, the percentage of HY-TCR-positive cells in female Id1/HY-TCR double-transgenic mice was found to be comparable to that in male HY-TCR transgenic mice. In contrast, a significantly lower percentage was observed in female HY-TCR transgenic mice (Fig. 7C).
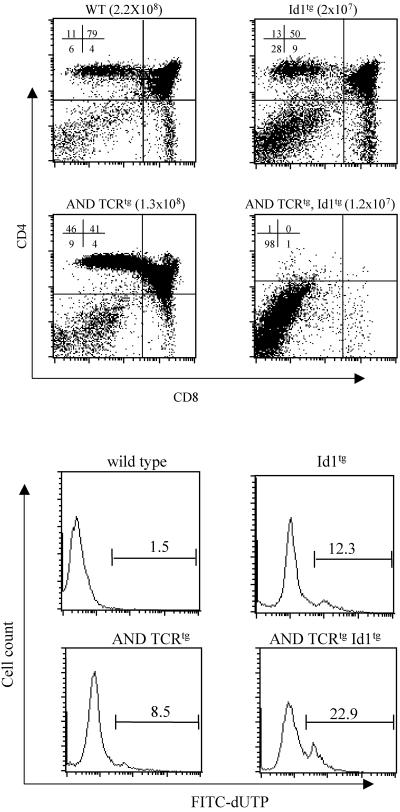
We also examined the effect of Id1 on MHC class II-restricted positive selection by using AND-TCR transgenic mice (22). The AND-TCR recognizes a peptide fragment from pigeon cytochrome c bound to MHC class II-Ek. CD4+ thymocytes are positively selected in AND transgenic mice. Indeed, 46% of total thymocytes were CD4+ in AND transgenic mice, whereas the percentage of CD4+ cells in wild-type mice was 11%. However, when Id1 transgenic mice were crossed with AND-TCR transgenic mice, no thymocytes beyond DN stages were found in transheterozygotes (Fig. 8A). The total cell number in the transheterozygotes is also lower compared to either AND or Id1 transgenic heterozygotes, a finding analogous to the situation in female Id1tg, HY-TCRtg mice. Consistently, examination of apoptosis of these thymocytes revealed that a significant increase in the percentage of apoptotic thymocytes from AND-TCRtg, Id1tg mice (22.9%) compared to that in Id1tg (12.3%) or AND-TCRtg (8.5%) mice (Fig. 8B). Taken together, these results indicate that expression of the Id1 gene turns signals for positive selection into pseudonegative selection, probably by exaggerating TCR signals or responses to the signals.
FIG. 8.
Effect of Id1 on thymocyte positive selection in AND transgenic mice. (A) Thymocytes from littermates with the indicated genotypes were stained with anti-CD4 and anti-CD8 antibodies. The numbers shown indicate the percentages of the subsets. The total thymic cellularity is shown in parentheses. Eight litters of mice were analyzed, and identical results were obtained each time. The data shown are representative of analyses for one such litter. (B) Detection of apoptosis. Total thymocytes from mice with the indicated genotypes were labeled with FITC-conjugated dUTP and analyzed by flow cytometry. The percentage of apoptotic cells is shown on top of the gate.
DISCUSSION
TCR formation in Id1 transgenic T cells.
