Abstract
Although Jun upregulation and activation have been established as critical to oncogenesis, the relevant downstream pathways remain incompletely characterized. In this study, we found that c-Jun blocks erythroid differentiation in primary human hematopoietic progenitors and, correspondingly, that Jun factors block transcriptional activation by GATA-1, the central regulator of erythroid differentiation. Mutagenesis of c-Jun suggested that its repression of GATA-1 occurs through a transcriptional mechanism involving activation of downstream genes. We identified the hairy-enhancer-of-split-related factor HERP2 as a novel gene upregulated by c-Jun. HERP2 showed physical interaction with GATA-1 and repressed GATA-1 transcriptional activation. Furthermore, transduction of HERP2 into primary human hematopoietic progenitors inhibited erythroid differentiation. These results thus define a novel regulatory pathway linking the transcription factors c-Jun, HERP2, and GATA-1. Furthermore, these results establish a connection between the Notch signaling pathway, of which the HERP factors are a critical component, and the GATA family, which participates in programming of cellular differentiation.
Aberrant overexpression or activation of Jun factors has been detected in malignancies of the liver, breast, colon, endometrium, ovaries, erythroblasts, cutaneous T cells, and hematopoietic stem cells (3, 4, 7, 12, 18, 39, 44, 46, 63, 69, 70). The upregulation or activation of Jun factors in most of these malignancies occurs as a consequence of dysregulation of the Ras-mitogen-activated protein kinase pathway, one of the most common targets in oncogenic transformation (60). However, recent findings indicate frequent genomic amplification of JunB in cutaneous T-cell lymphomas (39).
The phenotypic consequences of Jun overexpression include alterations in cellular proliferation, survival, and differentiation. In the case of proliferation, c-Jun has been shown to be critical for progression of murine embryonic fibroblasts through the G1 phase of the cell cycle, a function that appears not to depend on phosphorylation via Jun N-terminal kinase (JNK) (68). Heterodimers of c-Jun with ATF2 confer growth factor-independent proliferation on chicken embryo fibroblasts, while heterodimers of c-Jun with Fos confer anchorage-independent proliferation (58). In the case of cell survival, inducible gene deletion in murine hepatocytes demonstrates a key c-Jun-mediated survival function both in the setting of chemical carcinogenesis and in response to tumor necrosis factor alpha treatment (12). In addition, c-Jun protects murine embryonic fibroblasts from UV-induced cell death, a function that is dependent on JNK phosphorylation (68). In the case of differentiation, all members of the Jun family (c-Jun, JunB, and JunD) prevent the differentiation of murine erythroleukemic cells (44). Conversely, downregulation of endogenous c-Jun with antisense oligonucleotides renders differentiation-resistant murine erythroleukemic cells competent for erythroid differentiation (18). Similarly, in human erythroleukemic cells, antagonism of AP-1 with a dominant negative c-Jun mutant enhances erythroid differentiation (49). Jun factors also probably participate in the inhibition of erythroid differentiation by the constitutively active Ras 12V mutant in the erythropoietin-responsive murine erythroleukemic line SKT6 (40).
c-Jun exerts such pleiotropic effects in most cases through a transcriptional regulatory mechanism, that is, coordinated control of direct and indirect downstream target genes. One of the relevant target genes mediating the cell cycle effects of c-Jun is cyclin D1 (52). Transcriptional upregulation of cyclin D1 by c-Jun is required for progression through G1 by murine embryonic fibroblasts and also plays a role in the proliferative phenotype of human breast cancer cells (68, 69). A relevant target gene for some of the antiapoptotic effects of c-Jun is p53 (52). c-Jun repression of p53 protects murine hepatocytes from apoptosis during chemical carcinogenesis and during tumor necrosis factor alpha treatment (12).
The downstream genes mediating Jun inhibition of cellular differentiation have not yet been identified. In this study, we found that c-Jun blocked the erythroid differentiation of primary human hematopoietic progenitors and correspondingly repressed the function of the key erythroid transcription factor GATA-1. Overexpression of GATA-1 reversed the inhibition of erythroid differentiation by c-Jun. GATA-1 normally functions to program terminal erythroid differentiation, including coordinated upregulation of erythroid-specific genes and downregulation of cellular proliferation (53, 56). Mutational analysis of Jun factors indicated that repression of GATA-1 correlated with the ability of c-Jun to mediate transcriptional activation. In particular, dominant negative mutants of c-Jun potentiated rather than inhibited the function of GATA-1.
Through microarray analysis followed by functional screening, we identified the basic helix-loop-helix (bHLH) protein HERP2 as a factor upregulated by wild-type c-Jun and capable of GATA-1 repression. In addition to blocking function, HERP-2 showed physical interaction with GATA-1. Enforced expression of HERP2 inhibited erythroid differentiation of K562 cells and of primary human hematopoietic progenitors. These results therefore identify a novel pathway for Jun-mediated inhibition of cellular differentiation, in which c-Jun causes upregulation of the Notch effector HERP2, which in turn directly inhibits the erythroid factor GATA-1. This pathway may have relevance to Jun transformation of several hematopoietic cell types, such as early progenitors, erythro-megakaryocytic progenitors, and T cells, in which GATA factor function plays a critical role in cellular development. In addition, these results describe a novel mechanism for potential regulation of GATA factor function by the Notch signaling pathway.
MATERIALS AND METHODS
Plasmid constructs.
The expression vectors for Flag-tagged HERP2 and HERP1 were kindly provided by Larry Kedes and Tatsuya Iso (31). The expression vector for the HERP2 mutant lacking the bHLH domain (HERP2 ΔbHLH) was made by subcloning a PstI-HindIII fragment into pCMV-Tag2C. Expression constructs for wild-type GATA-1 and GATA-2 as well as for GATA-1 mutants employed the pEF vector and were generously supplied by Jane Visvader (59). The GATA-1 mutant lacking both zinc fingers (Δ200-290) was generated by overlap PCR followed by subcloning into pCMV5; for a positive control, PCR-derived wild-type, full-length GATA-1 was similarly subcloned into pCMV5. pcDNA3-Epas-1 was generously contributed by Steven McKnight and Richard Bruick (57). pCMV-cJunwt, pCMV-cJunΔTAD, pCMV-cJun-DBD-Cla linker, and pCMV-cJunΔLZ have been described previously (54). The GATA-responsive luciferase reporter plasmids αIIb-598 and αIIb-98 have also been described previously (45).
c-Jun retroviral constructs were derived from PCR fragments of full-length c-Jun, ΔTAD c-Jun (c-Jun mutant lacking a transcription activation domain), and cJun-DBD-Cla linker subcloned into EcoRI and XhoI sites of MSCV-IRES-GFP (43). The HERP2 retroviral construct was generated by subcloning an EcoRI-BamHI fragment of HERP2, with a blunted BamHI end, into the EcoRI and XhoI sites of MSCV-IRES-GFP, with a blunted XhoI end. The retroviral construct for stable, blasticidin S-selectable c-Jun expression in K562 cells, pLRT-c-Jun, has been described previously (36, 64). The full-length GATA-1 retroviral construct was generated by subcloning an XhoI fragment into the corresponding site of MSCV-IRES-GFP. Replacement of c-Jun TAD with VP16 TAD was accomplished by subcloning a PCR fragment of c-Jun ΔTAD into the NcoI and XbaI sites of pVP-HA2, a mammalian expression vector for VP16 fusions kindly provided by Richard Baer (27). For mammalian expression of glutathione S-transferase (GST)-GATA-1, full-length GATA-1 was ligated into the BamHI and NotI sites of pEBG, as described (14). The bHLH (amino acids 1 to 111) and Orange (amino acids 113 to 175) domains of HERP2 were subcloned as BamHI-blunt and BamHI-KpnI fragments, respectively, into pEBG. The short interfering RNA (siRNA) expression construct for HERP2 was assembled by inserting 5′-GGATCCCGTAATTGAGAAGCGCCGACGTTCAAGAGACGTCGGCGCTTCTCAATTATTTTTTCCAAAAGCTT-3′ (italics show the sense and antisense sequences of human HERP2 nucleotides 167 to 185) into the BglII and HindIII sites of pSUPER.
Retroviral transduction and cell culture.
Human CD34+ cells at greater than 98% purity were derived at the National Hematopoietic Cell Processing Core (NIH grant HL 66947) directed by Shelly Heimfeld (Fred Hutchinson Cancer Research Center). In brief, granulocyte colony-stimulating factor-mobilized peripheral blood mononuclear cells from normal donors underwent purification with CliniMACS magnetic beads (Miltenyi Biotec, Auburn, Calif.). All experiments with human cells were approved by the University of Virginia Human Investigations Committee. Prior to retroviral transduction, the CD34+ cells underwent stimulation for 48 h in serum-free medium consisting of Iscove's modified Dulbecco's medium supplemented with BIT 9500 (Stem Cell Technologies, Vancouver, Canada) with the following cytokines added: stem cell factor at 100 ng/ml, thrombopoietin at 100 ng/ml, FLT3 ligand at 100 ng/ml, and interleukin-3 at 20 ng/ml (all from Peprotech, Rocky Hill, N.J.). Supernatants containing retroviral particles, derived from transient transfection of Phoenix and FLYRD18 packaging cells (20, 32), were used to load retronectin-coated plates (PanVera, Madison, Wis.).
