Abstract
After each spliceosome cycle, the U4 and U6 snRNAs are released separately and are recycled to the functional U4/U6 snRNP, requiring in the mammalian system the U6-specific RNA binding protein p110 (SART3). Its domain structure is made up of an extensive N-terminal domain with at least seven tetratricopeptide repeat (TPR) motifs, followed by two RNA recognition motifs (RRMs) and a highly conserved C-terminal sequence of 10 amino acids. Here we demonstrate under in vitro recycling conditions that U6-p110 is an essential splicing factor. Recycling activity requires both the RRMs and the TPR domain but not the highly conserved C-terminal sequence. For U6-specific RNA binding, the two RRMs with some flanking regions are sufficient. Yeast two-hybrid assays reveal that p110 interacts through its TPR domain with the U4/U6-specific 90K protein, indicating a specific role of the TPR domain in spliceosome recycling. On the 90K protein, a short internal region (amino acids 416 to 550) suffices for the interaction with p110. Together, these data suggest a model whereby p110 brings together U4 and U6 snRNAs through both RNA-protein and protein-protein interactions.
Nuclear pre-mRNA splicing takes places in a large RNP complex, the spliceosome, which is assembled in an ordered multistep process. It consists of five small nuclear RNAs (the U1, U2, U4, U5, and U6 snRNAs) and more than 100 proteins, as recent proteomic analyses have determined (15, 16, 39). The spliceosome shows characteristic dynamics during assembly and splicing catalysis. For example, only the U2, U5, and U6 snRNAs participate in the catalytic center of the spliceosome, whereas the U1 and U4 snRNAs play essential roles only during the early assembly stages. After completion of the two-step splicing reaction and the release of mRNA and lariat products, the spliceosome disassembles into its components. Before entering a new cycle, at least some the components presumably must be reactivated. However, very little is known about this recycling phase of the spliceosome cycle.
The U4, U5, and U6 snRNAs enter the prespliceosome in the form of the 25S U4/U6.U5 tri-snRNP but are released from the spliceosome in their singular forms, the U4 and U6 snRNPs. These interact with each other to regenerate the U4/U6 di-snRNP, in which the two snRNAs are stably base paired (see Fig. 6C). The addition of the U5 snRNP generates the U4/U6.U5 tri-snRNP, which is integrated into the spliceosome. During each spliceosome cycle, the participating snRNAs undergo extensive structural rearrangements governed by specificprotein factors (for reviews, see references 7, 27, 28, and 33).
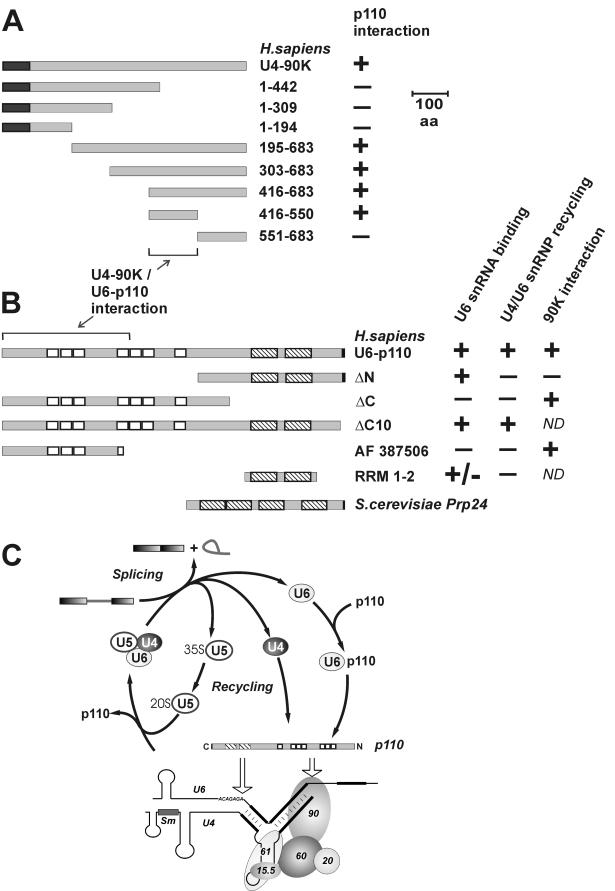
FIG. 6.
Summary of the domain organization and mutant derivatives of human U4-90K and U6-p110 proteins and model of U4/U6 snRNP recycling. (A) Human U4-90K wild-type protein (683 amino acids [aa]) and mutant derivatives containing amino acids 1 to 442, 1 to 309, 1 to 194, 195 to 683, 303 to 683, 416 to 683, 416 to 550, and 551 to 683 are schematically represented. The dark gray boxes indicate the N-terminal PWI domain. The abilities of 90K wild-type protein and mutant derivatives to interact with p110 in two-hybrid assays (Fig. 5C) and GST pull-down assays (Fig. 5D) are shown on the right (+ or −). Regions sufficient for the U4-90K or U6-p110 protein interactions are marked by the bracket and the arrow. (B) The following activities of p110 protein, of mutant derivatives ΔN, ΔC, ΔC10, and RRM 1-2, and of splice variant AF 387506 are summarized: U6 snRNA binding (Fig. 2), U4/U6 snRNP recycling (Fig. 4), and 90K protein interactions (Fig. 5). The p110 domain structure is schematically represented (RRMs, striped boxes; HAT or TPR domain, white boxes; conserved C-terminal region, black boxes). +, activity like that of full-length p110; +/−, low activity above the background; −, no activity; ND, not determined. For comparison, the domain organization of the p110-related Prp24 protein from the yeast S. cerevisiae is also shown. (C) Model of U4/U6 snRNP recycling and p110 interactions. The model of the U4 snRNP is that described by Nottrott et al. (24).
