The development of genetic profiles of invading plants can provide insight towards a better understanding of niche utilization. Recent advances in sequencing techniques have greatly facilitated genetic assessment and profiling. Here we review our recent studies on selected Australian Cucurbitaceae and Boraginaceae to demonstrate the application of genetic sequencing across a selection of invasive Australian plants.
Keywords: DNA barcoding, genetic profile, invasion history, plant invasion, sequencing technology
Abstract
Part of the challenge in dealing with invasive plant species is that they seldom represent a uniform, static entity. Often, an accurate understanding of the history of plant introduction and knowledge of the real levels of genetic diversity present in species and populations of importance is lacking. Currently, the role of genetic diversity in promoting the successful establishment of invasive plants is not well defined. Genetic profiling of invasive plants should enhance our understanding of the dynamics of colonization in the invaded range. Recent advances in DNA sequencing technology have greatly facilitated the rapid and complete assessment of plant population genetics. Here, we apply our current understanding of the genetics and ecophysiology of plant invasions to recent work on Australian plant invaders from the Cucurbitaceae and Boraginaceae. The Cucurbitaceae study showed that both prickly paddy melon (Cucumis myriocarpus) and camel melon (Citrullus lanatus) were represented by only a single genotype in Australia, implying that each was probably introduced as a single introduction event. In contrast, a third invasive melon, Citrullus colocynthis, possessed a moderate level of genetic diversity in Australia and was potentially introduced to the continent at least twice. The Boraginaceae study demonstrated the value of comparing two similar congeneric species; one, Echium plantagineum, is highly invasive and genetically diverse, whereas the other, Echium vulgare, exhibits less genetic diversity and occupies a more limited ecological niche. Sequence analysis provided precise identification of invasive plant species, as well as information on genetic diversity and phylogeographic history. Improved sequencing technologies will continue to allow greater resolution of genetic relationships among invasive plant populations, thereby potentially improving our ability to predict the impact of these relationships upon future spread and better manage invaders possessing potentially diverse biotypes and exhibiting diverse breeding systems, life histories and invasion histories.
Introduction
The seriousness of the global challenge due to invasive plant species has been increasingly recognized in the past two decades, while climate change and increased global trade have served to accelerate plant invasion (Beerling et al., 1995; Bunce and Ziska, 2000; Ding et al., 2008; McDonald et al., 2009; Clements and Ditommaso, 2011; Hyvönen et al., 2012; Gallagher et al., 2013; Singh et al., 2013; Clements et al., 2014; Seebens et al., 2015). In Australia, >3000 non-native plant species are now recorded as naturalized (Stow et al., 2014), and threats from these species are increasing exponentially. Many of these invaders have become noxious or weedy, with an estimated annual cost of over 4 billion AUD (Olson, 2006). For example, one invasive weed, bitou bush (Chrysanthemoides monilifera), is associated with population decline in 63 rare and threatened native plant species in New South Wales alone (Kohli et al., 2008). In addition, when present in agricultural lands, weed infestation contributes to the majority (34%) of total losses attributable to pests relative to all crop pests (Oerke, 2006).
In order to address the challenges associated with invasive weeds, systems of prediction are being developed, in terms of both associated theoretical frameworks that attempt to identify the major predictors of invasion (e.g. Daehler, 2003) and models that predict the extent of invasion. In particular models, potential regions of further invasion are identified by evaluating current home ranges and predicted ranges where species may invade based on climate change and other factors (e.g. Kriticos et al., 2005; Ebeling et al., 2008; McDonald et al., 2009). Many attempts have been made to predict the scope of future invasions, but information on critical aspects of invasive plant biology is often lacking, including the ability of species to evolve in response to selection pressures, such as climate change (Clements and DiTommaso, 2011, 2012).
Fundamental to the nature of a given species or individual organism is a plant's genetic identity. Gene regulation and environmental interactions determine the physiological nature of a plant as it develops from seeds and/or other types of propagules, which in turn determines its eco-physiological success and eventual impact on ecosystems and/or human economies. A common shortcoming in the management of invasive plants is the failure to recognize a given weed species as not only a single genetic identity but a collection of populations that may vary greatly across a variety of scales from local to regional to global (Meekins et al., 2001; Lavergne and Molofsky, 2007; Prentis et al., 2008). Therefore, although a species may be a single entity by definition, populations of a particular species may exhibit both genotypic and phenotypic variation. Thus, their successful management may be improved greatly by addressing specific genetic manifestations of the species resulting in phenotypic variation attributable to genetic variation and/or plasticity.
Genotypic and phenotypic diversity is also observed in invaders across all taxa, but it is important to highlight particular features of plants that are crucial in predicting the success of plant invasions. Plant breeding systems and life histories are therefore key considerations. In terms of breeding systems, plants may fall anywhere on a spectrum from obligate outcrossing to 100% selfing (Clements et al., 2004). Many plants forgo reproduction by seed as well, often making use of the advantages afforded by vegetative propagation from already vigorously growing plant parts. In terms of plant life histories, whether a plant is an annual or perennial or some intermediate of the two extremes can influence whether or not it is or could become a problematic invader (Meekins et al., 2001; Lavergne and Molofsky, 2007; Prentis et al., 2008).
Although we know much about the genetics of particular invasive species, there are still many gaps in our knowledge (Bock et al., 2015). For example, there remain important questions around what factors influence the primary sources of genetic variation, the role of genetic bottlenecks in potentially hindering the success of plants at the fringe of an invasion wave, and whether propagule load is more important than genetic diversity in promoting establishment; these are questions that may be answered by both genomic studies and studies of plant ecophysiology using model organisms (Bock et al., 2015).
In this review, we compare and contrast the genetic diversity of two models; Australian congeneric invaders representing the Cucurbitaceae and Boraginaceae. The Boraginaceae model compares two congeric invaders introduced to Australia in a similar time frame; one highly successful invader and the other a niche colonizer with similar morphological, chemical and biological features (Skoneczny et al., 2015; Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, under review; Zhu et al., 2016). The Cucurbitaceae model compares three related melons that appear to have been introduced to Australia via camel trading routes established in the 1800s, with genetic diversity among the three largely selfing species varying from existing as a single in genotype in Australia for prickly paddy melon (Cucumis myriocarpus) and camel melon (Citrullus lanatus) to the more heterogeneous populations of Citrullus colocynthis composed of two major introduced genotypes in Australia (Shaik et al., 2015). Two of the melon species (C. myriocarpus) and (C. lanatus) are annuals, whereas C. colocynthis is perennial. The experience of working with these two different plant families in Australia using similar genetic analysis methods enables us to draw some general conclusions on the value of such analyses in characterizing continental invasions by a variety of taxa. Thus, our overall objective is to examine how recent advancements in genetic characterization and sequence analysis can be applied successfully to invasive plants with varying life histories, breeding systems and invasion histories.
Recent advances in DNA sequencing for invasive plants
One key innovation in recent years is the development of DNA barcoding for rapidly characterizing invasive plant genetics. DNA barcoding can be defined as ‘a diagnostic technique in which short DNA sequence(s) can be used for species identification’ (Savolainen et al., 2005). DNA barcoding using the 648 bp region of the mitochondrial gene cytochrome c oxidase I is a well-accepted method of species identification in animals (Wiemers and Fiedler, 2007; Lahaye et al., 2008; Hollingsworth et al., 2009). Successful use of barcoding requires that genetic distance between species is larger than within-species distance. Its success also depends on monophyly of the species examined (Wiemers and Fiedler, 2007). Interestingly, species boundaries in plants are typically less pronounced than in animals. In some cases, up to 50% of plants show higher levels of gene tree paraphyly, and interspecific hybridization exacerbates this, often making fine-scale species distinction within plants difficult (Fazekas et al., 2009). Owing to the absence of a standard barcode region in plants, appropriate sampling and a careful choice of markers are essential prerequisites for correct plant species identification (Mort et al., 2007; Fazekas et al., 2009).