Complete inhibition of E-protein (E2A and HEB) function in thymocytes of Id1 homozygous transgenic mice results in developmental arrest at the DN1 stage (CD4− CD8− CD44+ CD25−) (23, 24). At this stage, TCR gene rearrangement has not begun. However, partial inhibition of E-protein function in Id1 heterozygous transgenic mice allows a fraction of T cells to mature beyond the DN stage. However, the number of total thymocytes remains very small in Id1 heterozygous mice, and the percentage of DP cells is significantly lower than that of wild-type mice. As demonstrated by a number of assays (23, 24), the reduced cellularity may, at least in part, be attributed to massive apoptosis occurring in the thymus. Examination of TCR gene rearrangement revealed that rearrangements of TCRβ and -α loci in apoptotic thymocytes occurred at similar frequencies as those in viable cells from Id1 transgenic or wild-type mice (23). We have now shown that percentages of productive rearrangements of the TCRβ and -α loci in apoptotic cells are similar as those in wild-type viable cells. These results suggest that Id1 transgenic apoptotic cells contained functional pre-TCR or TCR. The developmental defect seen in Id1 transgenic mice is thus unlikely due to inabilities to rearrange TCR genes, which would be predicted to arrest development at the DN3 stage. Instead, the profound developmental defects of Id1 transgenic mice include accumulation of DN1 cells and dramatic reduction of DP cells. Furthermore, no direct biochemical evidence are presented to demonstrate that these E proteins are involved in TCRβ gene rearrangement, even though E2A and HEB are shown to facilitate the rearrangement of immunoglobulin genes, as well as the TCRγ and -δ loci in lymphoid or nonlymphoid cells (6, 18, 43).
This notion is also consistent with the findings that expression of functionally rearranged TCR transgenes such as the HY- or AND-TCR genes fails to rescue T-cell development in Id1 transgenic mice. On the contrary, expression of the TCR transgenes further impairs T-cell development (Fig. 7 and 8). In addition, Id1 transgenic thymocytes are capable of differentiating to the DP stage in the absence of the RAG1 gene, suggesting Id1 expression at least partially creates an effect analogous to that generated by pre-TCR signaling. This may explain why the viable transgenic thymocytes have an unexpected lower percentage of productive TCRβ rearrangement; Id1 expression allows DN cells to bypass β selection. Furthermore, the absence of a functional TCRβ chain is probably advantageous for the survival of Id1 thymocytes because cells with functional TCR might be more prone to apoptosis due to hyperresponses to TCR signaling.
Impact on T-cell proliferation by Id1 expression.
Activation of naive CD4+ T cells normally depends on signals from the complex between TCR and peptide-MHC molecules, as well as those from the interaction between a costimulatory receptor and its ligand such as CD28 and CD80/CD86. In experimental systems, anti-CD3 treatment fails to trigger T-cell proliferation without costimulation. In contrast, Id1 transgenic CD4+ cells responded vigorously to the anti-CD3 antibody in the absence of costimulation from anti-CD28 or PMA. This remarkable characteristic suggests that expression of Id1 or inhibition of E protein function causes an amplification of TCR signaling such that costimulation is no longer needed for proliferation. Alternatively, Id1 expression in T cells may have an effect similar to that created by costimulation. The costimulation-independent proliferation of Id1 transgenic thymocytes is reminiscent of that seen in c-Cbl/Cbl-b double-knockout splenic CD4+ cells (11, 35). Cbl double-knockout splenic T cells proliferate in the absence of costimulation originating from CD28 or IL-2. In addition, lack of Cbl function leads to IL-2 production upon anti-CD3 treatment without anti-CD28 or even in CD28−/− T cells. It has been suggested that Cbl (particularly Cbl-b) negatively influences CD28-mediated costimulation (11). Spontaneous mutation of the SHP-1 gene in motheaten mice also results in enhanced potential to proliferate and secrete IL-2 by splenic T cells when stimulated with both anti-CD3 and IL-2 (20). In this regard, SHP-1, a protein tyrosine phosphatase, is thought to be responsible for dampening the signals originating from the TCR (38). Unlike Cbl- and SHP-1-deficient cells, Id1 transgenic thymocytes produce a very low level of IL-2 when stimulated with anti-CD3 alone but secrete five times more IL-2 than wild-type thymocytes upon treatment with both anti-CD3 and anti-CD28. Therefore, it appeared that the abilities of Id1 transgenic T cells to proliferate and secrete IL-2 were disconnected. The former does not require anti-CD28 costimulation but the latter does. This situation is similar to costimulation mediated by anti-CD5 or anti-CD9, which facilitates proliferation but not IL-2 production (48). The ability of Id1 transgenic thymocytes to proliferate vigorously is consistent with the finding that DN3 thymocytes in E47-deficient mice, which lack one of the proteins encoded by the E2A gene, are hyperproliferative as indicated by bromodeoxyuridine incorporation compared to their wild-type counterpart. This result suggests that E proteins suppress pre-TCR-stimulated proliferation (16).