Regarding the molecular organization of the mammalian U4/U6 snRNP, a hierarchical assembly pathway has been demonstrated; this pathway also is conserved in the related U4atac/U6atac snRNP of the minor spliceosome (24). Of the five specific protein components, the 15.5K protein initially recognizes the loop region of the U4 5′ stem-loop (23), followed by binding of the 61K protein. The subsequent integration of the 20K/60K/90K protein heterotrimer requires an intact U4/U6 stem II structure. In addition, the U4 snRNA is associated with the canonical Sm heptamer core, the U6 snRNA with the corresponding LSm heptamer (1, 21).
Only recently a 110-kDa protein (called p110 hereafter) has been identified in the mammalian system which is required for recycling of the U4/U6 snRNP from singular U4 and U6 snRNPs (3). In addition, p110 was detected in the U6atac snRNP and found it to function in recycling of the U4atac/U6atac snRNP (8). The human p110 protein (also referred to as SART3) had originally been characterized as a nuclear protein copurifying with U6 snRNA capping activity (14, 31). It associates only transiently with U6 snRNA, since it can be detected in the U6 and U4/U6 snRNPs but is absent from both the U4/U6.U5 tri-snRNP and the spliceosome. p110 is functionally related to the yeast splicing factor Prp24 (30), which has been demonstrated to function in U4/U6 RNA annealing and U4/U6 snRNP recycling (11, 28, 36). It is concentrated in the Cajal bodies, although it is also present in the nucleoplasm, suggesting that Cajal bodies represent the cellular site of U4/U6 snRNP recycling (34).
The domain structure of the human U6-p110 protein consists of a large N-terminal region with at least seven tetratricopeptide repeat (TPR) motifs, two RNA recognition motifs (RRMs) in the C-terminal half of U6-p110, and a stretch of 10 highly conserved amino acids at the C terminus (3, 26). A splice variant of the human protein has been reported as a full-length cDNA (AF 387506) (19) which arises through the usage of a different 5′ splice site in exon 7; this usage is expected to result in a truncated protein carrying only three of the seven TPR motifs.
The 34-amino-acid TPR domain provides a structural unit of two antiparallel α helices that assemble to a platform for specific protein-protein interactions (reviewed in reference 6). This function is supported by the determination of the structure of the Ser/Thr phosphatase PP5 protein (9). The domain present in p110 (and many other RNA-processing factors) deviates slightly from the TPR consensus domain and has been referred to as half a TPR (HAT) (25) or the crooked-neck-like TPR domain (22, 38). The TPR-related pentatricopeptide repeat motifs and the HEAT, armadillo, and ankyrin repeats are all members of a large family of tandemly arranged helical repeat units (13, 32).
The p110 domain organization is conserved in many other eukaryotes, including Caenorhabditis elegans, Arabidopsis thaliana, Schizosaccharomyces pombe, and Drosophila melanogaster (3, 26); of the orthologues, only the well-known yeast Prp24 protein deviates from this domain organization, in that it lacks the entire N-terminal half with the TPR domain. Therefore, we were particularly interested in the possible role of the conserved TPR domain in U4/U6 snRNP formation and recycling.
In this study, we have mapped the domains of p110 required for specific snRNA binding, U4/U6 snRNP recycling activity, and protein-protein interaction. In particular, we identified a novel interaction of p110 with the U4/U6 snRNP-specific 90K protein. We further demonstrate that under recycling conditions, p110 functions as an essential splicing factor. Our study yields new functional insights into how both RNA-protein and protein-protein contacts contribute to U4/U6 snRNP assembly.
MATERIALS AND METHODS
RNA analysis.
RNAs were separated by electrophoresis in denaturing polyacrylamide-urea gels (8% acrylamide, 0.42% bisacrylamide, 50% urea; Tris-borate-EDTA buffer) and visualized by silver staining or Northern blot analysis as previously described (2). Digoxigenin (DIG)-labeled probes directed against human U4 and U6 snRNAs were obtained by PCR with PCR DIG labeling mix (Roche), M13 forward and reverse primers, and SP6-U4 (37) or SP6-U6 (5) as a template, respectively.
Expression and purification of recombinant p110 protein and mutant derivatives, Coomassie blue staining, and Western blotting.
Full-length His6-p110 protein was expressed as described previously (3). For the glutathione S-transferase (GST) fusion of the protein, the GST open reading frame and the p110 coding sequence were amplified from pAcGHLT-A/p110nrb (14) and cloned between the RsrII and XhoI sites of vector pFASTBAC HTa (Invitrogen Life Technologies). The protein was expressed by using the Bac-To-Bac baculovirus expression system (Invitrogen) as described previously (3).
The following shortened forms of the p110 open reading frame (NM_014706) were constructed by PCR methods that introduced restriction sites and termination codons with the primers: ΔC, ΔC10, ΔN, AF 387506, and RRM 1-2 (see Results). The PCR products were cloned into vector pFASTBAC HTa, replacing the first methionine with the N-terminal His tag of the vector. Recombinant baculovirus for protein expression in SF21 cells was obtained by using the Bac-to-Bac baculovirus expression system. For purification of recombinant p110 mutant derivatives, cytoplasmic and nuclear extracts from infected SF21 cells were prepared (10, 18). Proteins were affinity selected under native conditions on Ni-nitrilotriacetic acid-agarose (QIAGEN), eluted with 250 mM imidazole, and dialyzed against buffer D (10). Proteins were separated in a sodium dodecyl sulfate (SDS)-8% polyacrylamide gel and analyzed by Coomassie blue staining or Western blotting with a polyclonal anti-p110 antibody (3). Proteins were transferred to a Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham Pharmacia Biotech), immunostained with anti-p110 antiserum (1:2,000 dilution) and anti-rabbit antibody-peroxidase (Roche; 1:20,000 dilution), and detected by enhanced chemiluminescence.
p110 immunodepletion from HeLa cell nuclear extracts.
