Abstract
Eukaryotic messenger RNAs containing premature stop codons are selectively and rapidly degraded, a phenomenon termed nonsense-mediated mRNA decay (NMD). Previous studies with both Caenohabditis elegans and mammalian cells indicate that SMG-2/human UPF1, a central regulator of NMD, is phosphorylated in an SMG-1-dependent manner. We report here that smg-1, which is required for NMD in C. elegans, encodes a protein kinase of the phosphatidylinositol kinase superfamily of protein kinases. We identify null alleles of smg-1 and demonstrate that SMG-1 kinase activity is required in vivo for NMD and in vitro for SMG-2 phosphorylation. SMG-1 and SMG-2 coimmunoprecipitate from crude extracts, and this interaction is maintained in smg-3 and smg-4 mutants, both of which are required for SMG-2 phosphorylation in vivo and in vitro. SMG-2 is located diffusely through the cytoplasm, and its location is unaltered in mutants that disrupt the cycle of SMG-2 phosphorylation. We discuss the role of SMG-2 phosphorylation in NMD.
Nonsense-mediated mRNA decay (NMD) rapidly and selectively degrades eukaryotic mRNAs containing premature translation termination codons (PTCs) (reviewed in references 27, 43, and 65). NMD likely improves the fidelity of gene expression by degrading aberrant mRNAs, thereby protecting cells from potentially deleterious consequences of their translation (9, 28, 53). The biological sources of NMD substrates are only partially understood. For example, unspliced, unproductively spliced, and aberrantly spliced mRNAs are degraded by NMD (29, 39, 48). NMD strongly influences the expression of certain genes for which mRNAs containing PTCs are a normal feature of their expression. For example, gene rearrangements of T-cell receptor genes often result in mRNAs that contain PTCs and are subjected to NMD (11).
Gene products required for NMD have been identified in fungi, nematodes, insects, and mammals (13, 16, 24, 49), and putative orthologs of these genes are evident in many sequenced eukaryotic genomes. A core group of three genes first identified in the yeast Saccharomyces cerevisiae (UPF1, NMD2/UPF2, and UPF3) (reviewed in reference 69) are required for NMD in all tested eukaryotes (3, 40, 45, 50, 57, 61). The conservation of both the sequence and function of such genes indicates that NMD is an evolutionarily ancient process and suggests that elements of the mechanism of NMD will be similar in most, or perhaps all, eukaryotes.
PTCs are distinguished from normal termination codons in both yeast and mammalian cells by the context of translation termination. Specific cis-acting elements mark the open reading frame of mRNAs and activate NMD when translation terminates upstream of the marks. The Upf proteins appear to be involved both in sensing the presence of downstream signals and in activating the degradation machinery. In yeast cells, the cis-acting elements are termed downstream sequence elements, which are thought to be present throughout coding sequences but not in 3′ untranslated regions (54). Hrp1p binds both downstream sequence elements and Upf1p and, in addition to other roles, is involved in NMD (26). In mammalian cells, cis-acting elements that mark open reading frames are exon junction complexes (EJCs), which are deposited near splice junctions in mature mRNAs (37). The EJC communicates with Upf proteins via interactions between Upf3 and the EJC components RNPS1 and Y14 (25, 36, 41). Upf3 interacts with Upf2, which interacts with Upf1 (40, 57). Although the marking of open reading frames is different in yeast cells and mammals, other aspects of NMD (for example, the roles of UPF proteins) are quite similar.
Upf1 is a key regulator of NMD (reviewed in reference 27) and is an ATPase/helicase whose catalytic activity is required for NMD (66). Upf1 interacts with downstream activators of NMD (26), with other Upf proteins (31), with translation release factors (17), and with components of either the 5′ or 3′ decay pathways (23, 42, 62). Upon premature termination, Upf1 activity is required to commence either 5′ or 3′ (or both) exonucleolytic degradation of substrate mRNAs (12, 38, 47, 62). Thus, Upf1 is uniquely positioned to regulate decay by communicating the presence of downstream information and activating subsequent steps of degradation. Important unanswered questions concerning Upf1 include how downstream activators of NMD influence its activity and how Upf1 in turn activates the 5′ or 3′ decay pathway.
Functions of at least seven genes are required for NMD in Caenorhabditis elegans. smg-2, smg-3, and smg-4 are orthologs of yeast UPF1, UPF2, and UPF3 (3, 50; S. L. Kuchma and P. Anderson, unpublished data). Four additional genes (smg-1, smg-5, smg-6, and smg-7), which are not evident in yeast cells, are required for NMD in C. elegans, Drosophila melanogaster, and mammalian cells (13, 19, 24, 49, 70). SMG-2 undergoes cycles of phosphorylation that appear to be required for NMD. Indeed, all known smg genes other than smg-2 affect the state of SMG-2 phosphorylation (50). SMG-1, SMG-3, and SMG-4 are needed to phosphorylate SMG-2 while SMG-5, SMG-6, and SMG-7 are needed to dephosphorylate SMG-2. SMG-5 is a component of a protein phosphatase 2A complex (PP2A) that interacts with SMG-2 in both C. elegans and mammalian cells (2, 13, 49). This SMG-5-PP2A complex is thought to be the SMG-2/human UPF1 (hUPF1)-specific phosphatase. Human SMG-1 phosphorylates hUPF1 both in vitro and in vivo (19, 70). Taken together, these observations suggest that regulation of SMG-2 via cycles of phosphorylation is central to NMD. We report here the molecular analysis of smg-1, its interactions with components of C. elegans NMD, and its role in SMG-2 phosphorylation.
MATERIALS AND METHODS
Strains and genetic methods.
Genetic methods for C. elegans were described by Brenner (7). Alleles used in this study were smg-1(r861, r879, r884, r887, r904, r910, and r913), smg-2(r908), smg-3(r930), smg-4(r1169), smg-5(r860), unc-54 (r293), and unc-119(ed3) (32, 52; this study).