Cells resuspended in retroviral supernatants supplemented with stem cell factor, thrombopoietin, FLT3 ligand, and interleukin-3 at prestimulation concentrations were then seeded onto the preloaded plates. Following supplementation of medium with 5 μg of protamine (Sigma, St Louis, Mo.) per ml, cells were subjected to spinoculation at 2,000 rpm for 90 min at room temperature. After an additional 2.5 h of incubation, the cells were supplemented with an equal volume of fresh retroviral supernatant with cytokines, followed by overnight incubation. This process was repeated 2 additional days before transferring cells to erythroid differentiation medium consisting of serum-free medium with 3 U of human erythropoietin (Amgen Inc., Thousand Oaks, Calif.) per ml and 25 ng of stem cell factor per ml. Transduction of K562 cells followed by purification of green fluorescent protein (GFP)-positive cells by flow sorting for two rounds was performed as previously described (14). K562 cells transduced with pLRT-c-Jun were grown in RPMI 1640 with 10% fetal bovine serum, 8 μg of blasticidin S per ml, and 1 μg of doxycycline per ml. These cells were superinfected with either the MSCV-IRES-GFP parent vector or the MSCV-GATA-1-IRES-GFP vector, followed by flow sorting for GFP-positive cells.
Flow cytometry and benzidine staining.
Erythroid differentiation of transduced cells was assessed by staining cells with allophycocyanine-conjugated anti- glycophorin A (GPA) or with phycoerythrin-conjugated anti-GPA (for costaining with allophycocyanine-anti-CD34 or allophycocyanine-anti-CD41) or with appropriately conjugated isotype-matched control antibody, as previously described (11) (BD Pharmingen, San Diego, Calif.). In the experiments in Fig. 9C, cells were costained for granulocytic, immaturity, and megakaryocytic markers with phycoerythrin-anti-CD13, allophycocyanine-anti-CD34, and allophycocyanine-anti-CD41, respectively, or conjugated isotype-matched control antibodies (BD Pharmingen). Cells were analyzed on a FACSCalibur instrument (Becton Dickinson, San Jose, Calif.) with FCSPress software (FCSPress, Cambridge, United Kingdom) for Fig. 1A, 1B, and 9B or FlowJo software (Treestar, Inc., Ashland, Oreg.) for Fig. 9C. Gating for GFP-positive cells was based on the use of control, nontransduced parallel cultures, and gating for antibody staining was based on isotype-matched control results.
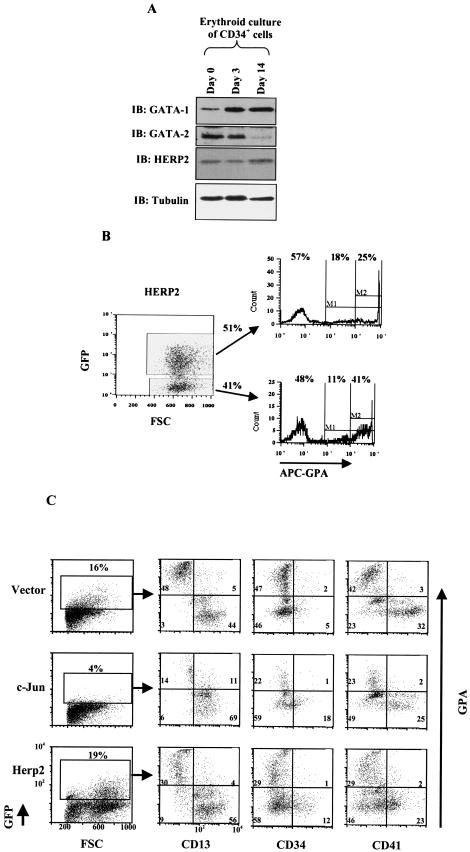
FIG.9.
Expression and function of HERP2 in erythroid differentiation of primary human CD34+ hematopoietic progenitor cells. (A) Expression of HERP2, GATA-1, and GATA-2 proteins in CD34+ cells cultured in erythroid medium for the indicated durations. Whole-cell lysates were analyzed by immunoblotting with the indicated antibodies. (B) HERP2-transduced CD34+ cells show impaired erythroid differentiation. Cells transduced with MSCV-HERP2-IRES-GFP were cultured in erythroid medium for 7 days, followed by flow cytometric analysis for the erythroid marker glycophorin A (GPA), as in Fig. 1. Results from parallel transduction with the control vector are shown in Fig. 1A. Similar results were obtained from two independent experiments. (C) c-Jun and HERP2 both interfere with erythroid maturation. CD34+ cells transduced with constructs encoding the vector, c-Jun, or HERP2 were cultured in erythroid medium for 7 days, followed by flow cytometric analysis with costaining for GPA plus CD13 (granulocytic lineage marker), GPA plus CD34 (immaturity marker), or GPA plus CD41 (megakaryocyte-associated antigen).
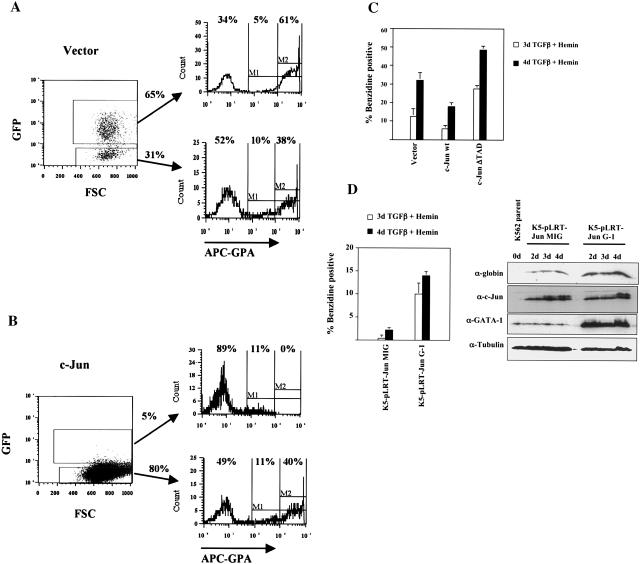
FIG. 1.
Expression of c-Jun inhibits erythroid differentiation of primary human CD34+ hematopoietic progenitors and of K562 cells. (A) Purified human CD34+ hematopoietic progenitor cells transduced with the control vector (MSCV-IRES-GFP) were cultured in erythroid medium for 7 days, followed by flow cytometric analysis for the erythroid marker glycophorin A (GPA). As shown, GFP-positive (transduced) and GFP-negative (nontransduced) populations were separately gated for analysis. FSC, forward angle light scatter. (B) CD34+ cells transduced with the c-Jun-encoding retrovirus (MSCV-c-Jun-IRES-GFP) were cultured in erythroid medium and analyzed for GPA expression as in A. (C) K562 cells transduced with the indicated retroviral constructs were subjected to erythroid induction as indicated, followed by benzidine staining for hemoglobin. Results represent the mean of three determinations ± standard error of the mean. (D) Overexpression of GATA-1 reverses c-Jun inhibition of erythroid differentiation. K562 cells stably overexpressing c-Jun were superinfected with either the control vector (K5-pLRT-Jun MIG) or a GATA-1-encoding retroviral vector (K5-pLRT-Jun G-1). Erythroid differentiation was induced with transforming growth factor β (TGFβ) plus hemin for the indicated durations, followed by either benzidine staining or immunoblot analysis of whole-cell lysates.
Benzidine staining of retrovirally transduced K562 cells induced to undergo erythroid differentiation with hemin (60 μM) (Sigma) plus transforming growth factor β (0.5 ng/ml) (R&D Systems, Minneapolis, Minn.) followed an established protocol (41). Cells showing either dim or bright benzidine staining were scored as positive by standard light microscopy.
Transient transfection and reporter assays.
K562 cells were cotransfected with the indicated expression constructs, luciferase reporters, and pCMV-βGal exactly as previously described (14). At 40 h posttransfection, cells were harvested for luciferase and β-galactosidase assays as described previously (14). In all cases, the results represent the mean ± standard error of the mean derived from three independent transfections, with use of β-galactosidase results to normalize for transfection efficiency. In addition, duplicate transfections were subjected to whole-cell lysis for immunoblot determination of protein expression levels. Trichostatin A (EMD Biosciences, San Diego, Calif.) was employed at a final concentration of 500 nM and compared with control cells treated with vehicle only (0.05% dimethyl sulfoxide).
Immunoblot and Northern blot analysis.
Immunoblot analysis of K562 whole-cell lysates was conducted as previously described (11, 14). GATA-1 was detected with rat monoclonal anti-GATA-1 (N6; Santa Cruz Biotechnology, Santa Cruz, Calif.). GATA-2 was detected with rabbit polyclonal anti-GATA-2 (SC-9008; Santa Cruz Biotechnology). Jun factors were detected with polyclonal rabbit anti-Jun (SC-44; Santa Cruz Biotechnology) (Fig. 4) or with polyclonal rabbit anti-c-Jun (9162; Cell Signaling, Beverly, Mass,) (Fig. 1D). The Flag epitope tag was detected with the M2 mouse monoclonal antibody (Sigma). Human HERP2 was detected with polyclonal rabbit anti-Hey1 (Abcam, Cambridge, Mass.). Tubulin and globin were detected as previously described (14).
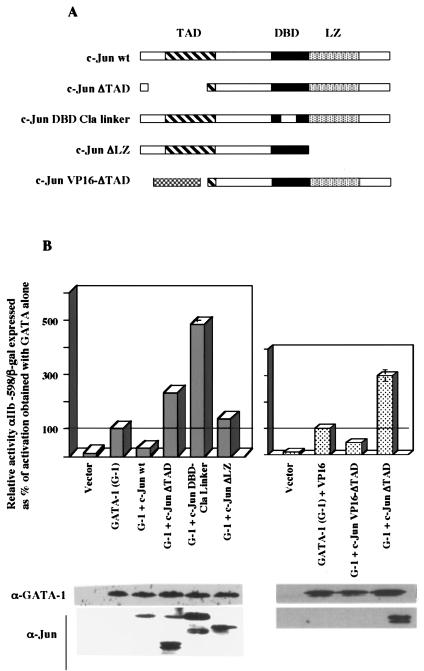
FIG. 4.