For immunodepletion of p110 from HeLa cell nuclear extracts, 200 μl of packed protein A-Sepharose CL-4B (Amersham) was equilibrated in buffer N100 (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.05% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) and incubated with rotation overnight at 4°C with 200 μl of polyclonal anti-p110 antiserum and 200 μl of buffer N100. Beads then were washed five times with 1 ml of buffer N100 each time, followed by the addition of nuclear extract (4C Biotec, Seneffe, Belgium) and incubation at 4°C for 1 h (450 μl of extract per 100 μl of packed beads). This depletion step was repeated twice. As a control, mock-depleted HeLa cell nuclear extracts were prepared in parallel without antiserum.
Analysis of RNA binding of p110 by coimmunoprecipitation.
Total RNA was purified from 400 μl of HeLa cell S100 extract and incubated with 2.5 μg of recombinant full-length p110 protein or the same molar amount of the mutant derivatives for 1 h at 30°C in a 250-μl reaction mixture (0.5 mM ATP, 20 mM creatine phosphate, 4.5 mM MgCl2, 50 mM KCl, and 80 U of RNasin [Promega] in buffer D). After the addition of 200 μl of buffer N100, p110 complexes were immunoprecipitated with immobilized anti-p110 antibodies. Coprecipitated RNAs were isolated after heating of the beads for 10 min at 80°C in proteinase K buffer (100 mM Tris-HCl [pH 8.0], 12.5 mM EDTA, 150 mM NaCl, 1% SDS) and were analyzed as described above.
In vitro splicing under recycling conditions in U4/U6 snRNP recycling assays.
Twofold reactions were prepared as described before (4, 17) with 30 μl of HeLa cell nuclear extract, mock-depleted extract, p110-immunodepleted extract, or p110-depleted extract supplemented with baculovirus-expressed recombinant p110 protein (500 ng per reaction). As a splicing substrate, 32P-labeled in vitro-transcribed MINX pre-mRNA (40) was added (10 ng per 25-μl reaction mixture, corresponding to 1× splicing substrate); for 5- and 10-fold excesses, additional unlabeled MINX pre-mRNA was added to final concentrations of 100 and 200 ng, respectively, per 25-μl reaction mixture. Splicing was performed at 30°C, and aliquots were taken after 45 and 90 min. After proteinase K treatment, RNAs were prepared by phenolization, followed by fractionation in a denaturating 8% polyacrylamide gel and autoradiography. Splicing efficiencies were quantitated by using TotalLabSoftware (Nonlinear Dynamics Limited).
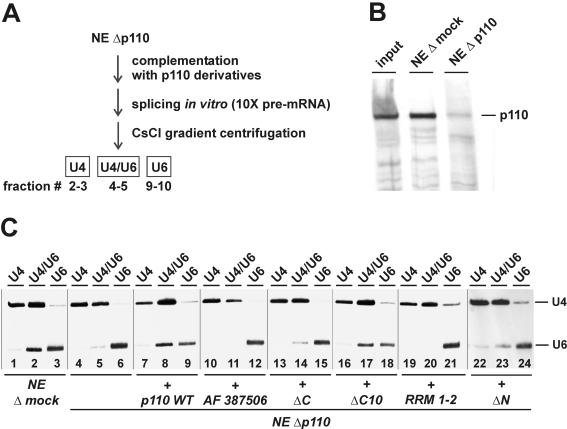
U4/U6 snRNP recycling assays were performed as described before (3). For testing of the p110 mutant derivatives, p110-depleted HeLa cell nuclear extracts were supplemented with 500 ng (per 25-μl reaction mixture) of recombinant p110 protein or the same molar amount of the mutant derivatives. In vitro splicing was performed under recycling conditions (10-fold excess of MINX splicing substrate) for 60 min at 30°C, and the entire reaction was fractionated by CsCl density gradient centrifugation as described previously (20) with a CsCl-buffer D solution at a density of 1.55 g/ml and containing 15 mM MgCl2. A total of 10 fractions were obtained, and fractions 2 and 3 (free U4), 4 and 5 (U4/U6), and 9 and 10 (free U6) were pooled. RNA was purified and analyzed in a denaturing 8% polyacrylamide gel, followed by Northern hybridization with DIG-labeled U4- and U6-specific probes.
Yeast two-hybrid and in vitro interaction assays.
Protein-protein interactions were assayed by using the Matchmaker-III two-hybrid system (Clontech). The open reading frames of the U4/U6-specific 15.5K, 61K, 20K, 60K, and 90K proteins as well as the full-length p110 protein, the p110 mutant derivatives ΔN and ΔC, and the splice variant AF 387506 were cloned into vectors pGBKT7 and pGADT7 (Clontech). The sequences encoding the full-length 90K protein and truncated versions containing amino acids 1 to 442, 1 to 309, 1 to 194, 195 to 683, 303 to 683, 416 to 683, 416 to 550, and 551 to 683 were introduced into vector pGADT7. After cotransformation of the constructs into yeast host strain AH109 by the lithium acetate method, cotransformants were selected for 5 days at 30°C on minimal synthetic dropout medium lacking the amino acids leucine and tryptophan. For assaying protein-protein interactions, cotransformants were replica plated on minimal synthetic dropout medium lacking leucine, tryptophan, and histidine and, for a more stringent selection, also lacking adenine. As positive controls, vectors pGADT7-T and pGBKT7-53 (Clontech) containing simian virus 40 large T antigen and p53 coding sequences, respectively, were used, and empty vectors served as negative controls.