Transformation rescue assay.
smg-1(r861) unc-54(r293) hermaphrodites were coinjected with DNA to be tested and marker plasmid pRF4[rol-6(su1006)] by using established methods (44). unc-54(r293); smg(+) animals are paralyzed, whereas unc-54(r293); smg(−) animals have normal motility (32). Heritable transformed lines were established and scored for motility.
Sequence and phylogenetic analysis.
Sequence and phylogenetic analysis was performed by using internet resources providing the following algorithms: CLUSTALW, Neighbor-Joining, and Puzzle (55, 60, 64). Amino acid sequences were aligned with CLUSTALW. Gapped positions and regions of uncertain alignment were omitted from further analysis. Trees were constructed by Neighbor-Joining and Puzzle, and confidence values were based on 1,000 bootstrap trees or puzzling steps.
Analysis of SMG-1 kinase-inactive mutants.
Constructs used in analysis of smg-1 kinase mutants included a copy of unc-119(+) and a wild-type (clone TR #430) or mutant (TR#431) genomic copy of smg-1 (NcoI-SalI fragment, nucleotides [nt] 13852 to 27566 of U97189); TR#431 was made by mutating nt 16626 (T to G) within a KpnI-BglII fragment (nt 16353 to 17293) by using QuikChange (Stratagene) and the appropriate primers and was then used to replace the corresponding piece of TR no. 430. Transgenic animals were generated by microparticle bombardment of smg-1(r861); unc-119(ed3) with either TR #430 or TR#431, and heritable lines were established as described previously (52). RNA isolation and rpl-12 reverse transcription-PCR were performed as described previously (48) with primers GTTGCGTCGGAGGAGAAGTCG (forward) and GATGATGTCGTGTGGGTGTTGTC (reverse).
Antibodies.
SMG-1 antisera were generated by immunizing two rabbits with recombinant, hexahistidine-tagged, partial-length SMG-1 (amino acids 477 to 903) purified from inclusion bodies (68) after expression in Escherichia coli (pET28a system; Novagen). Anti-SMG-1 antibodies were affinity purified as described previously (4) with a glutathione S-transferase-tagged partial-length SMG-1 encoding aa 507 to 817 (pGEX4T system; Amersham Pharmacia). Anti-SMG-1 antibodies were eluted with ActiSep (Sterogene), desalted, and concentrated.
Coimmunoprecipitation assays.
Mixed-stage animals were suspended in 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7.2) buffer containing protease and phosphatase inhibitors. Worms were homogenized by using a French pressure cell (12,000 lb/in2) and centrifuged at 21,000 × g for 25 min, retaining only the soluble supernatant fraction. Protein concentrations were measured and equalized with 20 mM MOPS (pH 7.2) before immunoprecipitation. Extracts were adjusted to 100 mM NaCl and incubated with antibody, followed by the addition of protein A-Sepharose (Amersham Pharmacia). Precipitated proteins were collected by centrifugation and washed six times with 20 mM MOPS (pH 7.2) and 100 mM NaCl. RNase-treated extracts were prepared by incubating extracts described above with 1 mg of RNase A/ml for 1 h on ice prior to immunoprecipitation.
Immunoprecipitation kinase assays.
SMG-2 was immunoprecipitated from total soluble protein extracts as described above. Washed immunoprecipitate pellets were further washed twice with kinase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2) with protease inhibitors and resuspended in a total volume of 100 μl of kinase buffer. [γ-32P]ATP (40 μCi) was added, and the reaction mixtures were incubated at room temperature for 60 min with gentle agitation. Samples containing wortmannin were incubated for 1 h at room temperature in 1 μM wortmannin (or mock control) prior to the addition of [γ-32P]ATP. Reactions were stopped by the addition of an equal volume of boiling sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, and products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were dried and visualized with a PhosphorImager (Molecular Dynamics).
Immunofluorescence microscopy.
Immunolocalization was performed by using standard procedures (15). Extruded gonads were fixed with methanol-acetone and washed in phosphate-buffered saline-Tween 20 buffer. Affinity-purified anti-SMG-2 antibody was precleared by treating the samples with an acetone powder generated from smg-2(r908) (a null allele). Mab414 antibody was added to cleared anti-SMG-2 antibody prior to staining. Images were obtained on an MRC1024 confocal microscope.
RESULTS
Positional cloning of smg-1.
We mapped smg-1 between dpy-14 and sem-4 on linkage group I. Among 47 crossovers between dpy-14 and sem-4, 14 occurred leftward and 33 occurred rightward of smg-1. We tested nearby cosmids for smg-1(+) function by using a transformation rescue assay (see Materials and Methods). Two cosmids, F56F2 and C14A12, fully rescued the NMD phenotype of smg-1(r861). We tested subclones of F56F2 or the overlapping cosmid C48B6 and identified a 13.7-kb NcoI-SalI fragment (nt 13852 to 27566 of cosmid C48B6) as the smallest tested fragment providing smg-1(+) function. This region is predicted to encode a single intact gene (WormBase transcript C48B6.6) and detects on Northern blots a single mRNA of approximately 7.8 kb in the wild type (data not shown).
We identified and sequenced a series of overlapping cDNA clones from oligo(dT)-primed or random-primed cDNA libraries that define a 7.6-kb full-length smg-1 mRNA. Multiple 3′ clones contained a poly(A) tract at their 3′ ends, and multiple 5′ clones contained SL1 at their 5′ ends. SL1 is a trans-spliced leader sequence present at the 5′ ends of many C. elegans transcripts (5). Thus, the assembled sequence (GenBank accession no. AF149821) represents full-length smg-1 mRNA. We examined all available smg-1 expressed sequence tag sequences and determined that each of them is colinear with our assembled cDNA sequence. smg-1 contains 42 exons, spans 12.6 kb of genomic DNA, and encodes a 2,322-aa protein. The intron and exon structure of smg-1 is described at http://www.wormbase.org.
We sequenced three independent smg-1 alleles. smg-1(r913) and smg-1(r910) are mutator-induced alleles that contain Tc1 insertions at TA dinucleotides 17105-17106 and 24620-24621 of cosmid C48B6, respectively. smg-1(r884) is an ethyl methanesulfonate-induced mutation changing codon 1941 (UGG; C48B6 nt 16697) to a UGA premature stop. An additional unsequenced allele, smg-1(r904), contains a Tc3 insertion in the vicinity of C48B6 nt 19600 based on Southern blot and PCR analyses.
