Abstract
Translation termination in eukaryotes is mediated by two release factors, eRF1 and eRF3. eRF1 recognizes each of the three stop codons (UAG, UAA, and UGA) and facilitates release of the nascent polypeptide chain. eRF3 is a GTPase that stimulates the translation termination process by a poorly characterized mechanism. In this study, we examined the functional importance of GTP hydrolysis by eRF3 in Saccharomyces cerevisiae. We found that mutations that reduced the rate of GTP hydrolysis also reduced the efficiency of translation termination at some termination signals but not others. As much as a 17-fold decrease in the termination efficiency was observed at some tetranucleotide termination signals (characterized by the stop codon and the first following nucleotide), while no effect was observed at other termination signals. To determine whether this stop signal-dependent decrease in the efficiency of translation termination was due to a defect in either eRF1 or eRF3 recycling, we reduced the level of eRF1 or eRF3 in cells by expressing them individually from the CUP1 promoter. We found that the limitation of either factor resulted in a general decrease in the efficiency of translation termination rather than a decrease at a subset of termination signals as observed with the eRF3 GTPase mutants. We also found that overproduction of eRF1 was unable to increase the efficiency of translation termination at any termination signals. Together, these results suggest that the GTPase activity of eRF3 is required to couple the recognition of translation termination signals by eRF1 to efficient polypeptide chain release.
Translation termination occurs when a stop codon enters the ribosomal A site and signals polypeptide chain release from the peptidyl-tRNA located in the ribosomal P site. This process is facilitated by two general groups of accessory proteins (15, 34, 43). The class I release factors recognize the stop codon in the A site and stimulate nascent peptide chain release. Prokaryotic organisms have two class I release factors with different stop codon specificities. RF1 recognizes UAA and UAG codons, while RF2 recognizes UAA and UGA codons. In contrast, eukaryotes possess a single class I release factor, eRF1, that decodes all three stop codons. The class II release factors in both prokaryotes and eukaryotes are GTP binding proteins that facilitate the termination process. RF3 is the bacterial class II release factor, while eRF3 functions as the eukaryotic class II release factor.
The yeast eRF1 protein (the product of the essential SUP45 gene) does not share significant sequence homology with its prokaryotic counterparts. However, like RF1 or RF2, it is thought to recognize the termination signal in the A site and stimulate peptide release through an interaction with the peptidyl transferase center located in the large ribosomal subunit. eRF1 is comprised of three distinct domains. Domain 1 includes the conserved amino acid motifs YxCxxxF (yeast amino acid residues 122 to 128) and TASNIKS (yeast amino acid residues 55 to 61), which have been implicated in stop codon binding (19, 31, 46). Domain 2 contains the conserved GGQ motif (yeast amino acid residues 180 to 182), which forms a highly exposed minidomain that is thought to play a crucial role in the interaction of eRF1 with the peptidyl transferase center (22, 45, 48). Interestingly, the GGQ motif is also found in prokaryotic RF1 or RF2 and presumably plays a similar role in those proteins (22). Finally, domain 3 of eRF1 mediates its association with eRF3 (14, 16, 20, 30, 41).
The eRF3 protein (the product of the essential SUP35 gene) also contains three distinct regions (Fig. 1A). The N and M regions occupy amino acid residues 1 to 253 and are dispensable for both translation termination and cell viability (52). In contrast, the C region is essential for viability and contains a GTPase fold (51, 52) (amino acid residues 254 to 479) similar to those found in all G proteins (49). Eukaryotic eRF3 and eubacterial RF3 share only limited sequence homology that is restricted to the 225-amino-acid region that comprises the GTPase fold. It has been shown that RF3 recycles the class I factor(s) from the posttermination complex in prokaryotes (17, 55, 56), and it was suggested that eRF3 carries out a similar function in eukaryotic cells (55). However, such a role has not been demonstrated. In the present study, we examined how mutations in the GTPase domain of eRF3 influence translation termination. Our results are not consistent with a mechanism in which eRF3 functions primarily to recycle eRF1. Instead, the results indicate that the GTPase activity of eRF3 acts to couple the recognition of translation termination signals by eRF1 to efficient polypeptide chain release.
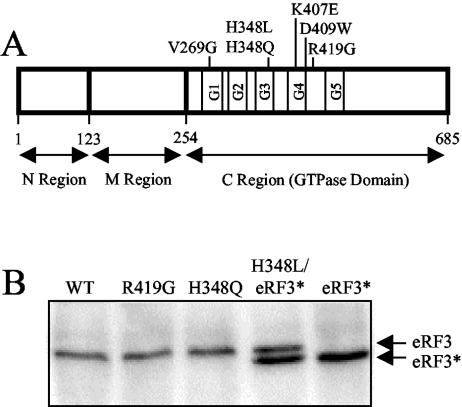
FIG. 1.
Diagram showing the location of eRF3 mutations analyzed in this study. (A) Schematic map of eRF3 showing the mutations introduced into the C-terminal GTPase domain. (B) Steady-state levels of wild-type (WT) and mutant forms of eRF3 as determined by Western blot analysis. Levels of eRF3 were measured in a sup35Δ strain (YBD498) expressing the indicated wild-type or mutant forms of eRF3 from a low-copy-number plasmid. Due to the inability of eRF3-H348L to support cell viability, it was coexpressed with a derivative of wild-type eRF3 (eRF3*) containing a small internal deletion (amino acids residues 21 to 67) in the N region. Twenty-five micrograms of total protein was loaded per lane.
MATERIALS AND METHODS
Strains and growth conditions.
The Saccharomyces cerevisiae strains used in this study were 614 (MATa leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 [psi−]), YDB498 (MATa leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 sup35::HIS3 [psi−]), YDB340 (MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-80 suc2-Δ901 [psi−]), and YDB447 (MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-80 suc2-Δ901 sup45::HIS3 [psi−]). Strains YDB498 and YDB447 were derived from 614 and YDB340, respectively, by using standard yeast genetic techniques (6). SMD is supplemented minimal medium (6) containing 2% glucose and other required nutritional supplements.
Gene disruptions.
Yeast strains YDB498 and YDB447 were generated by using a one-step gene replacement method. The SUP35 gene was disrupted by the removal of the entire open reading frame and the insertion of the HIS3 gene from Candida glabrata by using a PCR-based gene deletion approach (39) with plasmid pH 4 as a template (35). Yeast strain 614 carrying pPW12.1 (a plasmid carrying SUP35 with its own promoter in the vector YCp50) was transformed with the disruption fragment generated by PCR. His+ transformants were screened by PCR and tested for the ability to evict pPW12.1. The correct genomic integration event was then verified by Southern blot analysis. To generate the sup45Δ strain YDB447, the S. cerevisiae HIS3 gene was subcloned between the BglII and BamHI sites of SUP45. This removed the entire SUP45 gene except the first 28 nucleotides and the last 23 nucleotides of the open reading frame. The resulting plasmid was designated pDB532. An 1,800-bp NciI/XhoI fragment containing this knockout construct was then used to transform a diploid yeast strain generated by mating GT17 and GT197 (MATa/MATα ade1-14/ade1-14 his3-Δ200/his3-Δ200 trp1-289/trp1-289 leu2-3,112/leu2-3,112 ura3-52/ura3-52 LYS2/lys2 [psi−]). The SUP45/sup45::HIS3 diploid strain was transformed with pDB709 (carrying wild-type SUP45 in YCplac22) and induced to sporulate. The gene disruption was verified by Southern blotting, and the resulting haploid strain containing the sup45::HIS3 allele (complemented by the plasmid-based SUP45 gene) was backcrossed to YDB340 five times.
Plasmids.