The Consortium for the Barcode of Life (CBOL) recommended a two-marker-based system as a barcode for flowering plants, i.e. maturase K (matK) and ribulose-bisphosphate carboxylase gene (rbcL; Hollingsworth et al., 2009). Although this combination of gene regions works for some plants (Steven and Subramanyam, 2009), it may not be useful in others (Zhang et al., 2014). This failure can be attributed to low sequence polymorphism between species at rbcL and difficulty in sequence retrieval in the case of matK, for example, as seen in Zingiberaceae (Kress et al., 2005). Some studies have suggested that the matK region alone can potentially be used for plant barcoding, e.g. for species distinction in Annona, a genus belonging to pawpaw/sugar apple family Annonaceae (Lahaye et al., 2008; Hollingsworth et al., 2009; Larranaga and Hormaza, 2015). Molecular systematics and phylogeographic studies have also extensively used evolutionarily conserved chloroplast DNA (Parducci and Szmidt, 1999; Desplanque et al., 2000; Xu et al., 2001). The chloroplast genes, although uniparentally inherited and highly conserved, can be extremely useful for species and haplotype distinction in some cases. In Dendrobium species, 100% species resolution was observed by using the chloroplast psbA-trnH intergenic spacer (Yao et al., 2009).
For plants in the genus Citrullus, genetic diversity has also been determined by using chloroplast DNA and sequencing analysis of several non-coding regions (Dane and Bakhtiyarova, 2003; Dane et al., 2004; Dane and Liu, 2007). Relative to nuclear markers, maternally inherited chloroplast markers may sometimes be associated with low polymorphism, caused by slow evolution owing to a reduced rate of substitution at synonymous sites and also in non-coding inverted repeat sequences (Wolfe et al., 1987). Furthermore, chloroplast capture events and intraspecific hybridizations may cause selective sweeps, resulting in shared haplotype formation and incongruent gene trees, as noted in Australian populations of golden wattle (Acacia pycnantha; Ndlovu et al., 2013). This, in turn, can lead to failure in species identification when using a combined data set of multiple chloroplast genes, as was observed in some cases (Rosenthal et al., 2008; Twyford, 2014), such as willow (Salix spp.; Percy et al., 2014) and sweet chestnut fruit (Castanea spp.; Li and Dane, 2013).
The evolution of nuclear genes is independent from plastid DNA; therefore, nuclear regions, including the internal transcribed spacer region (ITS), may also be required for increased resolution (Chase et al., 2005) and hybridization testing (Chase et al., 2005; Zhang et al., 2013). The ITS from nuclear ribosomal DNA typically shows greater discriminatory power (Hollingsworth et al., 2011) and is easily amplified by using universal primers in some plant molecular studies. It has been successfully used for phylogenetic studies in some families, e.g. in the Euphorbiaceae (Pang et al., 2010). The ITS region was also used to infer phylogenetic relationships in Cucumis and Citrullus (Jarret and Newman, 2000; Garcia-Mas et al., 2004). Some limitations of ITS use include difficulty in obtaining the sequences and incomplete concerted evolution of the gene, leading to divergent paralogous copies within the same individual. Additionally, polymorphic sites need to be scored carefully (Mort et al., 2007; Hollingsworth et al., 2011).
Systematists have argued that dependence on a single sequenced region may result in a distorted picture of phylogenetic relationships, as incongruence has been observed between phylogenetic trees of nuclear and chloroplast origin (Fehrer et al., 2007); hence, phylogenetic inferences are now being made using multiple gene regions (Soltis and Soltis, 2004). Some researchers recommend using multiple markers from independent genomes, including a chloroplast and a nuclear gene together, for better taxon discrimination (Kress et al., 2005; Mort et al., 2007; Zhang et al., 2014). This helps to overcome the inherent inaccuracies of using single gene markers (Rubinoff et al., 2006; Mort et al., 2007). A combination of nuclear G3pdh and chloroplast ycf6-psbM regions was successfully used to distinguish species within Citrullus (Dane et al., 2007). This suggests that successful identification mainly depends on successful determination of a gene region or a combination of gene regions.
Numerous markers have also been used for plant genetic diversity and species identification studies during the last few decades, including Simple Sequence Repeat (SSR), Amplified Fragment Length Polymorphism (AFLP), Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD) and Inter Simple Sequence Repeat (ISSR). Compared with DNA barcoding, these markers can be cheaper and sometimes more polymorphic. However, they can also be impracticable because of erroneous results in scoring (electropherogram base calling; Devey et al., 2009). Successful use of marker-based systems for analysis of diversity requires subjective human judgment and editing, which can sometimes be overcome using PeakScanner and SPAGeDi software (Ley and Hardy, 2013). In addition, markers such as RAPDs and ISSRs are generally subject to reproducibility issues between laboratories (Gaskin et al., 2011). The cost of sequencing technology and analysis has recently been dramatically reduced. Complete plastid genome sequencing or even whole genome sequencing using next generation sequencing may eventually prove affordable, and these technologies will provide much useful information for those willing to work with large data sets and perform bioinformatics (Su et al., 2011).
Recently, we conducted DNA sequence analysis on five Australian invasive plant species in two families, the Cucurbitaceae and Boraginaceae. These species exhibit a variety of breeding systems, life histories and introduction histories across Australia (Table 1).
Table 1:
Comparison of the plants featured in DNA sequencing case studies of invasive species from Cucurbitaceae (data from Shaik et al., 2015) and Boraginaceae (data from Zhu et al., 2016) in Australia
| Plant taxa | Life cycle | Chloroplast haplotypes | Nuclear genotypes | Invasiveness in Australia | Breeding system in Australia |
|---|---|---|---|---|---|
| Cucurbitaceae | |||||
| Cucumis myriocarpus | Annual | 1 | 1 | H | SC |
| Citrullus lanatus | Annual | 1 | 1 | H | SC |
| Citrullus colocynthis | Perennial | 2 | 4 | H | SC |
| Boraginaceae | |||||
| Echium plantagineum | Annual | 12 | 2 | H | SC |
| Echium vulgare | Perennial | 2 | 4 | L | SC |
Invasiveness ratings: H, high; L, low. Breeding systems: SC, self-compatible; SI, self-incompatible.
Cucurbitaceae case study
Species profiles
Three cucurbitaceous invasive melons, camel melon [Citrullus lanatus (Thunb.) Matsum. and Nakai], prickly paddy melon (Cucumis myriocarpus L.) and colocynth melon [Citrullus colocynthis (L.) Schrad.] are currently distributed across Australia (see Fig. 1 for illustrations of the first two), invading crops, fallow lands and natural habitats (Leys et al., 1990; Parsons and Cuthbertson, 2001; Johnson et al., 2006b; Richardson et al., 2006). In Australia, wild melons were cited as one of the main summer fallow weed problems in a Grains Research & Development Corporation (GRDC) survey conducted in 2014 (Llewellyn et al., 2016). Their expansion is likely to continue unless adequate control strategies are implemented. The first two are annual vines that germinate during spring, fruit during summer and senesce during autumn (Parsons and Cuthbertson, 2001), whereas the third, colocynth, is a perennial vine. Australian summer weeds can result in up to 1 ton wheat yield loss per hectare if left uncontrolled, causing a loss of soil moisture of up to 50 mm that would otherwise have been useful for subsequent winter crops (Van Rees et al., 2011).
Figure 1:

Fruits of invasive Cucurbitaceae in Australia. The large fruit is Citrullus lanatus (camel melon), which has an average diameter of 7–10 cm. The smaller fruit is Cucumis myriocarpus (prickly paddy melon), which has an average diameter of 2–3 cm. The fruit of Citullus colocynthis (colocynth melon) is similar in size and appearance to that of C. lanatus, but C. colocynthis rind tends to have a mottled or mosaic pattern as opposed to the spotted or striped pattern seen on C. lanatus (Shaik et al., 2012).