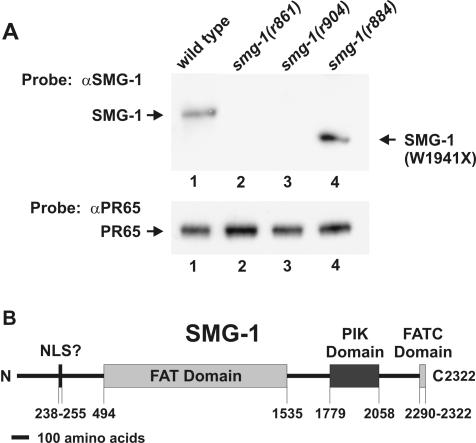
We prepared an affinity-purified anti-SMG-1 antibody directed against SMG-1 aa 477 to 903 (see Materials and Methods). This antibody recognizes a protein on Western blots whose mass is consistent with the cDNA prediction of 265 kDa. We examined a series of smg-1 mutants for expression of SMG-1-related proteins. Five tested alleles (r861, r879, r887, r904, and r910::Tc1) expressed no detectable SMG-1 (Fig. 1A, lanes 2 and 3 and data not shown). We conclude that smg-1 corresponds to transcript C48B6.6 and that the canonical allele smg-1(r861) is a null protein.
FIG. 1.
(A) Western blot of total protein from the wild type (lane 1) and three independent smg-1(−) mutants (lanes 2 to 4) probed with anti-SMG-1 (αSMG-1) antibody (top) and anti-PR65 (αPR65) antibody (bottom) as a loading control. (B) Domain structure of SMG-1. SMG-1 contains a FAT domain (pfam02259), a FATC domain (pfam02260), a PIK domain (pfam00454), and a putative nuclear localization signal (NLS?) at the indicated amino acid positions.
Two tested smg-1 alleles (r884 and r913::Tc1) express shortened SMG-1-related proteins whose estimated sizes (220 and 210 kDa, respectively) are consistent with premature termination caused by the DNA sequences described above (Fig. 1A, lane 4 and data not shown). The SMG-1-related proteins expressed by smg-1(r884) and smg-1(r913::Tc1) are nonfunctional, as the phenotypes of these mutants are indistinguishable from that of the null allele smg-1(r861).
smg-1 encodes a PIK-related protein kinase.
smg-1 cDNA encodes a protein of 2,322 aa (265 kDa). Four domains of SMG-1, diagrammed in Fig. 1B, are notable: (i) a putative bipartite nuclear localization signal (aa 238 to 255); (ii) a FAT (FRAP, ATM, and TRRAP) domain (aa 494 to 1535); (iii) a phosphatidylinositol kinase (PIK) domain (aa 1779 to 2058); and (iv) a FATC (FAT carboxyl terminus) domain (aa 2290 to 2322). The founding members of the PIK superfamily are lipid kinases, but the subfamily of PIK-related kinases are protein kinases (reviewed in reference 34). Phylogenetic analysis (see below) identifies SMG-1 as a probable protein kinase rather than a lipid kinase. Biochemical functions of the FAT and FATC domains are unknown, although they are found together in certain protein kinases of the PIK superfamily (6). The presence of SMG-1 FAT and FATC domains supports our conclusion that SMG-1 is a protein kinase rather than a lipid kinase.
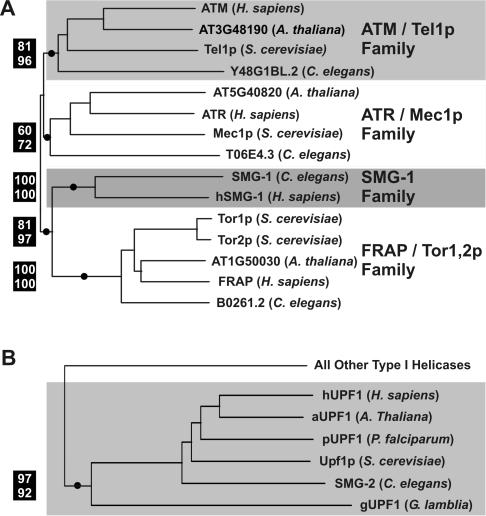
We constructed a phylogenetic tree consisting of all PIK superfamily members from C. elegans, human, yeast (Saccharomyces cerevisiae), and Arabidopsis thaliana. These species were selected as a proxy for higher eukaryotes. The most significant portions of the PIK superfamily tree are shown in Fig. 2A. Four families of protein kinases are identified. Kinases of the ATM/Tel1p, ATR/Mec1p, and FRAP/Tor1,2p families function predominantly in growth control, cell cycle regulation, and response to DNA damage (reviewed in reference 33). The four species we examined contain members of each of these subfamilies. In contrast, of the species we examined, only C. elegans and humans contain members of the SMG-1 family. SMG-1 family members are not found in either S. cerevisiae or Arabidopsis. Interestingly, the SMG-1 kinase family appears to be most closely related to the FRAP family of kinases (discussed below).
FIG. 2.
Phylogenetic analysis of SMG-1 and SMG-2. (A) Neighbor-joining tree of PIK-related protein kinases. (B) Neighbor-joining tree of the UPF1/SMG-2 subfamily of type I helicases. For both trees, confidence values are shown to the left of selected nodes marked with circles: percent bootstrap values by neighbor joining (top) and percent reliability values by quartet puzzling (bottom) are shown. Supported nodes (i.e., bootstrap values of >70%) indicate that sequences to the right of that node are more closely related to each other than any of them are to any other sequences included in the analysis.