The mechanism underlying costimulation-independent proliferation of Id1 transgenic T cells is not understood. We have previously shown that NF-κB transcription factors are dramatically activated in freshly isolated total thymocytes from Id1 transgenic mice (24). Since stimulation through CD28 is thought to cause activation of NF-κB (21), it is possible that activation of NF-κB by Id1 expression could substitute for costimulatory signals. Indeed, a peptide inhibitor (NBD) of NF-κB activation diminished the proliferation of Id1 transgenic CD4+ cells treated with anti-CD3 or both anti-CD3 and anti-CD28. This suggests that activation of NF-κB is necessary for proliferation. However, it is unlikely that costimulation-independent proliferation of Id1 transgenic cells is solely due to hyperactivation of NF-κB for the following two reasons. First, NBD was able to completely block the proliferative response of wild-type CD4+ cells stimulated with anti-CD3 and anti-CD28 but only diminished that of Id1 transgenic cells by ≤80%. Second, stimulation of wild-type cells with anti-CD3 activated NF-κB but failed to trigger their proliferation. Thus, it is possible that other transcription factors may act in concert with NF-κB in stimulating T-cell proliferation. Indeed, our preliminary data suggested that AP-1 transcription factors are also superactivated in Id1 transgenic thymocytes upon anti-CD3 or anti-CD3 plus anti-CD28 stimulation (data not shown). This is consistent with our previous finding in freshly isolated thymocytes where a higher AP-1 DNA-binding activity was detected in Id1 transgenic mice (24).
IL-2 is a cytokine known to facilitate T-cell proliferation in the presence of anti-CD3 (9, 38). We have shown that IL-2 level in anti-CD3-treated Id1 transgenic CD4+ cells is much lower than that in wild-type or Id1 transgenic cells treated with both anti-CD3 and anti-CD28 for 48 h (Fig. 5B). This is probably due to a powerful stimulatory effect by signaling from CD28 (40). However, we also found that the same low level of IL-2 was secreted by anti-CD3-treated Id1 transgenic cells but not by wild-type cells within the first 24 h of stimulation (data not shown). At this time point, cell proliferation had not occurred. Whether this low basal level of IL-2 is enough to support the proliferation of Id1 transgenic cells in the absence of costimulation remains to be investigated. However, it is possible that altered expression of other cytokines also contributes to costimulation-independent proliferation observed in Id1 transgenic thymocytes.
Impact on T-cell selection by Id1 expression.
The fate of CD4+ CD8+ DP thymocytes may be determined by the strength of signals (26). DP thymocytes expressing TCRs that have high affinities for peptide-MHC complex are negatively selected. On the other hand, those with TCRs that have moderate affinities are positively selected and allowed to mature to CD4+ or CD8+ thymocytes. However, Id1 expression appears to lower the threshold of TCR signaling such that signals from TCRs that normally mediate positive selection now cause apoptosis or pseudo-negative selection at the DP stage. The thymuses of female HY-TCR/Id1 transgenic mice resemble those of male HY-TCR transgenic mice. Pseudo-negative selection also appeared to occur in the thymuses of AND/Id1 double transgenic mice.
This effect of Id1 expression on T-cell selection is qualitatively consistent with that created by disruption of the E2A gene. In E2A-deficient mice, enhanced positive selection has been observed in either AND- or HY-TCR transgenic mice (5). Since both E2A and HEB are expressed in the thymus, disruption of either of the two genes does not completely eliminate the collective function of all E proteins. Thus, the effect of E2A deficiency is expected to be less than that caused by the expression of the Id1 protein, which inhibits both E2A and HEB proteins. We thus hypothesize that the moderate reduction in E-protein function due to E2A or HEB deficiency facilitates positive selection by slightly increasing TCR signaling strength, whereas a more complete elimination of E-protein function by Id1 dramatically amplifies TCR signaling and turns signals for positive selection into those for negative selection. It remains to be determined if this is achieved by alteration in the signaling events or by augmentation of the cellular responses to the same signals. One might then ask why then do a small number of DP and SP T cells accumulate in heterozygous Id1 transgenic mice carrying a natural repertoire of TCR receptors? Perhaps, these surviving T cells bear TCRs that generate weak signals that in the absence of Id1 expression would fail to promote positive selection and lead to death by neglect.