For immunoprecipitation assays, the coding sequences of the 90K and 60K proteins were in vitro transcribed with T7 RNA polymerase from the pGBKT7 constructs and translated with 35S-labeled methionine by using a TNT coupled reticulocyte lysate system (Promega). A total of 15 μl of translation reaction mixture was incubated for 2 h at 4°C in buffer N100 supplemented with 5 mM MgCl2 and 50 mM KCl or in buffer N100 supplemented with 500 ng of baculovirus-expressed full-length p110 protein. Complexes formed were precipitated with p110 antibodies prebound to protein A-Sepharose, followed by seven washes with 0.75 ml each of buffer N100. The bound proteins were eluted by being heated in protein sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.005% bromphenol blue, 5% β-mercaptoethanol), fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and visualized by autoradiography.
For assaying protein-protein interactions by GST pull-down assays, 90K mutant proteins were in vitro translated with the addition of 35S-labeled methionine by using a TNT T7 quick coupled transcription-translation system (Promega) according to the manufacturer's instructions. For each reaction, 1 μg of recombinant GST-p110 or a comparable molar amount of GST was added together with 100 μl of reaction buffer (20 mM HEPES-KOH [pH 7.5], 100 mM NaCl, 0.01% NP-40, 1 mM dithiothreitol). Glutathione-Sepharose 4 M beads (Amersham) were blocked for 2 h in reaction buffer supplemented with 1 μg of bovine serum albumin/μl, 200 ng of tRNA/μl, and 200 ng of glycogen/μl, washed in reaction buffer, and added to the proteins. After 1 h of incubation with head-over-tail rotation at room temperature, the beads were washed extensively with reaction buffer. The bound proteins were eluted by being heated in protein sample buffer, fractionated by SDS-PAGE, and visualized by autoradiography.
RESULTS AND DISCUSSION
p110 domain analysis shows that C-terminal RRMs are sufficient for specific U6 snRNA binding.
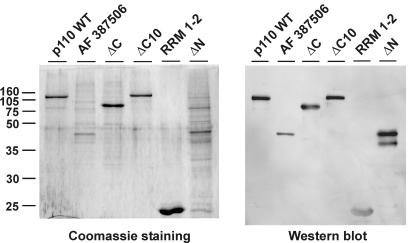
To delineate the domain structure of p110, we constructed a series of p110 mutant derivatives (Fig. 1; see Fig. 6B for a summary). Each carries an N-terminal His6 tag and was expressed and purified from baculovirus-transfected SF21 cells. Samples were analyzed after denaturing PAGE by Coomassie blue staining and Western blotting (Fig. 1). In addition to wild-type protein p110 (amino acids 2 to 963), p110 mutant derivatives ΔC, ΔC10, ΔN, and RRM 1-2 were expressed. p110 ΔC (amino acids 2 to 688) carries the entire TPR domain, including the putative nuclear localization signal region, but not the two RRMs and the C-terminal 10 amino acids. In p110 ΔC10 (amino acids 2 to 951), only the C-terminal 12 amino acids are missing; 10 of these are highly conserved (amino acids 952 to 961). p110 ΔN (amino acids 537 to 963) lacks the entire N-terminal TPR domain but contains the putative nuclear localization signal region, the two RRMs, and the C-terminal 10 amino acids. In addition, a minimal derivative that includes only the two RRMs (p110 RRM 1-2) was generated. Finally, we expressed a natural splice variant, AF 387506 (see above), which is truncated within the fourth TPR motif and contains amino acids 2 to 350, followed by 14 additional amino acids, RSTTESKGFGFICT.
FIG. 1.
Baculovirus expression of human U6-p110 mutant derivatives. The expression of wild-type U6-p110, splice variant AF 387506, and mutant proteins ΔC, ΔC10, RRM 1-2, and ΔN was detected by Coomassie blue staining (left panel) and by Western blotting (right panel). Molecular mass markers (in kilodaltons) are given on the left.
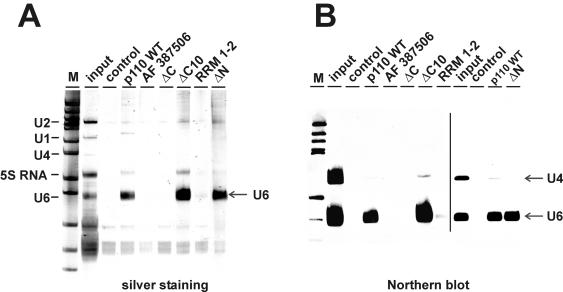
We tested the RNA binding specificity of these p110 derivatives by incubating each of them with total RNA prepared from an S100 extract and analyzing bound RNAs after immunoprecipitation with anti-p110 antibodies by using both silver staining (Fig. 2A) and Northern blotting with a probe specific for U4 and U6 snRNAs (Fig. 2B). Binding to U4 snRNA served as a negative control, since the total RNA preparation had been heat denatured before binding and the U4/U6 snRNA duplex is not stable under these conditions (data not shown). Our polyclonal anti-p110 antibody was able to recognize these different p110 derivatives with comparable efficiencies in solution (data not shown). The results showed that only two of the mutant proteins, p110 ΔC10 and p110 ΔN, were able to bind specifically U6 snRNA from total S100 extract RNA and with efficiencies similar to that of the wild-type protein. In contrast, p110 ΔC and the splice variant AF 387506 did not bind U6 snRNA detectably. The minimal construct with the two RRMs, p110 RRM 1-2, associated with U6 snRNA only at a very low efficiency.
FIG. 2.