Structural requirements for Jun-mediated inhibition of GATA-1; dominant negative c-Jun mutants potentiate GATA-1 function. (A) Diagrams of c-Jun mutants. TAD, transcription activation domain; DBD, DNA binding domain; LZ, leucine zipper. The VP16 ΔTAD mutant contains the herpes simplex virus VP16 TAD in place of the c-Jun TAD. (B) Analysis of transcriptional activation by GATA-1 with and without the indicated c-Jun mutants. Results ± standard error of the mean of three experiments are shown as relative reporter activity compared with activation by GATA-1 alone, normalized by β-galactosidase expression. Whole-cell lysates from a duplicate transfection were immunoblotted for GATA-1 or Jun factors.
Northern blot analysis of transduced K562 cells was carried out essentially as described previously (21); 20 μg of glyoxalated total cellular RNA, isolated with Trizol (Gibco-BRL, Carlsbad, Calif.), per sample was fractionated on 1.0% agarose gels, transferred to Zeta probe nylon membranes (Bio-Rad, Hercules, Calif.), and hybridized with the indicated 32P-labeled probes. Probes for HERP2 and EPAS-1 consisted of full-length cDNA inserts. The glyceraldehyde-3-phosphate dehydrogenase probe has been described (28).
Microarray analysis.
Total cellular RNA was extracted with a Qiagen RNeasy kit (Qiagen, Valencia, Calif.) from duplicate samples of flow-sorted K562 cells transduced with the following retroviral constructs: parent, wild-type c-Jun, c-Jun ΔTAD, and c-Jun DNA-binding domain (DBD)-Cla linker. Following synthesis of double-stranded cDNA, biotinylated antisense cRNA probes were transcribed in vitro. Fragmented cRNA probes were hybridized to the Affymetrix GeneChip human genome U133A and U133B arrays (Affymetrix, Santa Clara, Calif.), representing a total of ≈33,000 distinct genes, with the Affymetrix fluidics station. Fluorescence image analysis employed the Agilent GeneArray scanner in conjunction with Microarray Suite 5.0 software (Affymetrix).
Protein interaction assays.
Coimmunoprecipitations and mammalian GST pulldown assays were performed as recently detailed (14). For coimmunoprecipitation assays, extracts from transiently cotransfected 293T cells were precleared and incubated with M2 anti-Flag antibody. Immune complexes collected on protein G-agarose beads were washed and analyzed by immunoblotting for coprecipitated GATA-2 or GATA-1. For GST pulldown assays, extracts were incubated with glutathione-agarose beads to capture GST fusions, followed by extensive washing and immunoblot analysis for copurified proteins. For coimmunoprecipitation of endogenous GATA-1 and HERP2, 10 μg of either normal rat immunoglobulin G (Santa Cruz Biotechnology) or N6 rat anti-GATA-1 was added to 2 ml of K562 cellular extracts containing 12 mg of protein, followed by incubation overnight at 4°C. Immune complexes collected on Ultralink protein G beads (Pierce Biotechnology, Rockford, Ill.) were subjected to extensive washing and immunoblot detection with 40% of the immunoprecipitation sample loaded in each lane. Extraction and washing buffers both consisted of 50 mM Tris-HCl (pH 7.6), 200 mM Na Cl, 0.5% NP-40, 1 mM MgCl2, 50 μM Zn SO4, 10 mM NaF, 1 mM Na3VO4, and 1 tab of protease inhibitor cocktail (Roche, Indianapolis, Ind.) per 10 ml.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assay analysis of GATA-1 binding to DNA was performed essentially as described previously (10). The wild-type GATA binding site consisted of 5′-GATCTCCGGCAACTGATAAGGATTCCCTG-3′ as described before (10), and the mutant GATA binding site substituted C at the position of the italic G. Nuclear extracts from K562 cells transduced with parent vector or with HERP2-encoding retrovirus were isolated by the method of Andrews and Faller (2). For the results shown in Fig. 8D, 3 μg of nuclear extracts was incubated with 5 fmol of gel-purified double-stranded 32P-labeled probe (42), in a final volume of 20 μl. Binding reactions occurred on ice, with addition of probe subsequent to a 5-min preincubation. Unlabeled competitors were added 15 min after addition of probe, followed by an additional 30 min of incubation. The binding buffer resembled that of Crossley et al. except that KCl was used in place of potassium glutamate (10).
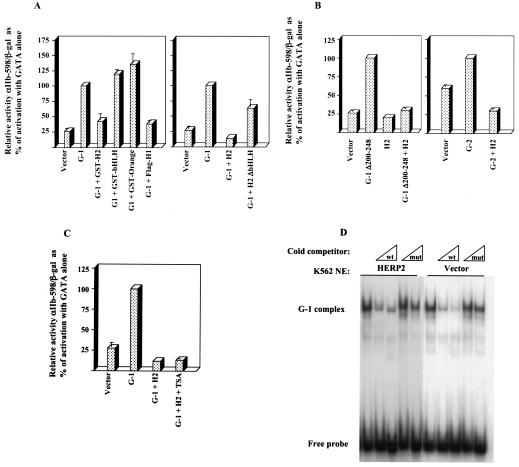
FIG. 8.
Characterization of the HERP2 repressive function. (A) Effects of various HERP factors on GATA-1 function. GST fusions contained full-length HERP2 (GST-H2) or isolated bHLH or orange domains. Also shown are results with Flag-HERP1 (Flag-H1) and a HERP2 truncation mutant lacking the bHLH domain (H2 ΔbHLH). Reporter assays were carried out as in Fig. 2. (B) HERP2 repression of GATA-1 lacking the N-finger (G-1 Δ200-248) and of GATA-2 (G-2). Reporter assays were carried out as in Fig. 2. (C) Histone deacetylase-independent repression of GATA-1 by HERP2. Transfectants were incubated with either vehicle control (0.05% dimethyl sulfoxide) or 500 nM trichostatin A (TSA). Reporter assays were carried out as in Fig. 2. (D) Lack of HERP2 influence on GATA-1 DNA binding. The electrophoretic mobility shift assay employed nuclear extracts (NE) from K562 cells transduced with either HERP2-encoding retrovirus or parent vector. Cells were treated for 48 h with 0.5 ng of transforming growth factor β per ml plus 60 μM hemin prior to harvest.
RESULTS
c-Jun blocks erythroid differentiation of both normal and leukemic human progenitor cells.
Previous work has demonstrated Jun-mediated inhibition of erythroid differentiation in Friend virus-transformed murine erythroleukemic cells (44). To ascertain the relevance of these findings to normal human erythroid differentiation, we examined the effects of enforced c-Jun expression in primary human hematopoietic progenitor cells undergoing erythroid differentiation. Purified human CD34+ progenitors cultured for 5 to 7 days in the presence of erythropoietin and stem cell factor show strong upregulation of the erythroid-specific marker glycophorin A (GPA) (55). We therefore transduced CD34+ cells with a control retrovirus vector (MSCV-IRES-GFP) or with a retrovirus encoding wild-type c-Jun (MSCV-cJun-IRES-GFP). After 7 days of culture in medium with erythropoietin plus stem cell factor, GFP-positive cells were analyzed for GPA expression by flow cytometry. As shown in Fig. 1A, 61% of vector-transduced cells were GPAbright (i.e., expressed high levels of GPA), characteristic of erythroid differentiation. By contrast, virtually none of the c-Jun-transduced cells were GPAbright (Fig. 1B). GFP-negative nontransduced cells in both cases showed similar expression of GPA, with ≈40% bright cells. The lower transduction efficiency associated with c-Jun was highly reproducible (seen in >6 different experiments), as was the inhibition of erythroid differentiation. The paucity of GFP-bright cells in Fig. 1B most likely reflects the inability of the CD34+ cells to tolerate c-Jun levels outside of a narrow range.
To determine whether c-Jun could similarly inhibit the differentiation of human erythroleukemic cells, K562 cells were transduced with constructs encoding the control vector, wild-type c-Jun, or a c-Jun mutant lacking a transcription activation domain (ΔTAD). After erythroid induction with transforming growth factor β plus hemin for 3 and 4 days, hemoglobin expression was quantitated by benzidine staining of cells. As shown in Fig. 1C, cells expressing wild-type c-Jun showed a ≈50% reduction in benzidine staining compared with control cells. Cells expressing c-Jun ΔTAD, on the other hand, showed significant enhancement of benzidine staining compared with control cells. These results confirm the ability of c-Jun to inhibit erythroid differentiation and show that a dominant negative mutant of c-Jun enhances erythroid differentiation.
The transcription factor GATA-1 functions as a central regulatory element in programming erythroid differentiation (8). We therefore examined whether overexpression of GATA-1 could reverse the inhibitory effects of c-Jun on erythroid differentiation. In Fig. 1D, control K562 cells overexpressing c-Jun (K562-pLRT-Jun MIG) showed minimal capacity for erythroid differentiation, as assessed by benzidine staining and globin expression at various points during induction. By contrast, GATA-1-transduced K562 cells overexpressing c-Jun (K562-pLRT-Jun G-1) showed recovery of erythroid differentiation in response to transforming growth factor β plus hemin. These results suggest that GATA-1 may serve as a target for the inhibitory effects of c-Jun on erythroid differentiation.