There has been confusion regarding identification of wild melons in Australia before flowering, at both the morphological and the taxonomic level. Herbicide control at the seedling stage is now recommended (Johnson et al., 2006a). However, clear identification of the cucurbitaceous species in question is challenging, as some herbicides do not fully control all three species (Johnson et al., 2006a). Wild melons ascribed to the species C. lanatus are also a prominent weed in other countries, including New Zealand and the USA (Parsons and Cuthbertson, 2001; Futch and Hall, 2003; Grichar et al., 2010; Abd El-Ghani et al., 2011). The other annual wild melon most common in Australia, C. myriocarpus, has also become naturalized in southern Europe and California (Grubben and Denton, 2004). In addition, the perennial wild melon species, C. colocynthis, is a weed in Australia and parts of Asia (Parsons and Cuthbertson, 2001; Dane et al., 2007; Burrows and Shaik, 2014).
DNA sequencing study insights
The two annual wild melon species share similar vegetative growth and produce yellow flowers; therefore, they are often confused. This is particularly so before fruit formation in the case of C. myriocarpus. The perennial species, C. colocynthis, is closely related, shares morphological similarity with C. lanatus and is often misidentified even on fruit formation. Interestingly, initial trials with chloroplastidic matK gene did not result in separation of these two congeneric species because their sequences were 100% similar (Shaik et al., 2011). However, a chloroplast gene (ycf6-psbM) and a nuclear gene (G3pdh intron region) based on Dane et al. (2007) proved useful in evaluating the interspecific and intraspecific variability among the three cucurbitaceous invasive species.
The results of extensive sampling across Australia showed that C. lanatus and C. myriocarpus were each represented by a single genotype and haplotype, indicating that the populations present were derived from a single introduction event or multiple introduction events of a single genotype (and subsequently selfing). Moderate levels of genetic diversity were present among Australian C. colocynthis, and this species sorted geographically into separate haplotypes found in eastern and western regions, suggesting at least two separate introductions from two different source populations (Shaik et al., 2015). These findings suggested that the two gene regions described above can be used to identify the invasives in question as C. myriocarpus subsp. myriocarpus for Australian prickly paddy melon and C. lanatus var. citroides for camel melon, previously described in the literature as the Australian wild melon C. lanatus var. lanatus (Shaik et al., 2011, 2012, 2015).
The findings of Shaik et al. (2015) suggest that an integrative approach, using both morphological characters and DNA-based methods, including sequence analysis for identification, is likely to be more successful than either approach alone. Based on the discovery that C. lanatus is a single genetic entity, it is likely that the Australian population can be controlled effectively by one efficacious method of control, barring any local variations in management required as a result of phenotypic differences. This is also thought to be the case with C. myriocarpus. However, populations of C. colocynthis may require differential methods of management should genotypic and phenotypic differences predominate among eastern and western populations.
Other well-described hypotheses that will not be discussed in detail in this review provide explanations of how populations with low genetic diversity can become invasive and include pre-adaptation (Dlugosch and Parker, 2007; Clark et al., 2013; Dostál et al., 2013), phenotypic plasticity and enhanced resource availability (Callaway and Aschehoug, 2000; Graebner et al., 2012; Stricker and Stiling, 2013), natural enemy release (Hinz et al., 2012) or a combination of factors (Geng et al., 2007; Eriksen et al., 2012; Vergeer and Kunin, 2013). Additional studies on the roles of breeding system and pollinator interactions may shed light on these successfully inbreeding invasive plants.
Boraginaceae case study
Species profiles
Australia has two exotic invasive Echium species: Paterson's curse (E. plantagineum; Fig. 2a) and viper's bugloss (E. vulgare; Fig. 2b). Both species originated in southern Europe and were introduced to Australia during the 19th century (Piggin, 1982; Klemow et al., 2002). The former soon became a serious weed after introduction, covering almost all biogeographical regions in southern Australia. Today, it is estimated to infest >33 million hectares, causing >250 million AUD in losses to the meat and wool industries (NRM South and the Southern Tasmanian Councils Authority, 2016). In contrast, E. vulgare, although more common across Europe, is a niche colonizer in Australia and is currently found in only a small subset of biogeographical regions across New South Wales, Victoria and Tasmania.
Figure 2:

(a) Inflorescence of Echium plantagineum (Paterson's curse). (b) Inflorescence of Echium vulgare (viper's bugloss). (c and d) Note that flower size in E. plantagineum (c) is typically larger and exhibits two protruding stamens, in contrast to smaller flower size in E. vulgare with a lack of protruding stamens (d).
Correct identification of these congeric Echium species has typically caused confusion in Australia, especially before anthesis. Prior to the 1950s, the common name ‘Paterson's curse’ was used for both species (Parsons, 1973). Piggin (1977) reported misidentification between the two in Australian herbaria collections, which contributes to confusion in tracking the dynamics of dispersal over time. The Australian introduction history of the highly invasive E. plantagineum is also not clear. Piggin (1982) suggested that E. plantagineum was introduced as an ornamental species from England; however, it is more likely that this species was introduced to Australia from Spain, potentially via South Africa, as a seed contaminant of hay through importation of Merino sheep in the late 18th century (Zhu et al., 2014b).
DNA sequencing study insights
The genetic diversity of E. plantagineum and E. vulgare was evaluated by sample collection from Queensland, New South Wales, Australian Capital Territory, Victoria, South Australia, Northern Territory and Western Australia. Results indicated that both Echium species were routinely identified and separated using any of four DNA regions under evaluation, which included one nuclear region ITS and three chloroplast regions (trnL intron, trnL-trnF spacer and psbA-trnH spacer; Zhu et al., 2014a). Echium plantagineum and E. vulgare possessed 12 and two haplotypes each, respectively, when separated using three chloroplast regions (Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, under review). The more successful invader, E. plantagineum, showed significantly higher levels of genetic diversity than did the less invasive E. vulgare, which supports the hypothesis that a certain level of genetic diversity is associated with success of invasion in herbaceous plants (Jose et al., 2013).
The relative pattern of introduction of Australian E. plantagineum was also observed through sequence analysis experimentation. The introduction of E. plantagineum was first reported historically in Albury (southern New South Wales), Gladstone (South Australia) and Western Australia in 1880, 1889 and 1881, respectively (Piggin, 1977; Kloot, 1982). Spatial-specific haplotypes were found near these sites, while western New South Wales, a buffer area between the South Australia and New South Wales introduction events, showed the greatest number of haplotypes detected in the study (Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, under review). These findings support the hypothesis that multiple introductions of E. plantagineum occurred across Australia. However, to unravel the pathway of E. plantagineum introduction to Australia further, additional investigation is required and is currently ongoing through evaluation of a global collection of samples.
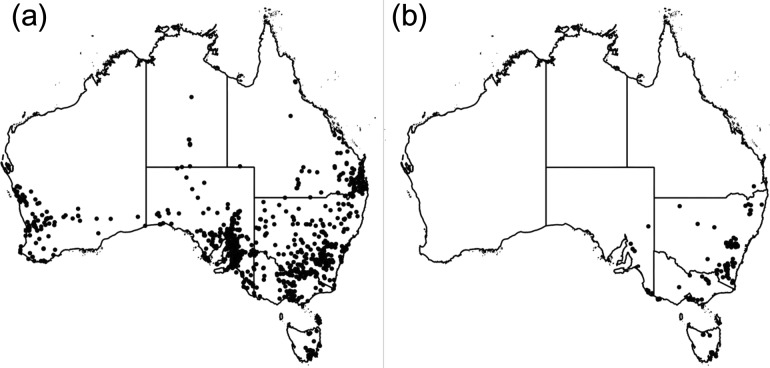
This study also highlights the limited genetic diversity found in Australian specimens of E. vulgare (Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, under review). Echium vulgare is restricted in its spread across Australia and is mainly found in the southeastern highlands (Fig. 3). As a perennial, it requires vernalization to induce flowering (Klemow et al., 2002), and is less drought tolerant when compared with E. plantagineum. Currently, E. vulgare is potentially under threat owing to its limited habitat as a niche colonizer. With exposure to a changing climate, its range may be further restricted in future years. In contrast, E. plantagineum, while already much more widely distributed than E. vulgare in Australia (Fig. 3), is predicted to become more invasive given its ability to withstand drought and its relatively high levels of genetic diversity, which may allow it successfully to adapt to recently changing environmental conditions across southern Australian biogeographical regions (Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, under review).