Mutational analysis of p110 protein-U6 snRNA binding in vitro. Total RNA from a HeLa cell S100 extract was incubated with recombinant full-length p110 protein or an equimolar amount of splice variant AF 387506 or mutant derivative ΔC, ΔC10, RRM 1-2, or ΔN. Ten percent of the RNA input was analyzed in parallel (input lanes). Control reactions were carried out in the absence of added p110 protein (control lanes). The p110-RNA complexes formed were immunoprecipitated with anti-p110 antibodies, and the coprecipitated RNAs were analyzed by denaturing gel electrophoresis and detected by silver staining (A) or by Northern blotting with U4- and U6-specific probes (B). The line in panel B separates autoradiograms from two separate experiments. The mobilities of the U1, U2, U4, U6, and 5S RNAs are marked on the left. Lanes M, pBR322/HpaII marker fragments (A) and DIG marker V (Roche) (B). The arrows indicate the positions of the U4 and U6 snRNAs.
We conclude that the RRMs are required for U6 snRNA binding; for efficient RNA binding, additional sequences flanking the RRMs are important (compare the U6 binding activities of p110 RRM 1-2 and p110 ΔN). However, the conserved C-terminal 10 amino acids and the TPR domain play no essential role in U6 snRNA binding (compare p110 wild-type, p110 ΔC10, and p110 ΔN).
Role of the p110 TPR domain in U4/U6 snRNP recycling.
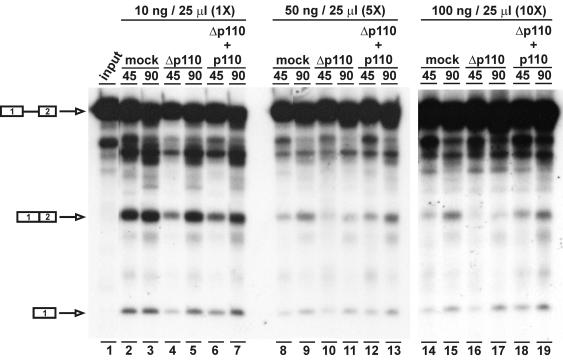
To establish in vitro recycling conditions, 32P-labeled MINX pre-mRNA (40) was spliced in vitro with a normal pre-mRNA concentration (10 ng per 25-μl reaction mixture) or 5- and 10-fold higher concentrations (Fig. 3). At the normal MINX pre-mRNA concentration, the MINX pre-mRNA:U4:U6 molar ratio corresponds to 1:5:10. In each case, reactions were carried out with three different nuclear extracts: mock-depleted extracts, p110-immunodepleted extracts, and p110-depleted extracts complemented with 500 ng of baculovirus-expressed recombinant p110 protein per 25-μl reaction mixture. Two time points (45 and 90 min) were assayed, and splicing efficiencies were quantitatively determined (expressed as the ratio of spliced mRNA to pre-mRNA). Under normal conditions, the MINX pre-mRNA was spliced efficiently and at comparable levels in the three extracts: splicing efficiencies at the 90-min time point in mock-depleted, p110-depleted, and p110-complemented extracts were 35, 34, and 32%, respectively (Fig. 3, left panel, lanes 3, 5, and 7). In contrast, at a fivefold higher pre-mRNA concentration (Fig. 3, middle panel), we consistently observed a large difference between the splicing activities in mock- and p110-depleted extracts (15 versus 6%); the addition of recombinant p110 protein fully restored the splicing activity level of the mock-depleted extract (to 18%), demonstrating that the splicing defect under these conditions is due to the specific depletion of p110 (lanes 9, 11, and 13). Finally, at a 10-fold higher pre-mRNA concentration (Fig. 3, right panel), splicing activity was almost undetectable in the p110-depleted extract (2% compared to 11% in the mock-depleted extract) but could be efficiently complemented by recombinant p110 protein to the level of the mock-depleted extract (15%) (lanes 15, 17, and 19).
FIG. 3.
U6-p110 is required for splicing in vitro only under recycling conditions. MINX pre-mRNA was spliced in vitro at concentrations of 10 ng (1×; lanes 2 to 7), 50 ng (5×; lanes 8 to 13), and 100 ng (10×; lanes 14 to 19) per 25-μl reaction in mock-depleted nuclear extracts (lanes 2, 3, 8, 9, 14, and 15), p110-depleted nuclear extracts (lanes 4, 5, 10, 11, 16, and 17), and p110-depleted nuclear extracts complemented with 500 ng of recombinant p110 (lanes 6, 7, 12, 13, 18, and 19). Input MINX pre-mRNA is shown in lane 1. Assays were carried out for 45 and 90 min. The positions of pre-mRNA, spliced product mRNA, and the first-exon intermediate are indicated on the left.
In summary, these findings demonstrate that p110 is required as a splicing factor only under conditions when relatively high levels of pre-mRNA are turned over, consistent with recent evidence that p110 is involved in U4/U6 snRNP recycling.
To identify which p110 domains are required for U4/U6 snRNP recycling, we tested each of the p110 derivatives under these conditions in a recently described in vitro complementation assay of U4/U6 snRNP recycling (3) (Fig. 4A shows a schematic representation). Briefly, in vitro splicing was carried out with an excess of pre-mRNA substrate and a p110-immunodepleted nuclear extract (Fig. 4B); a mock-depleted nuclear extract served as a control. After the splicing incubation, the base pairing status of U4 and U6 snRNAs was assessed through CsCl density gradient centrifugation (Fig. 4C). Thereby, free U4 (fractions 2 and 3) and U6 (fractions 9 and 10) were separated from the U4/U6 snRNP (fractions 4 and 5); the distribution of U4 and U6 snRNAs across the gradient was detected by Northern hybridization with U4- and U6-specific probes. p110 depletion resulted in a complete dissociation of the U4/U6 snRNP (Fig. 4C, compare lanes 4 to 6 with the mock-depleted reaction in lanes 1 to 3). In an analysis of the complementation reactions, only the wild-type protein (Fig. 4C, lanes 7 to 9) and p110 ΔC10 (lanes 16 to 18) were active in restoring the U4/U6 base-paired status. Neither p110 ΔC (Fig. 4C, lanes 13 to 15), RRM 1-2 (lanes 19 to 21), p110 ΔN (lanes 22 to 24), nor the splice variant AF 387506 (lanes 10 to 12) exhibited significant U4/U6 snRNP recycling activity above the level of the uncomplemented reaction (lanes 4 to 6).