Also, the effect of Id1 expression on T-cell selection is consistent with those caused by c-Cbl and SHP-1 deficiencies, which not only lead to hyperproliferative responses of CD4+ T cells but also facilitate positive selection in an HY-TCR transgenic background (36, 39). However, unlike Id1 expression, these deficiencies do not result in negative selection in HY-TCR transgenic mice. This may also be explained by quantitative differences in TCR signaling strength or response to TCR signals between Id1 transgenic mice and c-Cbl- or SHP-1-deficient mice. One indication may come from their proliferative responses. Although it is difficult to make direct comparisons, SHP-1-deficient CD4+ T cells display an enhanced proliferation in a costimulation-dependent manner. Although disruption of the c-Cbl gene alone has no effect on T-cell proliferation, c-Cbl/Cbl-b double-knockout cells do not proliferate as vigorously as Id1 transgenic cells in the absence of costimulation. Nevertheless, it appears that the hyperproliferative responses correlate with enhanced positive selection in these mouse models.
In summary, we have provided several lines of evidence to support the notion that E proteins play an important role in controlling the threshold of TCR signaling. Disruption of E-protein function by Id1 lowers the threshold, thus allowing costimulation-independent proliferation of CD4+ T cells and converting signals for positive selection into those for negative selection. Without proper control of the thresholds of pre-TCR and TCR signaling, developing T cells undergo apoptosis, which could, at least in part, contribute to the T-cell deficiency found in Id1 transgenic mice. The mechanisms by which E proteins control the threshold are not completely understood. Regulation of NF-κB activity may be one of the means but additional controls of cellular factors may also be involved.
Acknowledgments
We thank Dongsoo Kim for preparation of the genomic DNA from Id1 transgenic mice used in this study and Viji Dandapani for excellent assistance in cell sorting. We thank Yuanzheng Yang, Ying Zhao, and Mei-Ying Xiong for technical assistance. We are grateful to Linda Thompson and Darryll Dudley for critical reading of the manuscript.
The study was supported by grants from the National Institutes of Health to X.-H.S. (AI33597, CA77553, and RR15577 [COBRE award]).
REFERENCES
- 1.Acuto, O., and F. Michel. 2003. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 3:939-951. [DOI] [PubMed] [Google Scholar]
- 2.Alberola-Ila, J., K. A. Hogquist, K. A. Swan, M. J. Bevan, and R. M. Perlmutter. 1996. Positive and negative selection invoke distinct signaling pathways. J. Exp. Med. 184:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain, G., C. B. Cravatt, C. Loomans, J. Alberola-Ila, S. M. Hedrick, and C. Murre. 2001. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras- ERK MAPK cascade. Nat. Immunol. 2:165-171. [DOI] [PubMed] [Google Scholar]
- 4.Bain, G., I. Engel, E. C. Robanus Maandag, H. P. te Riele, J. R. Voland, L. L. Sharp, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, G., M. W. Quong, R. S. Soloff, S. M. Hedrick, and C. Murre. 1999. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 190:1605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bain, G., W. J. Romanow, K. Albers, W. L. Havran, and C. Murre. 1999. Positive and negative regulation of V(D)J recombination by the E2A proteins. J. Exp. Med. 189:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barndt, R., M. F. Dai, and Y. Zhuang. 1999. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J. Immunol. 163:3331-3343. [PubMed] [Google Scholar]
- 8.Barndt, R. J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant-negative mutation of HEB. Mol. Cell. Biol. 20:6677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bash, R. O., S. Hall, C. F. Timmons, W. M. Crist, M. Amylon, R. G. Smith, and R. Baer. 1995. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A pediatric oncology study. Blood 86:666-676. [PubMed] [Google Scholar]