The construct used to disrupt the SUP45 gene was made as follows. SUP45 from pUKC802 (51) was subcloned into the ClaI/XhoI sites of a pBluescript II KS(+) derivative (missing the BamHI site) (Stratagene), yielding pDB531. The final construct, pDB532, was obtained by replacing the BglII/BamH1 fragment from pDB531 with a BglII/BamH1 fragment from pJJ215 that includes the HIS3 gene (32).
Centromere-based plasmids were used to express wild-type and mutant forms of SUP35 in yeast. pDB663 contains the SUP35 gene under the control of its own promoter in a centromere-based vector. Site-directed mutations were introduced into SUP35 by using a QuikChange site-directed mutagenesis kit (Stratagene). Mutations H348Q (CAT to CAA), H348L (CAT to CTT), and R419G (CGT to GGT) were introduced into SUP35 (SUP35/pYES2). Finally, a StuI/NsiI fragment containing the mutated DNA was used to replace a StuI/NsiI fragment in pDB663. This process resulted in plasmids pDB670 (eRF3-H348Q), pDB697 (eRF3-H348L), and pDB783 (eRF3-R419G). Plasmid pDB734 was used to express eRF3*, which contained a small internal deletion (amino acid residues 21 to 67) within the N region of eRF3 in the centromeric plasmid YCp50. This shortened form of eRF3 was used in Western blot experiments to visualize the expression of eRF3 mutants that were unable to support yeast cell viability.
Plasmid pDB799 was used to express the GTPase domain of eRF3 (starting at methionine residue 254) in Escherichia coli for its subsequent purification. This construct, which contained a C-terminal His6 tag, was derived from pPW12.1 (54) and subcloned into the pET-3a vector (Novagen). To generate constructs for expression of the eRF3 mutants in E. coli, StuI/NsiI fragments from pDB670, pDB697, and pDB783 were used to replace the StuI/NsiI fragment of pDB799. This yielded pDB820 (eRF3-H348Q), pDB821 (eRF3-H348L), and pDB823 (eRF3-R419G). Plasmid pDB698 was also a pET3a derivative and was used to express an N-terminal His6-tagged version of eRF1 in E. coli. The gene encoding eRF1 (SUP45) was obtained from pJH109 (25). The centromeric plasmid pDB811 was used to express SUP45 from the CUP1 promoter. The CUP1 promoter was PCR amplified from YEp96 (26) and subcloned into SUP45-YCplac22. Similarly, the centromeric plasmid pDB824 was used to express SUP35 from the CUP1 promoter. Finally, the plasmid pDB773, which contained the SUP45 promoter and the SUP45 gene in the multicopy vector YEplac181, was used to overexpress eRF1.
Purification of proteins.
In preliminary experiments, we found that the expression of full-length His6-tagged eRF3 was relatively poor in E. coli BL21(DE3) cells, while a much greater amount of protein could be recovered from constructs that expressed a His6-tagged fragment with the GTPase domain alone. Consequently, each of these truncated eRF3 constructs (residues 254 to 685) of S. cerevisiae was expressed in BL21(DE3) cells and purified from the soluble fraction. Cells were lysed by sonication in lysis buffer (20 mM Tris-HCl [pH 8.0], 0.5 M KCl, 10% glycerol, 5 mM MgCl, 10% Nonidet P-40, 10 mM imidazole, 2 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml). A cleared lysate was obtained by centrifugation at 10,000 × g for 20 min. The supernatant was added to 250 μl (bed volume) of Ni-nitrilotriacetic acid resin (QIAGEN) equilibrated in wash buffer (20 mM Tris-HCl [pH 8.0], 0.5 M KCl, 10% glycerol, 5 mM MgCl, 10 mM imidazole). This slurry was incubated with shaking at 4°C for 1 h. The resin was then loaded on a Poly-Prep chromatography column (Bio-Rad) and washed. The protein was eluted in 9 ml of elution buffer (wash buffer plus 0.25 M imidazole). Peak fractions were concentrated by using a Centricon YM-30 concentrator (Amicon). The eRF1 protein was purified as described above for eRF3 with minor exceptions. MgCl2 was not present in the lysis and wash buffers. In general, the eRF1 and eRF3 proteins obtained by this procedure were nearly homogeneous as judged by Coomassie brilliant blue R staining.
Ribosome purification for GTPase assays.
Salt-washed 80S ribosomes were purified from the S. cerevisiae strain 614. Cells were resuspended in high-salt buffer (20 mM Tris-HCl, 1.0 M potassium acetate, 15 mM MgCl2, 1 mM dithiothreitol [pH 7.4]) supplemented with 1 mM PMSF and lysed by mechanical agitation with glass beads. The cleared lysate was loaded onto a 15% sucrose cushion (in high-salt buffer) and centrifuged at 100,000 × g for 5 h with an SW55Ti rotor (Beckman). The ribosomal pellet was resuspended in high-salt buffer and incubated overnight at 4°C. Salt-washed ribosomes were pelleted as before and resuspended in storage buffer (20 mM Tris-HCl, 30 mM NH4Cl, 15 mM MgCl2 [pH 7.5]) at −80°C until use.
GTPase assays.
The GTPase activity of wild-type and mutant forms of eRF3 was measured as described (18). The reaction mixture (12.5 μl) contained GTP (as indicated), [α-32P]GTP (0.1 μCi), and 3 pmol each of eRF3-ΔNM, eRF1, and salt-washed ribosomes in reaction buffer (20 mM Tris-HCl, 30 mM NH4Cl, 15 mM MgCl2, pH 7.5). GTPase assays were carried out at 25°C. The reaction was stopped by the addition of 1 μl of stop buffer (20 mM EDTA and 5% sodium dodecyl sulfate [SDS]). Five-microliter aliquots were spotted onto a polyethylenimine cellulose plate (Sigma) and resolved by thin-layer chromatography in 0.5 M LiCl and 2 M formic acid for 2 h. Plates were dried, and the reaction products were quantified by using a PhosphorImager (Molecular Dynamics). The rates of GTP hydrolysis were calculated for several GTP concentrations (1, 1.4, 2, 3, 5, 7, 15, and 20 μM), and the data were analyzed by using Lineweaver-Burk plots. The Vmax (the inverse of the y intercept) and the Km (negative inverse of the x intercept) for each set of reactions were calculated. The Kcat was derived from the Vmax (Kcat = Vmax/pmol eRF3).
Preparation of cell lysates and ribosomes for Western blot analysis.
After each strain was cultured for several generations, cell growth was terminated by the addition of 10% trichloroacetic acid. After a 30-min incubation on ice, the cells were collected by a brief centrifugation in a microcentrifuge. The cells were washed with ice-cold acetone, dried, resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% SDS) and lysed by mechanical agitation with glass beads. The samples were then boiled, cleared by a brief centrifugation in a microcentrifuge, and subjected to SDS-polyacrylamide gel electrophoresis and Western blotting.
For the isolation of ribosomes, cycloheximide (200 μg/ml) was added to the cultures and cells were harvested by centrifugation 15 min later. The pellet was resuspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM PMSF) supplemented with 200 μg of cycloheximide/ml. The cells were lysed by mechanical agitation with glass beads, and debris was removed by centrifugation at 12,000 × g for 15 min. The cell lysate was loaded onto a sucrose cushion (15% sucrose in lysis buffer), and ribosomes were obtained by centrifugation at 100,000 × g for 5 h with an SW55Ti rotor (Beckman).
Dual luciferase assays.
The dual luciferase reporters used to measure the efficiency of translation termination in yeast were adapted from plasmids previously used to monitor the efficiency of translation termination in mammalian cells (24, 27) as recently described (33). Dual luciferase assays were performed with a dual luciferase reporter assay system (Promega). Briefly, yeast strains were transformed with the indicated dual luciferase reporter plasmids. Approximately 104 cells from each strain were assayed for luminescence with a Berthold Lumat LB9507 luminometer. Assays were done in quadruplicate, and the data are expressed as the means ± the standard deviation. The percent readthrough in each strain is expressed as the firefly-Renilla luciferase activity (nonsense) divided by the firefly-Renilla luciferase activity (sense) multiplied by 100. For further details, see Keeling et al. (33).