FIG. 4.
Mutational analysis of p110 protein-U4/U6 snRNP recycling activity. (A) Schematic representation of the U4/U6 snRNP recycling assay (see the text and reference 3). NE Δp110, p110-depleted nuclear extract. (B) p110 immunodepletion of nuclear extracts. Aliquots of normal nuclear extracts (input lane) and nuclear extracts after mock depletion (NE Δmock lane) and after p110 depletion (NE Δp110 lane) were analyzed by Western blotting with anti-p110 antibodies. (C) U4/U6 snRNP recycling assay of p110 mutant derivatives. Splicing reactions were carried out with mock-depleted nuclear extracts (lanes 1 to 3), p110-depleted nuclear extracts (lanes 4 to 6), and p110-depleted nuclear extracts complemented with wild-type U6-p110 (lanes 7 to 9), splice variant AF 387506 (lanes 10 to 12), or mutant derivative ΔC (lanes 13 to 15), ΔC10 (lanes 16 to 18), RRM 1-2 (lanes 19 to 21), or ΔN (lanes 22 to 24). After CsCl gradient fractionation, the distribution of postspliceosomal U4, U4/U6, and U6 snRNPs was analyzed by Northern blotting with U4- and U6-specific probes and pooled fractions 2 and 3 (free U4), 5 and 6 (U4/U6), and 9 and 10 (free U6). The positions of the U4 and U6 snRNAs are marked on the right.
In conclusion, U4/U6 snRNP recycling activity in vitro requires both the N-terminal TPR domain and the C-terminal RRMs; however, the conserved C-terminal 10 amino acids are not essential (see Fig. 6B for a summary). These conclusions were surprising in light of recent studies with the yeast system that have revealed roles of the highly conserved C-terminal 10 amino acids in the interaction of Prp24 with the U6-specific LSm heptamer and in U4/U6 snRNP formation (26, 29). In contrast, the functional in vitro assays of the human protein reported here provided no evidence for an essential role of this C-terminal sequence. In particular, deletion of the C-terminal 10 amino acids did not detectably affect in vitro U4/U6 recycling activity (Fig. 4C). Possible explanations for this apparent discrepancy are (i) that there are some minor differences in the functional contributions of individual domains between the two systems, (ii) that our in vitro assay is not sensitive enough to detect the contribution of the LSm-p110 interaction, and (iii) that there are more built-in redundancies in the mammalian system than in the yeast system.
The inactivity of p110 ΔC in recycling can be explained by its U6 snRNA binding defect (Fig. 2). More importantly, the inactivity of p110 ΔN clearly dissects the U6 snRNA binding and U4/U6 snRNP recycling functions. Thus, the TPR domain fulfills a specific role in the U4/U6 recycling process, presumably through mediating protein-protein interactions (see below). Finally, the natural splice variant AF 387506, which retains only part of the TPR domain, was inactive in U4/U6 snRNP recycling.
p110 interacts through its TPR domain with a C-terminal region (amino acids 416 to 550) of the U4/U6 snRNP-specific 90K protein.
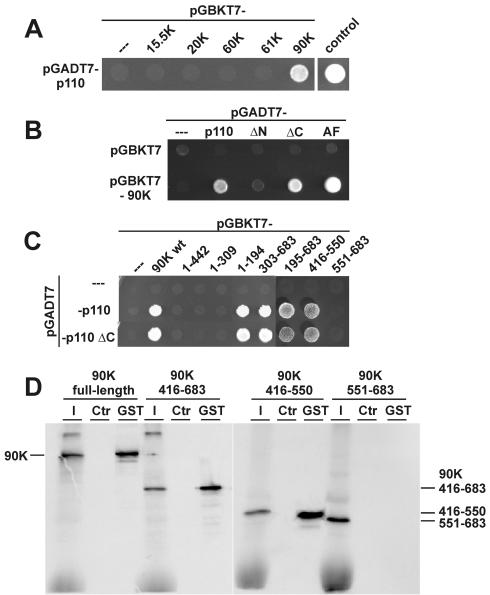
To investigate the role of the N-terminal TPR domain of p110, we performed yeast two-hybrid assays with the U4 and U4/U6 snRNP-specific 15.5K, 61K, 20K, 60K, and 90K proteins (Fig. 5A). Of these proteins, only the 90K protein showed an interaction with the full-length p110 protein. The T antigen-p53 interaction served as a positive control; cotransformation with pGBKT7 and pGADT7-p110 served as a negative control. Independently, we also twice recovered in a genomic yeast two-hybrid screen a full-length clone of the 90K protein, confirming the significance of this interaction (data not shown) and consistent with recent fluorescence resonance energy transfer measurements in HeLa cells (D. Stanek and K. M. Neugebauer, submitted for publication).