Figure 3:
Distribution of Echium plantagineum (Paterson's curse; a) and Echium vulgare (viper's bugloss; b) in Australia. Source of distribution data: Australia's Virtual Herbarium, 2015.
Value of DNA sequencing for a diverse array of invasive plants
Diverse breeding systems
Modes of reproduction and dispersal play a vital role in determining the genetic structure of a population of a particular species (Barrett et al., 2008; Petanidou et al., 2012). Many factors influence the genetic diversity of populations (Loveless and Hamrick, 1984), including ecological parameters. Genetic variation among populations is often solely dependent on the breeding system of the species (Schoen and Brown, 1991). Within-population genetic diversity is often reported to be low in the case of inbreeding populations and high in the case of outcrossing populations (Charlesworth and Charlesworth, 1995). In comparison, the among-population diversity was high (up to 51% of the total genetic diversity) in selfing and endemically distributed species. This is in direct contrast to outcrossing populations that were widely distributed geographically or those that were wind dispersed, where only a small proportion (~10%) of total genetic diversity was observed among populations (Hamrick et al., 1990, 1992; Hooper and Haufler, 1997).
For the Australian weeds discussed above, genetic characterization provided helpful insights into invasion history, although breeding systems varied widely in each case. Citrullus lanatus and C. myriocarpus reproduce by selfing and form large infestations of a single haplotype in the invaded range. The breeding system in Echium species features protandrous individual flowers, which cannot self-pollinate, but the possibility of fertilization by other flowers on the same plant renders them self-compatible (Klemow et al., 2002). Although E. plantagineum is self-compatible, outcrossing does occur via insect pollination, and outcrossing rates are generally high (Burdon and Brown, 1986; Burdon et al., 1988). The breeding system of E. vulgare is similar to that of E. plantagineum but exhibits slightly less incidence of outcrossing (Rademaker et al., 1999; Klemow et al., 2002) and predictably less genetic diversity in Australia, although other factors, such as a smaller available niche, are crucial when considering observed differences in distribution (Zhu X, Skoneczny D, Gopurenko D, Meyer L, Lepschi BJ, Weston PA, Callaway RM, Gurr GM, Weston LA. A tale of two plant invaders: comparison of the ecology and genetics of Echium plantagineum and E. vulgare in southern Australia. Scientific Reports, in preparation).
Diverse population genetics and invasion history
Assessment of genetic diversity can assist in pinpointing the origins, introduction history and invasion path of a particular species, and also point out invasion-prompting factors (Burrell et al., 2015). Little or no genetic variation has been noted in some invasive plant populations, including barbed goat grass (Aegilops triuncialis; Meimberg et al., 2006), cat's claw creeper (Macfadyena unguis-cati; Prentis et al., 2009), North American populations of perennial pepper weed (Lepidium latifolium; Gaskin et al., 2013) and giant reed (Arundo donax; Ahmad et al., 2008). Likewise, M. unguis-cati showed 27 chloroplast DNA haplotypes in its native range and only one haplotype in its invaded range (Sexton et al., 2002; Prentis et al., 2009). Sometimes invaded populations are far less diverse than their source populations, and such is the case in C. lanatus and C. myriocarpus in Australia (Shaik et al., 2015). In other cases, the level of genetic diversity in the non-native range can be similar to the native range, as is the case in Australian E. plantagineum when observed using isozyme marker studies (Burdon and Brown, 1986).
Alternatively, plant invaders can exhibit post-invasion genetic diversity (Jakobs et al., 2004) through mutation and novel chromosomal or ploidy changes, and also by hybridization and/or introgression with closely related congeners present in the invasive range (Prentis et al., 2009; Meyerson and Cronin, 2013; Ndlovu et al., 2013). The adaptability of such species can also be influenced by post-introduction genetic changes, including adaptive evolution through selection and genetic drift, resulting in the development of locally adapted ecotypes (Hahn et al., 2012; Wang et al., 2012; Oduor et al., 2016). For example, genes involved in stress responses were found to be over-expressed in annual ragweed (Ambrosia artemisiifolia) in its introduced range (Prentis and Pavasovic, 2013). Conversely, the genetically depauperate invasive populations of Japanese knotweed (Fallopia japonica) showed higher epigenetic variation (leading to phenotypic variation) than genetic variation (Richards et al., 2012). This demonstrates that a high level of genetic diversity in the invaded population is not always an essential prerequisite to invasion success.
It is also important to evaluate the invasive population's genetic make-up at both its native location and the invaded range, as careful study can provide information on the evolutionary processes that have occurred, as well as their role in invasion success (Hornoy et al., 2013), and invasion history (Le Roux et al., 2011), including the potential number of introductions (Meimberg et al., 2006). Greater knowledge can also assist in the reconstruction of introduction pathways (Novak and Mack, 2001; Le Roux et al., 2011; Hornoy et al., 2013; Kelager et al., 2013). Multiple introductions or a single introduction of multiple genotypes of a particular species to a location from diverse source populations can also result in enhanced genetic diversity in the invaded range, e.g. rugosa rose (Rosa rugosa) populations were diverse in the introduced European range, suggesting multiple introductions from their source populations in Japan (Simberloff, 2009; Hornoy et al., 2013; Kelager et al., 2013). In turn, this may result in the development of locally adapted ecotypes/genotypes through natural selection (Sexton et al., 2002; Prentis et al., 2008). Additionally, potential source populations of the invader can be identified (Clark et al., 2013; Kelager et al., 2013), and these populations may help further to locate associated natural enemies (Ellstrand and Schierenbeck, 2000; Ndlovu et al., 2013), which may later be useful as biological control agents (Goolsby et al., 2006).
Diverse management opportunities
Intraspecific diversity of an invasive plant species can have important implications for management; the genetic diversity among populations may be sufficiently great to warrant different control strategies. For example, in any invasive population, the presence of a mixture of resistant and non-resistant genotypes potentially impedes chemical and biological control (Burdon et al., 1981, 1984; Prentis et al., 2008). Hence, a genetically variable invasive plant population may be difficult to control because of naturally variable genotypes within the introduced population or the possibility of newly emerged resistant plants as a result of ongoing natural selection (Sterling et al., 2004). Such diverse populations may also show variable response to control by biocontrol agents (Bruckart et al., 2004). Knowledge of existing genetic variability in an invasive population provides further insight into the responses of weed populations to specific management strategies (Ward et al., 2008). Differential responses to the same management method have been observed in genetically diverse populations (Goolsby et al., 2006). Therefore an understanding of genetic diversity of invasive populations may help to predict the likelihood of successful management of invasive weeds, including the use of biocontrol programmes (Burdon and Marshall, 1981; Chapman et al., 2004; Gaskin et al., 2005). Knowledge of the source population of an invasive weed may also aid in finding potential biocontrol agents in the plant's native environment (Paterson et al., 2009).