RESULTS
Mutations within the GTPase domain of eRF3 compromise cell viability.
All G proteins share a structural core (the GTPase fold) that carries out GTP binding and hydrolysis (49). These GTPase folds are typically about 200 amino acids in length and consist of a central six-stranded β-sheet surrounded by α-helices. The nucleotide-binding site within this domain is formed by five highly conserved loops, designated G1 to G5 (Fig. 1A). The eukaryotic polypeptide chain release factor eRF3 belongs to a class of G proteins that includes the eukaryotic translation elongation factor eEF1A and its prokaryotic counterpart EF-Tu. The C-terminal GTPase domain of eRF3 exhibits significant sequence homology throughout the entire length of eEF1A (39% identity) and EF-Tu (31% identity). To test the hypothesis that the GTPase activity of eRF3 plays an important role in translation termination, a series of mutations were introduced into the GTPase domain of eRF3. The amino acid residues targeted for mutagenesis in eRF3 corresponded to mutations previously shown to alter the GTPase activities of either EF-Tu or eEF1A or to confer an “omnipotent suppressor” phenotype in eRF3 (2, 7, 11, 28, 36, 53, 57) (Fig. 1A).
Six mutations (V269G, H348L, H348Q, K407E, D409W, and R419G) were introduced into the yeast eRF3 gene (SUP35) by site-directed mutagenesis. The mutated genes were then cloned under SUP35 promoter control into a centromeric plasmid containing the TRP1 gene as the selectable marker. Initially, these plasmids were transformed into a yeast strain carrying a knockout of the genomic SUP35 gene (sup35Δ) whose viability was maintained by the presence of another plasmid carrying a wild-type copy of the SUP35 gene with the URA3 gene as the selectable marker. A plasmid shuffle procedure was then carried out to select cells that had lost the URA3 plasmid encoding wild-type SUP35. This was done by growth in medium containing 5-fluoroorotic acid (47). This compound is metabolized to the cytotoxic compound 5-fluorouracil in the presence of the wild-type URA3 gene (present on the plasmid expressing wild-type eRF3). Under these conditions, only cells that have spontaneously lost the URA3 plasmid can grow. The ability to form colonies indicates that the mutant eRF3 alone can support viability, while a complete absence of colonies indicates that the mutant form of eRF3 is unable to support cell viability. We found that the eRF3-H348Q and eRF3-R419G proteins were capable of supporting cell viability when present as the sole source of eRF3 in the cell, although the growth rate was reduced. In contrast, the eRF3-V269G, eRF3-H348L, eRF3-K407E, and eRF3-D409W proteins were unable to support cell growth in the absence of wild-type eRF3. These results are consistent with the previous conclusion that the GTPase domain of eRF3 is essential for cell viability (52). In addition, since most of these conserved residues were previously shown to be important for the GTP binding and/or hydrolysis in EF-Tu or eEF1A, these results strongly suggest that the basic features of GTP binding and hydrolysis are conserved between these translation factors. The recently published crystal structure of the C-terminal portion of eRF3 from Schizosaccharomyces pombe confirms the close structural similarities of the GTPase domains of these proteins (37).
Based on the results above, we chose the two mutant forms of eRF3 that maintained cell viability when present as the sole source of eRF3 in the cell (H348Q and R419G) and one mutant form (H348L) that was unable to support viability for further study. Initially, Western blot analysis was carried out to monitor the steady-state levels of these mutant forms of eRF3 (Fig. 1B). The two viable mutant proteins were expressed as the only form of eRF3 in the cells, while the eRF3-H348L mutant protein was coexpressed with a shortened form of wild-type eRF3 that was produced by an in-frame deletion of 47 amino acids (residues 21 to 67) within the nonessential N region (called eRF3*). We found that each of the mutant forms of eRF3 was present at a steady-state level similar to that of wild-type eRF3. This allowed us to correlate changes in GTP binding and/or hydrolysis resulting from these mutations with alterations in the efficiency of translation termination.
eRF3 mutations decrease the efficiency of translation termination in a distinct manner at different tetranucleotide termination signals.
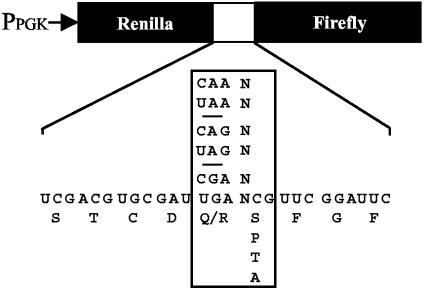
Previous studies have shown that a statistical bias exists in the nucleotides that surround naturally occurring stop codons in both prokaryotic and eukaryotic organisms (5). It was later shown that the identity of the first nucleotide following the stop codon influences the efficiency of translation termination by as much as 20-fold in eukaryotic systems, consistent with polypeptide chain release being mediated by a tetranucleotide termination signal (4, 40). Based on this information, we examined how mutations in the GTPase domain of eRF3 affected the efficiency of translation termination at each of the 12 possible tetranucleotide termination signals by using a dual luciferase reporter system (24, 27, 33) (Fig. 2). The dual luciferase reporter plasmids contain an upstream Renilla luciferase gene and a downstream firefly luciferase gene separated by different in-frame tetranucleotide termination signals. These constructs allowed us to monitor the translational readthrough of different termination signals by measuring firefly luciferase activity, while the Renilla luciferase activity served as an internal normalization control. These activities were then compared to Renilla and firefly luciferase activities assayed in the same strain with a sense codon in the readthrough position. This allowed us to determine the percent readthrough while carefully controlling for any differences in mRNA abundance or the efficiency of translation initiation.
FIG. 2.
Dual luciferase readthrough reporter constructs.
The eRF1 protein has been shown to interact directly with stop codons located in the ribosomal A site (8-10), while eRF3 is thought to play a more peripheral role in the termination process (55, 59). Mutations in the gene encoding eRF3 (SUP35) have frequently been reported to exhibit an omnipotent suppressor phenotype (23, 38), where readthrough is thought to increase at all stop codons. Therefore, we expected cells expressing the mutant forms of eRF3 to show an increased level of readthrough at each of the tetranucleotide termination signals. Yeast strains expressing either the eRF3-H348Q or the eRF3-R419G protein as the sole source of eRF3 were found to exhibit an increased level of readthrough compared to cells expressing wild-type eRF3, but the level of readthrough showed a strong bias toward certain tetranucleotide termination signals (Table 1).
TABLE 1.
Effect of eRF3 mutations on translation termination
| Stop signalb | % Readthrough fora:
|
Change (fold) (relative to wild type) forc:
|
|||
|---|---|---|---|---|---|
| Wild type | R419G | H348Q | R419G | H348Q | |
| UAAA | 0.30 ± 0.01 | 0.27 ± 0.05 | 0.27 ± 0.07 | 0.9 | 0.9 |
| UAAC | 0.45 ± 0.04 | 1.95 ± 0.15 | 1.52 ± 0.28 | 4.4 | 3.4 |
| UAAG | 0.30 ± 0.01 | 0.27 ± 0.04 | 0.36 ± 0.06 | 0.9 | 1.2 |
| UAAU | 0.25 ± 0.02 | 0.50 ± 0.05 | 0.24 ± 0.02 | 2.0 | 0.9 |
| UAGA | 0.16 ± 0.03 | 0.24 ± 0.03 | 0.26 ± 0.03 | 1.5 | 1.7 |
| UAGC | 0.33 ± 0.02 | 1.31 ± 0.17 | 1.03 ± 0.15 | 4.0 | 3.2 |
| UAGG | 0.26 ± 0.01 | 0.25 ± 0.01 | 0.24 ± 0.03 | 1.0 | 0.9 |
| UAGU | 0.12 ± 0.01 | 0.12 ± 0.01 | 0.14 ± 0.02 | 1.1 | 1.2 |
| UGAA | 0.49 ± 0.06 | 2.42 ± 0.10 | 2.90 ± 0.18 | 4.9 | 5.9 |
| UGAC | 1.40 ± 0.40 | 11.6 ± 1.00 | 23.9 ± 1.80 | 8.2 | 16.8 |
| UGAG | 0.85 ± 0.14 | 3.10 ± 1.00 | 3.90 ± 0.42 | 3.7 | 4.6 |
| UGAU | 0.21 ± 0.03 | 1.10 ± 0.2 | 0.94 ± 0.06 | 5.2 | 4.5 |
Percent readthrough is expressed as the mean ± standard deviation.