FIG. 5.
p110 interacts through its TPR domain with a C-terminal region (amino acids 416 to 550) of the U4/U6 snRNP-specific 90K protein. (A) Yeast two-hybrid assay of interactions between p110 and U4/U6-specific proteins. Yeast strain AH109 was transformed with plasmid pGBKT7-15.5K, pGBKT7-20K, pGBKT7-60K, pGBKT7-61K, or pGBKT7-90K in combination with plasmid pGADT7-p110. As controls, cotransformations were done with pGADT7-p110 and plasmid pGBKT7 as well as with plasmid pGADT7-T (containing simian virus 40 large T antigen) and plasmid pGBKT7-53 (containing p53). Protein-protein interactions were assayed by selection on minimal synthetic dropout medium lacking leucine, tryptophan, histidine, and adenine for 3 days at 30°C. (B) Yeast two-hybrid assay of interactions between p110 mutant derivatives and the U4/U6-specific 90K protein. Yeast strain AH109 was transformed with plasmid pGADT7-p110, pGADT7-ΔN, pGADT7-ΔC, or pGADT7-AF 387506 (AF) in combination with plasmid pGBKT7-90K. As controls, cotransformations were done with plasmid pGBKT7 or with plasmids pGADT7 and pGBKT7-90K. Selection was done for 3 days at 30°C on minimal synthetic dropout medium lacking leucine, tryptophan, and histidine (SD-LWH medium). (C) Yeast two-hybrid assay of interactions between p110 and U4/U6-specific 90K protein mutant derivatives. Yeast strain AH109 was transformed with plasmid pGADT7, pGADT7-p110, or pGADT7-ΔC. In addition, plasmid pGBKT7-90K or pGBKT7 constructs containing various regions of the 90K protein (amino acids 1 to 442, 1 to 309, 1 to 194, 303 to 683, 195 to 683, 195 to 683, 416 to 550, and 551 to 683) were cotransformed. Interactions were assayed for 3 days at 30°C on SD-LWH medium. wt, wild type. (D) Mutational analysis of the p110-90K protein interaction by GST pull-down assays. 35S-labeled full-length 90K protein and mutant derivatives containing amino acids 416 to 683, 416 to 550, and 551 to 683 were incubated with GST-p110 protein, followed by GST pull-down assays. Coprecipitated proteins were fractionated by SDS-PAGE and visualized by autoradiography. For each assay, 10% of the input (lanes I) and the precipitated material (lanes GST) are shown; control reactions contained only GST protein (lanes Ctr). The mobilities of the 90K full-length and mutant proteins are marked on the left and right.
The p110-90K protein interaction depended on the 90K protein component and was specific, since p110 ΔN gave a negative result; in contrast, both p110 ΔC and the natural splice variant AF 387506 gave a positive result for the 90K protein interaction (Fig. 5B). The specific association of p110 and the U4/U6-specific 90K protein was confirmed by a coimmunoprecipitation assay (data not shown). An in vitro-translated 90K protein could—after incubation with recombinant p110 protein—be efficiently coimmunoprecipitated with anti-p110 antibodies; the specificity of the interaction was demonstrated by the fact that an in vitro-translated 60K protein, another U4/U6-specific protein, was not coimmunoprecipitated.
Mutational analysis of the 90K protein (Fig. 5C) showed in two-hybrid assays that the amino-terminal half of the 90K protein did not interact with p110 (see constructs with amino acids 1 to 442, 1 to 309, and 1 to 194). In contrast, C-terminal fragments of the 90K protein were positive for the p110 interaction (see constructs with amino acids 303 to 683 and 194 to 683). This interaction domain of the 90K protein could be further shortened to C-terminal amino acids 416 to 683. Finally, this region was subdivided into two halves, amino acids 416 to 550 and 551 to 683; only the first half was able to interact with p110, whereas the latter half was negative in this assay. When the p110 mutant derivative ΔC was used instead of full-length p110, identical results were obtained, consistent with the TPR domain mediating the interaction between p110 and the 90K protein.
These conclusions from two-hybrid assays were subsequently confirmed by in vitro GST pull-down assays (Fig. 5D). GST-p110 (full length) efficiently interacted with the in vitro-translated, 35S-labeled 90K protein as well as the two 90K protein derivatives with C-terminal amino acids 416 to 683 and 416 to 550 but not the 90K protein derivative with C-terminal amino acids 551 to 683.
In summary, we conclude that the C-terminal part of p110 containing two RRMs and the highly conserved C-terminal sequence are dispensable for the 90K protein interaction. In contrast, the N-terminal TPR domain and the three N-terminal HAT motifs alone appear to be sufficient for 90K protein binding. As no other protein motifs are predicted in this N-terminal part of p110, the results strongly suggest that the HAT motifs mediate the specific interaction with the 90K protein. There seems to be some redundancy in the interaction, since the number of TPR motifs varies at least between three and seven in the known orthologues (3, 26). These data would also be consistent with our finding that the three N-terminal TPR motifs present in the splice variant AF 387506 appear to suffice for the 90K protein interaction (Fig. 5B). The latter finding raises the interesting question as to whether the splice variant may act as a negative regulator of p110 function, competing for the 90K protein interaction but lacking the RNA binding and recycling functions.
In the 90K protein, a short internal region near the C terminus (amino acids 416 to 550) that suffices for the p110 interaction was identified (Fig. 6A and B show a summary of the interaction data). This region provides a novel bridge between the U6 snRNA-bound p110 and the U4/U6-specific protein components and most likely plays an important role in U4/U6 snRNP assembly (Fig. 6C). In addition, the p110 interaction defines a new interface of the 90K protein, different from the amino-terminal PWI domain of the 90K protein (amino acids 3 to 76), which recognizes nucleic acids (35), and different from the central region, which mediates the interaction with the 60K protein (12). We also note that the 90K protein—within the 20K/60K/90K complex—contacts the U6 snRNA in the stem II region (24), and this event may stabilize the newly formed U4/U6 base-paired structure.