Jun factors repress the transcriptional function of GATA factors.
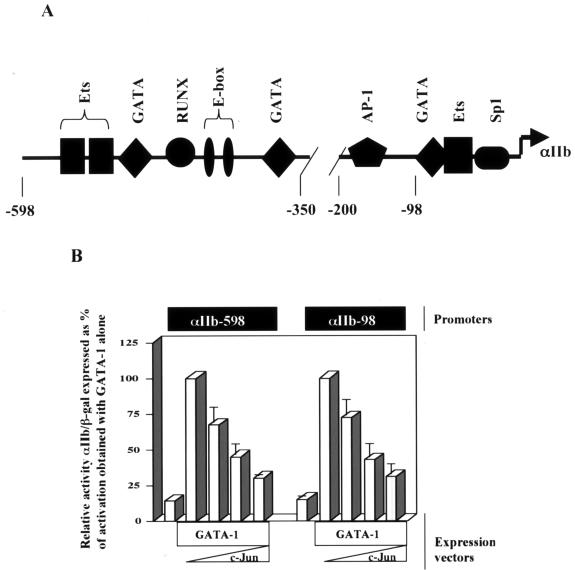
We examined the effects of c-Jun on the ability of GATA-1 to activate a GATA-responsive reporter construct in human erythroleukemic cells. The GATA-responsive αIIb promoter, with key transcription factor binding sites, is shown in Fig. 2A. c-Jun displayed a dose-dependent ability to repress GATA-1 transactivation of the αIIb promoter (−598); use of a minimized GATA-responsive reporter (−98) lacking any recognizable AP-1 binding sites demonstrated that the repressive effect did not require DNA binding by c-Jun adjacent to GATA-1 (Fig. 2B).
FIG. 2.
c-Jun inhibits transcriptional activation by GATA-1. (A) Map of the GATA-responsive αIIb promoter, showing positions of binding sites based on use of the TESS and TFSearch programs as previously described (14). (B) GATA-1 repression by c-Jun does not depend on cis-acting AP-1 sites. The potency of Jun-mediated repression of GATA-1 transcriptional activation was compared using two GATA-responsive reporter constructs, one possessing an AP-1 binding site (αIIb-598) and one lacking an AP-1 binding site (αIIb-98), as depicted in the diagram in A; 2 μg of the GATA-1 expression construct (pEF-GATA-1) was cotransfected with 0.2, 0.5, or 1.0 μg of the c-Jun expression construct (RSV-c-Jun). Results ± standard error of the mean of three experiments are shown as relative reporter activity compared with activation by GATA-1 alone, normalized to β-galactosidase expression.
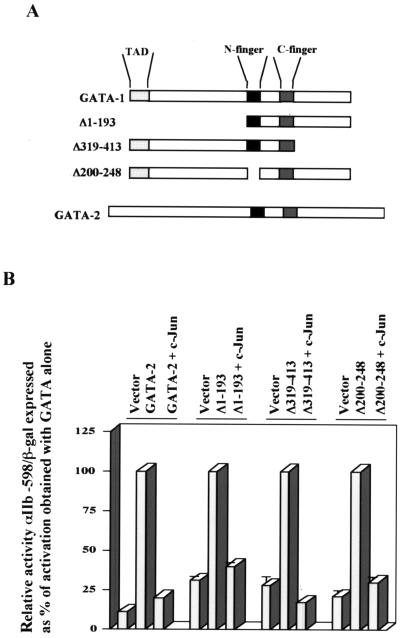
To determine whether specific domains within GATA-1 were targeted by Jun repression, GATA-1 deletion mutants as well as wild-type GATA-2 were analyzed (Fig. 3A). Notably, FOG1, a zinc finger cofactor of GATA proteins, can repress GATA-1 function through interaction with the N-terminal GATA finger (GATA-1 amino acids 200 to 248) (17). The results in Fig. 3B show that c-Jun retains repressive capacity for GATA-2 as well as for the GATA-1 deletion mutants, including GATA-1 Δ200-248. Similar results were obtained with GATA-1 Δ200-248 in the context of the minimized GATA-responsive reporter αIIb-98 (not shown). Therefore, Jun targets a structure that is conserved in GATA-1 and GATA-2 but most likely does not act through a FOG-dependent pathway.
FIG. 3.
c-Jun-mediated transcriptional repression operates through a FOG-independent mechanism. (A) Diagrams of GATA-1, GATA-2, and GATA-1 deletion mutants. (B) Analysis of transcriptional activation by the indicated GATA factors with and without c-Jun. Results ± standard error of the mean of three experiments are shown as relative reporter activity compared with activation by GATA-1 alone, normalized by β- galactosidase expression. For each GATA factor, the activation obtained with that factor alone was set at 100%.
Jun repression of GATA depends on the transcriptional activation functions of c-Jun.
To determine further structural requirements, domain-inactivating and domain-swapping mutants of c-Jun were analyzed for repression of GATA-1 (Fig. 4A). As shown in Fig. 4B, loss of transcriptional activation (ΔTAD) or loss of DNA binding (DBD-Cla linker) functions both converted c-Jun from a repressor to a potentiator of GATA-1-mediated activation. Loss of leucine zipper dimerization (ΔLZ) eliminated all effects of c-Jun on GATA-1. The potentiation of GATA-1 function by the c-Jun ΔTAD mutant correlates with the enhancement of erythroid differentiation by the same mutant, as illustrated in Fig. 1C. Replacement of the c-Jun TAD with the classic herpes simplex virus VP16 TAD restored the capacity for GATA-1 repression (Fig. 4B). On immunoblot, the VP16-Jun fusion protein showed evidence of marked instability, consistent with the recently described role of the VP16 TAD as a degron, i.e., a signaling sequence for ubiquitylation and degradation (51). Results similar to those in Fig. 4B were also obtained with the minimized GATA-responsive reporter αIIb-98 (not shown). Taken together, the results with the c-Jun mutants suggest a mechanism for repression that involves transcriptional activation of a Jun target gene, which in turn would mediate the repression of GATA factor function.
HERP2, a bHLH transcription factor, is upregulated by wild-type but not mutant c-Jun.
To identify downstream genes which might mediate GATA repression by c-Jun, microarray analysis was used to compare human erythroleukemic cells transduced with constructs encoding the vector, wild-type c-Jun, c-Jun ΔTAD, or c-Jun DBD-Cla linker. With the Affymetrix human genome U133A and U133B chips, representing approximately 33,000 distinct human genes, replicate samples were screened to identify transcription-regulatory genes upregulated by wild-type c-Jun but not by the vector or c-Jun mutants. Particular attention was given to genes showing ≥2-fold upregulation by wild-type c-Jun as well as detection P values of ≤0.05. Two genes clearly fit these criteria, EPAS-1 (also known as HIF-2α), which was upregulated ≈4-fold, and HERP2 (also known as Hesr1, Hey1, HRT1, and CHF2), which was upregulated ≈3-fold. EPAS-1, a member of the bHLH-pas family of transcription factors, shows preferential expression in endothelial cells and functions as part of the hypoxia response pathway (57). HERP2 is a member of the bHLH-orange domain family of transcription factors and has been implicated in the Notch signaling pathway (29-31).
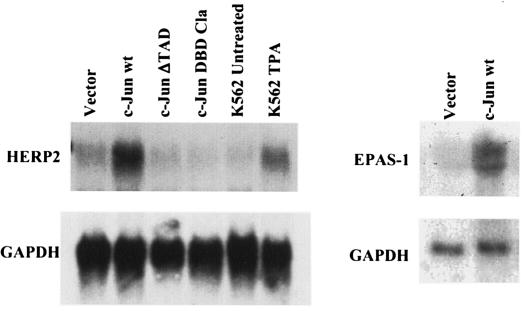
Northern blot analysis showed that wild-type c-Jun expression was associated with ≈5-fold upregulation of HERP2 mRNA, while the c-Jun mutants caused no detectable upregulation of HERP2 (Fig. 5). Immunoblot analysis confirmed equivalent levels of wild-type and DBD-Cla linker mutant c-Jun proteins in the transduced cells (not shown). K562 cells treated with tetradecanoyl phorbol acetate also showed weak upregulation of HERP2 mRNA, correlating with an upregulation of endogenous c-Jun expression (not shown) (1). Probing of Northern blots for EPAS1 also confirmed its upregulation by wild-type c-Jun (Fig. 5). Therefore, HERP2, previously identified as a downstream target of Notch signaling, and EPAS-1, previously identified as a participant in the hypoxia pathway, both represent novel targets of AP-1 signaling.
FIG. 5.
Expression of wild-type c-Jun is associated with upregulation of the hairy-enhancer-of-split-related bHLH factor HERP2. HERP2 mRNA levels in transduced cell lines expressing wild-type (wt) or mutant c-Jun as indicated were analyzed by Northern blot analysis, probing with HERP2 cDNA. Also analyzed were parental K562 with and without tetradecanoyl phorbol acetate (TPA) treatment (25 nM, 48 h). The membrane was stripped and reprobed with labeled glyceraldehhyde-3-phosphate dehydrogenase (GAPDH) cDNA. The right panel shows EPAS-1 and GAPDH mRNA levels in transduced cell lines, as indicated.
HERP2 represses GATA-mediated transcriptional activation and K562 erythroid differentiation in a manner similar to c-Jun.