All measurements were carried out with the [psi−] strain YDB498 (sup35Δ) carrying pDB663 (wild-type eRF3), pDB783 (eRF3-R419G), or pDB670 (eRF3-H348Q).
Changes in relative readthrough >2-fold are underlined.
The level of readthrough in strains expressing eRF3-H348Q or eRF3-R419G was increased at each of the four UGA(N) tetranucleotides (ranging from 3.7- to 16.8-fold) relative to a strain expressing wild-type eRF3 from the same plasmid. In contrast, an increase in readthrough attributable to these mutant release factors was either subtle or nonexistent at most UAGN or UAAN tetranucleotide stop signals. The major exceptions were the UAGC and UAAC tetranucleotides, where increases in readthrough of 3.2- to 4.4-fold were observed. Overall, the most dramatic increase in readthrough caused by the eRF3-R419G and eRF3-H348Q mutant proteins was observed at the UGAC termination signal, where readthrough was 8.2- and 16.8-fold higher, respectively, than the strain expressing wild-type eRF3.
Defects in the GTPase activity of eRF3 correlate with increased levels of readthrough.
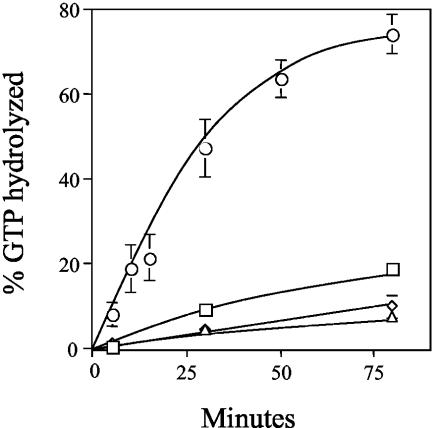
We next used an in vitro eRF3 GTPase assay to compare the biochemical properties of the wild-type and mutant forms of eRF3. In previous studies, a similar assay (with purified Xenopus laevis, rabbit, or human components) was used to show that both eRF1 and ribosomes are required to activate the GTPase activity of eRF3, suggesting that together they act as GTPase-activating proteins for eRF3 (18, 21). Here, we reconstituted a similar assay system to monitor GTP hydrolysis by eRF3 using purified yeast components. His6-tagged versions of full-length yeast eRF1 and the GTPase domain of yeast eRF3 (amino acids 254 to 685, hereafter referred to as eRF3-ΔNM) were purified from an E. coli expression system, while salt-washed ribosomes were purified from yeast cells. The GTPase domain of yeast eRF3 was used because the corresponding region of eRF3 from both X. laevis and humans was found to hydrolyze GTP in a manner similar to that of full-length eRF3 (18, 21). In addition, the GTPase domain of yeast eRF3 fully supports cell viability when provided as the sole source of eRF3 in yeast (52). In preliminary experiments, we found that none of the individual components alone (wild-type eRF3-ΔNM, eRF1, or ribosomes) showed significant GTPase activity (data not shown). Similarly, the pairwise combination of any of the three components was unable to stimulate more than a basal level of GTPase activity (Fig. 3). Only when all three components were present together in the reaction was the GTPase activity of wild-type eRF3-ΔNM activated. These results indicate that this yeast eRF3 GTPase assay system has the same requirements as a previously reported eRF3 assay that used components purified from other species (18).
FIG. 3.
eRF3 GTPase assays. GTPase assays were carried out with various combinations of purified eRF3-ΔNM, eRF1, and salt-washed ribosomes. The graph indicates the GTP hydrolysis associated with 3 pmol each of eRF3-ΔNM and eRF1 (triangles), eRF3-ΔNM and ribosomes (squares), eRF1 and ribosomes (diamonds), and eRF3-ΔNM, eRF1, and ribosomes (circles).
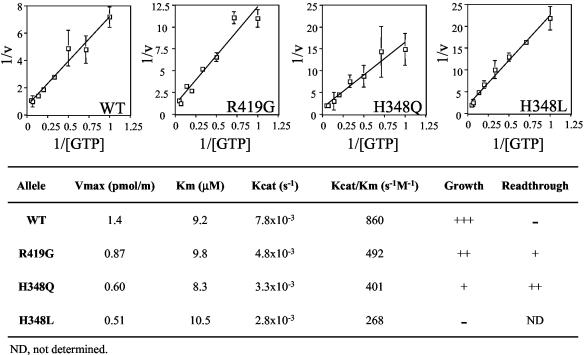
We next assayed the rate of GTP hydrolysis of the wild-type and mutant forms of eRF3 in the presence of different GTP concentrations. Kinetic parameters were determined by using Lineweaver-Burk plots, and these values were then compared to the efficiency of translation termination (Fig. 4). First, we found that a decrease in the efficiency of GTP hydrolysis (Kcat/Km) correlated with an increase in the readthrough of translation termination signals. This finding suggests that the GTPase activity of eRF3 plays an important role in the termination process. Second, we found that mutations that caused a reduction in the efficiency of GTP hydrolysis also caused a reduced growth rate. The H348L-eRF3 mutant, which showed the largest reduction in the efficiency of GTP hydrolysis of all the mutant proteins assayed, was unable to support cell viability as the sole source of eRF3. These results confirm that the GTPase activity of eRF3 is important for both efficient translation termination and cell viability. Furthermore, they indicate that a reduction in the efficiency of GTP hydrolysis leads to quite distinct effects on the efficiency of translation termination at different tetranucleotide termination signals.
FIG. 4.
Kinetic analysis of the GTPase activity of wild-type (WT) and mutant forms of eRF3. Reactions contained 3 pmol each of eRF1, eRF3-ΔNM, and ribosomes.
A reduction in the steady-state level of eRF1 does not cause selective readthrough at different termination signals.
It was recently shown that bacterial RF3 functions to maintain an adequate pool of free RF1 and RF2 available for translation termination by recycling these class I release factors following polypeptide chain release (17, 55, 56). In addition, it was suggested that eRF3 probably carries out a similar function during eukaryotic translation termination (55). This model predicts that mutations in eRF3 should lead to a reduction in the steady-state level of eRF1 available to recognize newly formed termination complexes. If correct, this model also predicts that the tetranucleotide-dependent readthrough documented above should also be observed if the pool of eRF1 available for termination is decreased by a means other than inefficient eRF1 recycling.