We performed a similar phylogenetic analysis of SMG-2. We constructed a phylogenetic tree consisting of all type I helicases from the representative eukaryotes C. elegans, humans, yeast, Arabidopsis, Plasmodium falciparum, and Giardia lamblia. Although the positions of both Plasmodium and Giardia within the eukaryotic tree are uncertain, Giardia is believed to have diverged from other eukaryotes very early (reviewed in reference 1). Figure 2B illustrates a portion of the superfamily tree and identifies a monophyletic group that includes the known orthologs SMG-2 (C. elegans), Upf1p (yeast), and hUPF1 (human). This group also includes putative SMG-2 orthologs in Arabidopsis (aUPF1), Plasmodium (pUPF1), and Giardia (gUPF1). The presence of a putative SMG-2 ortholog in Arabidopsis is not surprising, given that NMD is known to occur in plants (22). Putative SMG-2 orthologs in Plasmodium and the basal eukaryote Giardia, however, are significant and suggest that NMD has an ancient origin among the eukaryotes.
SMG-1 kinase activity is required for NMD.
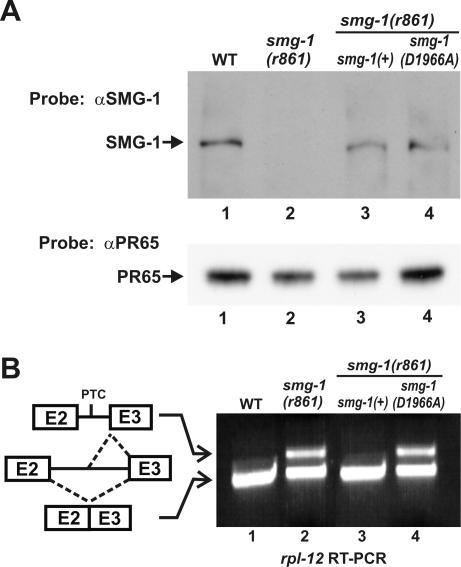
We tested whether the putative kinase activity of SMG-1 is required for NMD by constructing a kinase-inactive variant and determining whether it supplies smg-1(+) function when expressed as the sole source of SMG-1 in vivo. We mutated a highly conserved aspartate (SMG-1 position 1966) to alanine in an smg-1 genomic clone [smg-1(D1966A)], a substitution known to inactivate the catalytic activity of many different protein kinases, including PIK-type kinases (8, 56, 63, 70). We established transgenic lines expressing either smg-1(+) or smg-1(D1966A) in an smg-1(r861) host and verified with Western blots that stable SMG-1 protein was expressed in each line (Fig. 3A, lanes 1, 3, and 4).
FIG. 3.
The kinase domain of SMG-1 is required for NMD. (A) Western blots of total protein from the wild type (WT) (lane 1), smg-1(r861) (lane 2), and smg-1(r861) expressing either SMG-1(+) or SMG-1(D1966A) (lanes 3 and 4) probed with anti-SMG-1 (αSMG-1) (upper panel) or anti-PR65 (αPR65) antibodies (lower panel) as a loading control. (B) rpl-12 reverse transcription (RT)-PCR assay of the same strains analyzed in panel A. Alternative splicing of rpl-12 pre-mRNA is illustrated, with exons 2 and 3 indicated as numbered boxes. Intron 2 is alternatively spliced to yield in-frame mRNA unaffected by NMD (lower mRNA) and a PTC-containing mRNA that is degraded by NMD (upper mRNA).
We then tested transgenic lines for rescue of the smg-1(r861) NMD phenotype by monitoring steady-state abundance of endogenous rpl-12 mRNA. rpl-12 expresses two alternatively spliced mRNAs (48) (Fig. 3B). An unproductively spliced mRNA, which contains PTCs, is abundant in smg(−) strains but absent (or nearly so) in smg(+) strains. A productively spliced mRNA is abundant in both smg(−) and smg(+) strains and serves as an internal control. The ratio of unproductively to productively spliced rpl-12 mRNA reflects the function of NMD. As shown in Fig. 3B, expression of SMG-1(D1966A) fails to rescue the NMD defect of smg-1(r861) (lanes 1, 2, and 4) while expression of SMG-1(+) fully rescues the defect (lanes 1 to 3). We conclude that a functional SMG-1 kinase is likely required for NMD. We cannot rule out the possibility, however, that SMG-1(D1966A) affects not only the SMG-1 kinase but also the additional functions of SMG-1 that are unrelated to its kinase activity and essential for NMD.
In vitro phosphorylation of SMG-2 requires SMG-1 and is negatively regulated by SMG-5.
SMG-1 is required to phosphorylate SMG-2 in vivo (50). Human SMG-1, furthermore, phosphorylates hUPF1 (the ortholog of SMG-2) in vitro and likely also in vivo (19, 70). We investigated whether SMG-2 phosphorylation could be reconstituted in partially purified preparations by immunoprecipitating SMG-2 and testing whether copurifying kinase(s) phosphorylate SMG-2 in vitro. SMG-2 phosphorylation was assayed by resuspending immunoprecipitation pellets in buffers favorable for PIK-related kinases, incubating them with [γ-32P]ATP, and monitoring the transfer of the radiolabel to SMG-2. We additionally tested roles of other smg genes in this assay.
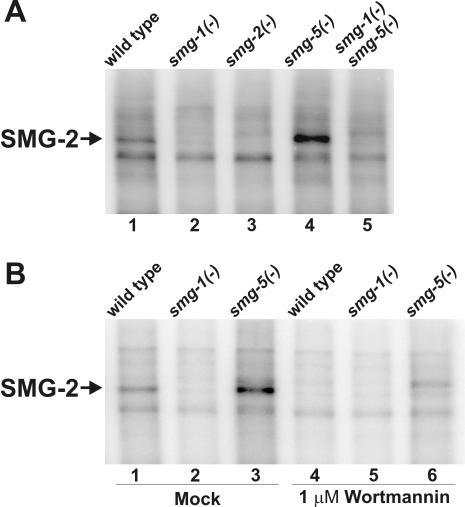
A protein of approximately 120 kDa, which we conclude is SMG-2, is efficiently radiolabeled in such immunoprecipitation kinase assays. Three lines of evidence indicate that this labeled protein is SMG-2: (i) the phosphorylated protein migrates at the known size of SMG-2 (Fig. 4A, lane 1); (ii) phosphorylation of the 120-kDa protein is not observed in assays of smg-2(r908), a smg-2-null allele (lane 3); and (iii) phosphorylation of the 120-kDa protein requires smg-1 function (lanes 2 and 5). We conclude that SMG-2 coimmunoprecipitates with an active kinase and that in vitro SMG-2 phosphorylation requires SMG-1.