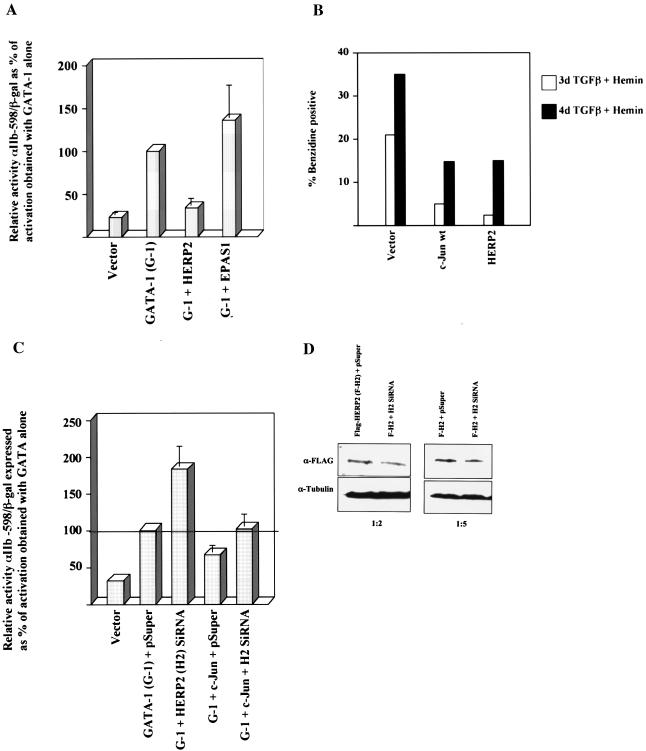
To test for functional effects of HERP2 and EPAS-1 on GATA-1, cells were cotransfected with the relevant expression constructs and a GATA-responsive reporter plasmid. As shown in Fig. 6A, HERP2 repressed GATA-1-mediated transcriptional activation. By contrast, EPAS-1 had no detectable effect on GATA-1 function. HERP2 repression of GATA-1 also occurred with the minimized GATA-responsive reporter αIIb-98, which lacks any identifiable E-box sites (not shown).
FIG. 6.
HERP2 inhibits transcriptional activation by GATA-1 and erythroid differentiation of K562 cells, and HERP2 siRNA interferes with c-Jun-mediated inhibition and augments GATA-1 function. (A) Analysis of transcriptional activation by GATA-1 with and without either HERP2 or EPAS-1. Reporter assays were carried out as described for Fig. 2. (B) K562 cells transduced with the indicated retroviral constructs were subjected to erythroid induction followed by benzidine staining for hemoglobin. (C) Analysis of transcriptional activation by GATA-1 with and without siRNA for HERP2 and with and without c-Jun; 2 × 106 K562 cells were transfected with 2 μg of pEF-GATA-1, 3 μg of pSUPER or pSUPER-siHERP2.2, 1 μg of pCMV-c-Jun, 1.5 μg of αIIb-598-Luc, and 0.5 μg of pCMV-βgal, as indicated. After 72 h of incubation, cells were harvested for standard luciferase and β-galactosidase assays. Results shown represent three independent experiments ± standard error of the mean. (D) Demonstration of Flag-HERP2 knockdown by the HERP2 siRNA construct. K562 cells cotransfected with pcDNA3.1(−)-Flag-HERP2 plus either the pSUPER vector or pSUPER-siHERP2.2 were analyzed by immunoblotting (IB) for Flag or tubulin. Transfections were carried out with 5 μg of DNA per 2 × 106 cells. Ratios below the panels indicate the relative amounts (in micrograms) of the Flag-HERP2 and siRNA vectors, respectively.
If the repressive effects of c-Jun on GATA-1 are mediated by HERP2, then knocking down endogenous HERP2 expression should augment GATA-1 function and interfere with c-Jun repression of GATA-1. As shown in Fig. 6C, cotransfection of a HERP2 siRNA vector augmented GATA-1 function by almost twofold and completely reversed the inhibitory of effect of c-Jun on GATA-1. Cotransfection experiments confirmed the ability of the HERP2 siRNA vector to knock down expression of a Flag-HERP2 transgene by ≈2-fold (Fig. 6D). The augmentation of GATA-1 function by HERP2 siRNA resembles the enhancement of GATA-1 transcriptional activation seen with the dominant negative c-Jun mutants in Fig. 4B. To compare the effects of HERP2 and c-Jun on an endogenous differentiation program, K562 cells transduced with the vector, c-Jun, or HERP2 constructs were analyzed for erythroid differentiation in response to transforming growth factor β plus hemin. Compared with control cells, c-Jun- and HERP2-transduced cells showed similar reductions in benzidine-positive (hemoglobin expressing) cells at days 3 and 4 of erythroid induction (Fig. 6B). Therefore, the repressive effects of c-Jun are recapitulated by HERP2.
Physical interaction of HERP2 with GATA factors.
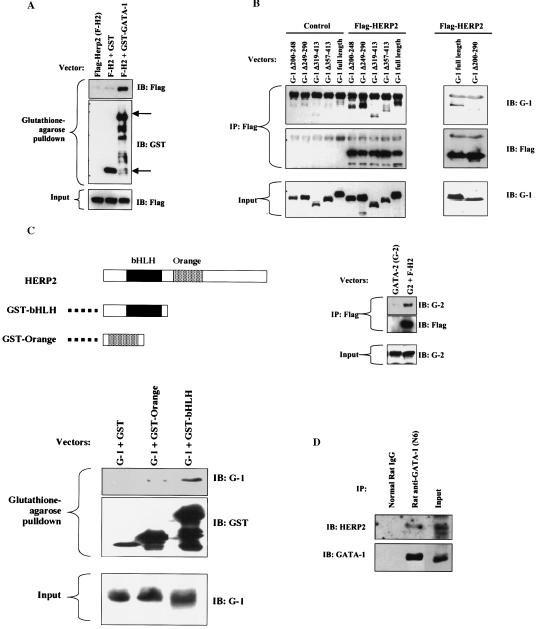
To address the physical basis for HERP2 repression of GATA-1, we conducted protein interaction assays. Flag-tagged HERP2, coexpressed in mammalian cells with either GST alone or GST-GATA-1, showed association with GST-GATA-1 but not GST in a pulldown assay (Fig. 7A). To map binding sites, a panel of GATA factors were coexpressed with Flag-HERP2, followed by immunoprecipitation with anti-Flag. Coprecipitation was detected by immunoblotting with anti-GATA antibodies. As shown in Fig. 7B, deletion of either zinc finger (Δ200-248 or Δ249-290) did not eliminate GATA-1 binding to HERP2. However, deletion of both zinc fingers (Δ200-290) abolished GATA-1 binding to HERP2. Carboxy-terminal truncations of GATA-1 (Δ319-413 and Δ357-413) weakened but did not eliminate HERP2 binding. GATA-2, whose homology to GATA-1 is largely restricted to the zinc fingers, also showed coprecipitation with Flag-HERP2. These results suggest that HERP2 can bind GATA factors through either of the two zinc fingers, following a pattern of GATA interaction observed previously with the Ets transcription factor Fli-1 (13). The carboxy-terminal tail of GATA-1 also appears to contribute to the efficiency of HERP2 binding, but its effects are likely to be indirect, i.e., through maintenance of conformational stability.
FIG. 7.
HERP2 interacts physically with GATA factors. (A) Physical interaction of HERP2 and GATA-1. 293T cells cotransfected with the indicated expression vectors were subjected to glutathione-agarose pulldown followed by immunoblotting with anti-Flag to detect Flag-HERP2 or with anti-GST to detect GST and GST-GATA-1. The upper arrow indicates the position of GST-GATA-1, and the lower arrow indicates the position of GST. (B) Physical interaction of HERP2 and GATA factors. 293T cells cotransfected with the indicated expression vectors were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting with antibodies to GATA-1 (IB: G-1), GATA-2 (IB: G-2), or Flag, as indicated. The GATA mutants consist of the following deletions: N-finger (Δ200-248), C-finger (Δ249-290), carboxy-terminal regions (Δ319-413 and Δ357-413), and both fingers (Δ200-290). (C) Physical interaction of GATA-1 with isolated HERP2 domains. A diagram of HERP2 and of GST fusion proteins is shown. Cotransfections, pulldowns, and immunoblotting were performed as indicated. (D) Physical association of endogenous GATA-1 and HERP2. Parental K562 cellular extracts were subjected to immunoprecipitation with 10 μg of either normal rat immunoglobulin G or rat anti-GATA-1 (N6) monoclonal antibody. Immunoprecipitates were immunoblotted with rabbit anti-HERP2 followed by stripping and reprobing with rabbit anti-GATA-1. The input consisted of 20 μg of cellular extracts.
To identify binding sites in HERP2, GST fusions of HERP2 domains (shown in Fig. 7C) were coexpressed in mammalian cells with full-length, wild-type GATA-1. Copurification on glutathione-agarose was determined by immunoblotting for GATA-1. GST fusions were employed because of the small size and instability of Flag-tagged HERP2 domains. Paradoxically, the full-length HERP2 became destabilized when fused to GST (not shown). As shown in Fig. 7C, the bHLH domain of HERP2 clearly interacted with GATA-1. By comparison, the orange domain of HERP2 showed minimal interaction with GATA-1, suggesting that the bHLH domain constitutes the major binding region.
Finally, coprecipitation experiments addressed whether endogenous GATA-1 and HERP2 formed a complex in K562 cells. As shown in Fig. 7D, immunoprecipitation of endogenous GATA-1 from K562 cells was associated with specific coprecipitation of endogenous HERP2. These results suggest that this physical interaction is not an artifact of enforced expression.