To test this possibility, we expressed the SUP45 gene under the control of the CUP1 promoter in a strain lacking the genomic copy of the SUP45 gene (sup45Δ). Gene expression from the CUP1 promoter is determined by the amount of copper in the growth medium, and trace amounts are normally sufficient for a partial induction of transcription. Pilot experiments demonstrated that the basal level of eRF1 expression driven by the CUP1 promoter during growth in standard SMD medium could be decreased further by the addition of EDTA (data not shown). When the growth medium was supplemented with 50 μM EDTA, we found that expression of eRF1 from the CUP1 promoter was reduced approximately twofold below the level of eRF1 produced from the SUP45 promoter at its normal genomic locus (Fig. 5). We found that this reduction in the eRF1 level coincided with a general increase in readthrough at all tetranucleotide termination signals that ranged from 3.3- to 12.5-fold (Table 2). In contrast, the experiments described above demonstrated that the eRF3-H348Q mutant led to an increase of 3.2- to 16.8-fold in readthrough at half of the termination signals but little or no increase in readthrough at the remainder. These striking differences in the pattern of suppression suggest that the termination signal-dependent readthrough associated with the eRF3 GTPase mutations is fundamentally different from the general increase in readthrough associated with a depletion of eRF1.
FIG. 5.
Reduced steady-state level of eRF1 following its expression from the CUP1 promoter as determined by Western blot analysis. Reduced expression of eRF1 was driven from the CUP1 promoter (PCUP1) in a sup45Δ strain, while the control strain expressed eRF1 from its natural promoter (PSUP45) at its genomic locus. Both strains were grown in SMD medium containing 50 μM EDTA to specifically reduce eRF1 expression from the CUP1 promoter. Twenty-five micrograms of total protein was loaded per lane.
TABLE 2.
Effect of eRF1 depletion on translation termination
| Stop signalb | % Readthrough fora:
|
Change (fold) | |
|---|---|---|---|
| PSUP45 | PCUP1 | ||
| UAAA | 0.19 ± 0.02 | 0.94 ± 0.05 | 5 |
| UAAC | 0.37 ± 0.06 | 2.8 ± 0.27 | 7.5 |
| UAAG | 0.22 ± 0.02 | 1.2 ± 0.07 | 5.3 |
| UAAU | 0.28 ± 0.01 | 0.97 ± 0.10 | 3.4 |
| UAGA | 0.18 ± 0.01 | 0.60 ± 0.05 | 3.3 |
| UAGC | 0.49 ± 0.02 | 2.0 ± 0.18 | 4.1 |
| UAGG | 0.13 ± 0.01 | 0.80 ± 0.04 | 6.5 |
| UAGU | 0.12 ± 0.02 | 0.70 ± 0.07 | 5.9 |
| UGAA | 0.37 ± 0.04 | 4.6 ± 0.50 | 12.5 |
| UGAC | 1.3 ± 0.12 | 11.7 ± 0.95 | 9 |
| UGAG | 0.55 ± 0.03 | 1.9 ± 0.23 | 3.5 |
| UGAU | 0.24 ± 0.05 | 1.2 ± 0.2 | 5 |
Percent readthrough is expressed as the mean ± standard deviation.
All measurements were carried out using the [psi−] strains YDB340 (wild type) or YDB447 (sup45Δ) carrying pDB811, which expresses eRF1 from the CUP1 promoter.
Depletion of eRF3 does not prevent eRF1 from associating with the ribosome.
The elongation factors EF-Tu and eEF1A are GTPases that bind and deliver aminoacyl-tRNAs to the ribosomal A site in prokaryotes and eukaryotes, respectively. The GTPase domain of eRF3 shares extensive sequence homologies with those elongation factors, and recent studies have suggested that eRF1 is a functional mimic of a tRNA molecule (48). Since our readthrough data indicated that the GTPase domain mutants of eRF3 cause a context-dependent readthrough phenotype, we next considered the possibility that a reduction in the GTPase activity of eRF3 could manifest these termination defects by inefficiently targeting eRF1 to the ribosome.
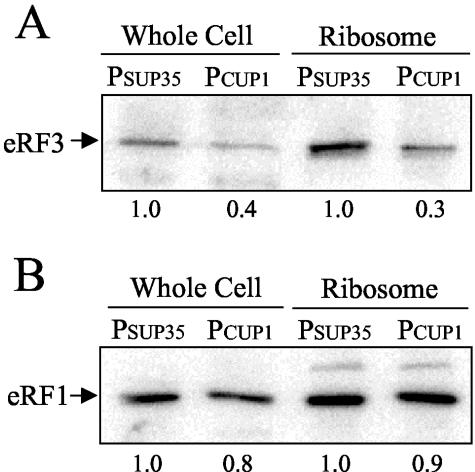
Previous studies have shown that eRF1 and eRF3 are present in roughly equimolar amounts in the cell, and the majority of these factors are associated with ribosomes at any given time (13, 50). Furthermore, eRF1 and eRF3 have been shown to interact both in vitro and in vivo, and the simultaneous overproduction of both factors is required to increase the efficiency of translation termination in yeast (51). One possible explanation for these results is that eRF3 is required to efficiently target eRF1 to ribosomes. This model predicts that a partial depletion of eRF3 should result in a corresponding decrease in the concentration of eRF1 bound to ribosomes. To test this possibility, we utilized the CUP1 promoter expression system to reduce eRF3 expression in a yeast strain that lacked the genomic SUP35 gene (sup35Δ). We found that the growth of this strain with the SUP35 gene under CUP1 promoter control in SMD medium supplemented with 50 μM EDTA resulted in a two- to threefold decrease in the level of eRF3 in both total cell extracts and ribosomal pellets compared to the normal level of eRF3 expressed from the SUP35 promoter at its genomic locus (Fig. 6A). However, we did not observe any significant difference in the concentration of eRF1 in ribosomal pellets prepared from these cells (Fig. 6B). While these results do not address possible differences in the rate of eRF1 targeting to ribosomes, they indicate that eRF3 does not play a significant role in determining the steady-state level of eRF1 bound to ribosomes.
FIG. 6.
Reduced steady-state level of eRF3 does not alter the amount of eRF1 associated with ribosomes. Western blots indicating total and ribosome-associated eRF3 levels (A) and total and ribosome-associated eRF1 levels (B) are shown. Twenty-five micrograms of total protein was loaded per lane. Reduced expression of eRF3 was driven from the CUP1 promoter (PCUP1) in a sup35Δ strain, while the control strain expressed eRF3 from its natural promoter (PSUP35) at its genomic locus. Both strains were grown in SMD medium containing 50 μM EDTA to specifically reduce eRF3 expression from the CUP1 promoter.
To determine the effect of eRF3 depletion on the efficiency of translation termination, we next examined the efficiency of translation termination at each of the 12 tetranucleotide termination signals. We again observed a general elevation of readthrough at each of these termination signals as a consequence of eRF3 depletion, with increases ranging from 5.0- to 16.4-fold (Table 3). These results indicate that a modest depletion of eRF3 causes a general decrease in the efficiency of translation termination even when a normal level of eRF1 is bound to the ribosome. Furthermore, these results demonstrate that a limitation of eRF3 is associated with a readthrough phenotype distinct from that observed with the eRF3 GTPase mutants.
TABLE 3.
Effect of eRF3 depletion on translation termination
| Stop signalb | % Readthrough fora:
|
Change (fold) | |
|---|---|---|---|
| PSUP35 | PCUP1 | ||
| UAAA | 0.16 ± 0.02 | 0.78 ± 0.02 | 5.0 |
| UAAC | 0.15 ± 0.01 | 1.2 ± 0.1 | 8.2 |
| UAAG | 0.13 ± 0.02 | 0.71 ± 0.12 | 5.6 |
| UAAU | 0.08 ± 0.01 | 0.79 ± 0.07 | 9.6 |
| UAGA | 0.10 ± 0.02 | 0.64 ± 0.1 | 6.4 |
| UAGC | 0.20 ± 0.03 | 1.7 ± 0.17 | 8.3 |
| UAGG | 0.13 ± 0.03 | 1.0 ± 0.15 | 8.1 |
| UAGU | 0.06 ± 0.01 | 0.30 ± 0.03 | 5.4 |
| UGAA | 0.17 ± 0.01 | 1.3 ± 0.14 | 7.7 |
| UGAC | 0.35 ± 0.01 | 5.3 ± 0.5 | 15.1 |
| UGAG | 0.11 ± 0.02 | 1.8 ± 0.06 | 16.4 |
| UGAU | 0.07 ± 0.02 | 0.96 ± 0.2 | 13.3 |
Percent readthrough is expressed as the mean ± standard deviation.