In the case of five invasive Australian weeds described above, genetic characterization allows managers to approach each of the species differently based on the degree of variation present. Chemical control is generally used for all of these species, except E. vulgare (Johnson et al., 2006a). However, a uniform chemical control technique is recommended for populations of the two annual melon species in Australia and is currently efficacious, possibly because these species are related and are also genetically uniform (i.e. C. lanatus and C. myriocarpus). For the other three species, control tactics should logically be designed to account for regional or local differences, especially if it is shown that responses to specific controls vary among genotypes. For example, different ecotypes of a biological agent currently being tested for use against knotweed (Fallopia spp.) in North America have been shown to favour particular Fallopia species, and there are three closely related target species, including Fallopia × bohemica, which form a hybrid swarm (Grevstad et al., 2013; Clements et al., 2016). In E. plantagineum, the success of biocontrol agents in Australia was clearly associated with long-term regional adaptation of each biocontrol organism; however, genetic differences among regional plant populations may also influence biocontrol (Weston et al., 2012), but this requires further investigation.
Conclusions
Comparisons among the five taxa evaluated in these case studies (Table 1) reveal a variety of patterns in species and population genetic diversity, dependent on invasion and life history and breeding systems, with implications for strategic management approaches. Invasive plants with varying levels of genetic diversity can provide important models with which to study plant invasion success. DNA sequencing technologies provide precise and clear information related to the identity of invasive plant species, along with information on genetic diversity and phylogeographic history. New sequencing technologies are also likely to continue to allow greater resolution of genetic relationships among invasive plant populations, thus improving our understanding of mechanisms driving successful invasion.
Acknowledgements
The authors acknowledge Dominik Skoneczny for providing E. vulgare images in Fig. 2.
Funding
Funding support for this work was provided by Charles Sturt University, the Australian Research Council Discovery Program DP130104346 (awarded to LA Weston, G Gurr and R Callaway) and Trinity Western University. We are also grateful to Curt Daehler for chairing the EMAPi (Ecology and Management of Alien Plant Invasions) 2015 conference and encouraging the publication of this review based on a presentation by DRC at the EMAPi meeting.
References
- Abd El-Ghani M, Abo El-Kheir M, Abdel-Dayem M, Abd El-Hamid M (2011) Vegetation analysis and soil characteristics of five common desert climbing plants in Egypt. Turk J Bot 35: 561–580. [Google Scholar]
- Ahmad R, Liow PS, Spencer DF, Jasieniuk M (2008) Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat Bot 88: 113–120. [Google Scholar]
- Barrett SCH, Colautti RI, Eckert CG (2008) Plant reproductive systems and evolution during biological invasion. Mol Ecol 17: 373–383. [DOI] [PubMed] [Google Scholar]
- Beerling DJ, Huntley B, Bailey JP (1995) Climate and the distribution of Fallopia japonica: use of an introduced species to test the predictive capacity of response surfaces. J Veg Sci 6: 269–282. [Google Scholar]
- Bock DG, Caseys C, Cousens RD, Hahn MA, Heredia SM, Hübner S, Turner KG, Whitney KD, Rieseberg LH (2015) What we still don't know about invasion genetics. Mol Ecol 24: 2277–2297. [DOI] [PubMed] [Google Scholar]
- Bruckart W, Cavin C, Vajna L, Schwarczinger I, Ryan FJ (2004) Differential susceptibility of Russian thistle accessions to Colletotrichum gloeosporioides. Biol Control 30: 306–311. [Google Scholar]
- Bunce JA, Ziska LH (2000) Crop ecosystem responses to climatic change: crop/weed interactions In Reddy KR, Hodges HF, eds, Climate Change and Global Crop Productivity. CABI, New York, NY, USA, pp 333–348. [Google Scholar]
- Burdon J, Brown A (1986) Population genetics of Echium plantagineum L. target weed for biological control. Aust J Biol Sci 39: 369–378. [Google Scholar]
- Burdon JJ, Marshall DR (1981) Biological control and the reproductive mode of weeds. J Appl Ecol 18: 649–658. [Google Scholar]
- Burdon JJ, Groves RH, Cullen JM (1981) The impact of biological control on the distribution and abundance of Chondrilla juncea in south-eastern Australia. J Appl Ecol 18: 957–966. [Google Scholar]
- Burdon JJ, Groves RH, Kaye PE, Speer SS (1984) Competition in mixtures of susceptible and resistant genotypes of Chondrilla juncea differentially infected with rust. Oecologia 64: 199–203. [DOI] [PubMed] [Google Scholar]
- Burdon J, Jarosz A, Brown A (1988) Temporal patterns of reproduction and outcrossing in weedy populations of Echium plantagineum. Biol J Linn Soc 34: 81–92. [Google Scholar]
- Burrell AM, Pepper AE, Hodnett G, Goolsby JA, Overholt WA, Racelis AE, Diaz R, Klein PE (2015) Exploring origins, invasion history and genetic diversity of Imperata cylindrica (L.) P. Beauv. (Cogongrass) in the United States using genotyping by sequencing. Mol Ecol 24: 2177–2193. [DOI] [PubMed] [Google Scholar]
- Burrows GE, Shaik RS (2014) Comparative developmental anatomy of the taproot of the cucurbitaceous vines Citrullus colocynthis (perennial), Citrullus lanatus (annual) and Cucumis myriocarpus (annual). Aust J Bot 62: 537–45. [Google Scholar]
- Callaway RM, Aschehoug ET (2000) Invasive plants versus their new and old neighbors: a mechanism for exotic invasion. Science 290: 521–523. [DOI] [PubMed] [Google Scholar]
- Chapman H, Robson B, Pearson ML (2004) Population genetic structure of a colonising, triploid weed, Hieracium lepidulum. Heredity 92: 182–188. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B (1995) Quantitative genetics in plants: the effect of the breeding system on genetic variability. Evolution 49: 911–920. [DOI] [PubMed] [Google Scholar]
- Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V (2005) Land plants and DNA barcodes: short-term and long-term goals. Philos Trans R Soc Lond B Biol Sci 360: 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LV, Evans KJ, Jasieniuk M (2013) Origins and distribution of invasive Rubus fruticosus L. agg. (Rosaceae) clones in the western United States. Biol Invasions 15: 1331–1342. [Google Scholar]
- Clements D, DiTommaso A (2011) Climate change and weed adaptation: can evolution of invasive plants lead to greater range expansion than forecasted. Weed Res 51: 227–240. [Google Scholar]
- Clements DR, DiTommaso A (2012) Predicting weed invasion in Canada under climate change: evaluating evolutionary potential. Can J Plant Sci 92: 1013–1020. [Google Scholar]
- Clements DR, DiTommaso A, Jordan N, Booth BD, Cardina J, Doohan D, Mohler CL, Murphy SD, Swanton CJ (2004) Adaptability of plants invading North American cropland. Agric Ecosyst Environ 104: 379–398. [Google Scholar]
- Clements DR, DiTommaso A, Hyvönen T (2014) Ecology and management of weeds in a changing climate In Chauhan BS, Mahajan G (eds) Recent Advances in Weed Management. Springer, pp 13–37. [Google Scholar]
- Clements DR, Larsen T, Grenz J (2016) Knotweed management strategies in North America with the advent of widespread hybrid Bohemian knotweed, regional differences, and the potential for biocontrol via the psyllid Aphalara itadori Shinji. Invasive Plant Sci Manag 9: 60–70. [Google Scholar]
- Daehler CC. (2003) Performance comparisons of co-occurring native and alien invasive plants: implications for conservation and restoration. Annu Rev Ecol Evol Syst 34: 183–211. [Google Scholar]