Characterization of the HERP2 repressive function.
To map repressive function, a panel of HERP factors were analyzed for effects on GATA-mediated transactivation. As shown in Fig. 8A, neither the HERP2 bHLH domain nor the orange domain in isolation could repress GATA-1 despite the ability of the bHLH domain to bind GATA-1. HERP1, whose homology to HERP2 is largely confined to the bHLH and orange domains (30), retained the capacity for GATA-1 repression. Deletion of the bHLH domain from HERP2 eliminated most but not all of the repressive function. Immunoblot analysis confirmed equivalent expression levels for full-length HERP2 and HERP2 ΔbHLH (not shown). These results indicate that the bHLH domain, while not sufficient, is necessary for efficient repression of GATA-1 by HERP2.
To determine whether HERP2 exerted repression in a FOG-dependent manner, HERP2 was coexpressed with GATA-1 Δ200-248, which lacks the N-finger involved in recruitment of FOG-1. As illustrated in Fig. 8B, HERP2 efficiently repressed the function of GATA-1 Δ200-248, providing evidence for a FOG-independent mechanism of repression, as was observed for c-Jun (Fig. 3B). These results also correlate with the ability of HERP2 to bind GATA-1 Δ200-248. In addition, HERP2 displayed repression of GATA-2 transactivation (Fig. 8B), as was observed with c-Jun (Fig. 3B), and further correlating with HERP2 binding to GATA factors (Fig. 7A).
Additional experiments addressed whether HERP2 repression of GATA-1 depended on histone deacetylase function or occurred by interference with DNA binding. Notably, trichostatin A, an inhibitor of class I and II histone deacetylase enzymes, had no effect on HERP2 repression of GATA-1 (Fig. 8C). In addition, immunoprecipitation of GATA-1 followed by enzymatic analysis for histone deacetylase activity showed no difference in GATA-1-associated histone deacetylase activity in parent vector- versus HERP2-transduced K562 cells (not shown).
Gel shift assays with nuclear extracts from K562 cells transduced with vector or with HERP2-encoding retrovirus showed no influence of HERP2 on GATA-1 DNA binding (Fig. 8D) despite the ability of HERP2 to block erythroid differentiation in these cells (Fig. 6B). Several measures were taken which ruled out subtle effects of HERP2 on GATA-1 DNA binding: comparison of nuclear extracts from uninduced as well as induced K562 cells, titration of nuclear extracts, and use of dissociation gel shift assays as previously described (9) (data not shown). Chromatin immunoprecipitation examining known DNA binding sites for GATA-1 in K562 cells (β-globin HS2A, -B, and -C) as described by Horak et al. (26), similarly showed no effect of HERP2 on GATA-1 DNA binding (not shown).
Expression and function of HERP2 in erythroid differentiation of normal human progenitor cells.
If the relative levels of HERP2 and GATA-1 determine the transcriptional activity of GATA-1, we would expect modulation of this ratio in the normal course of erythroid differentiation from progenitor cells. Furthermore, interference with this ratio, through the overexpression of HERP2, would be expected to impair erythroid differentiation, possibly retaining progenitors in a more primitive state. Figure 9A illustrates the expression patterns of GATA-1, GATA-2, and HERP2 during the course of erythroid differentiation of normal human CD34+ hematopoietic progenitor cells. Notably, GATA-1 protein levels underwent marked upregulation at days 3 and 14, while HERP2 levels remained constant at days 0, 3, and 14. Therefore, the ratios of HERP2 to GATA-1 dropped significantly during the course of normal human erythroid differentiation. GATA-2 levels showed a slight decline at day 3 and a marked decline at day 14.
Perturbation of the HERP2-GATA-1 ratio by HERP2 overexpression in primary human CD34+ hematopoietic progenitor cells, as predicted, significantly impaired erythroid differentiation (Fig. 9B). CD34+ cells transduced with HERP2-encoding retrovirus (as well as with control vector, shown in Fig. 1A) were subjected to 7 days of erythroid culture, followed by flow cytometric analysis for GPA expression, exactly as described for Fig. 1A. The HERP2-transduced progenitors (GFP positive) clearly showed diminished expression of GPA compared with vector-transduced cells (Fig. 1A, GFP-positive cells) and compared with the GFP-negative cells in Fig. 9B. In particular, HERP2-transduced cells failed to manifest the characteristic discreet peak of GPAbright cells normally associated with erythroid differentiation and displayed more of an ill-defined spectrum of cells with low to intermediate levels of GPA. Interestingly, the transduction efficiency of CD34+ cells by HERP2-encoding retrovirus was consistently much higher than that by c-Jun-encoding retrovirus. This difference may reflect a toxicity associated with c-Jun expression not shared by HERP2 expression. Similar results, showing HERP2 impairment of erythroid differentiation in primary human progenitors, have been obtained in two independent experiments.
To characterize the nature of the erythroid inhibition exerted by c-Jun and HERP2, transduced CD34+ cells cultured for 7 days in erythroid medium were subjected to multiparameter flow cytometry. We examined the coexpression of GPA with CD13, a marker of the granulocytic lineage, with CD34, a marker of immaturity, and with CD41, a marker associated with the megakaryocytic lineage. Vector-transduced cells showed three discrete populations, consisting of erythroid (GPA++ CD13− CD41−), granulocytic (GPA− CD13+ CD41−), and megakaryocytic (GPA− CD13−/+ CD41+) lineages (Fig. 9C). Very few immature GPA− CD13− and GPA− CD34+ cells were identified in the vector-transduced population. Transduction with either c-Jun or HERP2 constructs was associated with an increase in the immature compartment, characterized by increased numbers of both GPA− CD13− and GPA− CD34+ cells. In addition, both c-Jun and HERP2 transduction was associated with an absolute decrease in GPA+ cells, and HERP2 transduction was associated with a downward shift in GPA intensity within the GPA+ compartment. Both c-Jun- and HERP2-transduced cells also showed diminished CD41 expression, possibly reflecting impaired megakaryocytic differentiation. Neither c-Jun nor HERP2 caused a significant shift in the intensity of CD13 expression within the granulocytic compartment.
Taken together, these results suggest that in primary human hematopoietic progenitors, c-Jun and HERP2 impair erythroid and possibly also megakaryocytic differentiation, at least in part through retaining cells in a primitive or immature state. In contrast to their effects on the erythroid lineage, neither c-Jun nor HERP2 appeared to have a significant impact on the granulocytic lineage in these culture conditions.
DISCUSSION
AP-1 transcription factors play a critical role in cellular transformation, contributing to multiple features of the malignant phenotype through direct and indirect upregulation of target genes. In this study, we show that Jun factors may block GATA-1 function through the activation of a novel target gene, HERP2. As GATA-1 function is required for terminal erythroid differentiation, the ability of HERP2 to bind and repress GATA-1 is most likely responsible for the inhibition of erythroid differentiation by HERP2 observed in Fig. 6B and 9B. Our findings indicate that one of the oncogenic effects of Jun factors, inhibition of differentiation, could occur in erythroblasts through a novel regulatory pathway: Jun factors→HERP2—|GATA factors.
An interesting question raised by our model is how HERP2 can block erythroid differentiation if it also inhibits the function of GATA-2, which is thought to support early progenitor survival and proliferation and possibly oppose the activity of GATA-1 (6, 24). This question actually has implications for the mechanisms of virtually all GATA binding factors, the majority of which bind to the highly conserved zinc fingers. For example, PU.1 binds and represses both GATA-1 and GATA-2, with the net phenotypic result of erythroid blockade (33, 47, 48, 62, 71). Conversely, CBP binds and potentiates both GATA-1 and GATA-2, with the net phenotypic result of enhanced erythroid differentiation (5). Thus, our results and those of previous studies suggest that GATA cofactors may be able to target an overlapping proerythroid function shared by GATA-1 and GATA-2 (e.g., transactivation of the GATA-1 gene). Indeed, recent results have demonstrated the existence in vivo of such a shared proerythroid function (19, 33).
How c-Jun induces HERP2 mRNA upregulation is not clear. The human HERP2 promoter contains a perfect AP-1 consensus site at position −2478, TGACTCA (GenBank accession number AJ277506) (37). Accordingly, we isolated from a bacterial artificial chromosome clone a portion of the human HERP2 promoter encompassing nucleotides −2516 to +35. Subcloned into a luciferase reporter vector, this promoter fragment showed no activation in response to c-Jun expression in transient-transfection assays in K562 cells (data not shown). Therefore, additional cis-acting elements within the HERP2 locus may be necessary, or HERP2 may be activated in an indirect manner by c-Jun expression, possibly at the level of mRNA stabilization.
Multiple lines of evidence support the concept that HERP2 may function downstream of c-Jun in the inhibition of GATA-1 function. Most compellingly, siRNA for HERP2 blunted the ability of c-Jun to repress GATA-1 function; furthermore, HERP2 siRNA augmented GATA-1 function in a manner reminiscent of the dominant negative c-Jun mutants (Fig. 6C and 4). Both c-Jun and HERP2 repressed GATA-1 in an N-finger-independent manner, with the GATA-1 ΔN-finger mutant retaining HERP2 binding (Fig. 3B, 7B, and 8B). Thus, c-Jun and HERP2 both appear to regulate GATA-1 in a FOG-independent manner. Both c-Jun and HERP2 also displayed inhibition of erythroid differentiation in K562 cells (Fig. 1C and 6B). Interestingly, in the primary human CD34+ progenitors, c-Jun inhibited erythroid differentiation more thoroughly than HERP2, possibly due to a HERP2-independent toxic effect of c-Jun on primary erythroid cells. Nevertheless, both c-Jun and HERP2 exerted a similar tendency to retain erythroid progenitors in an immature state (Fig. 9C).