All measurements were carried out using the [psi−] strains 614 (wild type) or YDB498 (sup35Δ) carrying pDB824, which expresses eRF3 from the CUP1 promoter.
Overproduction of eRF1 does not cause an increase in the efficiency of translation termination.
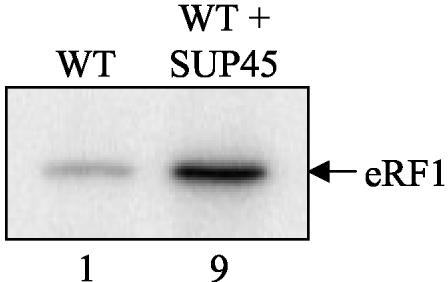
The bacterial recycling model for RF3 function predicts that RF1 or RF2 is alone sufficient to mediate polypeptide chain release (17, 55, 56). Consistent with this prediction, the overproduction of only RF1 is sufficient to increase the efficiency of translation termination at UAG-containing termination signals (12). Similarly, if eRF1 alone was sufficient to facilitate polypeptide chain release, an increase in the steady-state level of eRF1 in the presence of a normal level of eRF3 should increase the efficiency of translation termination. To test this possibility, we overproduced eRF1 in yeast cells by introducing a multicopy plasmid that expressed the SUP45 gene under the control of its own promoter. Western blot analysis indicated that this plasmid resulted in a ninefold increase in the steady-state level of eRF1 (Fig. 7). We then introduced the dual luciferase reporter constructs and assayed the efficiency of translation termination at each of the 12 tetranucleotide termination signals. We found that eRF1 overproduction did not significantly change the efficiency of translation termination at most signals (Table 4). Importantly, we did not observe an increase in the efficiency of termination at any signal. Instead, excess eRF1 appeared to cause a small decrease in termination efficiency at the UGAC and UGAG signals. These results indicate that the overproduction of eRF1 alone is insufficient to increase the efficiency of translation termination in vivo.
FIG. 7.
Introduction of a multicopy plasmid expressing the SUP45 gene from its own promoter leads to an increased steady-state level of eRF1. WT, wild type.
TABLE 4.
Effect of eRF1 overexpression on translation termination
| Stop signalb | % Readthrough fora:
|
Change (fold) | |
|---|---|---|---|
| Wild type | High-copy-no. eRF1 | ||
| UAAA | 0.18 ± 0.03 | 0.17 ± 0.02 | 0.94 |
| UAAC | 0.16 ± 0.05 | 0.15 ± 0.02 | 0.94 |
| UAAG | 0.13 ± 0.01 | 0.14 ± 0.01 | 1.1 |
| UAAU | 0.11 ± 0.01 | 0.12 ± 0.02 | 1.1 |
| UAGA | 0.09 ± 0.01 | 0.09 ± 0.01 | 0.96 |
| UAGC | 0.18 ± 0.02 | 0.18 ± 0.03 | 1.0 |
| UAGG | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.96 |
| UAGU | 0.05 ± 0.01 | 0.06 ± 0 | 1.2 |
| UGAA | 0.21 ± 0.03 | 0.21 ± 0.01 | 1.0 |
| UGAC | 0.34 ± 0.04 | 0.53 ± 0.02 | 1.5 |
| UGAG | 0.13 ± 0.02 | 0.26 ± 0.02 | 2.0 |
| UGAU | 0.05 ± 0 | 0.06 ± 0.01 | 1.1 |
Percent readthrough is expressed as the mean ± standard deviation.
All measurements were carried out using the [psi−] strain 614 with or without pDB773, which expresses eRF1 from the SUP45 promoter.
DISCUSSION
The objective of this study was to gain insights into eRF3 function during eukaryotic translation termination. Recent studies have shown that bacterial RF3 functions primarily to recycle the class I release factors RF1 and RF2 from the termination complex following polypeptide chain release (17, 55, 56). It was shown that nucleotide exchange (GDP→GTP) by RF3 facilitates RF1 and RF2 dissociation from the ribosome following nascent chain release, while GTP hydrolysis mediates the subsequent departure of RF3 from the posttermination complex. We reasoned that we could determine whether eRF3 facilitates a similar recycling mechanism during eukaryotic translation termination through the use of eRF3 mutants that exhibit defects in GTP binding and/or hydrolysis. Based on the bacterial model, mutants that reduced the rate of nucleotide exchange should also reduce eRF1 release from the termination complex following polypeptide chain release. This should lead to a reduction in the pool of eRF1 available for recruitment to subsequent termination complexes. In contrast, mutants that reduced the rate of GTP hydrolysis should be defective in the release of eRF3 from the ribosome. This should deplete the level of eRF3 available to bind subsequent termination complexes, which should again ultimately reduce the recycling of eRF1. Our kinetic analysis indicated that the eRF3-H348Q and eRF3-R419G mutants belonged to this latter class, since both exhibited defects in the rate of GTP hydrolysis (Kcat) while retaining normal Km values.
Since the mutations characterized in the present study were predicted to reduce the pool of eRF1 available to form new termination complexes according to the recycling model, we asked whether a partial depletion of eRF1 by another means could cause a similar pattern of termination signal-dependent readthrough of stop codons. Significantly, we found that a moderate (twofold) reduction in the level of eRF1 resulted in an increase in readthrough at all 12 tetranucleotide termination signals. This result differed significantly from the finding that eRF3 mutations caused readthrough at some stop signals but not at others. This difference in the pattern of readthrough was not due simply to a more severe depletion of eRF1 in this experiment, since eRF1 depletion increased readthrough at the UGAC termination signal by 9-fold, while readthrough increased 17-fold at the same termination signal in a strain expressing eRF3-H348Q.
The recycling model for bacterial RF3 function predicts that an increase in the concentration of RF1 alone should increase the efficiency of translation termination. Notably, in vivo studies have shown this prediction to be correct (12). In contrast, it was reported that the overproduction of either eRF1 or eRF3 alone in S. cerevisiae is unable to increase the efficiency of translation termination, while the overproduction of both factors together increases the termination efficiency (referred to as antisuppression) (51). Using a dual luciferase readthrough system, we found that a ninefold overproduction of eRF1 alone did not lead to antisuppression at any of the 12 tetranucleotide termination signals. These results clearly suggest that eRF1 alone is not capable of mediating translation termination in vivo. In fact, eRF1 overproduction reduced the efficiency of termination at the UGAG and UGAC signals, two of the weakest tetranucleotide termination signals. This suggests that excess eRF1 may act as a mild competitor for the formation of productive termination complexes.
eRF1 and eRF3 associate to form a stable complex (14, 16, 44, 51), although it is not known whether complex formation is a prerequisite for the efficient binding of either factor to ribosomes. To address this possibility, we also examined the effect of reducing the steady-state level of eRF3 in vivo. We found that a normal level of eRF1 was associated with ribosomes even when they contained two- to threefold less eRF3 than normal. This indicates that the formation of a complex between eRF1 and eRF3 is not required to deliver eRF1 to ribosomes. Moreover, we found that a reduction in the steady-state level of eRF3 caused a general decrease in the efficiency of translation termination at all tetranucleotide termination signals. Again, these results are distinct from those obtained with the eRF3 mutants that exhibited a reduced efficiency of translation termination at a distinct subset of termination signals. These findings lead us to conclude that eRF3 limitation is not the primary defect resulting from the eRF3 GTPase mutations analyzed in our study.