- 10.Casanova, J. L., P. Romero, C. Widmann, P. Kourilsky, and J. L. Maryanski. 1991. T-cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T-cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 174:1371-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, Y. J., H. K. Kole, K. Brown, M. Naramura, S. Fukuhara, R. J. Hu, I. K. Jang, J. S. Gutkind, E. Shevach, and H. Gu. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403:216-220. [DOI] [PubMed] [Google Scholar]
- 12.Dudley, E. C., H. T. Petrie, L. M. Shah, M. J. Owen, and A. C. Hayday. 1994. T-cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity 1:83-93. [DOI] [PubMed] [Google Scholar]
- 13.Dutton, R. W., L. M. Bradley, and S. L. Swain. 1998. T-cell memory. Annu. Rev. Immunol. 16:201-223. [DOI] [PubMed] [Google Scholar]
- 14.Engel, I., C. Johns, G. Bain, R. R. Rivera, and C. Murre. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194:733-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel, I., and C. Murre. 2001. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 16.Engel, I., and C. Murre. 2004. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 23:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehling, H. J., and H. von Boehmer. 1997. Early alpha beta T-cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 9:263-275. [DOI] [PubMed] [Google Scholar]
- 18.Goebel, P., N. Janney, J. R. Valenzuela, W. J. Romanow, C. Murre, and A. J. Feeney. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heemskerk, M. H., B. Blom, K. Oda, A. P. Stegmann, A. Q. Bakker, K. Weijer, P. C. Res, and H. Spits. 1997. Inhibition of T cell and promotion of natural killer cell development by the dominant-negative helix loop helix factor Id3. J. Exp. Med. 186:1597-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, K. G., F. G. LeRoy, L. K. Borysiewicz, and R. J. Matthews. 1999. TCR signaling thresholds regulating T cell development and activation are dependent upon SHP-1. J. Immunol. 162:3802-3813. [PubMed] [Google Scholar]
- 21.Kane, L. P., J. Lin, and A. Weiss. 2002. It's all Rel-ative: NF-κB and CD28 costimulation of T-cell activation. Trends Immunol. 23:413-420. [DOI] [PubMed] [Google Scholar]
- 22.Kaye, J., M. L. Hsu, M. E. Sauron, S. C. Jameson, N. R. Gascoigne, and S. M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341:746-749. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D., X. C. Peng, and X. H. Sun. 1999. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol. Cell. Biol. 19:8240-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D., M. Xu, L. Nie, X. C. Peng, E. Jimi, R. E. Voll, T. Nguyen, S. Ghosh, and X. H. Sun. 2002. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-κB activities. Immunity 16:9-21. [DOI] [PubMed] [Google Scholar]
- 25.Kisielow, P., H. Bluthmann, U. D. Staerz, M. Steinmetz, and H. von Boehmer. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333:742-746. [DOI] [PubMed] [Google Scholar]
- 26.Kisielow, P., and H. von Boehmer. 1990. Negative and positive selection of immature thymocytes: timing and the role of the ligand for alpha beta T cell receptor. Semin. Immunol. 2:35-44. [PubMed] [Google Scholar]
- 27.Kruisbeek, A. M., M. C. Haks, M. Carleton, A. M. Michie, J. C. Zuniga-Pflucker, and D. L. Wiest. 2000. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol. Today 21:637-644. [DOI] [PubMed] [Google Scholar]
- 28.Lafferty, K. J., I. S. Misko, and M. A. Cooley. 1974. Allogeneic stimulation modulates the in vitro response of T cells to transplantation antigen. Nature 249:275-276. [DOI] [PubMed] [Google Scholar]