FIG. 4.
Phosphorylation of SMG-2 in vitro requires SMG-1 and is sensitive to wortmannin. (A) SMG-2 immunoprecipitation kinase assays. smg-1(r861), smg-2(r908), and smg-5(r860) are null alleles of the respective genes. (B) SMG-2 immunoprecipitation kinase assays in the presence (lanes 4 to 6) or absence (lanes 1 to 3) of 1 μM wortmannin.
Kinases of the PIK superfamily, including human SMG-1, are specifically inhibited by wortmannin (19, 70). If SMG-1 is the SMG-2 kinase described above, phosphorylation of SMG-2 in vitro should be inhibited by wortmannin. We therefore incubated SMG-2 immunoprecipitated pellets with buffer alone (Fig. 4B, lanes 1 to 3) or with 1 μM wortmannin (lanes 4 to 6) prior to the addition of [γ-32P]ATP in immunoprecipitation kinase assays. SMG-2-specific kinase activity was absent in wortmannin-treated samples. We conclude that the SMG-2 kinase observed in vitro is a PIK-type kinase.
SMG-5 is a component of a PP2A complex that is required for SMG-2 dephosphorylation in vivo (2, 49). The SMG-2 kinase activity that we detect in vitro is regulated by smg-5. In vitro phosphorylation of SMG-2 is increased in immunoprecipitation pellets of smg-5(r860) (Fig. 4A, lane 4) and is eliminated in smg-5(−) smg-1(−) double mutants (lane 5). Thus, in vitro phosphorylation of SMG-2 recapitulates the important aspects of SMG-2 phosphorylation observed in vivo. Previous studies demonstrate that SMG-5 interacts with and is coimmunoprecipitable with SMG-2 in crude extracts (2). Thus, the elevated SMG-2 kinase activity in smg-5(−) mutants is likely a direct effect of the absence of SMG-5 in the SMG-2 immunoprecipitation pellets.
SMG-1 interacts with SMG-2.
If SMG-1 is the SMG-2 kinase, as suggested above, then the two proteins may interact as part of a stable complex. Such interactions have been described for hSMG-1 and hUPF1 (70). We therefore investigated whether SMG-1 interacts physically with SMG-2 in crude extracts by using coimmunoprecipitation assays. Our strategy was to immunoprecipitate SMG-2 (or SMG-1) from crude extracts and to examine the pellets for the presence of SMG-1 (or SMG-2) by using antibodies described above and previously (50). We additionally tested smg-3, smg-4, and smg-5 mutants for effects on SMG-1-SMG-2 interactions. smg-3 and smg-4 are required in vivo for SMG-2 phosphorylation, whereas smg-5 is required for SMG-2 dephosphorylation (50).
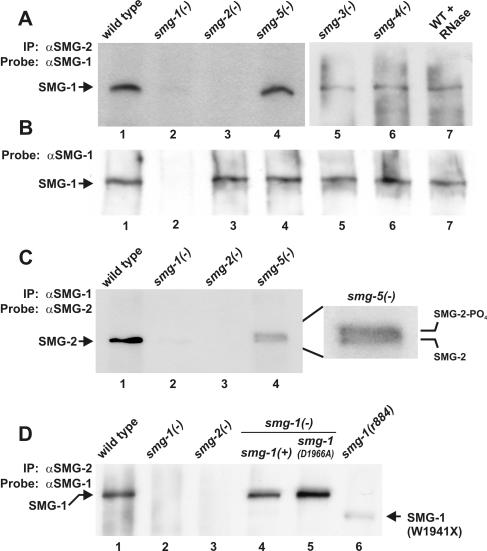
SMG-1, which migrates as a 265-kDa protein, copurifies with immunoprecipitated SMG-2 from wild-type extracts (Fig. 5A, lane 1), including those that have been treated with RNase A (lane 7). SMG-1 is absent from the immunoprecipitation pellets of control strain smg-2(r908) (lane 3; r908 is a protein-null allele), even though its expression is normal in smg-2(r908) (Fig. 5B, lane 3). SMG-1 additionally copurifies with immunoprecipitated SMG-2 from extracts of smg-3(r930) and smg-4(r1169) (Fig. 5A, lanes 5 and 6). We conclude that SMG-1 interacts with SMG-2 either directly or indirectly, that the SMG-1-SMG-2 interaction is not bridged by RNA, and that phosphorylation of SMG-2 is not required for its interaction with SMG-1.
FIG. 5.
SMG-1 interacts with SMG-2. (A) Proteins immunoprecipitated (IP) by anti-SMG-2 (αSMG-2) antibodies were electrophoresed and probed on Western blots with anti-SMG-1 (αSMG-1) antibodies. WT, wild type. (B) Western blots of strains used in Fig. 5A probed with anti-SMG-1. (C) Proteins immunoprecipitated by anti-SMG-1 antibodies were electrophoresed and probed on Western blots with anti-SMG-2 antibodies. (D) Proteins immunoprecipitated by anti-SMG-2 antibodies were electrophoresed and probed on Western blots with anti-SMG-1 antibodies. SMG-1(+) and SMG-1(D1966A) are expressed from transgenes in the strains of lanes 4 and 5, respectively. smg-1(r861), smg-2(r908), smg-3(r930), smg-4(r1169), and smg-5(r860) are null alleles of the respective genes.
Reciprocal immunoprecipitations (Fig. 5C) confirm that SMG-2, which migrates as a 120-kDa protein, copurifies with SMG-1 (lane 1). Previous controls demonstrate that SMG-2 is expressed normally in smg-1(r861), a negative control (50). Both hyperphosphorylated and hypophosphorylated isoforms of SMG-2 coimmunoprecipitate with SMG-1 from extracts of smg-5(r860) (lane 4). SMG-5 is a component of an SMG-2 phosphatase complex (2, 49), and hyperphosphorylated SMG-2 accumulates to abnormally high levels in smg-5 mutants (50). The ratio of SMG-2 isoforms copurifying with SMG-1 (lane 4) is similar to that observed in vivo with smg-5 mutants (50). These results confirm the SMG-1-SMG-2 interaction and indicate again that the state of SMG-2 phosphorylation does not strongly influence its interactions with SMG-1.