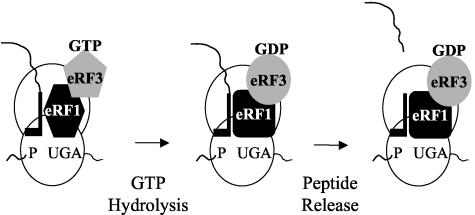
It is thought that stop codon recognition is mediated by eRF1 (3, 8-10, 19, 31). The results of this study indicate that the signal-dependent defects in translation termination associated with the eRF3 GTPase mutations are not caused by a depletion of either eRF1 or eRF3. This result strongly suggests that eRF3 does not function simply to recycle eRF1. Another model proposed by Frolova et al. suggests that the GTP-bound form of eRF3 controls the positioning of eRF1 toward the termination signal and the peptidyl tRNA, while the GDP-bound form promotes the release of both eRF1 and eRF3 from the ribosome (18). While this model is consistent with some of our results, it does not adequately explain how mutations that alter the rate of GTP catalysis can influence stop signal decoding in a signal-dependent manner. To accommodate the results of our study, we propose a revised model in which the GTPase activity of eRF3 assists in stop signal decoding by eRF1 prior to nascent chain release (Fig. 8). In this model, a stable interaction between eRF1 and a stop codon in the ribosomal A site serves to stimulate GTP hydrolysis by eRF3. This event then activates eRF1 in some way so that it can efficiently stimulate nascent chain release. If eRF1 binds to each stop signal with a distinct affinity, its ability to activate GTP hydrolysis by eRF3 may be slower at low-affinity signals (such as UGAC). We propose that the slower kinetics of GTPase activation at low-affinity stop signals remain sufficient to activate efficient nascent chain release by eRF1 under normal conditions. However, in the presence of eRF3 mutants that exhibit a reduced rate of GTP hydrolysis, the incidence of release factor dissociation from the termination complex could become more frequent. This would lead to a decrease in termination efficiency at a subset of termination signals, as was observed in the present study.
FIG. 8.
Model illustrating how GTP hydrolysis by eRF3 may enhance the decoding of termination signals during translation termination.
This model is also consistent with the results of other recent studies of eukaryotic translation termination. In vitro cross-linking experiments using termination complexes generated with mRNA containing 4-thiouridine at position 1 of the A site codon have shown that eRF1 can be cross-linked directly to the first nucleotide of stop codons (8-10). However, it was found that eRF1 also cross-linked to the UGG (tryptophan) codon much more readily than to other sense codons. This finding suggests that it may be difficult for eRF1 alone to distinguish the UGG codon from bona fide stop codons (9). Based on those cross-linking results, it was hypothesized that eRF3 may help eRF1 discriminate between UGG and certain stop codons. In light of this prediction, it was particularly striking that we observed high levels of readthrough with both the eRF3-H348Q and eRF3-R419G mutants at each of the four UGA-containing tetranucleotides (see the UGAA, UGAC, UGAG, and UGAU termination signals in Table 1). These results suggest that the UGA termination signals are the most difficult for eRF1 to productively decode in vivo when the GTPase activity of eRF3 is compromised. At each of the three triplet stop codons, we also observed a significantly higher level of readthrough in strains expressing the eRF3 GTPase mutants when a C residue was present at position 4 of the tetranucleotide termination signal (see the UGAC, UAGC, and UAAC termination signals in Table 1). This finding suggests that eRF1 may also recognize tetranucleotide termination signals that contain a C residue in position 4 less efficiently than other termination signals.
Our experiments that examined the effects of the eRF3 GTPase mutants and the consequences of eRF1 overproduction suggest that GTP hydrolysis by eRF3 is required to stimulate nascent chain release. Based on these results, we propose that eRF3 is recruited to the ribosome in the GTP-bound form, and the subsequent hydrolysis of this nucleotide is important for eRF1 to induce nascent chain release. This aspect of our model is consistent with a recent publication describing the crystal structure of a C-terminal derivative of eRF3 (amino acid residues 196 to 662) from S. pombe (37). In that study, it was found that eRF3 is unable to bind GDP in the presence of 2 mM Mg2+. While eRF3 could tightly bind GDP in the complete absence of Mg2+ (Kd = 3.8 μM), binding was severely reduced when the Mg2+ concentration was increased to only 0.3 mM Mg2+. In contrast, eRF3 bound the GTP analogue GDPNP well (Kd = 200 to 300 μM) in the presence of 2 mM Mg2+. Since the cytoplasmic Mg2+ concentration in eukaryotes is thought to be 0.5 to 1.0 mM (1, 58), these results suggest that the large majority of cellular eRF3 will exist in the GTP-bound state at any given time. This makes it highly unlikely that nucleotide exchange (GDP→GTP) by eRF3 will play the same critical role in eukaryotic translation termination that it does in bacteria.
It may seem unlikely that class II release factors would carry out such different functions during translation termination in prokaryotes compared to eukaryotes. However, these proteins do not represent a phylogenetically well-conserved component of the translational machinery. In fact, it has been proposed that RF3 has a common origin with EF-G, while eRF3 originated from the same ancestor protein as eEF1A (29, 42). These distinct origins are also consistent with the observation that bacterial RF3 is dispensable for growth, while yeast eRF3 is essential for cell viability. In addition, little or no sequence homology is found between prokaryotic and eukaryotic class I release factors, while the eukaryotic and Archaea class I release factors share ∼30% sequence identity (48). The observation that no genes encoding a homologue of RF3 (or eRF3) have been found in several sequenced Archaea genomes is consistent with the notion that these organisms may also have developed an alternative strategy to recycle their class I release factors. In eubacteria, RF1 decodes UAG and UAA stop codons, while RF2 decodes UGA and UAA stop codons (34). This division of labor in stop codon recognition may provide an adequate level of accuracy during the termination process. In contrast, eukaryotes have only one class I release factor that must properly decode UAG, UAA, and UGA stop codons and distinguish them from near-cognate codons, particularly the UGG codon (15, 34, 43). This may necessitate an additional step during the termination process that is facilitated by eRF3.
Acknowledgments
We thank John Atkins, Yury Chernoff, Mike Culbertson, James Friesen, Mark Hochstrasser, Mick Tuite, Jonathan Weissman, and Ming Du for providing strains and reagents. We also thank Peter Prevelige, Kim Keeling, and other Bedwell lab members for critically reading the manuscript.
This study was supported by NIH grant GM 68854 (D.M.B.).
REFERENCES
- 1.Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell, 4th ed., p. 616. Garland Science, New York, N.Y.