Does the fact that the entire TPR domain is absent in the yeast Prp24 protein reflect a yeast-specific feature of the U4/U6 snRNP recycling mechanism? This scenario appears to be unlikely, because—except for Prp24 and p110—all known protein constituents of the U4/U6 snRNP are highly conserved between the yeast and mammalian systems. As discussed by Rader and Guthrie (26), the 90K protein-TPR domain interaction may in fact be conserved in Saccharomyces cerevisiae, but with the TPR domain residing in a polypeptide other than Prp24. This suggestion is supported by the fact that the C terminus of the 90K protein, which interacts with p110 in the human system, is conserved in the yeast Prp3 protein.
How do these findings change our model of the U4/U6 snRNP assembly pathway? In contrast to the yeast protein, the mammalian p110 protein carries only two RRMs homologous to the highly conserved second and third RRMs of Prp24. As proposed by Rader and Guthrie (26), the additional RRMs of Prp24, 1 and 4, may recognize U4 snRNA and thereby promote the U4/U6 interaction. Perhaps this U4 interaction has shifted from an RNA-protein interaction (such as in yeast Prp24 RRMs 1 and 4 with the U4 snRNA) in the mammalian and other systems to a protein-protein interaction (p110 TPR domain with the 90K protein). What makes p110 leave the reassembled U4/U6 snRNP? Most likely this signal is provided by the U5 snRNP association, which results in U4/U6.U5 tri-snRNP formation. It will be important in further studies to identify this signal, with the aim of fully describing the structural rearrangements that accompany the recycling and assembly of functional snRNPs.
Acknowledgments
We thank G. Makarov for sharing the results of snRNA quantitation in HeLa cell nuclear extracts and Karla Neugebauer for communicating unpublished results and discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (grants Lu 294/12-1 and SFB 523/A8 to R.L. and grants Bi 316/9-1 and Bi 316/9-2 to A.B.).
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, M., and A. Bindereif. 1999. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 27:3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, M., S. Schreiner, A. Damianov, R. Reddy, and A. Bindereif. 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 21:2724-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindereif, A., and M. R. Green. 1987. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 6:2415-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindereif, A., T. Wolff, and M. R. Green. 1990. Discrete domains of human U6 snRNA required for the assembly of U4/U6 snRNP and splicing complexes. EMBO J. 9:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blatch, G. L., and M. Lässle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 7.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 8.Damianov, A., S. Schreiner, and A. Bindereif. 2004. Recycling of the U12-type spliceosome requires p110, a component of the U6atac snRNP. Mol. Cell. Biol. 24:1700-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A. K., P. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghetti, A., M. Company, and J. Abelson. 1995. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA 1:132-145. [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Santos, J. M., A. Wang, J. Jones, C. Ushida, J. Liu, and J. Hu. 2002. Central region of the human splicing factor Hprp3p interacts with Hprp4p. J. Biol. Chem. 277:23764-23772. [DOI] [PubMed] [Google Scholar]
- 13.Groves, M. R., and D. Barford. 1999. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9:383-389. [DOI] [PubMed] [Google Scholar]
- 14.Gu, J., S. Shimba, N. Nomura, and R. Reddy. 1998. Isolation and characterization of a new 110 kDa human nuclear RNA-binding protein (p110nrb). Biochim. Biophys. Acta 1399:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krainer, A. R., T. Maniatis, B. Ruskin, and M. R. Green. 1984. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36:993-1005. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K. A., A. Bindereif, and M. R. Green. 1988. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech. 5:22-31. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y., J. Li, B. O. Kim, B. S. Pace, and J. J. He. 2002. HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J. Biol. Chem. 277:23854-23863. [DOI] [PubMed] [Google Scholar]
- 20.Lücke, S., T. Klöckner, Z. Palfi, M. Boshart, and A. Bindereif. 1997. Trans-mRNA splicing in trypanosomes: cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 16:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayes, A. E., L. Verdone, P. Legrain, and J. D. Beggs. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean, M. R., and B. C. Rymond. 1998. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 18:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nottrott, S., H. Urlaub, and R. Lührmann. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 21:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preker, P. J., and W. Keller. 1998. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23:15-16. [DOI] [PubMed] [Google Scholar]
- 26.Rader, S. D., and C. Guthrie. 2002. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8:1378-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghunathan, P. L., and C. Guthrie. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8:847-855. [DOI] [PubMed] [Google Scholar]
- 28.Raghunathan, P. L., and C. Guthrie. 1998. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279:857-860. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, D. E., S. W. Stevens, and J. Abelson. 2002. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA 8:1011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon, K. W., and C. Guthrie. 1991. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 5:773-785. [DOI] [PubMed] [Google Scholar]
- 31.Shimba, S., and R. Reddy. 1994. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J. Biol. Chem. 269:12419-12423. [PubMed] [Google Scholar]
- 32.Small, I. D., and N. Peeters. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25:46-47. [DOI] [PubMed] [Google Scholar]
- 33.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 34.Stanek, D., S. D. Rader, M. Klingauf, and K. M. Neugebauer. 2003. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 160:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szymczyna, B. R., J. Bowman, S. McCracken, A. Pineda-Lucena, Y. Lu, B. Cox, M. Lambermon, B. R. Graveley, C. H. Arrowsmith, and B. J. Blencowe. 2003. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 17:461-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidaver, R. M., D. M. Fortner, L. S. Loos-Austin, and D. A. Brow. 1999. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics 153:1205-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wersig, C., and A. Bindereif. 1990. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 18:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, K., D. Smouse, and N. Perrimon. 1991. The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes Dev. 5:1080-1091. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 40.Zillmann, M., M. L. Zapp, and S. M. Berget. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]