- Dane F, Bakhtiyarova R (2003) Diagnostic chloroplast DNA haplotypes to distinguish cultivated from citron type watermelon. Cucurbit Genetics Coop Rpt 26: 36–39. [Google Scholar]
- Dane F, Liu J (2007) Diversity and origin of cultivated and citron type watermelon (Citrullus lanatus). Genet Resour Crop Evol 54: 1255–1265. [Google Scholar]
- Dane F, Lang P, Bakhtiyarova R (2004) Comparative analysis of chloroplast DNA variability in wild and cultivated Citrullus species. Theor Appl Genet 108: 958–966. [DOI] [PubMed] [Google Scholar]
- Dane F, Liu J, Zhang C (2007) Phylogeography of the bitter apple, Citrullus colocynthis. Genet Resour Crop Evol 54: 327–336. [Google Scholar]
- Desplanque B, Viard F, Bernard J, Forcioli D, Saumitou-Laprade P, Cuguen J, Van Dijk H (2000) The linkage disequilibrium between chloroplast DNA and mitochondrial DNA haplotypes in Beta vulgaris ssp. maritima (L.): the usefulness of both genomes for population genetic studies. Mol Ecol 9: 141–154. [DOI] [PubMed] [Google Scholar]
- Devey DS, Chase MW, Clarkson JJ (2009) A stuttering start to plant DNA barcoding: microsatellites present a previously overlooked problem in non-coding plastid regions. Taxon 58: 7–15. [Google Scholar]
- Ding J, Mack RN, Lu P, Ren M, Huang H (2008) China's booming economy is sparking and accelerating biological invasions. BioScience 58: 317–324. [Google Scholar]
- Dlugosch KM, Parker IM (2007) Molecular and quantitative trait variation across the native range of the invasive species Hypericum canariense: evidence for ancient patterns of colonization via pre-adaptation. Mol Ecol 16: 4269–4283. [DOI] [PubMed] [Google Scholar]
- Dostál P, Dawson W, van Kleunen M, Keser LH, Fischer M (2013) Central European plant species from more productive habitats are more invasive at a global scale. Glob Ecol Biogeogr 22: 64–72. [Google Scholar]
- Ebeling SK, Welk E, Auge H, Bruelheide H (2008) Predicting the spread of an invasive plant: combining experiments and ecological niche model. Ecography 31: 709–719. [Google Scholar]
- Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants. Proc Natl Acad Sci USA 97: 7043–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen RL, Desronvil T, Hierro JL, Kesseli R (2012) Morphological differentiation in a common garden experiment among native and non-native specimens of the invasive weed yellow starthistle (Centaurea solstitialis). Biol Invasions 14: 1459–1467. [Google Scholar]
- Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, Newmaster SG, Hajibabaei M, Husband BC (2009) Are plant species inherently harder to discriminate than animal species using DNA barcoding markers. Mol Ecol Resour 9: 130–139. [DOI] [PubMed] [Google Scholar]
- Fehrer J, Gemeinholzer B, Chrtek J Jr, Brautigam S (2007) Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol Phylogenet Evol 42: 347–361. [DOI] [PubMed] [Google Scholar]
- Futch SH, Hall DW (2003) Identification of Vine Weeds in Florida Citrus. University of Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, Electronic Data Information Source, Gainesville, FL, USA.
- Gallagher R, Duursma DE, O'Donnell J, Wilson P, Downey P, Hughes L, Leishman M (2013) The grass may not always be greener: projected reductions in climatic suitability for exotic grasses under future climates in Australia. Biol Invasions 15: 961–975. [Google Scholar]
- Garcia-Mas J, Monforte AJ, Arúis P (2004) Phylogenetic relationships among Cucumis species based on the ribosomal internal transcribed spacer sequence and microsatellite markers. Plant Syst Evol 248: 191–203. [Google Scholar]
- Gaskin JF, Dao-Yuan Z, Bon M-C (2005) Invasion of Lepidium draba (Brassicaceae) in the western United States: distributions and origins of chloroplast DNA haplotypes. Mol Ecol 14: 2331–2341. [DOI] [PubMed] [Google Scholar]
- Gaskin JF, Bon M-C, Cock MJW, Cristofaro M, Biase AD, De Clerck-Floate R, Ellison CA, Hinz HL, Hufbauer RA, Julien MH et al. (2011) Applying molecular-based approaches to classical biological control of weeds. Biol Control 58: 1–21. [Google Scholar]
- Gaskin JF, Schwarzländer M, Hinz HL, Williams L, Gerber E, Rector BG, Zhang DY (2013) Genetic identity and diversity of perennial pepperweed (Lepidium latifolium) in its native and invaded ranges. Invasive Plant Sci Manag 6: 268–280. [Google Scholar]
- Geng Y-P, Pan X-Y, Xu C-Y, Zhang W-J, Li B, Chen J-K, Lu B-R, Song Z-P (2007) Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol Invasions 9: 245–256. [Google Scholar]
- Goolsby JA, De Barro PJ, Makinson JR, Pemberton RW, Hartley DM, Frohlich DR (2006) Matching the origin of an invasive weed for selection of a herbivore haplotype for a biological control programme. Mol Ecol 15: 287–297. [DOI] [PubMed] [Google Scholar]
- Graebner RC, Callaway RM, Montesinos D (2012) Invasive species grows faster, competes better, and shows greater evolution toward increased seed size and growth than exotic non-invasive congeners. Plant Ecol 213: 545–553. [Google Scholar]
- Grevstad F, Shaw R, Bourchier R, Sanguankeo P, Cortat G, Reardon RC (2013) Efficacy and host specificity compared between two populations of the psyllid Aphalara itadori, candidates for biological control of invasive knotweeds in North America. Biol Control 65: 53–62. [Google Scholar]
- Grichar WJ, Dotray PA, Baughman TA (2010) Postemergence weed control in peanut using reduced rate or combinations of imazapic and imazethapyr. Crop Manag 9. doi:10.1094/CM-2010-1110-01-RS. [Google Scholar]
- Grubben GJH, Denton OA (eds) (2004) Plant Resources of Tropical Africa 2: Vegetables. PROTA Foundation, Wageningen, pp 1–668. [Google Scholar]
- Hahn MA, Buckley YM, Müller-Schärer H (2012) Increased population growth rate in invasive polyploid Centaurea stoebe in a common garden. Ecol Lett 15: 947–954. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Godt M, Brown AH, Clegg MT, Kahler A, Weir B (1990) Allozyme diversity in plant species. Plant Popul Genet Breed Genet Resour: 43–63. [Google Scholar]
- Hamrick JL, Godt M, Sherman-Broyles S (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests 6:95–124. [Google Scholar]
- Hinz HL, Schwarzländer M, McKenney JL, Cripps MG, Harmon B, Price WJ (2012) Biogeographical comparison of the invasive Lepidium draba in its native, expanded and introduced ranges. Biol Invasions 14: 1999–2016. [Google Scholar]
- Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ et al. (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS One 6: e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper E, Haufler C (1997) Genetic diversity and breeding system in a group of neotropical epiphytic ferns (Pleopeltis; Polypodiaceae). Am J Bot 84: 1664. [PubMed] [Google Scholar]
- Hornoy B, Atlan A, Roussel V, Buckley YM, Tarayre M (2013) Two colonisation stages generate two different patterns of genetic diversity within native and invasive ranges of Ulex europaeus. Heredity 111: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvönen T, Luoto M, Uotila P (2012) Assessment of weed establishment risk in a changing European climate. Agric Food Sci 21: 348–360. [Google Scholar]
- Jakobs G, Weber E, Edwards PJ (2004) Introduced plants of the invasive Solidago gigantea (Asteraceae) are larger and grow denser than conspecifics in the native range. Divers Distrib 10: 11–19. [Google Scholar]
- Jarret RL, Newman M (2000) Phylogenetic relationships among species of Citrullus and the placement of C. rehmii De Winter as determined by internal transcribed spacer (ITS) sequence heterogeneity. Genet Resour Crop Evol 47: 215–222. [Google Scholar]
- Johnson A, Brooke G, Thompson R, Roberts K, Hertel K, Border N, McNee T, Sullivan P (2006. a) Weed control for cropping and pasture in central west NSW. Orange, N.S.W. : NSW Department of Primary Industries, 2006 viii, 94 p.