We investigated whether mutations that inactivate or truncate the SMG-1 kinase domain affect the SMG-1-SMG-2 interaction (Fig. 5D). SMG-1(D1966A) (discussed above) is a substitution known to inactivate the kinase activity of PIK-type kinases. SMG-1(W1941X) is a truncated form of SMG-1 expressed by the nonsense allele smg-1(r884), which removes the carboxyl terminus of SMG-1, including about half of the SMG-1 kinase domain (Fig. 1A). Both SMG-1(D1966A) and SMG-1(W1941X) copurify with immunoprecipitated SMG-2 (Fig. 5D, lanes 5 and 6). We conclude that an active or intact SMG-1 kinase domain is not required for interactions between SMG-1 and SMG-2, despite being required for NMD.
SMG-2 is localized to the cytoplasm.
An important unanswered question concerning NMD is when during gene expression the surveillance complex (Upf1p, Upf2p, Upf3p, and their associated proteins) is assembled on mRNAs. Yeast UPF3 and hUPF3 and possibly hUPF1 and hUPF2 shuttle between the nucleus and the cytoplasm (40, 45, 46, 57, 59). The significance of this shuttling and the functions of surveillance proteins in different compartments of the cell are presently unknown. More generally, concentrated foci of proteins involved in mRNA degradation have been described for both yeast and mammalian cells (58; reviewed in reference 67). The dynamics of these foci and their roles in mRNA turnover are poorly understood. In principle, phosphorylation of UPF1/SMG-2, which is required for NMD, may regulate its trafficking, either in the nucleus or the cytoplasm. We therefore investigated the intracellular location of SMG-2 in the wild type and in mutants that disrupt cycles of SMG-2 phosphorylation.
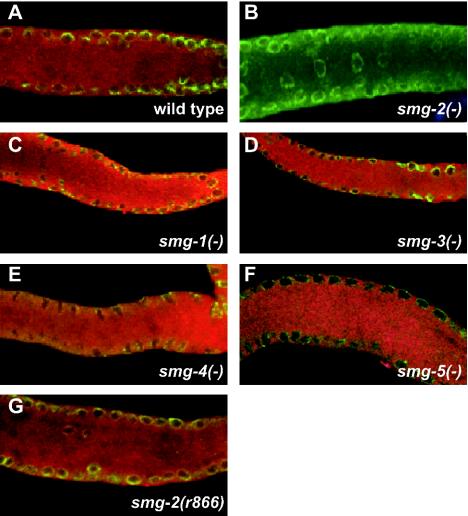
We stained fixed specimens with anti-SMG-2 antibody (50) and with anti-Mab414. Mab414 identifies the nuclear envelope (18), although it is unknown whether Mab414 marks the inner or outer layer (or both). Representative immunofluorescent images are shown in Fig. 6, with SMG-2 labeled with Texas Red and Mab414 labeled with fluorescein (green). SMG-2 is located diffusely throughout the cytoplasm in the wild type (Fig. 6A) and is eliminated in smg-2(r908) (Fig. 6B; r908 is a null allele). The staining of the wild type is somewhat granular in appearance, but the foci of the concentrated signal are not apparent. We detect no changes in the SMG-2 distribution in smg-1(r904), smg-3(r930), and smg-4(r1169) (Fig. 6C to E), all of which are required for SMG-2 phosphorylation and are null alleles of the affected gene (3; Kuchma and Anderson, unpublished). Similarly, SMG-2 is diffusely cytoplasmic in smg-5(r860) (Fig. 6F) and in smg-2(r866) (Fig. 6G). Phosphorylated SMG-2 accumulates to abnormally high levels in both of these mutants, due either to loss of the SMG-5-associated phosphatase (2) or to a missense mutation of SMG-2 itself (50). We conclude that neither SMG-1, SMG-3, SMG-4, nor SMG-5 is required for the gross localization of SMG-2 within cells.
FIG. 6.
Double labeling of wild-type and smg(−) mutant worm gonads with anti-SMG-2 antibody (red signal) and Mab414, an antibody that stains the nuclear envelope (green signal). Genotypes are wild type (A), smg-2(r908) (B), smg-1(r904) (C), smg-3(r930) (D), smg-4(r1169) (E), smg-5(r860) (F), and smg-2(r866) (G). With the exception of smg-2(r866), all smg alleles are null.
DISCUSSION
Seven smg genes are required for NMD in C. elegans (10, 53). We cloned smg-1 as part of an effort to characterize genes involved in NMD. Three lines of evidence demonstrate that the gene we describe is smg-1. First, clones of smg-1 restore NMD when introduced into smg-1(−) mutants. Second, Southern, Northern and Western blots of independent smg-1(−) mutants exhibit allele-specific abnormalities of smg-1 or its encoded mRNA and protein. Third, three sequenced smg-1(−) alleles contain mutations in the cloned gene. Most tested smg-1 mutants express no detectable protein, indicating that smg-1 mutations are loss-of-function alleles.
Cycles of SMG-2 phosphorylation are necessary for NMD in C. elegans, and SMG-1 appears to be the SMG-2 kinase. Phosphorylated SMG-2 is detected in vivo (19, 50, 70), and all other smg genes influence the state of its phosphorylation. SMG-5 is a component of a PP2A complex that interacts with SMG-2 and is required for NMD (2, 13, 49). Multiple lines of evidence indicate that C. elegans SMG-1 is the likely SMG-2 kinase: (i) smg-1 function is required to phosphorylate SMG-2 both in vivo and in vitro, (ii) smg-1 encodes a predicted protein kinase, (iii) an intact SMG-1 kinase domain is essential for NMD, (iv) wortmannin, an inhibitor of PIK-related kinases, inhibits smg-1-dependent phosphorylation of SMG-2 in vitro, and (v) SMG-1 and SMG-2 physically interact. Taken together, these observations suggest strongly that SMG-1 directly phosphorylates SMG-2. Our results indicate that the biochemical functions of C. elegans SMG-1 are similar to those of human SMG-1 (19, 70).