An additional correlation consisted of upregulation of endogenous HERP2 mRNA in response to tetradecanoyl phorbol acetate, a stimulus known to upregulate c-Jun in K562 cells (1). Notably, tetradecanoyl phorbol acetate treatment also causes downregulation of the erythroid marker GPA in K562 cells (11). The question arises how K562 cells may undergo megakaryocytic differentiation in response to tetradecanoyl phorbol acetate when the upregulation of HERP2 is expected to block GATA factor function. Important caveats are that we have not been able to document clear upregulation of HERP2 protein in response to tetradecanoyl phorbol acetate and that the megakaryocytic differentiation of K562 cells in response to tetradecanoyl phorbol acetate is only partial (11). Nevertheless, it is possible that different thresholds for HERP2 inhibition of GATA function exist in erythroid versus megakaryocytic differentiation. For example, GATA finger binding by Fli-1, known to occur during megakaryocytic differentiation (13), could compete for HERP2 binding to a similar domain. Alternatively, tetradecanoyl phorbol acetate induction of megakaryocytic differentiation in K562 cells may occur through a GATA-independent pathway.
HERP2 repression of GATA-1 occurs independently of histone deacetylase function and without affecting GATA-1 DNA binding. This repressive mechanism also does not depend on HERP2 binding to cis-acting E-boxes, as evidenced by the repression obtained with the −98 reporter lacking any potential E-box sites (Fig. 2A). Most likely, physical interaction between HERP2 and GATA-1 plays a role, but simple binding of HERP2 to GATA-1 does not suffice because the HERP2 bHLH domain can bind but cannot repress GATA-1 (Fig. 7C and 8A). The physical interaction and the repressive effects rely on conserved domains within HERP2 and GATA-1, namely, the bHLH and zinc finger regions, respectively. The orange domain within HERP2 also binds weakly to GATA-1 and may contribute to the repressive function (Fig. 7C and 8A).
Upregulation of HERP2 expression by c-Jun provides an additional example of the cross talk that may occur between Ras and Notch pathways during cellular transformation. c-Jun has long been recognized as a major downstream target of Ras signaling, and HERP2 was recently described as one of the most robust targets of physiologic Notch signaling (29, 30). In a previous example of Ras-mediated transformation of human cells in vitro, Notch-1 signaling appeared to lie downstream of a Ras-p38/mitogen-activated protein kinase pathway and to induce several features of the transformed phenotype (66). In a murine mammary tumor model, inhibition of Ras signaling impaired Notch-mediated transformation (16).
Our results identify HERP2 as a novel modulator of GATA factor function, with relative levels of HERP2 and GATA-1 influencing the differentiation outcome of erythroid progenitors. This regulatory relationship may explain several previous observations relating to the effect of Notch signaling on cellular differentiation and transformation. With regard to differentiation, Notch signaling has been shown to inhibit erythroid maturation in multiple cell line model systems (34, 35). That GATA-1 is the target for this inhibition is suggested by the inability of activated-Notch-transduced cells to downregulate GATA-2 mRNA upon erythroid induction (34). Downregulation of GATA-2 mRNA during erythroid development has been shown to depend critically on GATA-1 function (22). Thus, the ability of activated Notch to prevent GATA-2 downregulation during erythroid induction could reflect HERP factor interference with GATA-1 function.
With regard to transformation, aberrant Notch activation has played a documented role in cases of human T-cell precursor acute leukemia and mucoepidermoid carcinoma (15, 38, 67). Inhibition of GATA factor function by HERP factors may offer an explanation for some of the oncogenic features of Notch signaling. For example, GATA-1 has recently been identified as a true tumor suppressor gene, sustaining inactivating mutations in cases of human acute myeloid leukemia (25, 65) and normally functioning to restrain cellular proliferation during hematopoiesis (56, 61). It is therefore plausible that HERP2 interference with GATA-1 function could dysregulate the proliferation of hematopoietic progenitors or, by extension, that HERP factor interference with GATA-3 function could disrupt growth control during T-cell ontogeny. GATA-3 normally coordinates key developmental transitions at both pre-T and mature T-cell stages (23, 50, 72). Thus, functional regulation of GATA proteins by HERP factors could provide a strategy for physiologic control of GATA transcriptional activity during normal tissue development and for differentiation blockade during oncogenic transformation.
Acknowledgments
We acknowledge the generosity of Larry Kedes and Tatsuya Iso for providing the Flag-HERP2 and Flag-HERP1 expression vectors in pcDNA3.1(−); Steven McKnight and Richard Bruick for providing pcDNA3-Epas-1; Jane Visvader for providing GATA expression constructs; and Richard Baer for providing the VP16 expression vector. Thanks also go to Joanne Lannigan for assistance with flow cytometry, to Yongde Bao for assistance with microarray analysis, and to Shelly Heimfeld of the PEGT-HCPC. Finally, thanks go to Ying Ye for expert experimental assistance.
This work was supported in part by Public Health Service grants CA93735 (A.N.G.) and CA100057 (A.N.G.) from the National Cancer Institute and K08 HL04017 (F.K.R.) from the National Heart, Lung and Blood Institute.
REFERENCES
- 1.Alitalo, R., J. Partanen, L. Pertovaara, E. Holtta, L. Sistonen, L. Andersson, and K. Alitalo. 1990. Increased erythroid potentiating activity/tissue inhibitor of metalloproteinases and jun/fos transcription factor complex characterize tumor promoter-induced megakaryoblastic differentiation of K562 leukemia cells. Blood 75:1974-1982. [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. C. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamberger, A. M., K. Milde-Langosch, E. Rossing, C. Goemann, and T. Loning. 2001. Expression pattern of the AP-1 family in endometrial cancer: correlations with cell cycle regulators. J. Cancer Res. Clin. Oncol. 127:545-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauknecht, T., P. Angel, M. Kohler, F. Kommoss, G. Birmelin, A. Pfleiderer, and E. Wagner. 1993. Gene structure and expression analysis of the epidermal growth factor receptor, transforming growth factor-alpha, myc, jun, and metallothionein in human ovarian carcinomas. Classification of malignant phenotypes. Cancer 71:419-429. [DOI] [PubMed] [Google Scholar]
- 5.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with the transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briegel, K., K. C. Lim, C. Plank, H. Beug, J. D. Engel, and M. Zenke. 1993. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes Dev. 7:1097-1109. [DOI] [PubMed] [Google Scholar]
- 7.Burgess, G. S., E. A. Williamson, L. D. Cripe, S. Litz-Jackson, J. A. Bhatt, K. Stanley, M. J. Stewart, A. S. Kraft, H. Nakshatri, and H. S. Boswell. 1998. Regulation of the c-jun gene in p210 BCR-ABL transformed cells corresponds with activity of JNK the c-jun N-terminal kinase. Blood 92:2450-2460. [PubMed] [Google Scholar]
- 8.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 10.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delehanty, L. L., M. Mogass, S. L. Gonias, F. K. Racke, B. Johnstone, and A. N. Goldfarb. 2003. Stromal inhibition of megakaryocytic differentiation is associated with blockade of sustained Rap1 activation. Blood 101:1744-1751. [DOI] [PubMed] [Google Scholar]
- 12.Eferl, R., R. Ricci, L. Kenner, R. Zenz, J.-P. David, M. Rath, and E. F. Wagner. 2003. Liver tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell 112:181-192. [DOI] [PubMed] [Google Scholar]
- 13.Eisbacher, M., M. L. Holmes, A. Newton, P. J. Hogg, L. M. Khachigian, M. Crossley, and B. H. Chong. 2003. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol. Cell. Biol. 23:3427-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elagib, K. E., F. K. Racke, M. Mogass, R. Khetawat, L. L. Delehanty, and A. N. Goldfarb. 2003. RUNX-1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101:4333-4341. [DOI] [PubMed] [Google Scholar]
- 15.Ellisen, L. W., J. Bird, D. C. West, A. Lee Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald, K., A. Harrington, and P. Leder. 2000. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene 19:4191-4198. [DOI] [PubMed] [Google Scholar]
- 17.Fox, A. H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francastel, C., R. Groisman, C. M. Pfarr, and J. Robert-Lezenes. 1994. Antisense c-jun overcomes a differentiation block in a murine erythroleukemia cell line. Oncogene 9:1957-1964. [PubMed] [Google Scholar]
- 19.Fujiwara, Y., A. N. Chang, A. M. Williams, and S. H. Orkin. 2004. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood 103:583-585. [DOI] [PubMed] [Google Scholar]
- 20.Gatlin, J., M. W. Melkus, A. Padgett, P. F. Kelly, and J. V. Garcia. 2001. Engraftment of NOD/SCID mice with human CD34+ cells transduced by concentrated oncoretroviral vector particles pseudotyped with the feline endogenous retrovirus (RD114) envelope protein. J. Virol. 75:9995-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfarb, A. N., L. L. Delehanty, D. Wang, F. K. Racke, and I. M. Hussaini. 2001. Stromal inhibition of megakaryocytic differentiation correlates with blockade of signaling by protein kinase C-ɛ and ERK/MAPK. J. Biol. Chem. 276:29526-29530. [DOI] [PubMed] [Google Scholar]