- Johnson A, Preston C, Watts JH, Crossman ND (2006. b) Promoting best management practice for weed control in central west New South Wales. In 15th Australian weeds conference, Papers and Proceedings, Adelaide, South Australia, 24–28 September 2006: Managing weeds in a changing climate. Torrens Park, SA: Weed Management Society of South Australia, pp 403–405. [Google Scholar]
- Jose S, Singh HP, Batish DR, Kohli RK (2013) Invasive Plant Ecology. CRC Press, Hoboken. [Google Scholar]
- Kelager A, Pedersen J, Bruun H (2013) Multiple introductions and no loss of genetic diversity: invasion history of Japanese Rose, Rosa rugosa, in Europe. Biol Invasions 15: 1125–1141. [Google Scholar]
- Klemow KM, Clements DR, Threadgill PF, Cavers PB (2002) The biology of Canadian weeds. 116. Echium vulgare L. Can J Plant Sci 82: 235–248. [Google Scholar]
- Kloot PM. (1982) The naturalization of Echium plantagineum L. in Australia. Aust Weeds 1: 29–31. [Google Scholar]
- Kohli RK, Jose S, Singh HP, Batish DR (2008) Invasive Plants and Forest Ecosystems. CRC Press, Boca Raton, FL, USA. [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriticos D, Yonow T, McFadyen R (2005) The potential distribution of Chromolaena odorata (Siam weed) in relation to climate. Weed Res 45: 246–254. [Google Scholar]
- Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105: 2923–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larranaga N, Hormaza JI (2015) DNA barcoding of perennial fruit tree species of agronomic interest in the genus Annona (Annonaceae). Front Plant Sci 6: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J (2007) Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA 104: 3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Brown GK, Byrne M, Ndlovu J, Richardson DM, Thompson GD, Wilson JRU (2011) Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) in South Africa. Divers Distrib 17: 861–871. [Google Scholar]
- Ley AC, Hardy OJ (2013) Improving AFLP analysis of large-scale patterns of genetic variation – a case study with the Central African lianas Haumania spp (Marantaceae) showing interspecific gene flow. Mol Ecol 22: 1984–1997. [DOI] [PubMed] [Google Scholar]
- Leys AR, Amor RL, Barnett AG, Plater B (1990) Evaluation of herbicides for control of summer-growing weeds on fallows in south-eastern Australia. Anim Prod Sci 30: 271–279. [Google Scholar]
- Li X, Dane F (2013) Comparative chloroplast and nuclear DNA analysis of Castanea species in the southern region of the USA. Tree Genet Genomes 9: 107–116. [Google Scholar]
- Llewellyn R, Ronning D, Clarke M, Mayfield A, Walker S, Ouzman J (2016) Impact of Weeds in Australian Grain Production. Grains Research and Development Corporation, Kingston, ACT, Australia. [Google Scholar]
- Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 15: 65–95. [Google Scholar]
- McDonald A, Riha S, DiTommaso A, DeGaetano A (2009) Climate change and the geography of weed damage: analysis of US maize systems suggests the potential for significant range transformations. Agric Ecosyst Environ 130: 131–140. [Google Scholar]
- Meekins J, Ballard H Jr, McCarthy B (2001) Genetic variation and molecular biogeography of a North American invasive plant species (Alliaria petiolata, Brassicaceae). Int J Plant Sci 162: 161–169. [Google Scholar]
- Meimberg H, Hammond JI, Jorgensen CM, Park TW, Gerlach JD, Rice KJ, McKay JK (2006) Molecular evidence for an extreme genetic bottleneck during introduction of an invading grass to California. Biol Invasions 8: 1355–1366. [Google Scholar]
- Meyerson L, Cronin J (2013) Evidence for multiple introductions of Phragmites australis to North America: detection of a new non-native haplotype. Biol Invasions 15: 2605–2608. [Google Scholar]
- Mort ME, Archibald JK, Randle CP, Levsen ND, O'Leary TR, Topalov K, Wiegand CM, Crawford DJ (2007) Inferring phylogeny at low taxonomic levels: utility of rapidly evolving cpDNA and nuclear ITS loci. Am J Bot 94: 173–183. [DOI] [PubMed] [Google Scholar]
- Ndlovu J, Richardson DM, Wilson JRU, O'Leary M, Le Roux JJ (2013) Elucidating the native sources of an invasive tree species, Acacia pycnantha, reveals unexpected native range diversity and structure. Ann Bot (Lond) 111: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natural Resource Management and the Southern Tasmanian Councils Authority (2009). Paterson's curse. Weeds of Southern Tasmania Natural Resource Management and the Southern Tasmanian Councils Authority. Retrieved 25th January 2016, from http://www.nrmsouth.org.au/wp-content/uploads/2014/10/patersons_curse.pdf.
- Novak SJ, Mack RN (2001) Tracing plant introduction and spread: genetic evidence from Bromus tectorum (cheatgrass). BioScience 51: 114–122. [Google Scholar]
- Oduor AMO, Leimu R, van Kleunen M (2016) Invasive plant species are locally adapted just as frequently and at least as strongly as native plant species. J Ecol 104: 957–968. [Google Scholar]
- Oerke E-C. (2006) Crop losses to pests. J Agric Sci 144: 31–43. [Google Scholar]
- Olson LJ. (2006) The economics of terrestrial invasive species: a review of the literature. Agric Resour Econ Rev 35: 178. [Google Scholar]
- Pang X, Song J, Zhu Y, Xie C, Chen S (2010) Using DNA barcoding to identify species within Euphorbiaceae. Planta Med 76: 1784–1786. [DOI] [PubMed] [Google Scholar]
- Parducci L, Szmidt AE (1999) PCR-RFLP analysis of cpDNA in the genus Abies. Theor Appl Genet 98: 802–808. [Google Scholar]
- Parsons WT. (1973) Noxious Weed of Victoria . Inkata Press Proprietary Limited, Melbourne, VIC, Australia. [Google Scholar]
- Parsons WT, Cuthbertson EG (2001) Noxious Weeds of Australia, Ed 2 CSIRO Publishing, Collingwood, VIC, Australia. [Google Scholar]
- Paterson ID, Downie DA, Hill MP (2009) Using molecular methods to determine the origin of weed populations of Pereskia aculeata in South Africa and its relevance to biological control. Biol Control 48: 84–91. [Google Scholar]
- Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, Husband BC, Newmaster SG, Barrett SCH, Graham SW (2014) Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep. Mol Ecol 23: 4737–4756. [DOI] [PubMed] [Google Scholar]
- Petanidou T, Godfree RC, Song DS, Kantsa A, Dupont YL, Waser NM (2012) Self-compatibility and plant invasiveness: comparing species in native and invasive ranges. Perspect Plant Ecol Evol Syst 14: 3–12. [Google Scholar]
- Piggin CM. (1977) The herbaceous species of Echium (Boraginaceae) naturalized in Australia. Muelleria 3: 215–244. [Google Scholar]
- Piggin CM. (1982) The biology of Australian weeds. 8. Echium plantagineum L. J Aust Inst Agric Sci 48: 3–16. [Google Scholar]
- Prentis PJ, Pavasovic A (2013) Understanding the genetic basis of invasiveness. Mol Ecol 22: 2366–2368. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ (2008) Adaptive evolution in invasive species. Trends Plant Sci 13: 288–294. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Sigg DP, Raghu S, Dhileepan K, Pavasovic A, Lowe AJ (2009) Understanding invasion history: genetic structure and diversity of two globally invasive plants and implications for their management. Divers Distrib 15: 822–830. [Google Scholar]
- Rademaker M, De Jong T, Van Der Meijden E (1999) Selfing rates in natural populations of Echium vulgare: a combined empirical and model approach. Funct Ecol 13: 828–837. [Google Scholar]