Core components of NMD (UPF1, UPF2, and UPF3) are found in animals, plants, and fungi, but to date, components involved in the phosphorylation of UPF1/SMG-2 have only been described for animals (13, 19, 24, 49, 70). Human and Drosophila orthologs of smg-1, smg-5, smg-6, and smg-7 are known to be involved in NMD and in UPF1/SMG-2 phosphorylation, but yeast orthologs have not been described. Nevertheless, the phosphorylation of Upf1p in yeast cells was recently described (20). Yeast cells do not have an apparent smg-1 ortholog, but they do contain other members of the PIK-related family of kinases. Perhaps one of these kinases is the undiscovered Upf1p kinase. The subfamily of PIK-related kinases most closely related to SMG-1 is the FRAP/Tor1,2 subfamily (Fig. 2). A possibly similar example of regulation via phosphorylation has been described for the yeast FRAP/Tor1,2 subfamily. Yeast Tap42p is reversibly phosphorylated by the competing activities of a Tor1,2p kinase and a PP2A phosphatase (21, 35). Our phylogenetic analysis indicates that the SMG-1 and FRAP/Tor1,2 subfamilies are distinct. C. elegans and hSMG-1 are monophyletic, and C. elegans and humans contain other PIK-related kinases of the FRAP/Tor1,2 subfamily. If an undiscovered Upf1p kinase exists, an undiscovered Upf1p phosphatase must exist also. Unambiguous orthologs of SMG-5, SMG-6, or SMG-7 are not evident in yeast cells, but Nmd4p contains a PINc domain (14), as do C. elegans SMG-5 and SMG-6 (2; H. Shang and P. Anderson, unpublished data). A function for Nmd4p has not been established, but remarkably, it interacts with Upf1p in a yeast two-hybrid assay (30). Perhaps required cycles of UPF1/SMG-2 phosphorylation and dephosphorylation are more widespread than presently described.
An important unanswered question concerning NMD is the function of UPF1/SMG-2 phosphorylation and dephosphorylation. hUPF1 may shuttle between the nucleus and cytoplasm (46), and in principle, phosphorylation of UPF1/SMG-2 may regulate this or other trafficking events. We examined the intracellular location of SMG-2 in mutants that disrupt cycles of SMG-2 phosphorylation, but we observed no alterations in the mutants (Fig. 6). Our analysis included mutants defective for either SMG-2 phosphorylation [smg-1(−), smg-3(−), and smg-4(−)] or SMG-2 dephosphorylation [smg-5(−) and smg-2(r866)]. SMG-2 phosphorylation, therefore, appears to not be involved in overall localization of SMG-2 within the cell. Targeting of SMG-2 on a submicroscopic scale, for example, association with polysomes (51) or differential association with isoforms of UPF3/SMG-4 (49), would not have been detected by our methods.
What then does phosphorylation of SMG-2 regulate? UPF1/SMG-2 is the key regulator of NMD. It interacts with the translation termination machinery and with downstream signals of open reading frames. It functions to integrate signals from multiple sources and to activate either 5′ or 3′ decay pathways. Both phosphorylation and dephosphorylation of SMG-2 are required for NMD, but the steady-state level of phosphorylated SMG-2 is quite low (50). Assuming that a significant fraction of SMG-2 is actively engaged in NMD at steady state, its phosphorylation must be a transient event. Perhaps phosphorylation of SMG-2 regulates its ATPase/helicase activity, with either phosphorylation or dephosphorylation being the trigger that activates 5′ decapping or 3′ decay. Perhaps SMG-2 phosphorylation regulates assembly or dynamics of the numerous protein-protein interactions of the surveillance complex (49), or perhaps phosphorylation regulates disassembly of the surveillance complex and recycling of components for subsequent rounds of NMD. Overexpression of hSMG-1 increases the level of hUPF1 phosphorylation and increases the efficiency of NMD (51, 70), which is consistent with each of these models.
Acknowledgments
This work was supported by the University of Wisconsin Training Grant in Genetics, by a Howard Hughes Predoctoral Fellowship (to A.G.), and by a research grant (GM50933) from the NIH.
We thank Bob Barstead for the cDNA libraries, Sarah Crittenden for help with immunofluorescent microscopy, and Chris Malone for help with microparticle bombardment.