- 2.Anborgh, P. H., R. H. Cool, F. Gumusel, K. Harmark, E. Jacquet, A. Weijland, M. Y. Mistou, and A. Parmeggiani. 1991. Structure-function relationships of elongation factor Tu as studied by mutagenesis. Biochimie 73:1051-1059. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, G., H. A. Bell, D. W. Ritchie, G. Fullerton, and I. Stansfield. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonetti, B., L. Fu, J. Moon, and D. M. Bedwell. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251:334-345. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. M., P. A. Stockwell, C. N. Trotman, and W. P. Tate. 1990. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 18:6339-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, D., D. C. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Cavallius, J., and W. C. Merrick. 1998. Site-directed mutagenesis of yeast eEF1A. Viable mutants with altered nucleotide specificity. J. Biol. Chem. 273:28752-28758. [DOI] [PubMed] [Google Scholar]
- 8.Chavatte, L., L. Frolova, P. Laugaa, L. Kisselev, and A. Favre. 2003. Stop codons and UGG promote efficient binding of the polypeptide release factor eRF1 to the ribosomal A site. J. Mol. Biol. 331:745-758. [DOI] [PubMed] [Google Scholar]
- 9.Chavatte, L., S. Kervestin, A. Favre, and O. Jean-Jean. 2003. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s. EMBO J. 22:1644-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavatte, L., A. Seit-Nebi, V. Dubovaya, and A. Favre. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 21:5302-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cool, R. H., and A. Parmeggiani. 1991. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry 30:362-366. [DOI] [PubMed] [Google Scholar]
- 12.Crawford, D. J., K. Ito, Y. Nakamura, and W. P. Tate. 1999. Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3. EMBO J. 18:727-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didichenko, S. A., M. D. Ter-Avanesyan, and V. N. Smirnov. 1991. Ribosome-bound EF-1 alpha-like protein of yeast Saccharomyces cerevisiae. Eur. J. Biochem. 198:705-711. [DOI] [PubMed] [Google Scholar]
- 14.Ebihara, K., and Y. Nakamura. 1999. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA 5:739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrenberg, M., and T. Tenson. 2002. A new beginning of the end of translation. Nat. Struct. Biol. 9:85-87. [DOI] [PubMed] [Google Scholar]
- 16.Eurwilaichitr, L., F. M. Graves, I. Stansfield, and M. F. Tuite. 1999. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 32:485-496. [DOI] [PubMed] [Google Scholar]
- 17.Freistroffer, D. V., M. Y. Pavlov, J. MacDougall, R. H. Buckingham, and M. Ehrenberg. 1997. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 16:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolova, L., X. Le Goff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334-341. [PMC free article] [PubMed] [Google Scholar]
- 19.Frolova, L., A. Seit-Nebi, and L. Kisselev. 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frolova, L. Y., T. I. Merkulova, and L. L. Kisselev. 2000. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolova, L. Y., J. L. Simonsen, T. I. Merkulova, D. Y. Litvinov, P. M. Martensen, V. O. Rechinsky, J. H. Camonis, L. L. Kisselev, and J. Justesen. 1998. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 256:36-44. [DOI] [PubMed] [Google Scholar]
- 22.Frolova, L. Y., R. Y. Tsivkovskii, G. F. Sivolobova, N. Y. Oparina, O. I. Serpinsky, V. M. Blinov, S. I. Tatkov, and L. L. Kisselev. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlach, W. L. 1975. Genetic properties of some amber-ochre supersuppressors in Saccharomyces cerevisiae. Mol. Gen. Genet. 138:53-63. [DOI] [PubMed] [Google Scholar]
- 24.Grentzmann, G., J. A. Ingram, P. J. Kelly, R. F. Gesteland, and J. F. Atkins. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479-486. [PMC free article] [PubMed] [Google Scholar]
- 25.Himmelfarb, H. J., E. Maicas, and J. D. Friesen. 1985. Isolation of the SUP45 omnipotent suppressor gene of Saccharomyces cerevisiae and characterization of its gene product. Mol. Cell. Biol. 5:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochstrasser, M., M. J. Ellison, V. Chau, and A. Varshavsky. 1991. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 88:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard, M. T., B. H. Shirts, L. M. Petros, K. M. Flanigan, R. F. Gesteland, and J. F. Atkins. 2000. Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 48:164-169. [PubMed] [Google Scholar]
- 28.Hwang, Y. W., A. Sanchez, and D. L. Miller. 1989. Mutagenesis of bacterial elongation factor Tu at lysine 136. A conserved amino acid in GTP regulatory proteins. J. Biol. Chem. 264:8304-8309. [PubMed] [Google Scholar]
- 29.Inge-Vechtomov, S., G. Zhouravleva, and M. Philippe. 2003. Eukaryotic release factors (eRFs) history. Biol. Cell 95:195-209. [DOI] [PubMed] [Google Scholar]
- 30.Ito, K., K. Ebihara, and Y. Nakamura. 1998. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA 4:958-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, K., L. Frolova, A. Seit-Nebi, A. Karamyshev, L. Kisselev, and Y. Nakamura. 2002. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. USA 99:8494-8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, J. S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 33.Keeling, K. M., J. Lanier, M. Du, J. Salas-Marco, L. Gao, A. Kaenjak-Angeletti, and D. M. Bedwell. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kisselev, L., M. Ehrenberg, and L. Frolova. 2003. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 22:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitada, K., E. Yamaguchi, and M. Arisawa. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165:203-206. [DOI] [PubMed] [Google Scholar]
- 36.Knudsen, C. R., I. V. Kjaersgard, O. Wiborg, and B. F. Clark. 1995. Mutation of the conserved Gly94 and Gly126 in elongation factor Tu from Escherichia coli. Elucidation of their structural and functional roles. Eur. J. Biochem. 228:176-183. [PubMed] [Google Scholar]
- 37.Kong, C., K. Ito, M. A. Walsh, M. Wada, Y. Liu, S. Kumar, D. Barford, Y. Nakamura, and H. Song. 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell 14:233-245. [DOI] [PubMed] [Google Scholar]
- 38.Liebman, S. W., and M. Cavenagh. 1980. An antisuppressor that acts on omnipotent suppressors in yeast. Genetics 95:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.McCaughan, K. K., C. M. Brown, M. E. Dalphin, M. J. Berry, and W. P. Tate. 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 92:5431-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merkulova, T. I., L. Y. Frolova, M. Lazar, J. Camonis, and L. L. Kisselev. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443:41-47. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, Y., and K. Ito. 1998. How protein reads the stop codon and terminates translation. Genes Cells 3:265-278. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, Y., and K. Ito. 2003. Making sense of mimic in translation termination. Trends Biochem. Sci. 28:99-105. [DOI] [PubMed] [Google Scholar]
- 44.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1997. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol. 17:2798-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seit-Nebi, A., L. Frolova, J. Justesen, and L. Kisselev. 2001. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29:3982-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seit-Nebi, A., L. Frolova, and L. Kisselev. 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 9:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 48.Song, H., P. Mugnier, A. K. Das, H. M. Webb, D. R. Evans, M. F. Tuite, B. A. Hemmings, and D. Barford. 2000. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100:311-321. [DOI] [PubMed] [Google Scholar]
- 49.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66:639-678. [DOI] [PubMed] [Google Scholar]
- 50.Stansfield, I., G. M. Grant, Akhmaloka, and M. F. Tuite. 1992. Ribosomal association of the yeast SAL4 (SUP45) gene product: implications for its role in translation fidelity and termination. Mol. Microbiol. 6:3469-3478. [DOI] [PubMed] [Google Scholar]
- 51.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ter-Avanesyan, M. D., V. V. Kushnirov, A. R. Dagkesamanskaya, S. A. Didichenko, Y. O. Chernoff, S. G. Inge-Vechtomov, and V. N. Smirnov. 1993. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 7:683-692. [DOI] [PubMed] [Google Scholar]
- 53.Velichutina, I. V., J. Y. Hong, A. D. Mesecar, Y. O. Chernoff, and S. W. Liebman. 2001. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol. 305:715-727. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, P. G., and M. R. Culbertson. 1988. SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J. Mol. Biol. 199:559-573. [DOI] [PubMed] [Google Scholar]
- 55.Zavialov, A. V., R. H. Buckingham, and M. Ehrenberg. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107:115-124. [DOI] [PubMed] [Google Scholar]
- 56.Zavialov, A. V., L. Mora, R. H. Buckingham, and M. Ehrenberg. 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10:789-798. [DOI] [PubMed] [Google Scholar]
- 57.Zeidler, W., C. Egle, S. Ribeiro, A. Wagner, V. Katunin, R. Kreutzer, M. Rodnina, W. Wintermeyer, and M. Sprinzl. 1995. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur. J. Biochem. 229:596-604. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, A., T. P. Cheng, X. Y. Wu, B. T. Altura, and B. M. Altura. 1997. Extracellular Mg2+ regulates intracellular Mg2+ and its subcellular compartmentation in fission yeast, Schizosaccharomyces pombe. Cell. Mol. Life Sci. 53:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]