- Richards CL, Schrey AW, Pigliucci M (2012) Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol Lett 15: 1016–1025. [DOI] [PubMed] [Google Scholar]
- Richardson FJ, Richardson RG, Shepherd RCH (2006) Weeds of the South-East: an Identification Guide for Australia, Ed 2 R.G. and F.J. Richardson Publishers, Meredith, VIC, Australia. [Google Scholar]
- Rosenthal DM, Ramakrishnan AP, Cruzan MB (2008) Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol Ecol 17: 4657–4669. [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Cameron S, Will K (2006) Are plant DNA barcodes a search for the Holy Grail. Trends Ecol Evol 21: 1–2. [DOI] [PubMed] [Google Scholar]
- Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R (2005) Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philos Trans R Soc Lond B Biol Sci 360: 1805–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen DJ, Brown AH (1991) Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc Natl Acad Sci USA 88: 4494–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebens H, Essl F, Dawson W, Fuentes N, Moser D, Perg J, Pyšek P, van Kleunen M, Weber E, Winter M et al. (2015) Global trade will accelerate plant invasions in emerging economies under climate change. Glob Change Biol 21: 4128–4140. [DOI] [PubMed] [Google Scholar]
- Sexton JP, McKay JK, Sala A (2002) Plasticity and genetic diversity may allow salt cedar to invade cold climates in North America. Ecol Appl 12: 1652–1660. [Google Scholar]
- Shaik RS, Weston LA, Burrows GE, Gopurenko D (2011) A comparative phenological and genetic diversity analysis of two invasive weeds, camel melon (Citrullus lanatus (Thunb.) Matsum. and Nakai var. lanatus) and prickly paddy melon (Cucumis myriocarpus L.), in inland Australia In Adkins S, ed, 23rd Asian-Pacific Weed Science Society Conference. The Sebel Cairns, Cairns, Queensland, Australia, pp 510–518. [Google Scholar]
- Shaik RS, Gopurenko D, Burrows GE, Urwin NAR, Lepschi BJ, Hildebrand SM, Weston LA (2012) Identification of the invasive weeds, camel melon, prickly paddy melon and colocynth in Australia – a morphological and molecular approach In Eldershaw V, ed, 18th Australasian Weeds Conference. Weed Science Society of Victoria Inc., The Sebel and Citigate Albert Park, Melbourne, Victoria, Australia, pp 73–77. [Google Scholar]
- Shaik RS, Gopurenko D, Urwin NAR, Burrows GE, Lepschi BJ, Weston LA (2015) Population genetics of invasive Citrullus lanatus, Citrullus colocynthis and Cucumis myriocarpus (Cucurbitaceae) in Australia: inferences based on chloroplast and nuclear gene sequencing. Biol Invasions 17: 2476–2490. [Google Scholar]
- Simberloff D. (2009) The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst 40: 81–102. [Google Scholar]
- Singh RP, Prasad PV, Reddy KR (2013) Impacts of changing climate and climate variability on seed production and seed industry. Adv Agron 118: 49–110. [Google Scholar]
- Skoneczny D, Weston PA, Zhu X, Gurr GM, Callaway RM, Weston LA (2015) Metabolomic profiling of pyrrolizidine alkaloids in foliar of two Echium spp. invaders in Australia – a case of novel weapons. Int J Mol Sci 16: 26721–26737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS (2004) Amborella not a ‘basal angiosperm’? Not so fast. Am J Bot 91: 997–1001. [DOI] [PubMed] [Google Scholar]
- Sterling TM, Thompson DC, Abbott LB (2004) Implications of invasive plant variation for weed management. Weed Technol 18: 1319–1324. [Google Scholar]
- Steven GN, Subramanyam R (2009) Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol Ecol Resour 9: 172–180. [DOI] [PubMed] [Google Scholar]
- Stow A, Maclean N, Holwell GI (2014) Austral Ark. Cambridge University Press, Cambridge and London, UK. [Google Scholar]
- Stricker BK, Stiling P (2013) Seedlings of the introduced invasive shrub Eugenia uniflora (Myrtaceae) outperform those of its native and introduced non-invasive congeners in Florida. Biol Invasions 15: 1973–1987. [Google Scholar]
- Su Z, Ning B, Fang H, Hong H, Perkins R, Tong W, Shi L (2011) Next-generation sequencing and its applications in molecular diagnostics. Expert Rev Mol Diagn 11: 333–343. [DOI] [PubMed] [Google Scholar]
- Twyford AD. (2014) Testing evolutionary hypotheses for DNA barcoding failure in willows. Mol Ecol 23: 4674–4676. [DOI] [PubMed] [Google Scholar]
- Van Rees H, Jackson D, Nuske K (2011) Summer Weeds: Counting the Costs for a Climate-Changed Future . The Rural Industries Research and Development Corporation (RIRDC), Barton, ACT, Australia. [Google Scholar]
- Vergeer P, Kunin WE (2013) Adaptation at range margins: common garden trials and the performance of Arabidopsis lyrata across its northwestern European range. New Phytol 197: 989–1001. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen G, Zan Q, Wang C, Su Y-j (2012) AFLP genome scan to detect genetic structure and candidate loci under selection for local adaptation of the invasive weed Mikania micrantha. PLoS One 7: e41310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Reid SD, Harrington J, Sutton J, Beck KG (2008) Genetic variation in invasive populations of yellow toadflax (Linaria vulgaris) in the western United States. Weed Sci 56: 394–399. [Google Scholar]
- Weston PA, Weston LA, and Hildebrand S (2012) Environmental impact on biocontrol agents and secondary chemistry of Paterson's curse (Echium plantagineum). In V. Eldershaw, ed, Proceedings of the 18th Australasian Weeds Conference, pp. 203–207. [Google Scholar]
- Wiemers M, Fiedler K (2007) Does the DNA barcoding gap exist? – a case study in blue butterflies (Lepidoptera: Lycaenidae). Front Zool 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DH, Abe J, Kanazawa A, Gai JY, Shimamoto Y (2001) Identification of sequence variations by PCR-RFLP and its application to the evaluation of cpDNA diversity in wild and cultivated soybeans. Theor Appl Genet 102: 683–688. [Google Scholar]
- Yao H, Song J-Y, Ma X-Y, Liu C, Li Y, Xu H-X, Han J-P, Duan L-S, Chen S-L (2009) Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region. Planta Med 75: 667–669. [DOI] [PubMed] [Google Scholar]
- Zhang D, Duan L, Zhou N (2014) Application of DNA barcoding in Roscoea (Zingiberaceae) and a primary discussion on taxonomic status of Roscoea cautleoides var. pubescens. Biochem Syst Ecol 52: 14–19. [Google Scholar]
- Zhang W, Fan XH, Zhu SF, Zhao H, Fu LZ (2013) Species-specific identification from incomplete sampling: applying DNA barcodes to monitoring invasive Solanum plants. PLoS One 8: e55927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Meyer L, Gopurenko D, Weston PA, Gurr GM, Callaway RM, Lepschi BJ, Weston LA (2014. a) Selection of DNA barcoding regions for identification and genetic analysis of two Echium invaders in Australia: E. plantagineum and E. vulgare. In Baker M., ed, Proceedings of the 19th Australasian Weeds Conference, Hobart, Australia, pp 396–400.
- Zhu X, Ryan B, Sokolov DV, Gurr GM, Weston LA (2014. b) Isohexenylnaphthazarins in primary and secondary roots of Paterson's curse (Echium plantagineum) In Rogers M., Sánchez-Moreiras A., eds, 7th World Congress on Allelopathy, Vigo, Spain, p 74. [Google Scholar]
- Zhu X, Skoneczny D, Weidenhamer JD, Mwendwa JM, Weston PA, Gurr GM, Callaway RM, Weston LA (2016) Identification and localization of bioactive naphthoquinones in the roots and rhizosphere of Paterson's curse (Echium plantagineum), a noxious invader. J Exp Bot 67: 3777–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]