REFERENCES
- 1.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders, K. R., A. Grimson, and P. Anderson. 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronoff, R., R. Baran, and J. Hodgkin. 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene 268:153-164. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Peled, M., and N. V. Raikhel. 1996. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241:140-142. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. [DOI] [PubMed] [Google Scholar]
- 6.Bosotti, R., A. Isacchi, and E. L. L. Sonnhammer. 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25:225-227. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, E. J., P. A. Beal, C. T. Keith, J. Chen, T. B. Shin, and S. L. Schreiber. 1995. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature 377:441-446. [DOI] [PubMed] [Google Scholar]
- 9.Cali, B. M., and P. Anderson. 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260:176-184. [DOI] [PubMed] [Google Scholar]
- 10.Cali, B. M., S. L. Kuchma, J. Latham, and P. Anderson. 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter, M. S., J. Doskow, P. Morris, S. Li, R. P. Nhim, S. Sandstedt, and M. F. Wilkinson. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270:28995-29003. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. Y., and A. B. Shyu. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 23:4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu, S. Y., G. Serin, O. Ohara, and L. E. Maquat. 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG-5 and SMG-7 that functions in the dephosphorylation of Upf1. RNA 9:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clissold, P. M., and C. P. Ponting. 2000. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10:R888-R890. [DOI] [PubMed] [Google Scholar]
- 15.Crittenden, S. L., and J. Kimble. 1999. Confocal methods for Caenorhabditis elegans. Methods Mol. Biol. 122:141-151. [DOI] [PubMed] [Google Scholar]
- 16.Culbertson, M. R., and P. F. Leeds. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13:207-214. [DOI] [PubMed] [Google Scholar]
- 17.Czaplinski, K., M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng, H. A. Perlick, M. D. Ter-Avanesyan, and S. W. Peltz. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis, L. I., and G. Blobel. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45:699-709. [DOI] [PubMed] [Google Scholar]
- 19.Denning, G., L. Jamieson, L. E. Maquat, E. A. Thompson, and A. P. Fields. 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 276:22709-22714. [DOI] [PubMed] [Google Scholar]
- 20.De Pinto, B., R. Lippolis, R. Castaldo, and N. Altamura. 2004. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol. Microbiol. 51:1129-1142. [DOI] [PubMed] [Google Scholar]
- 21.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 22.Dolferus, R., D. Van Den Bossche, and M. Jacobs. 1990. Sequence analysis of two null-mutant alleles of the single Arabidopsis Adh locus. Mol. Gen. Genet. 224:297-302. [DOI] [PubMed] [Google Scholar]
- 23.Dunckley, T., and R. Parker. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork, and E. Izaurralde. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gehring, N. H., G. Neu-Yilik, T. Schell, M. W. Hentze, and A. E. Kulozik. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11:939-949. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, C. I., M. J. Ruiz-Echevarria, S. Vasudevan, M. F. Henry, and S. W. Peltz. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5:489-499. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez, C. I., A. Bhattacharya, W. Wang, and S. W. Peltz. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274:15-25. [DOI] [PubMed] [Google Scholar]
- 28.Hall, G. W., and S. Thein. 1994. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood 83:2031-2037. [PubMed] [Google Scholar]
- 29.He, F., S. W. Peltz, J. L. Donahue, M. Rosbash, and A. Jacobson. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 90:7034-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, F., and A. Jacobson. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437-454. [DOI] [PubMed] [Google Scholar]
- 31.He, F., A. H. Brown, and A. Jacobson. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgkin, J., A. Papp, R. Pulak, V. Ambros, and P. Anderson. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoekstra, M. F. 1997. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr. Opin. Genet. Dev. 7:170-175. [DOI] [PubMed] [Google Scholar]
- 34.Hunter, T. 1995. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell 83:1-4. [DOI] [PubMed] [Google Scholar]
- 35.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, V. N., N. Kataoka, and G. Dreyfuss. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293:1832-1836. [DOI] [PubMed] [Google Scholar]
- 37.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, B. P., R. E. Green, and S. E. Brenner. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 100:189-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121-1131. [DOI] [PubMed] [Google Scholar]
- 41.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836-1839. [DOI] [PubMed] [Google Scholar]
- 42.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maquat, L. E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5:89-99. [DOI] [PubMed] [Google Scholar]
- 44.Mello, C., and A. Fire. 1995. DNA transformation. Methods Cell Biol. 48:451-482. [PubMed] [Google Scholar]
- 45.Mendell, J. T., S. M. Medghalchi, R. G. Lake, E. N. Noensie, and H. C. Dietz. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20:8944-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendell, J. T., C. M. J. ap Rhys, and H. C. Dietz. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419-422. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell, P., and D. Tollervey. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-5′ degradation. Mol. Cell 11:1405-1413. [DOI] [PubMed] [Google Scholar]
- 48.Mitrovich, Q. M., and P. Anderson. 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14:2173-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohnishi, T., A. Yamashita, I. Kashima, T. Schell, K. R. Anders, A. Grimson, T. Hachiya, M. W. Hentze, P. Anderson, and S. Ohno. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12:1187-1200. [DOI] [PubMed] [Google Scholar]
- 50.Page, M. F., B. Carr, K. R. Anders, A. Grimson, P. Anderson. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pal, M., Y. Ishigaki, E. Nagy, and L. E. Maquat. 2001. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7:5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885-1897. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Echevarria, M. J., C. I. Gonzalez, and S. W. Peltz. 1998. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 17:575-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 56.Schu, P. V., K. Takegawa, M. J. Fry, J. H. Stack, M. D. Waterfield, and S. D. Emr. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260:88-91. [DOI] [PubMed] [Google Scholar]
- 57.Serin, G., A. Gersappe, J. D. Black, R. Aronoff, and L. E. Maquat. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21:209-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirley, R. L., M. J. Lelivelt, L. R. Schenkman, J. N. Dahlseid, and M. R. Culbertson. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111:3129-3143. [DOI] [PubMed] [Google Scholar]
- 60.Strimmer, K., and A. Von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 61.Sun, X., H. A. Perlick, H. C. Dietz, and L. E. Maquat. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 95:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi, S., Y. Araki, T. Sakuno, and T. Katada. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, S. S., D. R. Knighton, J. Zheng, J. M. Sowadski, C. S. Gibbs, and M. J. Zoller. 1993. A template for the protein kinase family. Trends Biochem. Sci. 18:84-89. [DOI] [PubMed] [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner, E., and J. Lykke-Andersen. 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115:3033-3038. [DOI] [PubMed] [Google Scholar]
- 66.Weng, Y., K. Czaplinski, and S. W. Peltz. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 16:5477-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wickens, M., and A. Goldstrohm. 2003. Molecular biology: a place to die, a place to sleep. Science 300:753-755. [DOI] [PubMed] [Google Scholar]
- 68.Williams, J. A., J. A. Langeland, B. S. Thalley, J. B. Skeath, and S. B. Carroll. 1995. Expression of foreign proteins in E. coli using plasmid vectors and purification of specific polyclonal antibodies, p. 15-58. In D. M. Glover and B. D. Hames (ed.), DNA cloning 2: expression systems. A practical approach. Oxford University Press, Oxford, England.
- 69.Wilusz, C. J., W. Wang, and S. W. Peltz. 2001. Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev. 15:2781-2785. [DOI] [PubMed] [Google Scholar]
- 70.Yamashita, A., T. Ohnishi, I. Kashima, Y. Taya, and S. Ohno. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15:2215-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]