- 29.Lenschow, D. J., T. L. Walunas, and J. A. Bluestone. 1996. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 14:233-258. [DOI] [PubMed] [Google Scholar]
- 30.Mallick, C. A., E. C. Dudley, J. L. Viney, M. J. Owen, and A. C. Hayday. 1993. Rearrangement and diversity of T cell receptor beta chain genes in thymocytes: a critical role for the beta chain in development. Cell 73:513-519. [DOI] [PubMed] [Google Scholar]
- 31.May, M. J., F. D'Acquisto, L. A. Madge, J. Glockner, J. S. Pober, and S. Ghosh. 2000. Selective inhibition of NF-κB activation by a peptide that blocks the interaction of NEMO with the IκB kinase complex. Science 289:1550-1554. [DOI] [PubMed] [Google Scholar]
- 32.Michie, A. M., and J. C. Zuniga-Pflucker. 2002. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin. Immunol. 14:311-323. [DOI] [PubMed] [Google Scholar]
- 33.Morrow, M. A., E. W. Mayer, C. A. Perez, M. Adlam, and G. Siu. 1999. Overexpression of the helix-loop-helix protein Id2 blocks T-cell development at multiple stages. Mol. Immunol. 36:491-503. [DOI] [PubMed] [Google Scholar]
- 34.Murre, C., A. Voronova, and D. Baltimore. 1991. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol. Cell. Biol. 11:1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naramura, M., I. K. Jang, H. Kole, F. Huang, D. Haines, and H. Gu. 2002. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR downmodulation. Nat. Immunol. 3:1192-1199. [DOI] [PubMed] [Google Scholar]
- 36.Naramura, M., H. K. Kole, R. J. Hu, and H. Gu. 1998. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. USA 95:15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nossal, G. J. 1994. Negative selection of lymphocytes. Cell 76:229-239. [DOI] [PubMed] [Google Scholar]
- 38.Plas, D. R., R. Johnson, J. T. Pingel, R. J. Matthews, M. Dalton, G. Roy, A. C. Chan, and M. L. Thomas. 1996. Direct regulation of ZAP-70 by SHP-1 in T-cell antigen receptor signaling. Science 272:1173-1176. [DOI] [PubMed] [Google Scholar]
- 39.Plas, D. R., C. B. Williams, G. J. Kersh, L. S. White, J. M. White, S. Paust, T. Ulyanova, P. M. Allen, and M. L. Thomas. 1999. Cutting edge: the tyrosine phosphatase SHP-1 regulates thymocyte positive selection. J. Immunol. 162:5680-5684. [PubMed] [Google Scholar]
- 40.Powell, J. D., J. A. Ragheb, S. Kitagawa-Sakakida, and R. H. Schwartz. 1998. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol. Rev. 165:287-300. [DOI] [PubMed] [Google Scholar]
- 41.Renauld, J. C. 2003. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol. 3:667-676. [DOI] [PubMed] [Google Scholar]
- 42.Rivera, R. R., C. P. Johns, J. Quan, R. S. Johnson, and C. Murre. 2000. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12:17-26. [DOI] [PubMed] [Google Scholar]
- 43.Romanow, W. J., A. W. Langerak, P. Goebel, I. L. Wolvers-Tettero, J. J. van Dongen, A. J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5:343-353. [DOI] [PubMed] [Google Scholar]
- 44.Ruzinova, M. B., and R. Benezra. 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13:410-418. [DOI] [PubMed] [Google Scholar]
- 45.Uematsu, Y., S. Ryser, Z. Dembic, P. Borgulya, P. Krimpenfort, A. Berns, H. von Boehmer, and M. Steinmetz. 1988. In transgenic mice the introduced functional T-cell receptor beta gene prevents expression of endogenous beta genes. Cell 52:831-841. [DOI] [PubMed] [Google Scholar]
- 46.Voll, R. E., E. Jimi, R. J. Phillips, D. F. Barber, M. Rincon, A. C. Hayday, R. A. Flavell, and S. Ghosh. 2000. NF-κB activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity 13:677-689. [DOI] [PubMed] [Google Scholar]
- 47.von Boehmer, H. 1994. Positive selection of lymphocytes. Cell 76:219-228. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, X. Y., Y. Yashiro-Ohtani, M. Nakahira, W. R. Park, R. Abe, T. Hamaoka, M. Naramura, H. Gu, and H. Fujiwara. 2002. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. J. Immunol. 168:3847-3854. [DOI] [PubMed] [Google Scholar]