- 22.Grass, J. A., M. E. Boyer, S. Pal, J. Wu, M. J. Weiss, and E. H. Bresnick. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA 100:8811-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Hoyos, G., M. K. Anderson, C. Wang, E. V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19:83-94. [DOI] [PubMed] [Google Scholar]
- 24.Heyworth, C., K. Gale, M. Dexter, G. May, and T. Enver. 1999. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev. 13:1847-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitzler, J. K., J. Cheung, Y. Li, S. W. Scherer, and A. Zipursky. 2003. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 101:4301-4304. [DOI] [PubMed] [Google Scholar]
- 26.Horak, C. E., M. C. Mahajan, N. M. Luscombe, M. Gerstein, S. M. Weissman, and M. Snyder. 2002. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. USA 99:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu, H.-L., I. Wadman, and R. Baer. 1994. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc. Natl. Acad. Sci. USA 91:3181-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussaini, I. M., L. R. Karns, G. Vinton, J. E. Carpenter, G. T. Redpath, J. J. Sando, and S. R. VandenBerg. 2000. Phorbol 12-myristate 13-acetate induces protein kinase Cη-specific proliferative response in astrocytic tumor cells. J. Biol. Chem. 275:22348-22354. [DOI] [PubMed] [Google Scholar]
- 29.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 30.Iso, T., V. Sartorelli, G. Chung, T. Shichinohe, L. Kedes, and Y. Hamamori. 2001. HERP, a new primary target of Notch regulated by ligand binding. Mol. Cell. Biol. 21:6071-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iso, T., V. Sartorelli, C. Poizat, S. Iezzi, H.-Y. Wu, G. Chung, L. Kedes, and Y. Hamamori. 2001. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21:6080-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 33.Kitajima, K., M. Masuhara, T. Era, T. Enver, and T. Nakano. 2002. GATA-2 and GATA-2/ER display opposing activities in the development and differentiation of blood progenitors. EMBO J. 21:3060-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumano, K., S. Chiba, K. Shimizu, T. Yamagata, N. Hosoya, T. Saito, T. Takahashi, Y. Hamada, and H. Hirai. 2001. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood 98:3283-3289. [DOI] [PubMed] [Google Scholar]
- 35.Lam, L. T., C. Ronchini, J. Norton, A. J. Capobianco, and E. H. Bresnick. 2000. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by Notch-1. J. Biol. Chem. 275:19676-19684. [DOI] [PubMed] [Google Scholar]
- 36.Leaner, V. D., I. Kinoshita, and M. J. Birrer. 2003. AP-1 complexes containing cJun and JunB cause cellular transformation of Rat1a fibroblasts and share transcriptional targets. Oncogene 22:5619-5629. [DOI] [PubMed] [Google Scholar]
- 37.Maier, M. M., and M. Gessler. 2000. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275:652-660. [DOI] [PubMed] [Google Scholar]
- 38.Maillard, I., and W. S. Pear. 2003. Notch and cancer: best to avoid the ups and downs. Cancer Cell 3:203-205. [DOI] [PubMed] [Google Scholar]
- 39.Mao, X., G. Orchard, D. M. Lillington, R. Russell-Jones, B. D. Young, and S. J. Whittaker. 2003. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphomas. Blood 101:1513-1519. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki, T., K.-I. Aisaki, Y. Yamamura, M. Noda, and Y. Ikawa. 2000. Induction of erythroid differentiation by inhibition of Ras/ERK pathway in a Friend murine leukemia cell line. Oncogene 19:1500-1508. [DOI] [PubMed] [Google Scholar]
- 41.Orkin, S. H., F. I. Harosi, and P. Leder. 1975. Differentiation in erythroleukemic cells and their somatic hybrids. Proc. Natl. Acad. Sci. USA 72:98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer, L. A., B. Gaston, and R. A. Johns. 2000. Normoxic stabilization of hypoxia-inducible factor-1 expression and activity: redox-dependent effect of nitrogen oxides. Mol. Pharmacol. 58:1197-1203. [DOI] [PubMed] [Google Scholar]
- 43.Persons, D. A., J. A. Allay, E. R. Allay, R. A. Ashmun, D. Orlic, S. M. Jane, J. M. Cunningham, and A. W. Nienhuis. 1999. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 93:488-499. [PubMed] [Google Scholar]
- 44.Prochownik, E. V., M. J. Smith, K. Snyder, and D. Emeagwali. 1990. Amplified expression of three jun family members inhibits erythroleukemia differentiation. Blood 76:1830-1837. [PubMed] [Google Scholar]
- 45.Racke, F. K., D. Wang, Z. Zaidi, J. Kelley, J. Visvader, J.-W. Soh, and A. N. Goldfarb. 2001. A potential role for protein kinase C-ɛ in regulating megakaryocytic lineage commitment. J. Biol. Chem. 276:522-528. [DOI] [PubMed] [Google Scholar]
- 46.Rangatia, J., R. K. Vangala, S. M. Singh, A. A. Peer Zada, A. Elsasser, A. Kohlmann, T. Haferlach, D. G. Tenen, W. Hiddeman, and G. Behre. 2003. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPα DNA binding via leucine zipper domain interaction. Oncogene 22:4760-4764. [DOI] [PubMed] [Google Scholar]
- 47.Rekhtman, N., K. S. Choe, I. Matushansky, S. Murray, T. Stopka, and A. I. Skoultchi. 2003. PU. 1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol. Cell. Biol. 23:7460-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU. 1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosson, D., and T. G. O'Brien. 1998. AP-1 activity affects the levels of induced erythroid and megakaryocytic differentiation of K562 cells. Arch. Biochem. Biophys. 352:298-305. [DOI] [PubMed] [Google Scholar]
- 50.Rothenberg, E. 2002. T-lineage specification and commitment: a gene regulation perspective. Semin. Immunol. 14:431-440. [DOI] [PubMed] [Google Scholar]
- 51.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 52.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 53.Shivdasani, R. A., and S. H. Orkin. 1996. The transcriptional control of hematopoiesis. Blood 87:4025-4039. [PubMed] [Google Scholar]
- 54.Smith, L. M., S. C. Wise, D. T. Hendricks, A. L. Sabichi, T. Bos, P. Reddy, P. H. Brown, and M. J. Birrer. 1999. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive, and hormone resistant phenotype. Oncogene 18:6063-6070. [DOI] [PubMed] [Google Scholar]
- 55.Southcott, M. J. G., M. J. A. Tanner, and D. J. Anstee. 1999. The expression of human blood group antigens during erythropoiesis in a cell culture system. Blood 93:4425-4435. [PubMed] [Google Scholar]
- 56.Takahashi, S., T. Komeno, N. Suwabe, K. Yoh, O. Nakajima, S. Nishimura, T. Kuroha, T. Nagasawa, and M. Yamamoto. 1998. Role of GATA-1 in proliferation and differentiation of definitive erythroid and megakaryocytic cells in vivo. Blood 92:434-442. [PubMed] [Google Scholar]
- 57.Tian, H., R. E. Hammer, A. M. Matsumoto, D. W. Russell, and S. L. McKnight. 1998. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 12:3320-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Dam, H., S. Huguier, K. Kooistra, J. Baguet, E. Vial, A. J. van der Eb, P. Herrlich, P. Angel, and M. Castellazzi. 1998. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 12:1227-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visvader, J. E., M. Crossley, J. Hill, S. H. Orkin, and J. M. Adams. 1995. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Mol. Cell. Biol. 15:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt, P. K. 2001. Jun, the oncoprotein. Oncogene 20:2365-2377. [DOI] [PubMed] [Google Scholar]
- 61.Vyas, P., F. A. Norris, R. Joseph, P. W. Majerus, and S. H. Orkin. 2000. Inositol polyphosphate 4-phosphatase type 1 regulates cell growth downstream of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 97:13696-13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walsh, J. C., R. P. DeKoter, H.-J. Lee, E. D. Smith, D. W. Lancki, M. F. Gurish, D. S. Friend, R. L. Stevens, J. Anastasi, and H. Singh. 2002. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 17:665-676. [DOI] [PubMed] [Google Scholar]
- 63.Wang, H., M. Birkenbach, and J. Hart. 2000. Expression of Jun family members in human colorectal adenocarcinoma. Carcinogenesis 21:1313-1317. [PubMed] [Google Scholar]
- 64.Watsuji, T., Y. Okamoto, N. Emi, Y. Katsuoka, and M. Hagiwara. 1997. Controlled gene expression with a reverse tetracycline-regulated retroviral vector (RTRV) system. Biochem. Biophys. Res. Commun. 234:769-773. [DOI] [PubMed] [Google Scholar]
- 65.Wechsler, J., M. Greene, M. A. McDevitt, J. Anastasi, J. E. Karp, M. M. Le Beau, and J. D. Crispino. 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32:148-152. [DOI] [PubMed] [Google Scholar]
- 66.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, W. M. Kast, and L. Miele. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8:979-986. [DOI] [PubMed] [Google Scholar]
- 67.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wisdom, R., R. S. Johnson, and C. Moore. 1999. C-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wulf, G. M., A. Ryo, G. G. Wulf, S. W. Lee, T. Niu, V. Petkova, and K. P. Lu. 2001. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20:3459-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuen, M. F., P. C. Wu, V. C. Lai, J. Y. Lau, and C. L. Lai. 2001. Expression of c-Myc, c-Fos, and c-Jun in hepatocellular carcinoma. Cancer 91:106-112. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU. 1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]
- 72.Zheng, W., and R. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587-596. [DOI] [PubMed] [Google Scholar]